Abstract
The cap-binding complex (CBC) binds to the cap structure of mRNA to protect it from exonucleases as well as to regulate downstream post-transcriptional events, translational initiation and nonsense-mediated mRNA decay. However, its role in regulation of the upstream transcriptional events such as initiation or elongation remains unknown. Here, using a formaldehyde-based in vivo cross-linking and chromatin immunoprecipitation assay in conjunction with transcriptional, mutational and co-immunoprecipitational analyses, we show that CBC is recruited to the body of yeast gene, and then stimulates the formation of pre-initiation complex (PIC) at several yeast promoters through its interaction with Mot1p (modifier of transcription). Mot1p is recruited to these promoters, and enhances the PIC formation. We find that CBC promotes the recruitment of Mot1p which subsequently stimulates PIC formation at these promoters. Furthermore, the formation of PIC is essential for recruitment of CBC. Thus, our study presents an interesting observation that an mRNA binding factor exhibits a reciprocal synergistic effect on formation of PIC (and hence transcriptional initiation) at the promoter, revealing a new pathway of eukaryotic gene regulation in vivo.
INTRODUCTION
Gene regulation, which turns on and off the expression of specific RNA polymerase II (RNAPII) genes in response to environmental changes during cell differentiation and development, occurs widely at the level of transcriptional activation which is initiated by the promoter-specific activators. A typical activator contains a promoter-targeting region, which is often a sequence-specific DNA binding domain, and a distinct activation domain. A variety of studies indicates that activator interacts directly with one or more components of the transcription machinery to stimulate the assembly of general transcription factors (GTFs) such as TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH and RNAPII for formation of the pre-initiation complex (PIC) at the core promoter of RNAPII gene to initiate transcription which is followed by elongation, and subsequently termination to generate mRNA (1–11).
The mRNA has a characteristic 5′-end consisting of a 7-methylguanosine cap structure attached by a 5′-5′ phosphotriester linkage to the first encoded nucleotide. The cap-binding complex (CBC) binds to the cap structure to protect mRNA from 5′ → 3′ exonucleases. CBC is composed of two different proteins, namely, Cbp20p and Cbp80p. Cbp20p has a structure common to many RNA binding proteins and is tightly bound to Cbp80p in a way that leaves the RNA binding surface of Cbp20p exposed. Such an RNA binding surface, which is highly conserved in evolution from yeast to human, first binds to the cap, and then the flexible ends of Cbp20p keep the cap in place by folding over it with the help of Cbp80p (12–17). Thus, neither Cbp20p nor Cbp80p has been biochemically found to bind to the capped RNA alone (18). The homologs of both CBC proteins have been identified in all eukaryotes examined so far (16–23).
Both yeast and human CBC play analogous roles in pre-mRNA splicing. The efficiency with which U1 small nuclear ribonucleoprotein particle (snRNP) binds to the cap-proximal 5′ splice site is stimulated by CBC, and thus, the rate of recognition and splicing of the cap-proximal intron is enhanced (18,20,24–26). Further, formaldehyde-based in vivo cross-linking and chromatin immunoprecipitation (ChIP) studies have demonstrated that CBC is necessary for co-transcriptional spliceosome assembly at an intron-containing gene in yeast (27), indicating an essential link between CBC and spliceosome assembly in vivo. CBC has also been implicated recently in pre-mRNA splicing and alternative splicing in Arabidopsis thaliana (28,29). Like splicing, the 3′-end formation of pre-mRNA is regulated by CBC in vertebrates (30–36). For example, the depletion of CBC from HeLa cell nuclear extract strongly reduces the endonucleolytic cleavage step of the cleavage and polyadenylation process at the 3′-end of pre-mRNA. Unlike vertebrates, yeast strains lacking CBC do not biochemically exhibit defects in the 3′-end formation of pre-mRNA (26,37). However, a recent study (38) has demonstrated the role of CBC in regulation of the 3′-end formation of yeast pre-mRNA in vivo. Like RNA splicing and 3′-end formation, the export of the capped RNAs is stimulated by CBC (39–41). For example, CBC in vertebrates mediates the effect of the cap structure in the export of U snRNA, providing a direct evidence for the involvement of a cellular RNA-binding factor in the transport of RNA to cytoplasm. Further, Shen et al. (42) have demonstrated the genetic and physical interactions between CBC and Npl3p (an mRNA export factor) in yeast, indicating the role of CBC in mRNA export. Recently, Nojima et al. (43) have also demonstrated the interaction of CBC with RNA export factor in HeLa cells for stimulation of mRNA. Thus, besides protecting mRNA from the exonucleases, CBC also regulates the downstream post-transcriptional events such as RNA splicing, 3′-end formation and export (18,20,24–45). Further, CBC is involved in controlling translational initiation and nonsense-mediated mRNA decay (46–51). Intriguingly, the recent studies have also implicated CBC in processing primary microRNAs, and regulating histone H2B ubiquitylation, RNA interference and cell proliferation (22,52–55). Together, these studies reveal that CBC plays crucial roles in diverse biological processes to maintain normal cellular functions.
Although the function of CBC in regulation of downstream post-transcriptional events is well-documented, it is not yet known whether CBC plays any role in controlling the upstream transcriptional events such as initiation or elongation. Here, using a ChIP assay in conjunction with transcriptional, mutational and co-immunoprecipitational analyses, we demonstrate in Saccharomyces cerevisiae that CBC stimulates the PIC formation (and hence transcriptional initiation) at several promoters via its interaction with Mot1p (modifier of transcription), thus providing a novel function of CBC in eukaryotic gene regulation.
MATERIALS AND METHODS
Plasmids
The plasmid pFA6a-13Myc-KanMX6 (56) was used for genomic myc-epitope tagging of the proteins of interest. The plasmid pFA6a-3HA-His3MX6 (56) was used for genomic HA-epitope tagging of the proteins of interest. The plasmids, namely pRS416 and pRS413, were used in the PCR-based gene disruption.
Yeast strains
Yeast (S. cerevisiae) strains harboring temperature-sensitive (ts) mutation in SPT15 and its isogenic wild-type equivalent were obtained from the Struhl laboratory (Kevin Struhl, Harvard Medical School). The ceg1-ts (ceg1-63; YSB230) and cet1-ts (cet1-438; YSB717) mutants and their isogenic wild-type equivalents (YSB242 and YSB540, respectively) were obtained from the Buratowski laboratory (Stephen Buratowski, Harvard Medical School) (57,58). The mot1-ts (MY603) and its wild-type (MY2 or KY804) equivalent were obtained from the Collart laboratory (Martin Collart, CMU, Switzerland). FY67 and FY1097 strains were obtained from the Winston laboratory (Fred Winston, Harvard Medical School) (59,60). The strains MY3 (Cbp80p-myc in spt15-ts) and SGY177 (Cbp80p-myc in wild-type W303a strain) were generated by insertion of multiple myc-epitope tags at the original chromosomal locus of CBP80 in spt15-ts and W303a, respectively (56). The endogenous CBP20 gene of SGY177 was disrupted using a PCR-based gene knockout method (61) to generate NSY14 (cbp20Δ::URA3, Cbp80p-myc). Similarly, the endogenous CBP80 gene of W303a was disrupted to generate NSY15 (cbp80Δ::HIS3). Multiple myc-epitope tags were added at the original chromosomal loci of CBP20, RPB3, SRB4, SPT20, MOT1, RAD3 and SNF2 in NSY15 to generate NSY27 (cbp80Δ::HIS3, Cbp20p-myc), NSY30 (cbp80Δ::HIS3, Rpb3p-myc), PBY9 (cbp80Δ::HIS3, Srb4p-myc), PBY6 (cbp80Δ::HIS3, Spt20p-myc), SLY3 (cbp80Δ::HIS3, Mot1p-myc), ASY2 (cbp80Δ::HIS3, Rad3p-myc) and ASY10 (cbp80Δ::HIS3, Snf2p-myc), respectively. Similarly, multiple myc-epitope tags were added at the original chromosomal loci of CBP20, RPB3, SRB4, SPT20, MOT1, RAD3 and SNF2 in W303a to generate NSY26 (Cbp20p-myc), NSY17 (Rpb3p-myc), PBY8 (Srb4p-myc), ASY10 (Spt20p-myc), SLY2 (Mot1p-myc), ASY41 (Rad3p-myc) and ASY39 (Snf2p-myc), respectively. Multiple HA-epitope tags were added at the original chromosomal locus of CBP20 in SLY2 and SLY3 to generate SLY8 (Cbp20p-HA and Mot1p-myc) and SLY9 (Cbp20p-HA, Mot1p-myc; cbp80Δ::HIS3). The strains bearing HA-tagged Cbp80p (SLY14a) and Mot1p (SLY13a) were generated by separately adding multiple HA-epitope tags in the chromosomal loci of CBP80 and MOT1 in W303a. Multiple myc-epitope tags were added to MOT1 in FY67 (SPT20 wild-type), FY1097 (Δspt20), and SPT15 wild-type and ts mutant strains to generate GDY7, GDY8, GDY5 and GDY6, respectively.
Growth media
For the ChIP studies at ADH1, both the wild-type and deletion mutant strains were grown in YPD (yeast extract-peptone plus 2% dextrose) up to an optical density at 600 nm (OD600) of 1.0 at 30°C prior to formaldehyde-based in vivo cross-linking. For the studies at the GAL1 gene in the wild-type and deletion mutant strains, yeast cells were first grown in YPR (yeast extract-peptone plus 2% raffinose) up to an OD600 of 0.9, and then transferred to YPG (yeast extract-peptone plus 2% galactose) for various induction time periods at 30°C prior to formaldehyde-based cross-linking. However, the spt15-ts and mot1-ts mutant strains and their isogenic wild-type equivalents were grown in YPG at 23°C up to an OD600 of 0.85, and then transferred to 37°C for 1 h before cross-linking. For cet1-ts and ceg1-ts mutant strains and their wild-type equivalents, yeast cells were grown in YPG at 30°C up to OD600 of 0.85, and then transferred to 37°C for 1 h (for cet1-ts and its isogenic wild-type strains) or 4 hr (ceg1-ts and its isogenic wild-type strains) prior to cross-linking. Similar growth conditions were used for the GAL7, GAL10 and GAL2 genes. For the studies at HSP26 and other heat-shock genes (e.g. SSA3 and SSA4), yeast strains were grown in YPD at 23°C up to OD600 of 0.9, and then switched to 39°C for 30 min prior to cross-linking. The CUP1 gene was induced by 1 mM CuSO4 for 15 min in synthetic complete medium (yeast nitrogen base and complete amino acid mixture plus 2% dextrose) at 30°C.
ChIP assay
The ChIP assay was performed as described previously (62–65). Briefly, yeast cells were treated with 1% formaldehyde, collected and resuspended in lysis buffer. Following sonication, cell lysates (400 µl lysate from 50 ml of yeast culture) were precleared by centrifugation, and then 100 µl lysate was used for each immunoprecipitation. Immunoprecipitated protein–DNA complexes were treated with proteinase K, the cross-links were reversed, and then the DNA was purified. Immunoprecipitated DNA was dissolved in 20 µl TE 8.0 (10 mM Tris–HCl, pH 8.0, and 1 mM EDTA), and 1 µl of immunoprecipitated DNA was analyzed by polymerase chain reaction (PCR). PCR reactions contained [α-32P]dATP (2.5 µCi for each 25-µl reaction) and the PCR products were detected by autoradiography after separation on a 6% polyacrylamide gel. As a control, ‘input’ DNA was isolated from 5 µl lysate without going through the immunoprecipitation step and was suspended in 100 µl TE 8.0. To compare PCR signal arising from the immunoprecipitated DNA with the input DNA, 1 µl of input DNA was used in PCR analysis. Serial dilutions of the input and IP DNAs were used to assess the linear range of PCR amplification as described previously (66). The PCR data presented in this article are within the linear range of PCR analysis.
For analysis of Mot1p and Snf2p recruitment, we modified the above ChIP protocol as follows (65). Lysate (800 µl) was prepared from 100 ml of yeast culture. Lysate (400 µl) was used for each immunoprecipitation (using 10 µl of anti-HA or anti-myc antibody and 100 µl of protein A/G plus agarose beads from Santa Cruz Biotechnology Inc.), and immunoprecipitated DNA sample was dissolved in 10 µl TE 8.0 for PCR analysis. In parallel, the PCR for ‘input’ DNA was performed using 1 µl DNA that was prepared by dissolving purified DNA from 5 µl lysate in 100 µl TE 8.0.
Primer-pairs used for PCR analysis were as follows:
| ADH1(UAS): | 5′-GAGTTTCCGGGTGTACAATATGG-3′ |
| 5′-CTATTGTATATCTCCCCTCCGC-3′ | |
| ADH1(Core): | 5′-GGTATACGGCCTTCCTTCCAGTTAC-3′ |
| 5′-GAACGAGAACAATGACGAGGAAACAAAAG-3′ | |
| ADH1(ORF1): | 5′-CTGGTTACACCCACGACGGTTCTT-3′ |
| 5′-GCAGACTTCAAAGCCTTGTAGACG-3′ | |
| ADH1(ORF2): | 5′-CGGTAACAGAGCTGACACCAGAGA-3′ |
| 5′-ACGTATCTACCAACGATTTGACCC-3′ | |
| GAL1(UAS): | 5′-CGCTTAACTGCTCATTGCTATATTG-3′ |
| 5′-TTGTTCGGAGCAGTGCGGCGC-3′ | |
| GAL1(Core): | 5′-ATAGGATGATAATGCGATTAGTTTTTTAGCCTT-3′ |
| 5′-GAAAATGTTGAAAGTATTAGTTAAAGTGGTTATGCA-3′ | |
| GAL1(ORF1): | 5′-CAGTGGATTGTCTTCTTCGGCCGC-3′ |
| 5′-GGCAGCCTGATCCATACCGCCATT-3′ | |
| GAL1(ORF2): | 5′-CAGAGGGCTAAGCATGTGTATTCT-3′ |
| 5′-GTCAATCTCTGGACAAGAACATTC-3′ | |
| CUP1(Core): | 5′-TCTTCTAGAAGCAAAAAGAGCGATG-3′ |
| 5′-CGCTGAACATTTTATGTGATGATTG-3′ | |
| GAL2 (Core): | 5′-GCTAAAATGTGGAGATAGGATAAGT-3′ |
| 5′-TGAATAAGGTGCATAATGAAGAGCA-3′ | |
| GAL7 (Core): | 5′-CTATGTTCAGTTAGTTTGGCTAGC-3′ |
| 5′-TTGATGCTCTGCATAATAATGCCC-3′ | |
| GAL10 (Core): | 5′-GCTAAGATAATGGGGCTCTTTACAT-3′ |
| 5′-TTTCACTTTGTAACTGAGCTGTCAT-3′ | |
| HSP26 (Core): | 5′-AATAGGACCTCCATTAGTTAGAGAT-3′ |
| 5′-TGGACTGTTAAATGACATGTTAATTTGTTTAG-3′ | |
| SSA3 (Core): | 5′-ATATTGCACAATTGGAAACGAATGG-3′ |
| 5′-AGCGTTTAGTACCTATTCTATCCGT-3′ | |
| SSA4 (Core): | 5′-GGAAGCACCAAGAAAAAAGGAAG-3′ |
| 5′-GTGGTTTTTATTCGAAAGTTGTGGAGAAAG-3′ | |
| Chrom-V: | 5′-GGCTGTCAGAATATGGGGCCGTAGTA-3′ |
| 5′-CACCCCGAAGCTGCTTTCACAATAC-3′ |
Autoradiograms were scanned and quantitated by the National Institutes of Health image 1.62 program. Immunoprecipitated (IP) DNAs were quantitated and presented as the ratio of IP to input. Chrom-V, a non-transcribed region in chromosome V.
We have performed each set of the ChIP experiments multiple times, and consistent results were obtained. Most of our ChIP data in this manuscript are presented in the form of a histogram or line with error bars. Some of the ChIP data are presented as autoradiogram. In such presentation of autoradiogram, the quantitation of that particular autoradiogram is shown below the band, as has also been shown in our previous studies (62,65,66).
Whole-cell extract preparation and analysis
For analysis of global levels of TBP, Rpb1p and Rpb3p in the CBP80 deletion mutant and its isogenic wild-type equivalent, the yeast strains were grown in 50 ml YPR up to an OD600 of 0.9, and then induced for 60 min in YPG. The harvested cells were lysed and sonicated to prepare the whole-cell extract with solubilized chromatin following the protocol as described previously for the ChIP assay (62–65). The whole-cell extract was run on SDS-polyacrylamide gel, and then analyzed by western blot. The anti-TBP (obtained from Michael R. Green, University of Massachusetts Medical School), anti-Rpb1 (8WG16; Covance) and anti-myc (9E10; Santacruz Biotechnology Inc.) antibodies against TBP, Rpb1p and myc-tagged Rpb3p, respectively, were used for western blot analysis.
Total mRNA preparation
The total mRNA was prepared from yeast cell culture as described by Peterson et al. (67). Briefly, 10 ml yeast culture of a total OD600 of 1.0 in YPD was harvested, and then was suspended in 100 µl RNA preparation buffer (500 mM NaCl, 200 mM Tris–HCl, 100 mM Na2 EDTA and 1% SDS) along with 100 µl phenol/chloroform/isoamyl alcohol and 100 µl volume-equivalent of glass beads (acid washed; Sigma). Subsequently, yeast cell suspension was vortexed with a maximum speed (10 in VWR mini-vortexer; Cat. No. 58816-121) for five times (30 s each). Cell suspension was put in ice for 30 s between pulses. After vortexing, 150 µl RNA preparation buffer and 150 µl phenol/chloroform/isoamyl alcohol were added to yeast cell suspension followed by vortexing for 15 s with a maximum speed on VWR mini-vortexer. The aqueous phase was collected following 5 min centrifugation at a maximum speed in microcentrifuge machine. The total mRNA was isolated from aqueous phase by ethanol precipitation.
Primer extension analysis
Primer extension analysis was performed as described previously (64). The primer used for analysis of GAL1 mRNA was as follows:
GAL1: 5′-CCTTGACGTTACCTTGACGTTAAAGTATAGAGG-3′
Formaldehyde-based in vivo cross-linking and co-immunoprecipitation assay
The co-immunoprecipitation assay was performed as described previously (68). Briefly, yeast strains carrying myc-tagged Mot1p and HA-tagged Cbp20p in the CBP80 deletion and wild-type strains were grown in YPG up to an OD600 of 1.0, and then cross-linked by formaldehyde. The whole-cell extract was prepared by lysing and sonicating the cross-linked yeast cells. Immunoprecipiation was performed using an anti-HA antibody and protein A/G plus agarose beads. Anti-Flag was used as a non-specific antibody. After immunoprecipitaion, the agarose beads were washed as in the ChIP assay. The washed A/G plus agarose beads were boiled in the SDS-PAGE loading buffer, and supernatant was analyzed by SDS-PAGE and western blot. The anti-myc antibody was used in the western blot analysis. The input is 1.25% of the lysate that was used for immunoprecipitation.
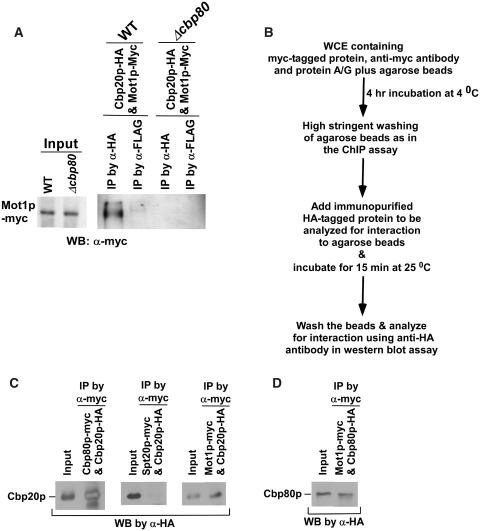
Immunopurification of Cbp20p and Cbp80p
The yeast strains expressing HA-tagged Cbp20p or Cbp80p were grown in 100 ml YPD up to an OD600 of 1.0, and then were harvested. Subsequently, 800 µl WCE was prepared from the culture of each strain. Immunoprecipitation was performed using anti-HA antibody attached to sepharose beads (mono-HA.11 clone 16B12 monoclonal antibody affinity matrix, AFC-101P; Covance Inc.) for 4 h at 4°C. 400 µl WCE and 100 µl sepharose beads (∼50 µl bed volume) were used for each immunoprecipitation. The sepharose beads following immunoprecipitation was washed under high stringent washing conditions as in the ChIP assay (62–65), but 0.5 M NaCl, instead of 1 M NaCl, was used in the second and third washes of the beads. Then the beads were equilibrated by buffer E (50 mM Tris-base, 250 mM NaCl, 1% NP-40, 1 mM EDTA; pH 8.5) before elution of HA-tagged protein by HA peptide. The immobilized protein (HA-tagged Cbp20p/Cbp80p) on sepharose beads was eluted by incubating the beads in two bed volumes (100 µl) of buffer E containing HA peptide with a final concentration of 1 mg/ml. The beads were incubated for 30 min at 25°C on rotor. Elution was performed three times. Buffer E (100 µl) with HA peptide was used for each elution. The eluted HA-tagged protein was analyzed by silver staining and western blot. An anti-HA antibody was used as a primary antibody in the Western blot analysis.
Protein interaction assay in vitro
To analyze the interaction of Cbp20p with Mot1p, the yeast strain expressing myc-tagged Mot1p was grown in 100 ml YPG up to an OD600 of 1.0, and subsequently 800 µl WCE was prepared. WCE (400 µl) was used for immunoprecipitation as in the ChIP assay using 10 µl anti-myc antibody and 100 µl protein A/G plus agarose beads. The immobilized Mot1p on beads was thoroughly washed under high stringent washing conditions as in the ChIP assay (62–65), and then the washed beads with immobilized Mot1p was incubated with immunopurified Cbp20p in buffer E containing HA peptide and aprotinin for 15 min at 25 °C. Subsequently, the beads were washed by buffer W (50 mM Tris-base, 2 mg/ml BSA, 250 mM NaCl, 1% NP-40, 1 mM EDTA; pH 8.5) containing aprotinin for four times (1 ml each time). Last wash was performed using buffer E containing aprotinin. The washed beads were then boiled in SDS–PAGE loading buffer, and the supernatant was subsequently analyzed by SDS–PAGE and western blot to determine the interaction between Cbp20p and Mot1p. An anti-HA antibody was used in the western blot analysis. Likewise, the interaction of Cbp80p with Mot1p was analyzed using myc-tagged Mot1p and immunopurified Cbp80p. Similar experimental protocol was also used to analyze the interaction of Cbp20p with Cbp80 and Spt20p. For analyzing the interaction of Cbp20p with Cbp80p, myc-tagged Cbp80p and immunopurified Cbp20 were used. Myc-tagged Spt20p and immunopurified Cbp20p were used to analyze Cbp20p–Spt20p interaction.
RESULTS AND DISCUSSION
CBC is co-transcriptionally recruited to the coding sequences of the active genes
To determine the role of CBC in regulation of transcriptional initiation or elongation, we first analyzed its association with an active gene, GAL1, using a ChIP (62–65) assay. In this direction, both the Cbp20p and Cbp80p components of CBC were tagged by multiple myc-epitopes in their chromosomal loci. Different sets of specific primer-pairs [(69), ‘Materials and Methods’ section] targeted to the UAS (upstream activating sequence), core promoter and two different locations (ORF1 and ORF2; towards the 5′- and 3′-ends of ORF, respectively) of the open reading frame (ORF) or coding sequence (bottom panel; Figure 1A) were used for PCR analysis of the immunoprecipitated DNA samples. Figure 1A (Top panel) shows that both the Cbp20p and Cbp80p components of CBC were predominantly recruited to the GAL1 coding sequence. However, these components were not recruited to the GAL1 promoter (top panel, Figure 1A). This was expected since CBC recruitment is dependent on nascent mRNA following transcriptional initiation and subsequent elongation. Consistently, we show that CBC (Cbp80p) was not recruited to GAL1 (right panel; Figure 1B) when RNAPII (Rpb1p, the largest subunit of RNAPII) was absent at the GAL1 core promoter in the ts mutant strain of SPT15 (that encodes TBP, TATA-box binding protein) (left panel; Figure 1B). Thus, CBC is co-transcriptionally recruited to GAL1 in an RNAPII-dependent manner.
Figure 1.
Analysis of the recruitment of CBC to the GAL1 and ADH1 genes. (A) Both the Cbp20p and Cbp80p components of CBC are predominantly recruited to the GAL1 ORF. Yeast strains expressing myc-tagged Cbp20p or Cbp80p were grown at 30°C in YPG. The ChIP assay was performed as previously described (62–65). Primer-pairs [(69), ‘Materials and Methods’ section] located in the UAS, core promoter and two different locations of ORF (ORF1 and ORF2) of GAL1 were used for PCR analysis of the immunoprecipitated DNA samples. Immunoprecipitation was performed using a mouse monoclonal antibody against the c-myc epitope-tag (9E10; Santa Cruz Biotechnology Inc.). The ratio of immunoprecipite over the input in the autoradiogram is indicated below each band. IP, immunoprecipitate. (B) Recruitment of CBC to the GAL1 ORF is dependent on TBP. Wild-type and ts mutant strains of TBP (spt15-ts) expressing myc-tagged Cbp80p were first grown in YPG at 23°C to an OD600 of 0.85, and then transferred to 37°C for 1 h prior to formaldehyde-based in vivo cross-linking. The mouse monoclonal antibody 8WG16 (Covance) against Rpb1p-CTD (carboxy terminal domain) was used. (C) Cbp20p and Cbp80p are mutually dependent for their recruitment to GAL1. Wild-type and mutant strains were grown in YPR up to an OD600 of 0.9, and then switched to YPG for 90 minutes at 30°C prior to cross-linking. (D) Western blot analysis. The wild-type and deletion mutant strains expressing myc-tagged Cbp20p/Cbp80p were grown as in panel C. The whole-cell extracts from the wild-type and deletion mutant strains were prepared as in the ChIP assay (62–65), and were analyzed by western blot using anti-myc and anti-TBP antibodies against myc-tagged Cbp20p/Cbp80p and TBP, respectively. (E) CBC is predominantly recruited to the ADH1 ORF. Primer pairs [(69), ‘Materials and Methods’ section] located in the UAS, core promoter and two different locations of ORF (ORF1 and ORF2) of ADH1 were used for PCR analysis of the immunoprecipitated DNA samples. (F) Recruitment of CBC to the ADH1 ORF is dependent on RNAPII as well as TBP. Wild-type and ts mutant strains of TBP (spt15-ts) and RPB1 expressing myc-tagged Cbp80p were first grown in YPD at 23°C to an OD600 of 0.85, and then transferred to 37°C for 1 h prior to formaldehyde cross-linking. Immunoprecipitations were performed as described in panels A and B. (G) Recruitments of Cbp20p and Cbp80p to ADH1 are dependent on each other.
Next, we asked whether the recruitment of Cbp20p and Cbp80p are mutually dependent. To address this question, we analyzed the recruitment of Cbp20p and Cbp80p to the GAL1 ORF in the cbp80Δ and cbp20Δ strains, respectively. If these proteins are functionally dependent on each other in recognizing the cap-structure of mRNA, Cbp20p and Cbp80p will not be recruited to the GAL1 ORF in the cbp80Δ and cbp20Δ strains, respectively. Indeed, the recruitment of Cbp20p was completely lost in cbp80Δ (left panel; Figure 1C). Similarly, Cbp80p was not recruited to the GAL1 ORF in cbp20Δ (right panel; Figure 1C). However, the stabilities of Cbp20p and Cbp80p were not altered in cbp80Δ and cbp20Δ, respectively (Figure 1D). Thus, Cbp20p and Cbp80p are dependent on each other for their recruitment to GAL1 in vivo. To determine whether Cbp20p and Cbp80p are also co-transcriptionally recruited to other active genes in a mutually dependent manner, we performed similar experiments at a constitutively active gene, ADH1. We find that CBC is predominantly recruited to the ADH1 coding sequence in an RNAPII-dependent manner (Figure 1E and F). Further, Cbp20p and Cbp80p are dependent on each other for their recruitment to ADH1 (Figure 1G). Thus, Cbp20p and Cbp80p are co-transcriptionally recruited to the active genes in a mutually dependent manner in vivo.
CBC stimulates the association of RNAPII with the GAL1 coding sequence
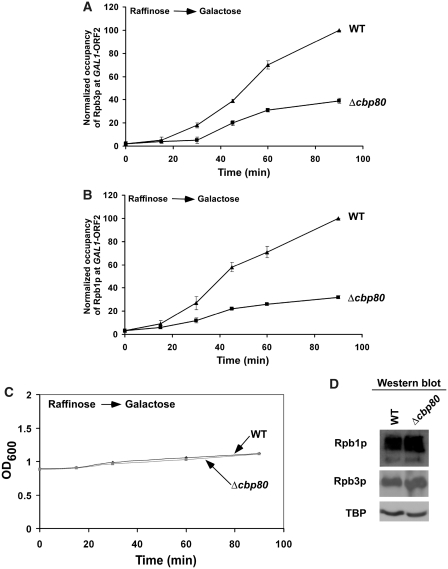
Our data (Figure 1) demonstrate that CBC is co-recruited with elongating RNAPII at the coding sequence of the active gene. Similarly, Gorneman et al. (27) show that the splicing factors are co-transcriptionally recruited to the active genes. Interestingly, co-transcriptionally recruited splicing factors have stimulatory effects on transcriptional elongation (70,71). We thus hypothesized that, like splicing factors, CBC might play an important role in transcriptional elongation, since it is co-transcriptionally recruited to the coding sequence (Figure 1A and E). To test this hypothesis, we analyzed the association of RNAPII with the coding sequence of the galactose-inducible GAL1 gene in the CBP80 deletion mutant and its isogenic wild-type equivalent, using a ChIP assay. Figure 2A shows that the association of the Rpb3p subunit of RNAPII with the GAL1 coding sequence in the wild-type yeast cells was gradually increased within 90 min upon switching the carbon source in the growth media from raffinose (non-inducing) to galactose (inducing). Interestingly, the association of Rpb3p with the GAL1 coding sequence was significantly reduced in Δcbp80 (Figure 2A). Similar reduction in the association of the Rpb1p subunit of RNAPII was also observed at the GAL1 coding sequence in Δcbp80 (Figure 2B). However, the reduced association of RNAPII with the GAL1 ORF in Δcbp80 could be due to slower growth of the mutant strain within 90 min of induction. To address this issue, we monitored the optical densities at 600 nm (OD600) of the mutant and wild-type yeast cells within 90 min of induction. Figure 2C shows that the OD600 of the mutant cells at different induction time points within 90 min was almost same as those of the wild-type cells. However, the mutant strain grew slowly as compared to the wild-type equivalent at later induction time points (Supplementary Figure S1), consistent with previous studies (26,42). We find that the Δcbp80 strain grew slowly after 90 min following the switch of the carbon source in the growth media from raffinose to galactose (Supplementary Figure S1A). Thus, the Δcbp80 strain grew slowly as compared to the wild-type equivalent in galactose-containing growth medium (Supplementary Figure S1B). However, the significant defect in the growth phenotype in Δcbp80 was not observed within 90 min following initial measurement of OD600 (Supplementary Figure S1B). Together, these growth curve analyses revealed that the Δcbp80 strain does not show significant defective growth phenotype within 90 min following induction. Hence, the reduced association of RNAPII with the GAL1 coding sequence in absence of CBC in Figure 2A and B is not attributed to the growth defect in the Δcbp80 strain within 90 min induction time-period. However, the reduced association of RNAPII with the GAL1 ORF in Δcbp80 might be due to decrease in the global levels of RNAPII components. To test this possibility, we analyzed the global levels of the Rpb1p and Rpb3p subunits of RNAPII in the Δcbp80 strain and its isogenic wild-type equivalent. Figure 2D shows that the global levels of Rpb1p and Rpb3p were not altered in Δcbp80. Similarly, the TBP level that was monitored as a loading control was not changed in the mutant strain (Figure 2D). Thus, the results presented in Figure 2 indicate the functional role of CBC in regulation of RNAPII association with the GAL1 coding sequence (and hence transcriptional elongation).
Figure 2.
The rate of association of RNAPII with the GAL1 coding sequence is significantly reduced in the absence of CBC. (A) Analysis of the rate of association of Rpb3p with the GAL1 coding sequence in Δcbp80 and its isogenic wild-type equivalent. The wild-type and CBP80 deletion mutant strains expressing myc epitope-tagged Rpb3p were first grown in raffinose-containing growth media (YPR) up to an OD600 of 0.9, and then shifted to galactose-containing growth media (YPG) for different time periods before treatment with formaldehyde. Immunoprecipitation was performed using a mouse monoclonal antibody against the c-myc epitope-tag. The specific primer pair targeted to the GAL1 ORF2 (Figure 1A) was used for PCR analysis of the immunoprecipitated DNA samples. The ratio of immunoprecipite over the input in the autoradiogram was measured. The maximum ratio was normalized to 100. The normalized ratio (represented as normalized occupancy) was plotted as a function of induction time. (B) Analysis of the rate of association of Rpb1p with the GAL1 coding sequence in Δcbp80 and its isogenic wild-type equivalent. The wild-type and deletion mutant strains were grown, cross-linked and immunoprecipitated as in panel A. (C) Growth analysis of the CBP80 wild-type and deletion mutant strains following induction in YPG. Both the wild-type and mutant strains were grown in raffinose-containing growth medium up to an OD600 of 0.9, and then switched to galactose-containing growth medium for 90 min. The OD600 of both the wild-type and mutant strains were measured in the galactose-containing growth medium within 90 min. (D) Western blot analysis. The wild-type and CBP80 deletion mutant strains were grown as in panel A, but induced for 60 min in YPG. The whole-cell extracts from both the wild-type and deletion mutant strains were prepared as in the ChIP assay (62–65). The whole-cell extracts were analyzed by western blot using anti-myc, anti-Rpb1 and anti-TBP antibodies against myc-tagged Rpb3p, Rpb1p and TBP, respectively.
CBC facilitates the rate of PIC formation at the GAL1 promoter
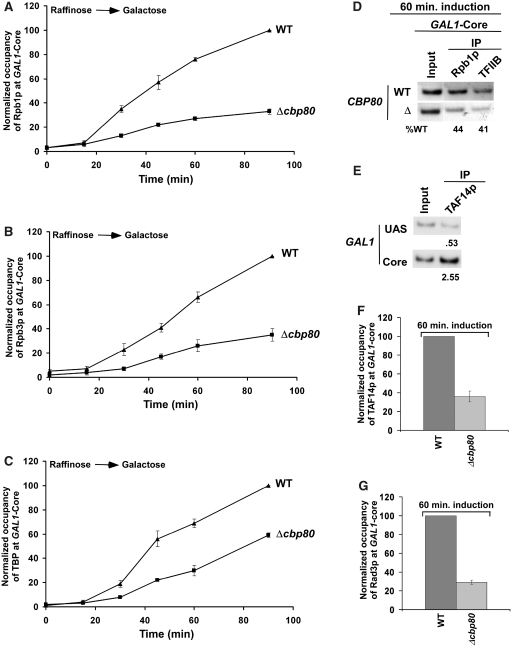
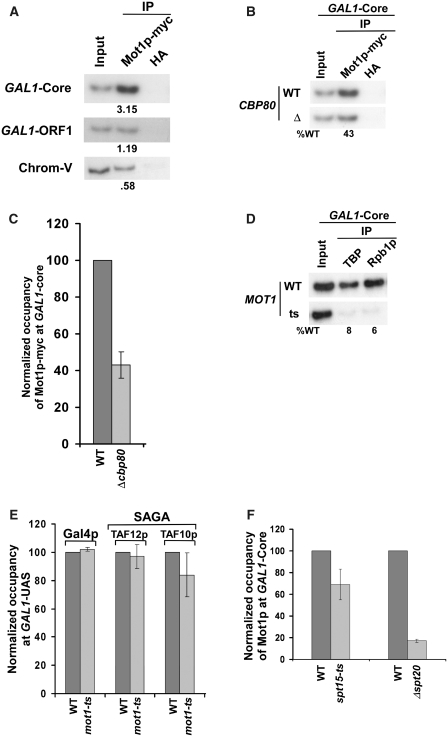
In Figure 2, we show that association of elongating RNAPII with GAL1 is significantly decreased in the absence of CBC. However, such an observation raised the possibility that the formation of PIC by GTFs (e.g. TBP, TFIIB, TFIIF, TFIIH, etc.) as well as RNAPII at the GAL1 core promoter might be decreased in the absence of CBC, hence lowering the association of RNAPII with the coding sequence. To test this possibility, we analyzed the rate of recruitment of RNAPII to the GAL1 core promoter following induction in the CBP80 deletion mutant and its isogenic wild-type equivalent. Interestingly, we find that the rate of recruitment of RNAPII (Rpb1p and Rpb3p) to the GAL1 core promoter was significantly reduced in Δcbp80 (Figure 3A and B). Similarly, CBC altered the rate of recruitment of TBP to the GAL1 core promoter (Figure 3C). Likewise, the recruitment of TFIIB, another component of the PIC assembly, was also reduced at the GAL1 core promoter in Δcbp80 following 60 min induction (Figure 3D). Next, we analyzed the recruitment of TAF14p component of TFIIF at the GAL1 core promoter in the CBP80 deletion mutant. TAF14p is also a component of the TFIID and SWI/SNF complexes. TFIID is recruited to the core promoter of the TAF-dependent genes (62,72) while the SWI/SNF complex is recruited to the UAS in an activator-dependent manner (73–75; see below Figure 4D). GAL1 is a TAF-independent gene (72), and thus, TAFs of the TFIID complex are not recruited to the GAL1 core promoter (62,72) while the TAF components of SAGA are recruited to the GAL1 UAS, but not core promoter (62). We find that TAF14p was predominantly recruited to the GAL1 core promoter as a component of TFIIF (Figure 3E). Further, we show that the recruitment of TAF14p to the GAL1 core promoter was significantly decreased in Δcbp80 (Figure 3F). Similarly, another component of the PIC assembly, TFIIH, was not efficiently recruited to the GAL1 core promoter in Δcbp80 (Figure 3G). We added the myc epitope at the C-terminal of Rad3p as a representative component of TFIIH that is recruited to the core promoter. Rad3p is predominantly recruited to the core promoter as a component of TFIIH (data not shown). However, Rad3p is also recruited to the site of DNA lesion as a component of nuclear excision repair machinery. Thus, the association of Rad3p with the GAL1 core promoter in the absence of DNA lesion is attributed to TFIIH. Taken together, our data demonstrate that CBC stimulates the PIC formation at the core promoter, and thus, the association of RNAPII with the coding sequence is significantly decreased in the absence of CBC.
Figure 3.
The stimulation of the PIC formation by CBC in vivo. (A) Analysis of the rate of recruitment of Rpb1p at the GAL1 core promoter in Δcbp80 and its isogenic wild-type equivalent. The wild-type and CBP80 deletion mutant strains were grown, cross-linked and immunoprecipitated as in Figure 2A and B. The specific primer pair targeted to the GAL1 core promoter was used for PCR analysis of the immunoprecipitated DNA samples. (B) Analysis of the rate of recruitment of Rpb3p at the GAL1 core promoter in Δcbp80 and its isogenic wild-type equivalent. (C) Analysis of the rate of recruitment of TBP at the GAL1 core promoter in Δcbp80 and its isogenic wild-type equivalent. Immunoprecipitations were performed using an anti-TBP antibody. (D) Analysis of recruitment of TFIIB at the GAL1 core promoter following 60 min induction in YPG. The wild-type and CBP80 deletion mutant strains were first grown in YPR up to an OD600 of 0.9 at 30°C, and then shifted to YPG for 60 min before treatment with formaldehyde. The mouse monoclonal antibody against TFIIB (obtained from Danny Reinberg, UMDNJ) was used. (E) TAF14p is predominantly recruited to the GAL1 core promoter as a component of TFIIF. Immunoprecipitation was carried out using a polyclonal antibody against TAF14p (obtained from Michael R. Green, UMass Medical School). (F) Recruitment of TAF14p (TFIIF) to the GAL1 core promoter is significantly decreased in the CBP80 deletion mutant strain. Both the wild-type and CBP80 deletion mutant strains were grown and cross-linked as in panel D. (G) Recruitment of Rad3p, a component of TFIIH, to the GAL1 core promoter is significantly decreased in the CBP80 deletion mutant strain. Both the wild-type and CBP80 deletion mutant strains were grown and cross-linked as in panel D.
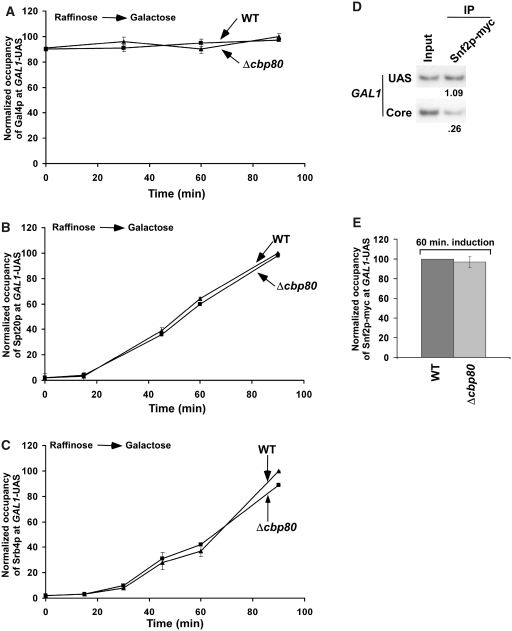
Figure 4.
CBC does not alter the recruitment of Gal4p, SAGA, Mediator and SWI/SNF to the GAL1 UAS. (A) Analysis of the rate of recruitment of Gal4p to the GAL1 UAS in Δcbp80 and its isogenic wild-type equivalent. The wild-type and CBP80 deletion mutant strains were grown, cross-linked and immunoprecipitated as in Figure 2A. A mouse monoclonal antibody against the DNA binding domain of Gal4p (RK5C1; Santa Cruz Biotechnology Inc.) was used. The specific primer-pair targeted to the GAL1 UAS was used for PCR analysis of the immunoprecipitated DNA samples. (B) Analysis of the rate of recruitment of SAGA (Spt20p-myc) to the GAL1 UAS in Δcbp80 and its isogenic wild-type equivalent. (C) Analysis of the rate of recruitment of Mediator (Srb4p-myc) to the GAL1 UAS in Δcbp80 and its isogenic wild-type equivalent. (D) Snf2p, a component of the SWI/SNF complex, is predominantly recruited to the GAL1 UAS. The yeast strain expressing myc epitope-tagged Snf2p was grown and cross-linked as in Figure 1A. Immunoprecipitation was performed using a modified ChIP protocol as described in the ‘Materials and Methods’ section. (E) The recruitment of Snf2p at the GAL1 UAS was not altered in the CBP80 deletion mutant strain. Both the wild-type and CBP80 deletion mutant strains expressing myc epitope-tagged Rad3p were grown and cross-linked as in Figure 3D.
CBC does not alter the recruitment of activator, coactivator or chromatin remodeling complex at the GAL1 UAS
The formation of PIC at the core promoter is regulated by activator and/or other factors (such as SAGA, Mediator and SWI/SNF) which are associated with the UAS (64,73–75). Thus, the rate of PIC formation at the GAL1 core promoter might be regulated by CBC through alteration of the recruitment of activator Gal4p, SAGA, Mediator or SWI/SNF at the UAS. To test this possibility, we analyzed the recruitment of Gal4p, SAGA, Mediator and SWI/SNF to the GAL1 UAS in the Δcbp80 strain and its isogenic wild-type equivalent following induction in galactose-containing growth medium. Figure 4A–C demonstrates that the rates of recruitment of Gal4p, SAGA (Spt20p) and Mediator (Srb4p) to GAL1 UAS were not altered in Δcbp80. In addition, the recruitment of the SWI/SNF complex (Snf2p) to the GAL1 UAS remained invariant in the absence of CBC (Figure 4D and E). Thus, CBC stimulates the PIC formation (and hence transcriptional initiation) without altering the recruitment of Gal4p, SAGA, Mediator or SWI/SNF at the active GAL1 gene in vivo. However, the slower rate of PIC formation at the GAL1 promoter in Δcbp80 could be due to alteration in the global levels of Gal3p and Gal80p which interact with the Gal4p activation domain, hence regulating the activity of Gal4p (76). If this occurs, the rate of recruitment of SAGA, Mediator or SWI/SNF would have decreased as the recruitment of these factors depends on the Gal4p activation domain (64,73–75). However, we rule out this possibility, since the recruitment of SAGA, Mediator or SWI/SNF to the Gal4p activation domain was not altered in Δcbp80 (Figure 4B, C and E).
Disassembly of PIC at GAL1 following Gal4p inactivation or turnover
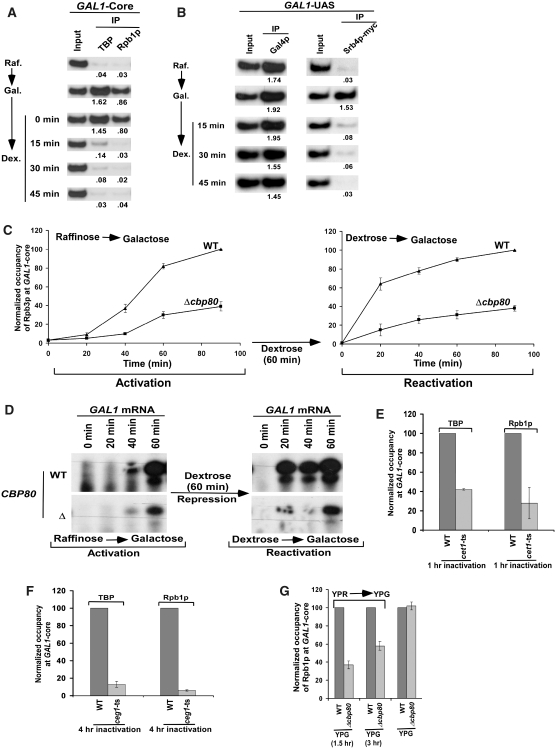
Previous in vitro studies (77,78) have demonstrated that GTFs are assembled at the promoter to form PIC for transcriptional initiation. Following initiation, TFIIB, TFIIF and RNAPII dissociate from PIC to trigger elongation, leaving behind a stable ‘scaffold’ complex consisting of TBP/TFIID, TFIIA, TFIIH, TFIIE and Mediator for reinitiation of transcription (78). Such a scaffold complex is stabilized in the presence of activator Gal4p(DBD)-VP16, but not Gal4p(DBD)-AH (78). However, according to this scaffold formation model in vitro (77,78), the rate of PIC formation at the GAL1 core promoter would not have altered in the subsequent rounds of transcription by CBC. To address this issue, we analyzed in vivo whether the stable scaffold is present at the GAL1 core promoter following inactivation of the Gal4p activator by the negative regulator, Gal80p. In this direction, we first monitored the recruitment of TBP and Rpb1p components of the PIC assembly at the GAL1 core promoter in raffinose-containing growth medium where Gal80p binds to the activation domain of Gal4p (64), hence preventing the PIC formation (Figure 5A). The yeast cells grown in raffinose-containing growth medium were then transferred to galactose-containing growth medium where Gal80p does not remain bound to the Gal4p activation domain (79,80), and hence PIC is assembled at the GAL1 core promoter (Figure 5A). Subsequently, these yeast cells were transferred back to dextrose (non-inducing) or raffinose-containing growth media. If the stable scaffold complex is formed at the core promoter, TBP would remain associated with the GAL1 promoter when Gal4p is inactivated by Gal80p following the transfer of the carbon source in the growth media from galactose to raffinose or dextrose. Interestingly, TBP was almost lost from the GAL1 promoter within 15 min after switching the carbon source from galactose to dextrose (Figure 5A) or raffinose (data not shown). Similar results were also obtained for another scaffold component, the Mediator complex (Srb4p), at the GAL1 promoter (Figure 5B). However, the recruitment of activator Gal4p was not altered within 45 min after switching the carbon source from galactose to dextrose (Figure 5B). Thus, the scaffold complex for transcriptional reinitiation does not seem to exist at the GAL1 core promoter when the activator is inactivated. Consistent with our in vivo data, Yudkovsky et al. (78) have demonstrated in vitro that the formation of the scaffold complex is stabilized by certain activation domain such as VP16. If the activator Gal4p stabilizes the scaffold complex for transcriptional reinitiation, how does CBC alter the rate of PIC formation at the GAL1 core promoter? The fact that CBC stimulates the rate of PIC formation raises the possibility that activator turnover leads to the formation of new PIC. Such a new PIC formation would be further stimulated by CBC that is bound to mRNA of the previous transcription cycle. In support of this possibility, activator or Gal4p turnover has been reported for transcriptional activation by several studies (81–87), indicating the recruitment of fresh activator or Gal4p in each cycle of transcription. Such recruitment of fresh activator in each transcription cycle will lead to the formation of new PIC, since the scaffold complex does not exist at the core promoter in the absence of activator. Thus, activator inactivation or turnover leads to the disassembly of PIC in vivo.
Figure 5.
The ChIP analysis for the recruitment of Gal4p, Mediator (Srb4p), TBP and RNAPII (Rpb1p) to the GAL1 promoter in the raffinose, galactose and dextrose-containing growth media. (A and B) Disassembly of PIC in the absence of active Gal4p. Yeast cells were first grown in raffinose-containing growth medium (Raf.) up to an OD600 of 0.8, and then transferred to galactose-containing growth medium (Gal.) for 90 min induction, and finally transferred to dextrose-containing growth medium (Dex.) for different time periods as mentioned in the left panel. The recruitment of TBP, RNAPII (Rpb1p), Mediator (Srb4p) and Gal4p was analyzed in raffinose, galactose and dextrose-containing growth media. The input DNA was diluted by 5-folds for the analysis of Mediator (Srb4p) recruitment. (C) The ChIP assay to analyze the recruitment of RNAPII (Rpb3p-myc) under the activated and reactivated conditions in Δcbp80 and its wild-type equivalent. The yeast cells were first grown in raffinose-containing growth medium, and then switched to galactose-containing growth medium for 90 min (activated state). These cells were then transferred to dextrose-containing growth medium for 60 min (repressed state), and finally switched to galactose-containing growth medium for 90 min (reactivated state). The recruitment of RNAPII was analyzed at the GAL1 core promoter under activated and reactivated conditions. (D) Primer extension analysis. Both the wild-type and CBP80 deletion mutant strains were grown under activated and reactivated conditions as in panel C. The total mRNA was isolated and analyzed by primer extension assay. (E) The recruitment of TBP and RNAPII to the GAL1 core promoter is significantly impaired in the cet1-ts mutant strain. (F) The recruitment of TBP and RNAPII to the GAL1 core promoter is significantly impaired in the ceg1-ts mutant strain. (G) The steady-state level of RNAPII at the core promoter is not altered in the absence of CBC. Analysis of recruitment of RNAPII to the GAL1 core promoter in the CBP80 wild-type and deletion mutant strains following a long period of induction in YPG. Both the wild-type and deletion mutant strains were grown as in Figure 2A, but induced for 1.5 and 3 h in YPG or continuously in YPG.
CBC regulates the PIC formation at GAL1 following mRNA capping
Like CBC, the capping machinery also regulates transcriptional initiation (88,89). For example, the Cet1p component of the capping machinery has been shown in vitro to repress the reinitiation of transcription before putting the cap-structure to newly synthesized mRNA, thus providing a quality control in the same cycle of transcription (88). Similarly, Schroeder et al. (89) have demonstrated by the ChIP assay that Abd1p, another component of the capping machinery, regulates promoter clearance in the same cycle of transcription. Thus, the capping machinery that is recruited to RNAPII when it is phosphorylated by Kin28p of TFIIH at the core promoter, regulates reinitiation, promoter clearance and mRNA cap formation (88,89). Unlike the capping machinery, CBC does not seem to regulate the promoter clearance or PIC formation in the first round of transcription. If CBC regulates the promoter clearance, the rate of RNAPII accumulation will increase at the core promoter, but will subsequently decrease at the 3′ end of the coding sequence in the absence of CBC. However, Figure 3A and B shows that the rate of RNAPII recruitment to the GAL1 core promoter was significantly decreased in Δcbp80. Thus, CBC does not regulate the promoter clearance. Further, CBC is less likely to enhance the PIC formation in the same cycle of transcription, since it binds to the cap structure that is formed by the capping machinery following transcriptional initiation. In support of this model, Raha et al. (90) have demonstrated that HIV-1 Tat which binds to the TAR element of the newly synthesized mRNA stimulates transcription complex assembly at the promoter in the subsequent rounds of transcriptional initiation, and hence enhances transcription in a positive feedback mechanism. Further, the recruitment of CBC (Cbp80p) is impaired (right panel; Figure 1B) when PIC is not assembled at the promoter in the absence of TBP [left panel; Figure 1B and previous study (62)]. This is expected, since mRNA synthesis and its capping are dependent on transcriptional initiation for recruitment of CBC. Intriguingly, CBC also facilitates the PIC formation (Figures 3 and 4). Thus, PIC and CBC are intimately coupled via their reciprocal synergism. This conclusion is further corroborated by the slower activation and re-activation of the GAL1 transcription in the absence of CBC as presented in Figure 5C and D. In the activation and re-activation experiments, we first activated the GAL1 gene by switching the carbon source in the growth medium from raffinose to galactose for 90 min, and then repressed in dextrose-containing growth medium for 60 min followed by re-activation in galactose-containing growth medium for 90 min in the CBP80 deletion mutant and its isogenic wild-type equivalent (Figure 5C), consistent with the protocol followed by Peterson and colleagues (91). Under these activating, repressing and re-activating conditions, we analyzed the recruitment of RNAPII (Rpb3p-myc) at the GAL1 core promoter in the absence and presence of CBC. The rate of RNAPII recruitment at the GAL1 core promoter in the wild-type strain is significantly faster in the reactivated state as compared to that of the activated state (Figure 5C). This is expected due to transcriptional memory, consistent with the studies of Peterson and colleagues (91). However, the rates of RNAPII recruitment to the GAL1 core promoter in the Δcbp80 strain were significantly slowed down in both activated and re-activated states as compared to those of the wild-type equivalent. Strikingly, the recruitment rates of RNAPII at the GAL1 core promoter in Δcbp80 in both the activated and reactivated states were almost equivalent. Similarly, we analyzed GAL1 mRNA in Δcbp80 and its wild-type strain under activating and re-activating conditions. The GAL1 mRNA levels were consistent with the patterns of RNAPII recruitment in the CBP80 deletion mutant and its isogenic wild-type equivalent following activation and re-activation (Figure 5D). Thus, CBC stimulates the PIC formation (and hence GAL1 transcription) in both activated and reactivated states.
Based on its known activity to recognize the cap-structure of mRNA, it is anticipated that CBC regulates the PIC formation at GAL1 via its binding to the capped-mRNA structure. However, it might be quite possible that CBC regulates the PIC formation at GAL1 prior to mRNA capping. To address this issue, we analyzed the recruitment of TBP and RNAPII to the GAL1 core promoter in the ts mutant strains of the capping enzymes, Cet1p and Ceg1p. If CBC regulates the PIC formation prior to mRNA capping, the formation of PIC at GAL1 would not be significantly decreased in the strains bearing the ts mutants of Cet1p and Ceg1p. However, we found that the recruitment of TBP and RNAPII to the GAL1 core promoter was significantly impaired in cet1-ts and ceg1-ts mutants following 1 and 4 h ts inactivation at the non-permissive temperature (37°C), respectively (Figure 5E and F). Thus, CBC regulates the PIC formation at GAL1 following mRNA capping.
CBC does not alter the steady-state level of PIC formation at GAL1
We next asked whether the occupancy of RNAPII at the GAL1 core promoter in Δcbp80 could reach the wild-type level if given enough time for attaining the steady-state. To address this question, we analyzed the recruitment of RNAPII to the GAL1 core promoter in the CBP80 wild-type and deletion mutant strains following longer periods of induction in galactose-containing growth medium. Figure 5G shows that the occupancy of RNAPII (Rpb1p) at the GAL1 core promoter in Δcbp80 reached to the wild-type level following a long induction. Thus, the loss of CBC results in a slower initial rate of recruitment of RNAPII to the GAL1 promoter. However, when the steady-state is reached after a long period of induction, RNAPII occupancy at the GAL1 promoter in Δcbp80 became similar to the wild-type level. Thus, CBC stimulates the rate of PIC formation (and hence transcriptional initiation). However, its effect is not observed when the steady-state of transcriptional initiation is reached. Analogous to this observation, we have also demonstrated previously that the loss of Ctk1p (that phosphorylates serine-2 at the C-terminal domain of the Rpb1p subunit of RNAPII) significantly slows down the rate of association of RNAPII with GAL1, and alters histone H3 lysine 4 methylation (92). However, when the steady-state is reached, the occupancy of RNAPII with GAL1 in Δctk1 reached to the wild-type level (92). Further, we find that Rad26p [a factor implicated in the stimulation of transcriptional elongation of GAL genes (93,94)] facilitates initial association of RNAPII with GAL1, but does not alter the steady-state level of RNAPII association with GAL1 after a long period of induction [(95) and unpublished data, the Bhaumik laboratory]. Thus, like the functions of Ctk1p and Rad26p in transcription, CBC does not appear to transiently stimulate the rate of PIC formation at GAL1, rather it persistently contributes to the rate of transcriptional initiation.
CBC stimulates the PIC formation at GAL1 via its interaction with Mot1p
To understand the stimulatory role of CBC in formation of PIC, we next searched for the transcriptional initiation factor(s) that interacts with CBC. Previous tandem-affinity purification, mass spectrometry and bioinformatic studies (96) have revealed an interaction of Cbp80p with Mot1p that is essential for transcriptional initiation of GAL1 via nucleosome remodeling and TBP recruitment (97). Mot1p has also been shown to be required for transcription of ∼7% of the yeast genes, and such a dependence is positively correlated with the recruitment of TBP (98–100). Thus, CBC seems to stimulate the PIC formation via its interaction with Mot1p that regulates the recruitment of TBP. To test this hypothesis, we first analyzed the interaction of CBC with Mot1p in vivo, using a formaldehyde-base cross-linking and co-immunoprecipitation assay (68). In this direction, we tagged the CBP20 and MOT1 genes by the HA and Myc epitopes, respectively, in their chromosomal loci. Using this strain, we performed the co-immunoprecipitation assay which revealed the interaction of Mot1p with CBC in vivo (Figure 6A). However, such an interaction was impaired in the CBP80 deletion mutant strain (Figure 6A), thus supporting the role of cap-binding on the interaction of CBC with Mot1p in vivo. Next, we analyzed their interaction in vitro. In this direction, we immunopurified Cbp20p and Cbp80p by HA epitope-tagging (Supplementary Figures S2–S4), and analyzed their interaction as a positive control. The experimental strategy for this interaction analysis is schematically shown in Figure 6B. We find that Cbp20p interacts with Cbp80p (Figure 6C). As a negative control, we show that Cbp20p does not interact with the Spt20p component of SAGA under similar experimental conditions (Figure 6C). Thus, the data presented in Figure 6C demonstrate that our assay to analyze protein–protein interaction in vitro is working. Therefore, using similar experimental conditions, we analyzed the interaction of Mot1p with Cbp20p and Cbp80p. We find that Mot1p interacts with Cbp20p (Figure 6C) as well as Cbp80p (Figure 6D). Thus, our in vitro experiments demonstrate that Mot1p interacts with CBC, consistent with in vivo results (Figure 6A). However, whether CBC interacts directly with Mot1p under physiological conditions remains to be elucidated. Nonetheless, our data support the interaction between CBC and Mot1p (either directly or at least indirectly), and such interaction is further complemented functionally as presented below (both Δcbp80 and mot1-ts strains show similar phenotypes with regard to the PIC formation).
Figure 6.
CBC interacts with Mot1p. (A) The formaldehyde-based in vivo cross-linking and co-immunoprecipitation assay as discussed in the ‘Materials and Methods’ section; WB, western blot. (B) The schematic outlines of the experimental strategy to analyze protein–protein interaction in vitro. (C) The analysis of interaction of Cbp20p with Cbp80p, Mot1p and Spt20p. (D) The analysis of interaction of Cbp80p with Mot1p.
Since CBC interacts with Mot1p, the recruitment of Mot1p to the GAL1 core promoter could be significantly decreased in the absence of Cbp80p. To test this hypothesis, we analyzed the recruitment of Mot1p to the GAL1 core promoter in Δcbp80. We find that the recruitment of Mot1p to the GAL1 core promoter is significantly decreased in the CBP80 deletion mutant strain as compared to the wild-type equivalent (Figure 7A–C). Further, we show that Mot1p is essential for recruitment of TBP and RNAPII (Rpb1p) to the GAL1 core promoter (Figure 7D). However, Mot1p is dispensable for recruitment of Gal4p and SAGA at the GAL1 UAS (Figure 7E), while SAGA that is dependent on activator for its recruitment to the UAS (62,101) is essential for recruitment of Mot1p (Figure 7F), consistent with previous studies (102). Further, the recruitment of Mot1p at the GAL1 core promoter does not appear to be significantly altered in the ts mutant strain of TBP (spt15-ts) following 1 h inactivation at 37°C (Figure 7F) Thus, activator or SAGA stimulates the recruitment of Mot1p which subsequently enhances the recruitment of TBP, and hence, promotes the formation of PIC at the GAL1 core promoter. However, in the subsequent rounds of transcriptional initiation, the recruitment of Mot1p is stimulated by both activator as well as CBC bound to the cap-structure of mRNA of previous transcriptional initiation cycle. Such a stimulation of Mot1p recruitment enhances the PIC formation (and hence transcriptional initiation) at the GAL1 core promoter. Thus, a modest decrease in the association of Mot1p with the GAL1 core promoter is observed in the ts mutant strain of TBP (spt15-ts) following 1 h inactivation at 37°C (Figure 7F), since the spt15-ts mutant strain lowers recruitment of CBC (via reduced formation of PIC) [Figure 1B, (62)] which, in turn, reduces the recruitment of Mot1p. Alternatively, the modest decrease in the recruitment of Mot1p in the spt15-ts mutant strain could also be due to the loss of reciprocal co-operativity, since Mot1p is essential for recruitment of TBP at the GAL1 core promoter (Figure 7D).
Figure 7.
CBC stimulates the recruitment of Mot1p to the core promoter of GAL1. (A) Mot1p is predominantly recruited to the GAL1 core promoter. The yeast strain expressing myc epitope-tagged Mot1p was grown and cross-linked as in Figure 1A. Immunoprecipitation was performed following the modified ChIP protocol as described in ‘Materials and Methods’ section. The anti-HA antibody was used as a non-specific antibody. (B) CBC promotes Mot1p recruitment to the GAL1 core promoter. Both the wild-type and CBP80 deletion mutant strains carrying myc-tagged Mot1p were grown in raffinose-containing growth medium up to OD600 of 0.9, and then shifted to galactose-containing growth medium for 90 min prior to formaldehyde-based in vivo cross-linking. Immunoprecipitation was performed as in panel A. (C) The results at the GAL1 core promoter in panel B were presented in the form of a histogram. (D) Mot1p is essential for recruitment of TBP and RNAPII. Both the wild-type and ts mutant strains were grown in galactose-containing growth medium up to OD600 of 0.85 at 23°C, and then switched to 37°C for 1 h prior to cross-linking. Immunoprecipitations were performed as in Figures 2B and 3C. (E) Mot1p is not essential for recruitment of Gal4p and SAGA to the GAL1 UAS. The MOT1 wild-type and ts mutant strains were grown and cross-linked as in panel D. Immunoprecipitation was performed using the polyclonal antibodies against TAF10p and TAF12p (obtained from Michael R. Green, UMass Medical School). (F) Analysis of the roles of SAGA and TBP for recruitment of Mot1p to the GAL1 core promoter. The wild-type and ts mutant strain of TBP (spt15-ts) were grown and cross-linked as in panel D. The SPT20 deletion mutant and its isogenic wild-type strains were grown and cross-linked as in panel B. Immunoprecipitation was performed as in panel A.
Regulation of the PIC formation at other Mot1p-dependent genes by CBC
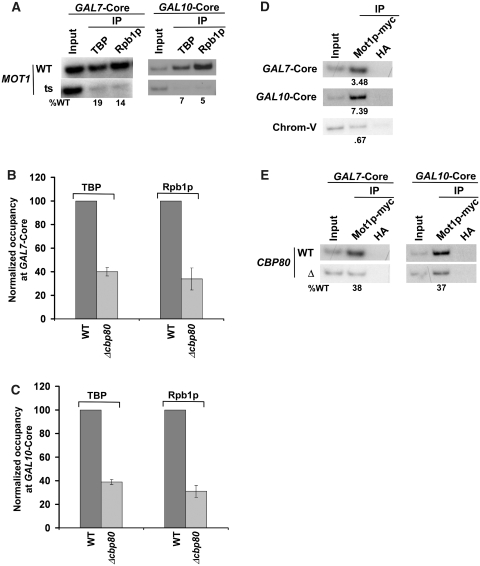
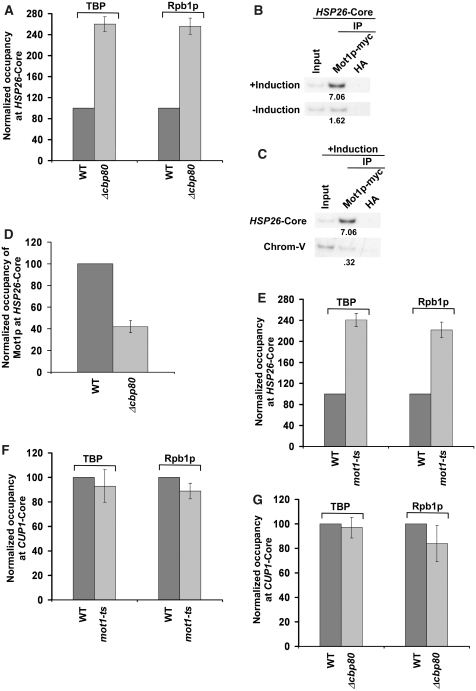
Mot1p promotes the transcription of a set of yeast genes including GAL1. We have demonstrated above that CBC stimulates the formation of the PIC at the GAL1 gene via its interaction with Mot1p. Next, we asked whether the formation of PIC at other genes that are positively regulated by Mot1p is also stimulated by CBC. In this direction, we first analyzed the formation of PIC at the GAL7 and GAL10 genes in the MOT1 wild-type and ts mutant strains. We find that Mot1p is essential for the PIC formation at the core promoters of GAL7 and GAL10 (Figure 8A). Next, we analyzed the PIC formation at the core promoters of these genes in the presence and absence of CBC. Our analysis revealed that the PIC formation at GAL7 and GAL10 was significantly impaired in the absence of CBC (Figure 8B and C). Further, we show that the recruitment of Mot1p to the GAL7 and GAL10 core promoters was significantly decreased in the absence of CBC (Figure 8D and E). Together, these results demonstrate that the absence of CBC lowers the recruitment of Mot1p by ∼2.5-fold which subsequently decreases the PIC formation by ∼2.5-folds (as Mot1p facilitates the PIC formation at these promoters). Thus, like that at GAL1, CBC promotes the PIC formation at GAL7 and GAL10. Similarly, CBC stimulates the PIC formation at the promoter of another gene, GAL2 (Supplementary Figure S5A) that requires Mot1p for facilitation of the PIC assembly (Supplementary Figure S5B). Based on the above results, one would expect the repressive role of CBC in formation of the PIC at the core promoter of a gene that is negatively regulated by Mot1p for the PIC formation. To test this possibility, we next analyzed the role of CBC in formation of the PIC at the core promoter of HSP26. The HSP26 gene was chosen on the basis of the previous studies (103) that demonstrated the role of Mot1p in repression of the HSP26 transcription. We find that CBC represses the PIC formation at HSP26 (Figure 9A), although the recruitment of Mot1p to the HSP26 core promoter is significantly decreased in the absence of CBC (Figure 9B–D). Further, we show that the formation of the PIC at HSP26 is enhanced in the mot1-ts mutant strain at the non-permissive temperature (Figure 9E). Thus, CBC exhibits its repressive role in formation of PIC at the gene that is negatively regulated by Mot1p. Likewise, CBC represses the PIC formation at the core promoters of other genes such as SSA3 and SSA4 (Supplementary Figure S6A and S6B) that are negatively regulated by Mot1p for the PIC assembly (Supplementary Figure S6C and S6D). As a control, we demonstrate that CBC does not alter the PIC formation at the Mot1p-independent gene, CUP1 (Figure 9F and G). Thus, CBC differentially controls the formation of PIC. Such a differential regulation is mediated via the interaction of CBC with Mot1p which controls Mot1p recruitment and subsequently the formation of PIC in vivo.
Figure 8.
CBC stimulates the PIC formation at the core promoters of the GAL7 and GAL10 genes. (A) Mot1p is essential for recruitment of TBP and RNA polymerase II at the GAL7 and GAL10 core promoters. Yeast cells were grown, cross-linked and immunoprecipitated as in Figure 7D. (B and C) Recruitment of TBP and RNA polymerase II at the GAL7 and GAL10 core promoters is significantly decreased in the CBP80 deletion mutant strain. Both the wild-type and CBP80 deletion mutant strains were grown, cross-linked and immunoprecipitated as in Figures 2A and B and 3C. (D) Mot1p is recruited to the GAL7 and GAL10 core promoters. Yeast cells were grown, cross-linked and immunoprecipitated as in Figure 7A. (E) The recruitment of Mot1p to the GAL7 and GAL10 core promoters is significantly decreased in the CBP80 deletion mutant strain. Both the wild-type and CBP80 deletion mutant strains were grown, cross-linked and immunoprecipitated as in Figure 7B.
Figure 9.
CBC represses the PIC formation at the core promoter of HSP26, but does not alter the PIC formation at the core promoter of CUP1. (A) CBC represses the PIC formation at the HSP26 core promoter. Immunoprecipitation was performed as in Figures 2B and 3C. (B and C) Mot1p is recruited to the HSP26 core promoter under inducible conditions. Immunoprecipitation was performed as in Figure 7A. (D) The recruitment of Mot1p at the HSP26 core promoter is significantly decreased in the CBP80 deletion mutant strain. (E) Mot1p represses the PIC formation at the HSP26 core promoter. Both the wild-type and ts mutant strains of MOT1 were grown in YPD at 23°C to 0.9, and then switched to 39°C for 30 min prior to cross-linking. (F) Mot1p does not alter the recruitment of TBP and RNA polymerase II at the CUP1 core promoter. Yeast cells were grown in synthetic complete medium at 23°C up to OD600 of 0.85, and then switched to 37°C for 45 min followed by 15 min induction using 1 mM CuSO4 before cross-linking. (G) CBC does not alter the recruitment of TBP and RNA polymerase II at the CUP1 core promoter.
CBC does not appear to regulate the PIC formation by indirect effects
We find that the formation of PIC at GAL1, GAL7, GAL10 and GAL2 is significantly decreased in the absence of CBC as presented above. Such a reduction in the PIC formation might be caused by indirect effects such as lower expression levels of the PIC components. According to this possibility, the reduced association of the PIC components would be observed at all promoters. However, we find the enhanced association of the PIC components at HSP26, SSA3 and SSA4, reduced PIC assembly at GAL1, GAL7, GAL10 and GAL2, and no alteration of the PIC formation at CUP1 in the absence of CBC (Figures 3, 8B and C, 9A and G; Supplementary Figures S5A, S6A and S6B). Further, we show that such a differential regulation of the PIC formation is mediated via Mot1p as discussed above. Thus, the regulation of PIC formation by CBC does not appear to be a result of altered expression levels of the PIC components. In support of this conclusion, we also show that the global levels of the PIC components such as TBP, Rpb1p and Rpb3p were not changed in the absence of CBC (Figure 2D). However, the global level of a protein may not be directly correlated with its targeted recruitment. For example, TBP is not recruited to the GAL1 core promoter in the absence of Gal4p while the global level of TBP is not altered in Δgal4 (62). Similarly, Tra1p is not recruited to the GAL1 UAS in the absence of Spt20p while its global level is not altered in Δspt20 (64).
It is quite likely that CBC regulates the PIC formation indirectly by altering the expression levels of other transcription factors that act early in the process of PIC formation. According to this model, the decreased association of activator or activator-target with the promoter would be observed in the absence of CBC. However, we do not find the altered recruitment of Gal4p, SAGA, Mediator and SWI/SNF to the GAL1 UAS in the absence of CBC (Figure 4). Rather, we specifically observed the decreased association of the PIC components with the GAL1 core promoter (Figure 3). We also find that CBC differentially regulates the PIC formation at the promoters as discussed above. These observations further support the fact that the role of CBC in formation of the PIC is not mediated by indirect effects such as altered expression levels of the PIC components or other transcription factors. Rather, it is mediated by a specific pathway (i.e. CBC-Mot1p interaction) as discussed above.
CONCLUSION
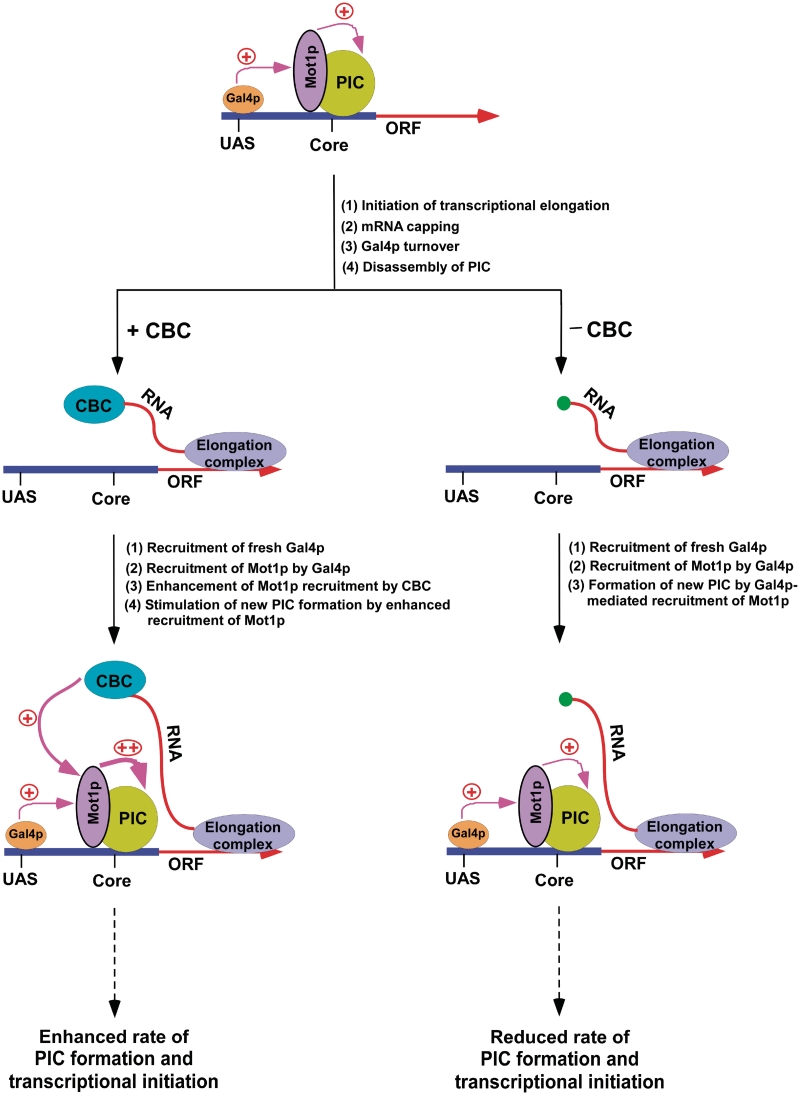
Here, we show that CBC facilitates the formation of PIC (and hence transcriptional initiation) at GAL1 via Mot1p following mRNA capping as schematically shown in Figure 10. Mot1p is recruited to the GAL1 core promoter. Such recruitment is dependent on Gal4p. Subsequent to its recruitment, Mot1p enhances the recruitment of TBP to the GAL1 core promoter, and consequently, promotes the PIC formation, hence leading to transcriptional initiation. Intriguingly, Gal4p-mediated recruitment of Mot1p to the GAL1 core promoter appears to be significantly enhanced in the subsequent rounds of transcriptional initiation by CBC that is bound to the cap-structure of mRNA of previous round of transcription. Such a stimulation of Mot1p recruitment by activator and CBC in the subsequent rounds of transcriptional initiation facilitates the PIC formation at the GAL1 core promoter, hence enhancing GAL1 transcription. However, Mot1p is recruited only by activator in the absence of CBC in the subsequent rounds of transcriptional initiation. Thus, the stimulation of Mot1p recruitment to the GAL1 core promoter is lost when CBC is not bound to the cap-structure of mRNA, lowering the efficiency of the PIC formation at the core promoter (and hence GAL1 transcription).
Figure 10.
A schematic model for the stimulatory role of CBC in formation of PIC (and hence transcriptional initiation) at the Gal4p-regulated genes. Gal4p is required for recruitment of Mot1p which is essential for the PIC formation at the Gal4p-regulated genes. Following PIC formation, mRNA is synthesized, and then CBC binds to the cap structure at the 5′-end of newly synthesized mRNA. The turnover of Gal4p destabilizes PIC. The fresh Gal4p is recruited to form new PIC in the subsequent round of transcription. CBC appears to promote the recruitment of Mot1p to the core promoter through its interaction with Mot1p in the subsequent rounds of transcription. Such stimulation of Mot1p recruitment by Gal4 as well as CBC enhances the rate of the PIC formation (and hence transcriptional initiation) in the subsequent rounds of transcription.
Although CBC is not recruited to the core promoter, it promotes the association of Mot1p with the core promoter. Similarly, we and others (62,101) have previously shown that SAGA which is recruited to the GAL1 UAS stimulates the PIC formation at the core promoter. Even though SAGA is not recruited to the GAL1 core promoter (62), it stimulates the PIC formation through its interaction with TBP (62,101,104). Likewise, CBC is not recruited to the core promoter, but it facilitates the formation of PIC through its interaction with Mot1p as schematically shown in Figure 10. Mot1p is recruited to the core promoter by activator. The CBC that is bound to mRNA interacts with Mot1p at the promoter. Such interaction stimulates the recruitment of Mot1p to the core promoter. Analogous to our observations, previous studies have also implicated multiple transcription factors/activators/enhanceosomes in stimulation of the transcription complex assembly at the promoter (and hence transcriptional activation/synergy) through multiple contacts (105–115).
How does CBC that is bound to the cap-structure of mRNA interact with Mot1p at the core promoter? Since mRNA is flexible, it can easily bend or loop to allow the interaction of CBC with Mot1p. Similarly, CBC has also been implicated to regulate mRNA export through its long-range interaction with Npl3p, an mRNA export factor (42). Likewise, CBC regulates RNA splicing and 3′-end formation (18,20,24–36,38). Similar long-range interactions among different proteins associated with the chromosome have also been demonstrated by chromosome conformation capture technique (116–118).
Mot1p has a positive effect on transcription of 7% yeast genes including GAL1 (97–100). Thus, it is likely that CBC might be facilitating the rate of PIC formation through its interaction with Mot1p at other genes that are positively regulated by Mot1p. Indeed, we show here that CBC stimulates the PIC formation at the core promoters of several other genes (such as GAL7, GAL10 and GAL2) that are positively regulated by Mot1p. Further, Mot1p has also been implicated in the repression of a subset of genes. Thus, based on our results, it is also anticipated that CBC might be playing an inhibitory role in formation of the PIC at the genes that are negatively regulated by Mot1p. In fact, our data demonstrate that CBC represses the PIC formation at the core promoter of a gene, HSP26 that is negatively regulated by Mot1p. Likewise, the PIC formation at other genes such as SSA3 and SSA4 is repressed by CBC, and these genes are negatively regulated by Mot1p. Further, we demonstrate that CBC does not alter the PIC formation at the core promoter of a Mot1p-independent gene, CUP1. Thus, CBC appears to interact with Mot1p in general in vivo, and subsequently, exhibits different phenotypes with regard to the PIC formation as dictated by Mot1p. Collectively, our results support that CBC differentially controls the PIC formation (and hence transcriptional initiation) via Mot1p in vivo, hence providing a novel regulatory mechanism of gene activation by CBC.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health grant (1R15GM088798-01); National Scientist Development Grant (0635008N) from American Heart Association; a Research Scholar Grant (06-52) from American Cancer Society; a Mallinckrodt Foundation award and several internal grants from Southern Illinois University. A.S. was supported by a predoctoral fellowship (0710187Z) from American Heart Association. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Michael R. Green and Danny Reinberg for antibodies; Stephen Buratowski, Kevin Struhl, Fred Winston and Martin Collart for yeast strains; Maheshi Udugama for technical assistance.
REFERENCES
- 1.Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988;335:683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- 2.Ptashne M, Gann AA. Activators and targets. Nature. 1990;346:329–331. doi: 10.1038/346329a0. [DOI] [PubMed] [Google Scholar]
- 3.Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 4.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 5.Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- 6.Hassan AH, Neely KE, Vignali M, Reese JC, Workman JL. Promoter targeting of chromatin-modifying complexes. Front Biosci. 2001;6:D1054–D1064. doi: 10.2741/hassan. [DOI] [PubMed] [Google Scholar]
- 7.Isogai Y, Tjian R. Targeting genes and transcription factors to segregated nuclear compartments. Curr. Opin. Cell Biol. 2003;15:296–303. doi: 10.1016/s0955-0674(03)00052-8. [DOI] [PubMed] [Google Scholar]
- 8.Green MR. Eukaryotic transcription activation: right on target. Mol. Cell. 2005;18:399–402. doi: 10.1016/j.molcel.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 9.Struhl K. Transcriptional activation: mediator can act after preinitiation complex formation. Mol. Cell. 2005;17:752–754. doi: 10.1016/j.molcel.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Venters BJ, Pugh BF. How eukaryotic genes are transcribed. Crit. Rev. Biochem. Mol. Biol. 2009;44:117–1141. doi: 10.1080/10409230902858785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sikorski TW, Buratowski S. The basal initiation machinery: beyond the general transcription factors. Curr. Opin. Cell Biol. 2009;21:344–351. doi: 10.1016/j.ceb.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazza C, Ohno M, Segref A, Mattaj IW, Cusack S. Crystal structure of the human nuclear cap binding complex. Mol. Cell. 2001;8:383–396. doi: 10.1016/s1097-2765(01)00299-4. [DOI] [PubMed] [Google Scholar]
- 13.Mazza C, Segref A, Mattaj IW, Cusack S. Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 2002;21:5548–5557. doi: 10.1093/emboj/cdf538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazza C, Segref A, Mattaj IW, Cusack S. Co-crystallization of the human nuclear cap-binding complex with a m7GpppG cap analogue using protein engineering. Acta Cryst. D. 2002;58:2194–2197. doi: 10.1107/s0907444902015445. [DOI] [PubMed] [Google Scholar]
- 15.Calero G, Wilson KF, Ly T, Rios-Steiner JL, Clardy JC, Cerione RA. Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nat. Struct. Biol. 2002;9:912–917. doi: 10.1038/nsb874. [DOI] [PubMed] [Google Scholar]
- 16.Worch R, Niedzwiecka A, Stepinski J, Mazza C, Jankowska-Anyszka M, Darzynkiewicz E, Cusack S, Stolarski R. Specificity of recognition of mRNA 5′ cap by human nuclear cap-binding complex. RNA. 2005;11:1355–1363. doi: 10.1261/rna.2850705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kierzkowski D, Kmieciak M, Piontek P, Wojtaszek P, Szweykowska-Kulinska Z, Jarmolowski A. The Arabidopsis CBP20 targets the cap-binding complex to the nucleus, and is stabilized by CBP80. Plant J. 2009;59:814–825. doi: 10.1111/j.1365-313X.2009.03915.x. [DOI] [PubMed] [Google Scholar]
- 18.Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj IW. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 19.Izaurralde E, Mattaj IW. RNA export. Cell. 1995;81:153–159. doi: 10.1016/0092-8674(95)90323-2. [DOI] [PubMed] [Google Scholar]
- 20.Colot H, Stutz F, Rosbash M. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev. 1996;10:1699–1708. doi: 10.1101/gad.10.13.1699. [DOI] [PubMed] [Google Scholar]
- 21.Visa N, Izaurralde E, Ferreira J, Daneholt B, Mattaj IW. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J. Cell Biol. 1996;133:5–14. doi: 10.1083/jcb.133.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S, Yang JY, Xu J, Jang IC, Prigge MJ, Chua NH. Two cap-binding proteins CBP20 and CBP80 are involved in processing primary MicroRNAs. Plant Cell Physiol. 2008;49:1634–1644. doi: 10.1093/pcp/pcn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geraldo N, Bäurle I, Kidou S, Hu X, Dean C. FRIGIDA delays flowering in Arabidopsis via a cotranscriptional mechanism involving direct interaction with the nuclear cap-binding complex. Plant Physiol. 2009;150:1611–1618. doi: 10.1104/pp.109.137448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis JD, Gorlich D, Mattaj IW. A yeast cap binding protein complex (yCBC) acts at an early step in pre-mRNA splicing. Nucleic Acids Res. 1996;24:3332–3336. doi: 10.1093/nar/24.17.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis JD, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj IW. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5' splice site. Genes Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- 26.Fortes P, Kufel J, Fornerod M, Polycarpou-Schwarz M, Lafontaine D, Tollervey D, Mattaj IW. Genetic and physical interactions involving the yeast nuclear cap-binding complex. Mol. Cell Biol. 1999;19:6543–6553. doi: 10.1128/mcb.19.10.6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gornemann J, Kotovic KM, Hujer K, Neugebauer KM. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol. Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 28.Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Rätsch G, Weigel D. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2008;105:8795–8800. doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raczynska KD, Simpson CG, Ciesiolka A, Szewc L, Lewandowska D, McNicol J, Szweykowska-Kulinska Z, Brown JW, Jarmolowski A. Involvement of the nuclear cap-binding protein complex in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 2010;38:265–267. doi: 10.1093/nar/gkp869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hart RP, McDevitt MA, Nevins JR. Poly(A) site cleavage in a HeLa nuclear extract is dependent on downstream sequences. Cell. 1985;43:677–683. doi: 10.1016/0092-8674(85)90240-5. [DOI] [PubMed] [Google Scholar]
- 31.Gilmartin GM, McDevitt MA, Nevins JR. Multiple factors are required for specific RNA cleavage at a poly(A) addition site. Genes Dev. 1988;2:578–587. doi: 10.1101/gad.2.5.578. [DOI] [PubMed] [Google Scholar]
- 32.Cooke C, Alwine JC. The cap and the 3′ splice site similarly affect polyadenylation efficiency. Mol. Cell. Biol. 1996;16:2579–2584. doi: 10.1128/mcb.16.6.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flaherty SM, Fortes P, Izaurralde E, Mattaj IW, Gilmartin GM. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc. Natl Acad. Sci. USA. 1997;94:11893–11898. doi: 10.1073/pnas.94.22.11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shatkin AJ, Manley JL. The ends of the affair: capping and polyadenylation. Nat. Struct. Biol. 2000;7:838–842. doi: 10.1038/79583. [DOI] [PubMed] [Google Scholar]
- 35.Balatsos NA, Nilsson P, Mazza C, Cusack S, Virtanen A. Inhibition of mRNA deadenylation by the nuclear cap binding complex (CBC) J. Biol. Chem. 2006;281:4517–4522. doi: 10.1074/jbc.M508590200. [DOI] [PubMed] [Google Scholar]
- 36.Nilsson P, Henriksson N, Niedzwiecka A, Balatsos NA, Kokkoris K, Eriksson J, Virtanen A. A multifunctional RNA recognition motif in poly(A)-specific ribonuclease with cap and poly(A) binding properties. J. Biol. Chem. 2007;282:32902–32911. doi: 10.1074/jbc.M702375200. [DOI] [PubMed] [Google Scholar]
- 37.Schwer B, Mao X, Shuman S. Accelerated mRNA decay in conditional mutants of yeast mRNA capping enzyme. Nucleic Acids Res. 1998;26:2050–2057. doi: 10.1093/nar/26.9.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong CM, Qiu H, Hu C, Dong J, Hinnebusch AG. Yeast cap binding complex impedes recruitment of cleavage factor IA to weak termination sites. Mol. Cell Biol. 2007;27:6520–6531. doi: 10.1128/MCB.00733-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj IW. A cap-binding protein complex mediating U snRNA export. Nature. 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- 40.Gorlich D, Kraft R, Kostka S, Vogel F, Hartmann E, Laskey RA, Mattaj IW, Izaurraide E. Importin provides a link between nuclear protein import and U snRNA export. Cell. 1996;87:21−32. doi: 10.1016/s0092-8674(00)81319-7. [DOI] [PubMed] [Google Scholar]
- 41.Hamm J, Mattaj IW. Monomethylated cap structures facilitate RNA export from the nucleus. Cell. 1990;63:109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- 42.Shen EC, Stage-Zimmermann T, Chui P, Silver PA. The yeast mRNA-binding protein Npl3p interacts with the cap-binding complex. J. Biol. Chem. 2000;275:23718–23724. doi: 10.1074/jbc.M002312200. [DOI] [PubMed] [Google Scholar]
- 43.Nojima T, Hirose T, Kimura H, Hagiwara M. The interaction between cap-binding complex and RNA export factor is required for intronless mRNA export. J. Biol. Chem. 2007;282:15645–15651. doi: 10.1074/jbc.M700629200. [DOI] [PubMed] [Google Scholar]
- 44.Narita T, Yung TM, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, Yamaguchi Y, Handa H. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol. Cell. 2007;26:349–365. doi: 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 45.Kuhn JM, Hugouvieux V, Schroeder JI. mRNA cap binding proteins: effects on abscisic acid signal transduction, mRNA processing, and microarray analyses. Curr. Top Microbiol. Immunol. 2008;326:139–150. doi: 10.1007/978-3-540-76776-3_8. [DOI] [PubMed] [Google Scholar]
- 46.Ishigaki Y, Li X, Sherin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 47.McKendrick L, Thompson E, Ferreira J, Morley SJ, Lewis JD. Interaction of eukaryotic translation initiation factor 4G with the nuclear cap-binding complex provides a link between nuclear and cytoplasmic functions of the m(7) guanosine cap. Mol. Cell. Biol. 2001;21:3632–3641. doi: 10.1128/MCB.21.11.3632-3641.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hosoda N, Kim YK, Lejeune F, Maquat LE. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol. 2005;12:893–901. doi: 10.1038/nsmb995. [DOI] [PubMed] [Google Scholar]
- 49.Svitkin YV, Herdy B, Costa-Mattioli M, Gingras AC, Raught B, Sonenberg N. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol. Cell. Biol. 2005;25:10556–10565. doi: 10.1128/MCB.25.23.10556-10565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell. Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 51.Pierrat OA, Mikitova V, Bush MS, Browning KS, Doonan JH. Control of protein translation by phosphorylation of the mRNA 5'-cap-binding complex. Biochem. Soc. Trans. 2007;35:1634–1637. doi: 10.1042/BST0351634. [DOI] [PubMed] [Google Scholar]
- 52.Hossain MA, Claggett JM, Nguyen T, Johnson TL. The cap binding complex influences H2B ubiquitination by facilitating splicing of the SUS1 pre-mRNA. RNA. 2009;15:1515–1527. doi: 10.1261/rna.1540409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gruber JJ, Zatechka DS, Sabin LR, Yong J, Lum JJ, Kong M, Zong WX, Zhang Z, Lau CK, Rawlings J, et al. Ars2 links the nuclear cap-binding complex to RNA interference and cell proliferation. Cell. 2009;138:328–339. doi: 10.1016/j.cell.2009.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sabin LR, Zhou R, Gruber JJ, Lukinova N, Bambina S, Berman A, Lau CK, Thompson CB, Cherry S. Ars2 regulates both miRNA- and siRNA- dependent silencing and suppresses RNA virus infection in Drosophila. Cell. 2009;138:340–351. doi: 10.1016/j.cell.2009.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nielsen AF, Gloggnitzer J, Martinez J. Ars2 and the cap-binding complex team up for silencing. Cell. 2009;138:224–226. doi: 10.1016/j.cell.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 56.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pingle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 57.Fresco LD, Buratowski S. Conditional mutants of the yeast mRNA capping enzyme show that the cap enhances, but is not required for, mRNA splicing. RNA. 1996;2:584–596. [PMC free article] [PubMed] [Google Scholar]
- 58.Takase Y, Takagi T, Komarnitsky PB, Buratowski S. The essential interaction between yeast mRNA capping enzyme subunits is not required for triphosphatase function in vivo. Mol. Cell. Biol. 2000;20:9307–9316. doi: 10.1128/mcb.20.24.9307-9316.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roberts SM, Winston F. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:3206–3213. doi: 10.1128/mcb.16.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roberts SM, Winston F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 62.Bhaumik SR, Green MR. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 2001;15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bhaumik SR, Green MR. Interaction of Gal4p with components of transcription machinery in vivo. Methods Enzymol. 2003;370:445–454. doi: 10.1016/S0076-6879(03)70038-X. [DOI] [PubMed] [Google Scholar]
- 64.Bhaumik SR, Raha T, Aiello DP, Green MR. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 2004;18:333–343. doi: 10.1101/gad.1148404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shukla A, Stanojevic N, Duan Z, Sen P, Bhaumik SR. Ubp8p, a histone deubiquitinase whose association with SAGA is mediated by Sgf11p, differentially regulates lysine 4 methylation of histone H3 in vivo. Mol. Cell. Biol. 2006;26:3339–3352. doi: 10.1128/MCB.26.9.3339-3352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bhaumik SR, Green MR. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 2002;22:7365–7371. doi: 10.1128/MCB.22.21.7365-7371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peterson CL, Kruger W, Herskowitz I. A functional interaction between the C-terminal domain of RNA polymerase II and the negative regulator SIN1. Cell. 1991;64:1135–1143. doi: 10.1016/0092-8674(91)90268-4. [DOI] [PubMed] [Google Scholar]
- 68.Hall DB, Struhl K. The VP16 activation domain interacts with multiple transcriptional components as determined by protein-protein cross-linking in vivo. J. Biol. Chem. 2002;277:46043–46050. doi: 10.1074/jbc.M208911200. [DOI] [PubMed] [Google Scholar]
- 69.Shukla A, Stanojevic N, Duan Z, Shadle T, Bhaumik SR. Functional analysis of H2B-Lys-123 ubiquitination in regulation of H3-Lys-4 methylation and recruitment of RNA polymerase II at the coding sequences of several active genes in vivo. J. Biol. Chem. 2006;281:19045–19054. doi: 10.1074/jbc.M513533200. [DOI] [PubMed] [Google Scholar]
- 70.Fong YW, Zhou Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001;414:929–933. doi: 10.1038/414929a. [DOI] [PubMed] [Google Scholar]
- 71.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu X-D. The splicing factor SC35 has an active role in transcriptional elongation. Nat. Struct. Mol. Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li X-Y, Bhaumik SR, Green MR. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science. 2000;288:1242–1244. doi: 10.1126/science.288.5469.1242. [DOI] [PubMed] [Google Scholar]
- 73.Yudkovsky N, Logie C, Hahn S, Peterson CL. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 1999;13:2369–2374. doi: 10.1101/gad.13.18.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peterson CL, Workman JL. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 75.Peterson CL, Logie C. Recruitment of chromatin remodeling machines. J. Cell. Biochem. 2000;78:179–185. doi: 10.1002/(sici)1097-4644(20000801)78:2<179::aid-jcb1>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 76.Acar M, Becskei A, van Oudenaarden A. Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435:228–232. doi: 10.1038/nature03524. [DOI] [PubMed] [Google Scholar]
- 77.Zawel L, Kumar KP, Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- 78.Yudkovsky N, Ranish JA, Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- 79.Leuther KK, Johnston SA. Nondissociation of GAL4 and GAL80 in vivo after galactose induction. Science. 1992;256:1333–1335. doi: 10.1126/science.1598579. [DOI] [PubMed] [Google Scholar]
- 80.Sil AK, Alam S, Xin P, Ma L, Morgan M, Lebo CM, Woods MP, Hopper JE. The Gal3p-Gal80p-Gal4p transcription switch of yeast: Gal3p destabilizes the Gal80p Gal4p complex in response to galactose and ATP. Mol. Cell. Biol. 1999;19:7828–7840. doi: 10.1128/mcb.19.11.7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Molinari E, Gilman M, Natesan S. Proteasome-mediated degradation of transcriptional activators correlates with activation domain potency in vivo. EMBO J. 1999;18:6439–6447. doi: 10.1093/emboj/18.22.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Muratani M, Tansey WP. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 2003;4:192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- 83.Métivier R, Penot G, Hübner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 84.Reid G, Hübner MR, Métivier R, Brand H, Denger S, Manu D, Beaudouin J, Ellenberg J, Gannon F. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol. Cell. 2003;11:695–707. doi: 10.1016/s1097-2765(03)00090-x. [DOI] [PubMed] [Google Scholar]
- 85.Lipford JR, Smith GT, Chi Y, Deshaies RJ. A putative stimulatory role for activator turnover in gene expression. Nature. 2005;438:113–116. doi: 10.1038/nature04098. [DOI] [PubMed] [Google Scholar]
- 86.Collins GA, Tansey WP. The proteasome: a utility tool for transcription? Curr. Opin. Genet. Dev. 2006;16:197–202. doi: 10.1016/j.gde.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 87.Bhaumik SR, Malik S. Diverse regulatory mechanisms of eukaryotic transcriptional activation by the proteasome complex. Crit. Rev. Biochem. Mol. Biol. 2008;43:419–433. doi: 10.1080/10409230802605914. [DOI] [PubMed] [Google Scholar]
- 88.Myers LC, Lacomis L, Erdjument-Bromage H, Tempst P. The yeast capping enzyme represses RNA polymerase II transcription. Mol. Cell. 2002;10:883–894. doi: 10.1016/s1097-2765(02)00644-5. [DOI] [PubMed] [Google Scholar]
- 89.Schroeder SC, Zorio DA, Schwer B, Shuman S, Bentley DA. Function of yeast mRNA cap methyltransferase, Abd1, in transcription by RNA polymerase II. Mol. Cell. 2004;13:377–387. doi: 10.1016/s1097-2765(04)00007-3. [DOI] [PubMed] [Google Scholar]
- 90.Raha T, Cheng SW, Green MR. HIV-1 Tat stimulates transcription complex assembly through recruitment of TBP in the absence of TAFs. PLoS Biol. 2005;3:e44. doi: 10.1371/journal.pbio.0030044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kundu S, Horn PJ, Peterson CL. SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster. Genes Dev. 2007;21:997–1004. doi: 10.1101/gad.1506607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wood A, Shukla A, Schneider J, Lee J, Stanton JD, Dzuiba T, Swanson SK, Florens L, Washburn MP, Wyrick J, et al. Ctk complex regulation of histone methylation by COMPASS. Mol. Cell. Biol. 2007;27:709–720. doi: 10.1128/MCB.01627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee SK, Yu SM, Prakash L, Prakash S. Requirement for yeast RAD26, a homologue of the human CSB gene, in elongation by RNA polymerase II. Mol. Cell. Biol. 2001;21:8651–8656. doi: 10.1128/MCB.21.24.8651-8656.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee SK, Yu SL, Prakash L, Prakash S. Yeast RAD26, a homolog of the human CSB gene, functions independently of nucleotide excision repair and base excision repair in promoting transcription through damaged bases. Mol. Cell. Biol. 2002;22:4383–4389. doi: 10.1128/MCB.22.12.4383-4389.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Malik S, Chaurasia P, Lahudkar S, Durairaj G, Shukla A, Bhaumik SR. Rad26p, a transcription-coupled repair factor, is recruited to the site of DNA lesion in an elongating RNA polymerase II-dependent manner in vivo. Nucleic Acids Res. 2009;38:1461–1477. doi: 10.1093/nar/gkp1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gavin AC, Bösche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 97.Topalidou I, Papamichos-Chronakis M, Thireos G, Tzamarias D. Spt3 and Mot1 cooperate in nucleosome remodeling independently of TBP recruitment. EMBO J. 2004;23:1943–1948. doi: 10.1038/sj.emboj.7600199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Andrau JC, Van Oevelen CJ, Van Teeffelen HA, Weil PA, Holstege FC, Timmers HT. Mot1p is essential for TBP recruitment to selected promoters during in vivo gene activation. EMBO J. 2002;21:5173–5183. doi: 10.1093/emboj/cdf485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Geisberg JV, Moqtaderi Z, Kuras L, Struhl K. Mot1 associates with transcriptionally active promoters and inhibits association of NC2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 2002;22:8122–8134. doi: 10.1128/MCB.22.23.8122-8134.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huisinga KL, Pugh BF. A TATA binding protein regulatory network that governs transcription complex assembly. Genome Biol. 2007;8:R46. doi: 10.1186/gb-2007-8-4-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Larschan E, Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.van Oevelen CJ, van Teeffelen HA, Timmers HT. Differential requirement of SAGA subunits for Mot1p and Taf1p recruitment in gene activation. Mol. Cell. Biol. 2005;25:4863–4872. doi: 10.1128/MCB.25.12.4863-4872.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dasgupta A, Juedes SA, Sprouse RO, Auble DT. Mot1-mediated control of transcription complex assembly and activity. EMBO J. 2005;24:1717–1729. doi: 10.1038/sj.emboj.7600646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Carey M, Lin YS, Green MR, Ptashne M. A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature. 1990;345:361–364. doi: 10.1038/345361a0. [DOI] [PubMed] [Google Scholar]
- 106.Lin YS, Carey M, Ptashne M, Green MR. How different eukaryotic transcriptional activators can cooperate promiscuously. Nature. 1990;345:359–361. doi: 10.1038/345359a0. [DOI] [PubMed] [Google Scholar]
- 107.Roeder RG. The complexities of eukaryotic transcription initiation: regulation of preinitiation complex assembly. Trends Biochem. Sci. 1991;16:402–408. doi: 10.1016/0968-0004(91)90164-q. [DOI] [PubMed] [Google Scholar]
- 108.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 109.Sauer F, Tjian R. Mechanisms of transcriptional activation: differences and similarities between yeast, Drosophila, and man. Curr. Opin. Genet. Dev. 1997;7:176–181. doi: 10.1016/s0959-437x(97)80126-8. [DOI] [PubMed] [Google Scholar]
- 110.Ptashne M, Gann A. Imposing specificity by localization: mechanism and evolvability. Curr. Biol. 1998;8:R812–R822. doi: 10.1016/s0960-9822(07)00508-8. [DOI] [PubMed] [Google Scholar]
- 111.Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 112.Nolis IK, McKay DJ, Mantouvalou E, Lomvardas S, Merika M, Thanos D. Transcription factors mediate long-range enhancer–promoter interactions. Proc. Natl Acad. Sci. USA. 2009;106:20222–20227. doi: 10.1073/pnas.0902454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Merika M, Thanos D. Enhanceosomes. Curr. Opin. Genet. Dev. 2001;11:205–208. doi: 10.1016/s0959-437x(00)00180-5. [DOI] [PubMed] [Google Scholar]
- 114.Näär AM, Beaurang PA, Robinson KM, Oliner JD, Avizonis D, Scheek S, Zwicker J, Kadonaga JT, Tjian R. Chromatin, TAFs, and a novel multiprotein coactivator are required for synergistic activation by Sp1 and SREBP-1a in vitro. Genes Dev. 1998;12:3020–3031. doi: 10.1101/gad.12.19.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Guermah M, Malik S, Roeder RG. Involvement of TFIID and USA components in transcriptional activation of the human immunodeficiency virus promoter by NF-kappaB and Sp1. Mol. Cell. Biol. 1998;18:3234–3244. doi: 10.1128/mcb.18.6.3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Miele A, Dekker J. Long-range chromosomal interactions and gene regulation. Mol. Biosyst. 2008;4:1046–1057. doi: 10.1039/b803580f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Miele A, Bystricky K, Dekker J. Yeast silent mating type loci form heterochromatic clusters through silencer protein-dependent long-range interactions. PLoS Genet. 2009;5:e1000478. doi: 10.1371/journal.pgen.1000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Miele A, Dekker J. Mapping cis- and trans- chromatin interaction networks using chromosome conformation capture (3C) Methods Mol. Biol. 2009;464:105–121. doi: 10.1007/978-1-60327-461-6_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.