Abstract
Argonaute (Ago) proteins are highly conserved between species and constitute a direct-binding platform for small RNAs including short-interfering RNAs (siRNAs), microRNAs (miRNAs) and Piwi interacting RNAs (piRNAs). Small RNAs function as guides whereas Ago proteins are the actual mediators of gene silencing. Although the major steps in RNA-guided gene silencing have been elucidated, not much is known about Ago-protein regulation. Here we report a comprehensive analysis of Ago2 phosphorylation in human cells. We find that the highly conserved tyrosine Y529, located in the small RNA 5′-end-binding pocket of Ago proteins can be phosphorylated. By substituting Y529 with a negatively charged glutamate (E) mimicking a phosphorylated tyrosine, we show that small RNA binding is strongly reduced. Our data suggest that a negatively charged phospho-tyrosine generates a repulsive force that prevents efficient binding of the negatively charged 5′ phosphate of the small RNA.
INTRODUCTION
Small non-coding RNAs such as miRNAs, endogenous short-interfering RNAs (endo-siRNAs) or Piwi interacting RNAs (piRNAs) form a specific class of non-coding RNAs with distinct functions in post-transcriptional gene regulation (1,2). MiRNAs are the best-characterized class of small non-coding RNAs in mammals. MiRNA genes are transcribed to primary miRNA transcripts (pri-miRNAs), which are processed to stem-loop structured miRNA precursors (pre-miRNAs). This processing event involves the action of the nuclear microprocessor complex containing the RNase III enzyme Drosha. In the cytoplasm, pre-miRNAs are further processed to short double-stranded (ds) RNA intermediates by the RNase III enzyme Dicer (3). After further processing and unwinding steps, one strand is incorporated into a miRNA–protein complex referred to as miRNP or miRNA-containing RNA-induced silencing complex (miRISC) (3). MiRNAs guide miRNPs to specific sites typically located in the 3′-untranslated region (UTR) of target mRNAs. Imperfect pairing of the miRNA with the target site leads to translational repression and/or mRNA degradation resulting in an efficient repression of gene expression. In contrast, perfect or nearly perfect pairing of a miRNA with its target RNA induces RNA interference (RNAi)-like cleavage (4–6).
Members of the Argonaute protein family represent the protein-binding partners of small RNAs (7). Argonaute proteins are typically composed of three distinct domains. The PAZ (Piwi–Argonaute–Zwille) domain recognizes the characteristic 2-nt 3′-overhangs generated by RNase III enzymes such as Dicer and Drosha and anchors the 3′-end of small RNAs (8). The PIWI (P-element-induced wimpy testes) domain folds similar to RNase H and it has been shown for some Argonaute proteins that the PIWI domain contains endonucleolytic activity (8). A third domain, termed MID domain because of its localization between the PAZ and the PIWI domain anchors the 5′-end of the small RNA (9,10). Thus, Argonaute proteins are highly specialized binding modules for functional small RNAs (11–13). Using RNAi reporter systems as well as in vitro RNA cleavage assays, it has been shown that Ago2 is the only member of the human Ago protein sub-family that possesses endonucleolytic cleavage activity although critical amino acids are conserved in other human Ago proteins as well (14,15). It is therefore still unclear what the exact functions of the individual human Ago proteins are.
Ago proteins interact with a conserved protein family generally referred to as the GW182 protein family (16–20). GW182 proteins have initially been reported as integral components of cytoplasmic processing bodies (P-bodies). P-bodies are only poorly understood protein–RNA aggregates that are enriched for enzymes that are important for RNA metabolism (21). Later on, it has been demonstrated that both miRNAs and Ago proteins localize to P-bodies as well (17,19,22–24). GW182 proteins are characterized by multiple glycine–tryptophan (GW) repeats that form multiple Ago interaction modules termed ‘Ago hooks’ (25). In human, three different GW182 homologs termed TNRC6A-C have been found (26). It has been demonstrated in mammals as well as in Drosophila that GW182 proteins interact with the poly(A)-binding protein PABP (27,28), thereby interfering with translational initiation (27).
To date, only little is known about how human small RNA-guided gene-silencing pathways are regulated. A number of post-translational modifications have been reported that suggest regulation of Argonaute function. Using mass spectrometry analysis, it has been found that human Ago2 is hydroxylated at proline-700 and that this post-translational modification influences Ago stability (29). Moreover, human Ago2 is phosphorylated at serine-387 leading to altered cellular localization (30). However, functional consequences of such phosphorylation events have not yet been reported. Therefore, we analyzed phosphorylation of human Ago proteins in detail and show that Ago proteins are phosphorylated at multiple sites. We find that Ago proteins are phosphorylated at a tyrosine residue in the MID domain that is critical for binding of the 5′-end of the small RNA. Furthermore, our data suggest that phosphorylation of this tyrosine inhibits small RNA binding. Therefore, we have identified a potential molecular switch that might allow for loading and unloading of Ago proteins with small RNAs.
MATERIALS AND METHODS
In vivo radiolabeling
HEK 293 cells grown on a 10 cm dish in DMEM-containing antibiotics but only 2% FBS were transfected by the calcium phosphate procedure as described before (31). Forty hours post-transfection, cells were rinsed with TBS and grown for further 4 h in the presence of 1.6 mCi ortho-[32P]-phosphate (Perkin Elmer). Total lysates were generated in the presence of sodium vanadate, a protein phosphotyrosyl phosphatase inhibitor.
In vitro RISC assays and quantification
Preparation of cap-32P-labeled RNA used as RISC substrate was described earlier (15). Assays were performed with 50% (v/v) immunoprecipitated protein coupled to anti-FLAG beads in a final volume of 25 µl in the presence of 40 mM KCl, 4 mM Mg2Cl, 5 mM DTT, 1 mM ATP, 0.2 mM GTP and 1.2 U RiboLock RNase inhibitor (Fermentas). Cleavage reactions were started by addition of the target RNA (1.0 Bq/cm2) and incubated at 30°C for 60 min. RNA extraction and analysis has been described before (31).
For kinetic studies a multiple of a 25 µl reaction was prepared and incubated at 30°C. After the time points indicated 25 µl were removed and immediately subjected to proteinase K digestion for subsequent RNA analysis by denaturing gel electrophoresis, while another 25 µl were removed and added to denaturing protein sample buffer for protein analysis. Signal intensities observed in the radiograph were quantified using ImageGauge software version 4.1 (Fuji). For each time point the intensities of the cleavage product as well as the one of the whole lane were determined and corrected for background. The product/total ratio was plotted against time.
The sequence of the exogenous, chemically 5′-modified small RNA used in Figure 6D is UCGAAGUAUUCCGCGUACGUdT. The small RNA was pre-incubated with the immunoprecipitated proteins for 20 min before the radioactively labeled substrate RNA was added and the RISC assay continued as described above.
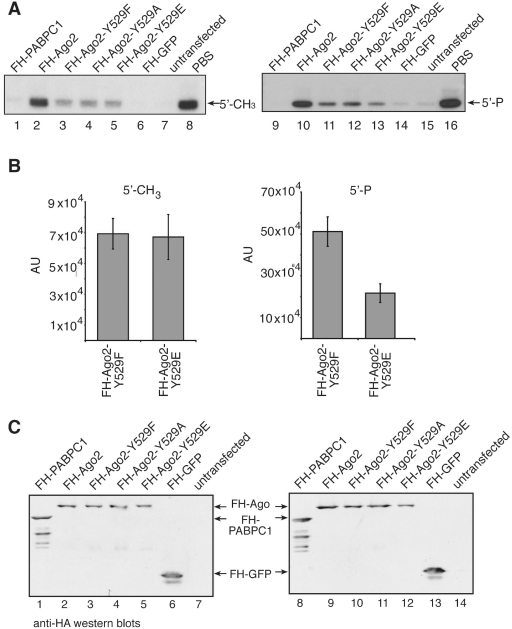
Figure 6.
Ago2 Y529 mutants bind 5′ methylated small RNAs equally well. (A) FH-PABPC1 (lanes 1 and 9) as an RNA-binding protein negative control, FH-Ago2 (lanes 2 and 10), FH-Ago2 Y529F (lanes 3 and 11), FH-Ago2 Y529A (lanes 4 and 12), FH-Ago2 Y529E (lanes 5 and 13) and FH-GFP as an unrelated negative control (lanes 6 and 14) were immunoprecipitated from HEK 293 cell lysates using anti-FLAG antibodies. Either a 5′ methylated (left panel) or 5′ phosphorylated (right panel) 21-nt single-stranded RNA was incorporated into the proteins. As control for unspecific degradation, PBS only was used (lanes 8 and 16). Tagged proteins were purified by immunoprecipitation and bound RNA was extracted for analysis by northern blotting using a probe specific to siR-21.27. (B) Quantification of the northern blot signals observed for Ago2 Y529F and Ago2 Y529E with a 5′ methylated (left panel) and a 5′ phosphorylated (right panel) 21-nt single-stranded RNA. Data used derive from two independent experiments. (C) Protein inputs that were used for the experiment shown in (A). (D) FH-Ago1 (lanes 2 and 10), FH-Ago2 (lanes 3 and 11), FH-Ago2 Y529F (lanes 4 and 12), FH-Ago2 Y529E (lanes 5 and 13), FH-Ago2 Y529A (lanes 6 and 14) were immunoprecipitated using anti-FLAG antibodies and subsequently incubated with a single stranded 5′ phosphorylated (lanes 2–8) or a 5′ methylated siRNA (lanes 10–16). After incubation a target RNA was added to each sample carrying a binding site of perfect complementarity to the siRNAs used. Cleavage products were analyzed by denaturing RNA PAGE. A western blot indicating the protein inputs is shown on the lower part of the figure.
In vitro Dicer assays
Pre-miR-27a was described before (32). In vitro transcription was performed in the presence of [32P]-UTP resulting in internally labeled pre-miR-27a. Immunoprecipitated proteins were incubated in a final volume of 20 µl in the presence of 5 mM ATP, 7.5 mM MgCl2, 4 U RiboLock RNase inhibitor (Fermentas) for 90 min at 37°C. RNA was phenol/chloroform extracted, ethanol precipitated and analyzed by denaturing gel electrophoresis.
Northern blotting
Northern blotting was performed as described earlier (33). Briefly, RNA isolated from immunoprecipitations was extracted and solved in formamide loading dye, separated by electrophoresis on 15% denaturing polyacrylamide gels and transferred to Hybond-N membrane (Amersham Bioscience) by semidry blotting. Membranes were dried, UV treated and incubated for 1 h at 80°C. After pre-hybridization the 5′-[32P]-labeled probe was added over night. Subsequently, the membrane was washed in 10 min intervals twice with 5× SSC; 1% (w/v) SDS and once with 1x SSC; 1% (w/v) SDS. Exposure to Kodak BioMax MS films (Kodak) was performed at −80°C using an intensifying screen (GE Healthcare). Probe for miR-19b: tcagttttgcatggatttgcaca; probe for miR-16: cgccaatatttacgtgctgcta; probe for miR-25: tcagaccgagacaagtgcaatg; probe for 21.27: cgtacgcggaatacttcgat (34).
Assembly of RISC using 5′-CH3- or 5′-P-modified RNAs and quantification
Immunoprecipitates of FH-tagged proteins were halved and subjected to RISC assembly in a total volume of 50 µl under the same buffer conditions as specified for the in vitro RISC assay. Either 1 pmol of 5′-P - 21.27 (=r(UCGAAGUAUUCCGCGUACGU)dT) or because of a weaker blot efficiency 6 pmol of 5′- CH3-21.27 single-stranded RNA were added to the immunoprecipitated proteins and incubated at 30°C for 30 min. Subsequently samples were washed twice with IP wash buffer (50 mM Tris–HCl, pH 7.5; 300 mM NaCl; 5 mM MgCl2; and 0,05% Nonidet P-40) followed by PBS and splitted again. One-half was subjected to protein analysis by western blotting, the other half to RNA analysis by northern blotting using a probe against siR-21.27 (34).
Signal intensities of two independent experiments were determinded as described above and corrected for background to finally plot the arithmetic mean against sample identity.
Immunoprecipitation of phosphotyrosine proteins
Monoclonal anti-phosphotyrosine antibodies or monoclonal anti-RMC antibodies were coupled to protein G sepharose over night. The matrix was subsequently incubated with 3 mg BSA for 1 h and washed with PBS before cytoplasmic extract from HEK 293 cells prepared in the presence of 10 mM sodium vanadat (inhibitor for protein phosphotyrosyl phosphatases) was added. After 4 h of slow rotation at 4°C beads were washed three times with IP wash buffer containing 1.2 M NaCl, twice with 0.6 M, once with PBS including 0.1% (v/v) Triton and finally with PBS only.
cDNA synthesis and qRT-PCR
RNA was extracted from 5% (v/v) of the input (500 µl) and from 50% of the immunoprecipitates by proteinase K digestion, phenol extraction and ethanol precipitation. RNA was treated with DNase I in a total volume of 18 µl for 30 min at 37°C followed by heat inactivation at 65°C. cDNA from 1 µl was synthesized using the first-strand-cDNA synthesis kit (Fermentas) and the random hexamers therein. Obtained cDNA was diluted to 100 µl and used for qRT-PCR, which was performed in quadruplicates. For each reaction 6.7 µl cDNA was added to MESA GREEN qPCR MasterMix Plus for SYBR Assay w/fluorescein (Eurogentec) and 0.2 µM primer in a total volume of 15 µl. Data were analyzed according to the 2(-delta delta C(T))-method (35). Gapdh was chosen for normalization.
RESULTS
Human Ago1 and Ago2 are phosphorylated in vivo
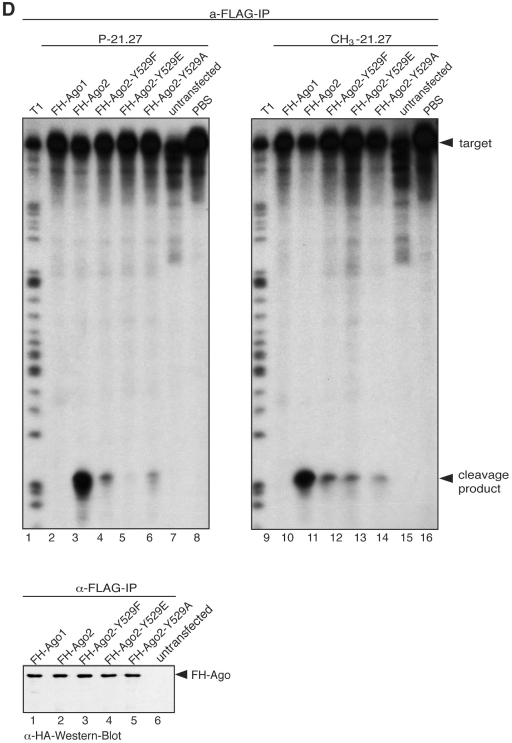
Many proteins are post-translationally modified. Phosphorylation, for example, can occur on serine, threonine and tyrosine side chains and is an important regulatory means for a wide range of human proteins. Therefore, we analyzed Ago1 and Ago2 phosphorylation in HEK 293 cells. Myc-Ago1 and myc-Ago2 were transiently transfected into HEK 293 cells followed by in vivo 32P labeling (Figure 1A). After labeling, cell lysates were generated and myc-Ago1/2 were immunoprecipitated using anti-myc antibodies. Because of the instability of protein phosphorylation, lysate preparation was carried out in the presence of phosphatase inhibitors. Immunoprecipitates were analyzed by autoradiography (left panel) or western blotting (right panel). Indeed, 32P was efficiently incorporated into myc-Ago1 as well as myc-Ago2 indicative of protein phosphorylation. We focused our further studies on Ago2 and analyzed specific phosphorylation sites by mass spectrometry. FLAG/HA-tagged Ago2 (FH-Ago2) was immunoprecipitated from HEK 293 cell lysates in the presence of phosphatase inhibitors and subsequently digested by trypsin. Digested peptides were enriched for phosphopeptides using TiO2 affinity purification and analyzed by nanoLC-ESI MS and MS/MS. We identified five phosphopeptides, which are listed in Figure 1B. Of note, phosphorylated serines (pS), threonines (pT) as well as tyrosines (pY) were found. Since tyrosine phosphorylation was found at very intriguing positions, we further validated our mass spec results, which were obtained from tagged Ago proteins. To rule out that the observed tyrosine phosphorylation resembles an Ago overexpression artifact, we immunoprecipitated endogenous proteins using an anti-phospho-tyrosine antibody and analyzed the immunoprecipitate for the presence of endogenous Ago2 by western blotting (Figure 1D). Indeed, Ago2 was readily co-immunoprecipitated suggesting that endogenous Ago2 is phosphorylated at the indicated tyrosine residues. In summary, our results demonstrate that human Ago2 is phosphorylated in vivo at a number of different amino acid residues.
Figure 1.
Human Ago1 and Ago2 can be phosphorylated in vivo. (A) Myc-Ago1/2 transfected HEK 293 cells were labeled with [32P] and myc-Ago1 (lanes 1 and 4), myc-Ago2 (lanes 2 and 5) or myc-GST (lanes 3 and 6) were immunoprecipitated using anti-myc antibodies. Radio-labeled proteins were visualized by autoradiography (left panel) and western blotting with anti-myc antibodies (right panel). (B) Phosphorylated Ago2 peptides detected by mass spectrometry. (C) Schematic representation of phosphorylated amino acid side chains on Ago2. (D) Tyrosine phosphorylated proteins were immunoprecipitated from HEK 293 lysate using the monoclonal anti-phospho-tyrosine antibody 4G10 (lane 2) or a control antibody (lane 3). Lane 1 shows 0.5% of the protein input that was used for the immunoprecipitations. Proteins were analyzed by western blotting using an anti-Ago2 antibody.
A highly conserved tyrosine located in the MID domain can be phosphorylated
We next analyzed the phosphorylated peptides in more detail (Figure 1C). We found three phosphorylated amino acid residues located in the PAZ domain (pS253, pT303, pT307), one in the PIWI domain (pS798), two in the L2 linker region (pS387, pY393) and one in the MID domain (pY529). The latter peptide 526-TPVpYAEVK-533, is part of the pocket that has been implicated in binding of the 5′ phosphate of the small RNA (9,10). Y529 is conserved not only in all other human Argonaute proteins but also in all other species that have been analyzed (Supplementary Figure S1, peptide 4). Therefore, we hypothesized that phosphorylation of Y529 could be a general regulatory mechanism for small RNA binding.
Ago2 Y529 mutants are functional and interact with Dicer and TNRC6B
To analyze the function of Ago2 Y529 in detail, we generated a number of different point mutants. It has been suggested that the aromatic ring of Y529 contributes to the binding of small RNAs by stacking interaction with the base of the 5′-terminal nucleotide of the small RNA (9,10). Furthermore, the hydroxyl group of Y529 forms a hydrogen bond with an oxygen atom of the 5′ phosphate. Therefore, we generated an Ago2 Y529F mutant that can perform stacking interaction with the first base but cannot be phosphorylated. To destroy stacking interaction as well, we replaced Y529 by alanine (Y529A). We further generated an Ago2 Y529Q mutant with a long aliphatic side chain. Finally, we replaced Y529 by a glutamate (Y529E). The Y529E mutant can neither form a hydrogen bond with the 5′ phosphate nor perform stacking interaction with the base of the 5′ terminal nucleotide. Furthermore, by mutating Y529 to E we placed a negatively charged side chain into the 5′-end-binding pocket thereby generating a negatively charged environment similar to a phosphorylated Y529.
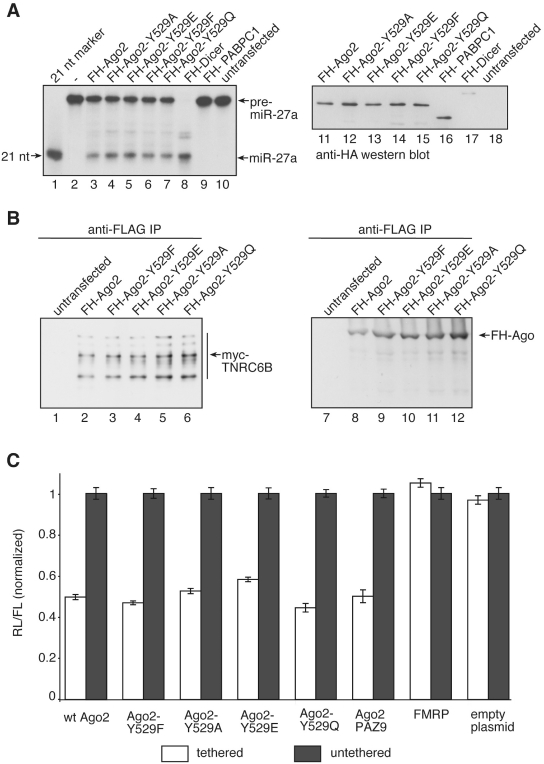
Substitution of specific amino acids can induce major structural changes followed by partial or complete unfolding. Therefore, we analyzed functionality of the Ago2 Y529 mutants. It has been shown before, that Ago proteins stably interact with Dicer. Therefore Dicer activity can be co-immunoprecipitated using antibodies against Ago proteins (Figure 2A) (17,36,37). We transfected FH-Ago2, FH-Ago2 Y529 mutants as well as FH-Dicer and immunoprecipitated the tagged proteins using anti-FLAG antibodies. Immunoprecipitates were incubated with 32P-labeled pre-miR-27a and Dicer cleavage products were analyzed by RNA gel electrophoresis. As expected, FH-Ago2 and FH-Dicer immunoprecipitates Dicer activity efficiently (lanes 3 and 8). Interestingly, all Ago2 Y529 mutants co-immunoprecipitated Dicer activity as efficiently as wild type (wt) Ago2 suggesting that the mutants are functional.
Figure 2.
Ago2 Y529 point mutants are functional. (A) [32P]-labeled pre-miR-27a was incubated with immunoprecipitated FH-Ago2 (lane 3), FH-Ago2 Y529A (lane 4), FH-Ago2 Y529E (lane 5), FH-Ago2 Y529F (lane 6), FH-Ago2 Y529Q (lane 7), FH-Dicer (lane 8), FH-PABPC1 (lane 9) or untransfected HEK 293 lysate (lane 10). Cleavage products were analyzed by denaturing RNA PAGE. Lane 1 shows a radio-labeled size marker. Lanes 11–18 show anti-HA western blots of the protein input. (B) Myc-TNRC6B was co-transfected with FH-Ago2 (lanes 2 and 8), FH-Ago2 Y529F (lanes 3 and 9), FH-Ago2 Y529E (lanes 4 and 10), FH-Ago2 Y529A (lanes 5 and 11) and FH-Ago2 Y529Q (lanes 6 and 12). Immunoprecipitation using anti-FLAG antibodies was performed and co-immunoprecipitated proteins were analyzed by western blotting against the myc-tag (left panel). The anti-HA western blot of the immunoprecipitates is shown to the right. In lanes 1 and 7, HEK 293 cell lysates containing only myc-TNRC6B were used. (C) Tethering experiments. Ago2, Ago2 Y529F, Ago2 Y529A, Ago2 Y529E, Ago2 Y529Q, Ago2 PAZ9 and FMRP as well as the empty plasmids as controls, all either λN-HA-(white bars) or only HA-conjugated (grey bars) were co-expressed with a renilla luciferase reporter that contained several boxB sites in its 3′-UTR. Proteins were tethered to the reporter by the λN-tag. Expression levels have been normalized to co-transfected firefly luciferase. For analysis data were normalized to the individual untethered controls.
It has been reported, that Ago2 Y529 is important for the interaction of Ago2 with TNRC6B (25). Therefore, we analyzed binding of TNRC6B to the Ago2 Y529 mutants that we have generated (Figure 2B). Myc-TNRC6B was transfected together with FH-Ago2 or FH-Ago2 Y529 mutants into HEK 293 cells. Ago2 complexes were immunoprecipitated using anti-FLAG antibodies and the immunoprecipitates were analyzed for the presence of myc-TNRC6B by anti-myc western blotting. Indeed, not only FH-Ago2 but also all Ago2 Y529 mutants interacted with myc-TNRC6B suggesting that Y529 is dispensable for interaction with TNRC6B.
Since miRNAs function as guides and Ago proteins artificially tethered to mRNAs repress gene expression (38), all Ago mutants were analyzed in tethering experiments using the λN–box B system (Figure 2C) (39). Consistently with the interaction data, tethered wt Ago2 as well as all mutants inhibited luciferase expression. Of note, the PAZ9 mutant that does not bind to small RNAs (23) can still repress luciferase expression when artificially tethered. These results further strengthen the assumption that the individual Ago2 Y529 mutants are functional and can be used for further analysis.
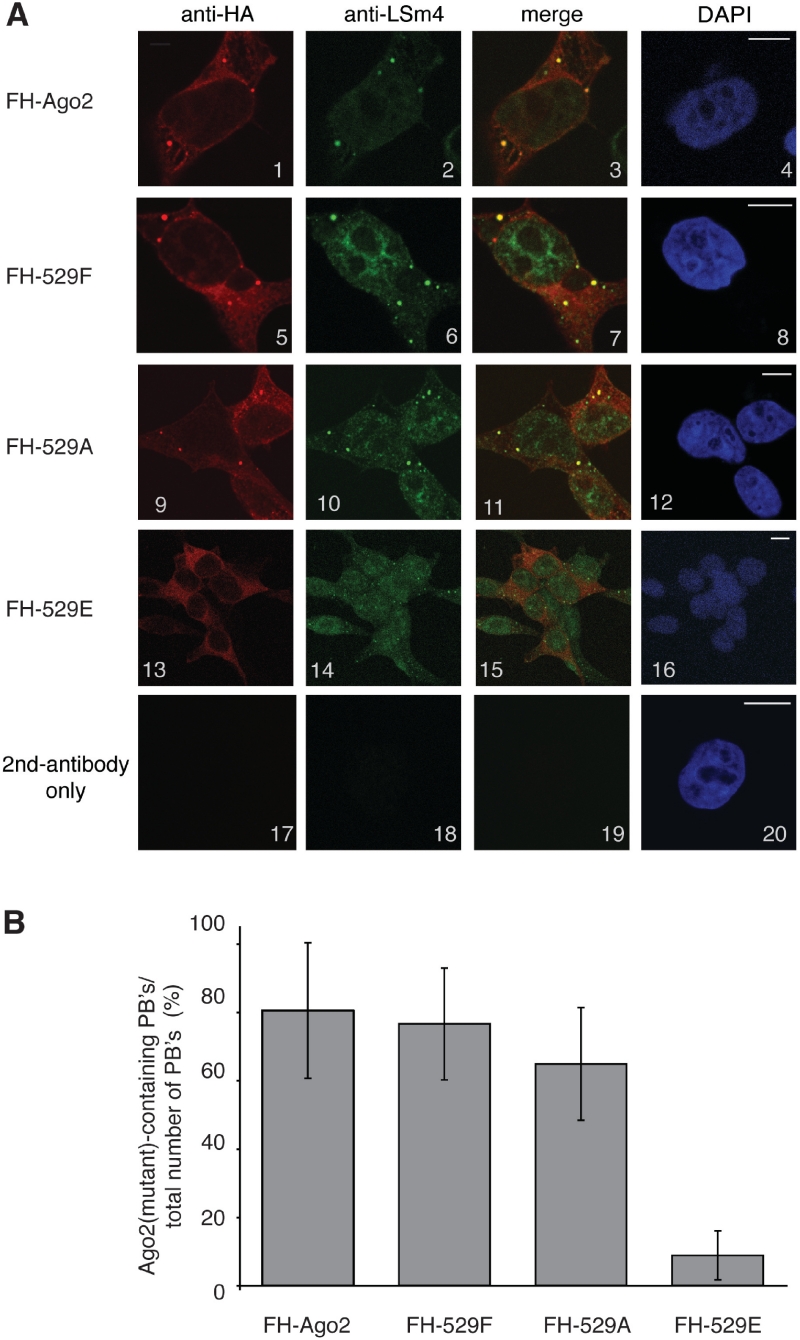
Strongly reduced P-body localization of Ago2 Y529E
It has been demonstrated before that Ago proteins as well as miRNAs localize to cytoplasmic P-bodies (17,19,22–24). We therefore analyzed sub-cellular localization of wt as well as Ago2 Y529 mutants (Figure 3). FH-Ago2 (panels 1–4), FH-Ago2 Y529F (panels 5–8), FH-Ago2 Y529A (panels 9–12) and FH-Ago2 Y529E (panels 13–16) were transfected and fixed cells were stained with antibodies against the HA-tag and LSm4, which served as P-body marker. Wt Ago2, the Ago2 Y529F and Ago2 Y529A mutants localized to P-bodies. However, the Ago2 Y529E mutant localized mainly to the diffuse cytoplasm and only a strongly reduced co-staining with the P-Body marker LSm4 was observed (Figure 3A and B). Our data suggest that Y529 itself is not required for P-body localization. However, the generation of a negatively charged environment in the 5′-end-binding pocket by substituting Y529 with E severely impairs P-body localization of Ago2.
Figure 3.
Cellular localization of Ago2 Y529 mutants. (A) FH-Ago2 (panels 1–4), FH-Ago2 Y529F (panels 5–8), FH-Ago2 Y529A (panels 9–12), FH-Ago2 Y529E (panels 13–16) were transfected into HEK 293 cells. Cells were stained with anti-HA antibodies (panels 1, 5, 9, 13) and antibodies against LSm4 (panels 2, 6, 10, 14). In panels 17–20, only the secondary antibody was used. Bars represent 10 µm. (B) Quantification of Ago2- or Ago2 Y529 mutant-positive P-bodies relative to the total number of counted P-bodies.
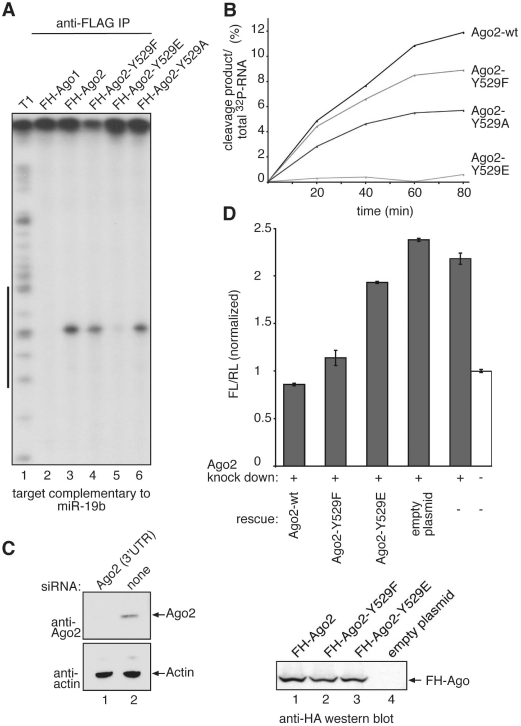
Mutation of Ago2 Y529 to E interferes with slicer activity in vitro and in vivo
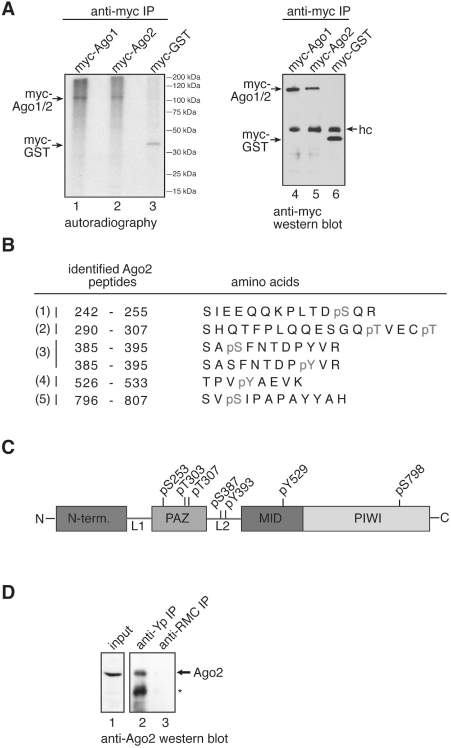
Ago2 is the only protein of the human Ago protein sub-family that cleaves target RNAs with perfect complementarity to the bound small RNA (14,31). Consequently, we analyzed the contribution of Y529 to the cleavage activity of Ago2 (Figure 4). FH-Ago2 as well as the FH-Ago2 Y529F, E and A mutants were transfected and immunoprecipitated using anti-FLAG antibodies. Isolated Ago complexes were incubated with a [32P]-cap-labeled target RNA complementary to endogenous miR-19b. Specific cleavage products were analyzed by denaturing RNA-gel electrophoresis (Figure 4A). Both the Y529F and A mutants cleaved the target RNA, although the cleavage activity was reduced compared to wt Ago2. Strikingly, the Y529E mutant showed very little cleavage activity indicating that carrying a negative charge in the 5′-end-binding pocket is incompatible with Ago2 slicer function in vitro. Time course experiments of wt Ago2 as well as the individual mutants revealed comparable results to the end-point measurements shown in Figure 4A (Figure 4B).
Figure 4.
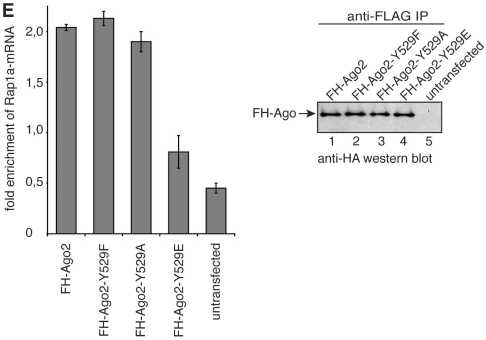
Cleavage activity of Ago2 Y529 mutants. (A) FH-Ago1 (lane 2) as negative control, FH-Ago2 (lane 3), FH-Ago2 Y529F (lane 4), FH-Ago2 Y529E (lane 5) and FH-Ago2 Y529A (lane 6) were immunoprecipitated from HEK 293 cell lysate and incubated with a [32P]-cap-labeled target RNA containing a perfect complementary target site for miR-19b. Specific cleavage products were analyzed by denaturing RNA PAGE. T1 indicates a partial nuclease T1 digestion of the target RNA (lane 1). The black bar to the left indicates the target site of miR-19b. (B) Kinetic analysis of Ago2 and Ago2-Y529 mutant-cleavage activity. Non-saturated radioactive signals were quantified using the ImageGauge software. (C) SiRNAs directed against the 3′-UTR of endogenous Ago2 were transfected into HeLa cells (lane 1) and knockdowns were analyzed by western blotting using anti-Ago2 (upper panel) or anti-actin antibodies (lower panel). (D) SiRNAs against the 3′-UTR of Ago2 (grey bars) or control siRNAs (white bar) were transfected into HeLa cells. After 48 h, knockdowns were rescued by transfecting wt Ago2, Ago2 Y529F, Ago2 Y529E or the empty plasmid together with a firefly luciferase reporter containing a perfectly complementary target site for endogenous miR-21. After 2 days, luciferase activity was measured. Firefly luciferase activity was normalized to renilla luciferase activity and the obtained values were normalized to the control without Ago2 knockdown (upper panel). Representative protein levels analyzed by anti-HA western blotting are shown in the lower panel. (E) FH-tagged Ago2 as well as the Ago2-Y529F, Ago2-Y529A and Ago2-Y529E mutants were immunoprecipitated from HEK 293 cell lysates using anti-FLAG antibodies. The co-immunoprecipitated miR-19b target Rap1a was analyzed by qRT-PCR for enrichment in the precipitates (left panel). Protein inputs that were used for the experiment are shown on the right.
Next, slicer activity was analyzed in vivo (Figure 4C and D). SiRNAs directed against the 3′-UTR of Ago2 were designed and used to knock down endogenous Ago2. Following Ago2 knockdown, HeLa cells were transfected with a reporter plasmid containing a cleavage site perfectly complementary to endogenous miR-21 within the 3′-UTR of firefly luciferase. As expected, relative firefly luciferase activity was elevated when Ago2 had been depleted. We then performed rescue experiments by co-transfecting siRNA-resistant wt Ago2 as well as the individual Ago2 Y529 mutants along with the reporter. Consistently with the in vitro data, wt Ago2 and Ago2 Y529F rescued silencing of the luciferase reporter (Figure 4D), while Ago2 Y529E did not reverse the Ago2 knockdown phenotype indicating that the Y529E mutant is cleavage incompetent in vivo as well.
In mammals, most miRNAs target only partially complementary sites and Ago2 contributes to translational repression as well as deadenylation-dependent mRNA decay. Therefore, we analyzed the interactions of wt Ago2 and the Ago2 Y529 mutants with an endogenous mRNA target. We have found that Rap1a expression is regulated by miR-19b in HEK cells (unpublished data) and this regulation requires stable interaction with Ago2-miR-19b complexes. FH-Ago2, FH-Ago2 Y529F, FH-Ago2 Y529A and FH-Ago2 Y529E constructs were transfected into HEK 293 cells and proteins were immunoprecipitated using anti-FLAG antibodies. The co-immunoprecipitated mRNAs were extracted and analyzed by qRT-PCR for enrichment of Rap1a after immunoprecipitation (Figure 4E). As expected, wt-Ago2 co-immunoprecipitates the Rap1a mRNA and similar effects were observed for the Y529F and Y529A mutants. However, the Ago2 Y529E mutant cannot interact with the endogenous mRNA target indicating that not only cleavage activity, but also mRNA interaction is impaired when a negatively charged environment is generated within the 5′ phosphate binding pocket of Ago2.
A negative charge in the 5′-end-binding pocket mimicking a phospho-tyrosine impairs small RNA binding
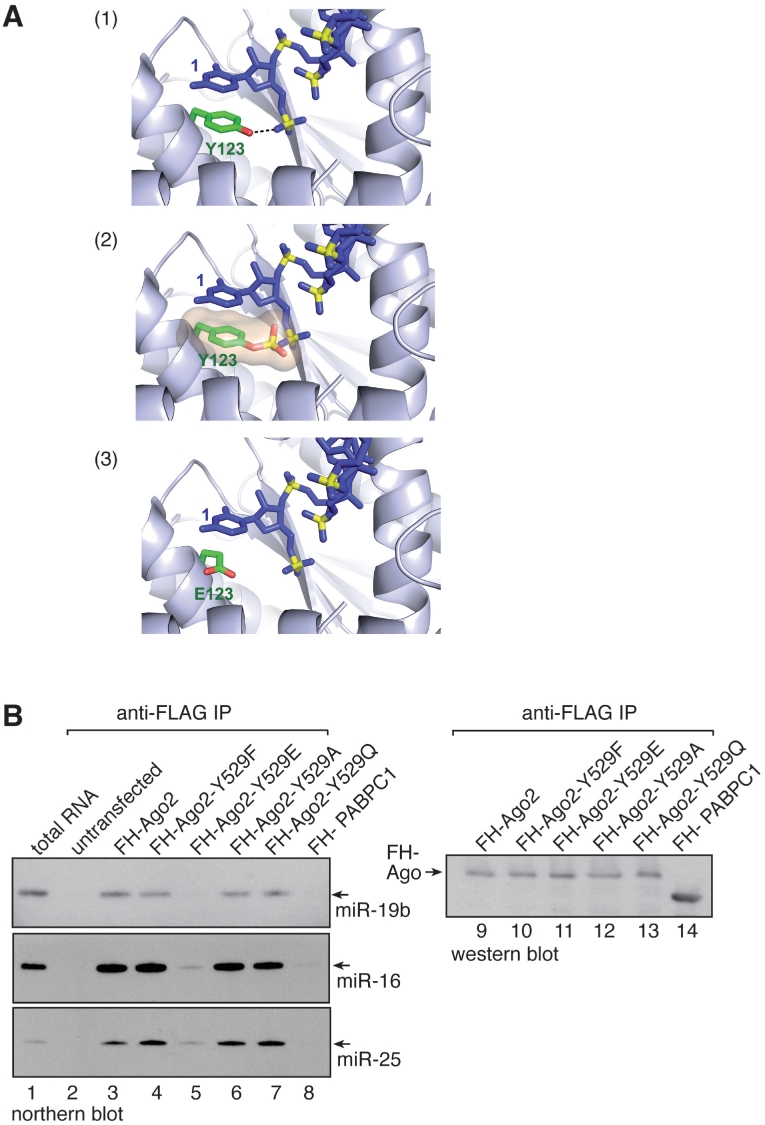
Based on the structure of the Archaeoglobus fulgidus Ago protein that contains the conserved tyrosine at position 123 we modeled a phospho-tyrosine into the 5′-end-binding pocket (Figure 5A). A phospho-tyrosine not only sterically interferes with the correct positioning of the 5′ phosphate of the small RNA but also repulses the negative charge of the small RNA’s 5′ phosphate (middle panel). According to the modeled structure, the Y529E mutant does not sterically hinder 5′ phosphate binding but its negative charge generates a repulsive force against the negatively charged 5′ phosphate of the small RNA (lower panel). Therefore, the effects observed with the Y529E mutant probably underestimate effects caused by pY529.
Figure 5.
Small RNA binding of Ago2 Y529E is strongly reduced. (A) Structure modeling of Archaeoglobus fulgidus Ago. Y123 was replaced by phospho-Y123 (panel 2) or E123 (panel 3). (B) FH-Ago2 (lane 3), FH-Ago2 Y529F (lane 4), FH-Ago2 Y529E (lane 5), FH-Ago2 Y529A (lane 6), FH-Ago2 Y529Q (lane 7) and FH-PABPC1 as negative control (lane 8) were transfected into HEK 293 cells and immunoprecipitated using anti-FLAG antibodies. Co-immunoprecipitated RNA was extracted and analyzed by northern blotting using probes specific to miR-19b (upper panel), miR-16 (middle panel) or miR-25 (lower panel). In lane 2, untransfected cell lysate was used for immunoprecipitation. Lane 1: 30 µg total RNA from HEK 293 cells was blotted. Lanes 9-14: anti-HA western blot of the immunoprecipitates used for RNA extraction.
The clear modeling data let to the conclusion that a small RNA carrying a 5′ phosphate can not efficiently interact with the 5′-end-binding pocket that contains a pY529 or 529E. Therefore, we analyzed direct miRNA binding to wt Ago2 or the individual Y529 mutants by northern blotting (Figure 5B). FH-Ago2 as well as the individual mutants were transfected into HEK 293 cells. Ago2 proteins were immunoprecipitated with anti-FLAG antibodies, associated RNA was extracted and analyzed by northern blotting using probes complementary to endogenous miR-19b (upper panel), miR-16 (middle panel) or miR-25 (lower panel). Wt Ago2 as well as Ago2 Y529F, Y529A and Y529Q efficiently co-immunoprecipitated the analyzed miRNAs. In agreement with the modeling data, Ago2 Y529E co-immunoprecipitated miRNAs only marginally.
Taken together, our data suggest that a negatively charged amino acid mimicking a phosphorylated tyrosine in the 5′ phosphate-binding pocket inhibits small RNA binding and as a consequence cleavage activity and P-body localization.
Ago2 Y529E interacts with 5′ methylated single stranded siRNAs
In order to further characterize the functional consequences of a negative charge within the 5′ end-binding pocket, we hypothesized that a small RNA without a 5′ phosphate should bind equally well to all Y529 mutants. We have demonstrated before that 5′ methylated single-stranded siRNAs are efficiently incorporated into RISC (34). FH-Ago2, FH-Ago2 Y529F, FH-Ago2 Y529A and FH-Ago2 Y529E were immunoprecipitated from HEK 293 cell lysates and incubated with 5′ methylated or 5′ phosphorylated single-stranded siRNAs (Figure 6A, left and right panels). Of note, single stranded siRNAs interact with wt Ago2 more efficiently compared to endogenous miRNAs, which binds equally well to the FH-Ago2 Y529F and FH-Ago2 Y529A mutants (Figure 5B). However, when comparing the FH-Ago2 Y529F and FH-Ago2 Y529E mutants we find no difference in binding when the 5′ methylated siRNA was used (Figure 6B, left panel). Binding of the 5′ phosphorylated siRNA to FH-Ago2 Y529E was significantly reduced (right panel), indicating that the FH-Ago2 Y529E mutant indeed generates a negatively charged environment that repulses the negatively charged 5′ phosphate of the siRNA. The protein input is shown in Figure 6C.
To further analyze the activity of the Ago2 mutants carrying 5′ phosphorylated or methylated single-stranded siRNAs, we performed in vitro cleavage assays (Figure 6D). FH-Ago2, FH-Ago2 Y529F, FH-Ago2 Y529E as well as FH-Ago2 Y529A were expressed in HEK 293 cells and isolated by anti-FLAG immunoprecipitation. The isolates were incubated with a 5′ phosphorylated or a 5′ methylated siRNA strand. After incubation a radio-labeled target RNA was added to the reaction and the cleavage products were analyzed by RNA PAGE. Consistently, with the small RNA-binding results shown in Figure 6A/B, the FH-Ago2 Y529E mutant showed very low-cleavage activity when loaded with a 5′ phosphorylated siRNA compared to FH-Ago2 Y529F. However, the cleavage activities of FH-Ago2 Y529F and FH-Ago2 Y529E were comparable when loaded with a 5′-methylated siRNA indicating that the FH-Ago2 Y529E mutant is generally active but cannot load a small RNA with a negatively charged phosphate group on its 5′-end.
DISCUSSION
Argonaute proteins are highly specialized binding modules for small RNAs and mediators of small RNA-guided gene silencing. However, not much is known about their regulation. Here, we found that human Ago proteins are phosphorylated at a number of different amino acids. Most importantly, one specific tyrosine, namely Y529 in human Ago2, located in the small RNA 5′-end-binding pocket of the MID domain can be phosphorylated in vivo. Of note, the amount of Ago2 phosphorylated on Y529 appears rather low in our experiments. However, we cannot exclude that this is due to experimental or technical reasons. Based on structural studies performed on archaeal or bacterial Ago proteins, it is known that this highly conserved tyrosine is located in close proximity to the 5′ phosphate of the small RNA that is generated by cleavage of long dsRNA precursors by the RNase III enzyme Dicer (9–12,40). Using structural modeling as well as a number of biochemical approaches, we found that placing a negative charge in position of the tyrosine side chain does not allow for binding of the 5′ phosphate of the small RNA. The phospho-tyrosine sterically hinders the docking of a second phosphate and the negative charge generates a repulsive force against another negatively charged group. Therefore, phosphorylation of the highly conserved tyrosine Ago2-Y529 within the 5′-end-binding pocket of the MID domain might function as a molecular switch that promotes or inhibits small RNA binding to Argonaute proteins.
Phosphorylation of Ago2 Y529 may have a number of biologically important implications. Specific kinases and phosphatases could turn Ago proteins on and off depending on cellular conditions such as stress-induced signaling, differentiation or cell-cycle regulation. Our future work clearly aims at the identification of signaling pathways that influence Ago phosphorylation under various cellular conditions. Interestingly, it has been suggested recently that Ago2 expression is controlled through MAP kinase (MAPK) signaling (41). The stability of Ago2 is influenced by the epidermal growth factor receptor (EGFR) as part of the MAPK pathway. The mechanistic details as well as the functional consequences behind this observation, however, remain elusive. It has further been reported before that Ago2 is phosphorylated at serine 387 and it has been shown that the p38 MAP-kinase pathway is required for this modification. Phosphorylation at S387 influences P-body localization of Ago2 and might serve as cellular regulator for Ago2 (30). It is becoming more and more apparent that phosphorylation of Ago proteins is a common means for regulating the activity of small RNA pathways. Detailed knowledge of such modifications may also help to better understand a number of diseases with abnormalities in miRNA function or may also help to improve RNAi for therapeutic applications.
Phosphorylation sites are typically located on the surface of proteins allowing for efficient accession of kinases and phosphatases. However, Ago2 Y529 is located in the 5′-end-binding pocket of the MID domain and not on the surface of Ago2. Three scenarios of how Y529 might be phosphorylated could be envisioned. First, highly specialized kinases or phosphatases that reach Y529 within the 5′-end-binding pocket for phosphorylation or de-phosphorylation could be responsible for the observed modification. Second, Ago proteins could undergo conformational changes leading to a relocation of Y529 to the protein surface, where Y529 would be more accessible for kinases or phosphatases. Different structural studies on the same Ago protein from Aquifex aeolicus suggest that the Ago structure is at least to some extend flexible (40,42,43). Third and most provocatively, the 5′ phosphate of the small RNA might be transferred directly to Ago2 Y529. Structural modeling studies (Figure 5A) suggest that the 5′ phosphate is positioned in close proximity to Y529. The biological consequences of such a transfer mechanism remains elusive. It is conceivable that phosphate transfer might occur during RISC turn over and Y529 phosphorylated Ago2 resembles a RISC break down intermediate. More functional experiments will be needed to clearly identify the biological function of Y529 phosphorylation of human Ago2.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (DFG Me 2064/2-1 and FOR855 DFG Me 2064/3-1 and 3-2, in part); European Union (LSHG-CT-2006-037900, SIROCCO, in part); Bavarian Genome Research Network (BayGene) and the Max-Planck-Society (in part). Funding for open access charge: University of Regensburg.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to Bernd Haas and Sabine Rottmüller for technical help. We thank Witek Filipowicz for plasmids pCIneo-NHA, pCIneo-HA and pRL-5boxB and Axel Ullrich for the generous gift of anti-phospho-tyrosine-antibody (4G10).
REFERENCES
- 1.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 4.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 5.Meister G. miRNAs get an early start on translational silencing. Cell. 2007;131:25–28. doi: 10.1016/j.cell.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 6.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 7.Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol. Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 9.Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 15.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Liu J, Rivas FV, Wohlschlegel J, Yates JR, 3rd, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005;7:1161–1166. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Luhrmann R, Tuschl T. Identification of novel argonaute-associated proteins. Curr. Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 18.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jakymiw A, Lian S, Eystathioy T, Li S, Satoh M, Hamel JC, Fritzler MJ, Chan EK. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- 20.Eystathioy T, Chan EK, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 22.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 25.Till S, Lejeune E, Thermann R, Bortfeld M, Hothorn M, Enderle D, Heinrich C, Hentze MW, Ladurner AG. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat. Struct. Mol. Biol. 2007;14:897–903. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- 26.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 27.Zekri L, Huntzinger E, Heimstadt S, Izaurralde E. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol. Cell Biol. 2009;29:6220–6231. doi: 10.1128/MCB.01081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol. Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qi HH, Ongusaha PP, Myllyharju J, Cheng D, Pakkanen O, Shi Y, Lee SW, Peng J. Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature. 2008;455:421–424. doi: 10.1038/nature07186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng Y, Sankala H, Zhang X, Graves PR. Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem. J. 2008;413:429–436. doi: 10.1042/BJ20080599. [DOI] [PubMed] [Google Scholar]
- 31.Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A Human snoRNA with MicroRNA-Like Functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 33.Rudel S, Flatley A, Weinmann L, Kremmer E, Meister G. A multifunctional human Argonaute2-specific monoclonal antibody. RNA. 2008;14:1244–1253. doi: 10.1261/rna.973808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen PY, Weinmann L, Gaidatzis D, Pei Y, Zavolan M, Tuschl T, Meister G. Strand-specific 5′-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity. RNA. 2008;14:263–274. doi: 10.1261/rna.789808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 38.Pillai RS, Artus CG, Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA. 2004;10:1518–1525. doi: 10.1261/rna.7131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell. 2003;11:939–949. doi: 10.1016/s1097-2765(03)00142-4. [DOI] [PubMed] [Google Scholar]
- 40.Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G, Chen HY, Dauter Z, Tuschl T, Patel DJ. Crystal Structure of A. aeolicus Argonaute, a site-specific DNA-Guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams BD, Claffey KP, White BA. Argonaute-2 expression is regulated by epidermal growth factor receptor and mitogen-activated protein kinase signaling and correlates with a transformed phenotype in breast cancer cells. Endocrinology. 2009;150:14–23. doi: 10.1210/en.2008-0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rashid UJ, Paterok D, Koglin A, Gohlke H, Piehler J, Chen JC. Structure of Aquifex aeolicus argonaute highlights conformational flexibility of the PAZ domain as a potential regulator of RNA-induced silencing complex function. J. Biol. Chem. 2007;282:13824–13832. doi: 10.1074/jbc.M608619200. [DOI] [PubMed] [Google Scholar]
- 43.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.