Abstract
Deoxyribonuclease I digestion of the deoxyoligodecamer 5'd(ATCGTACGAT)2(3') has been examined in detail to study the kinetic and structural properties of this enzyme substrate system in solution. In addition, these studies have defined, in general, those DNase I conditions to be used in future drug-DNA footprinting experiments. Special attention has been taken of those properties of DNase I that are critical for quantitation of ligand binding to small DNA fragments, and that aid in designing oligomers to be used in footprinting experiments. Enzyme activity was observed at all phosphodiester bonds in the decamer studied with varying affinity, except for the first four bonds at the 5' end of the oligomer. The DNA substrate concentration is always in excess, in order to achieve conditions of no more than one DNase I cleavage per DNA molecule. Reactions were controlled so that 65% or more of the initial amount of decamer substrate remained after DNase I digestion. It was observed that the rate of enzyme reactivity decreases with digestion time and is sensitive to the experimental conditions.
Full text
PDF



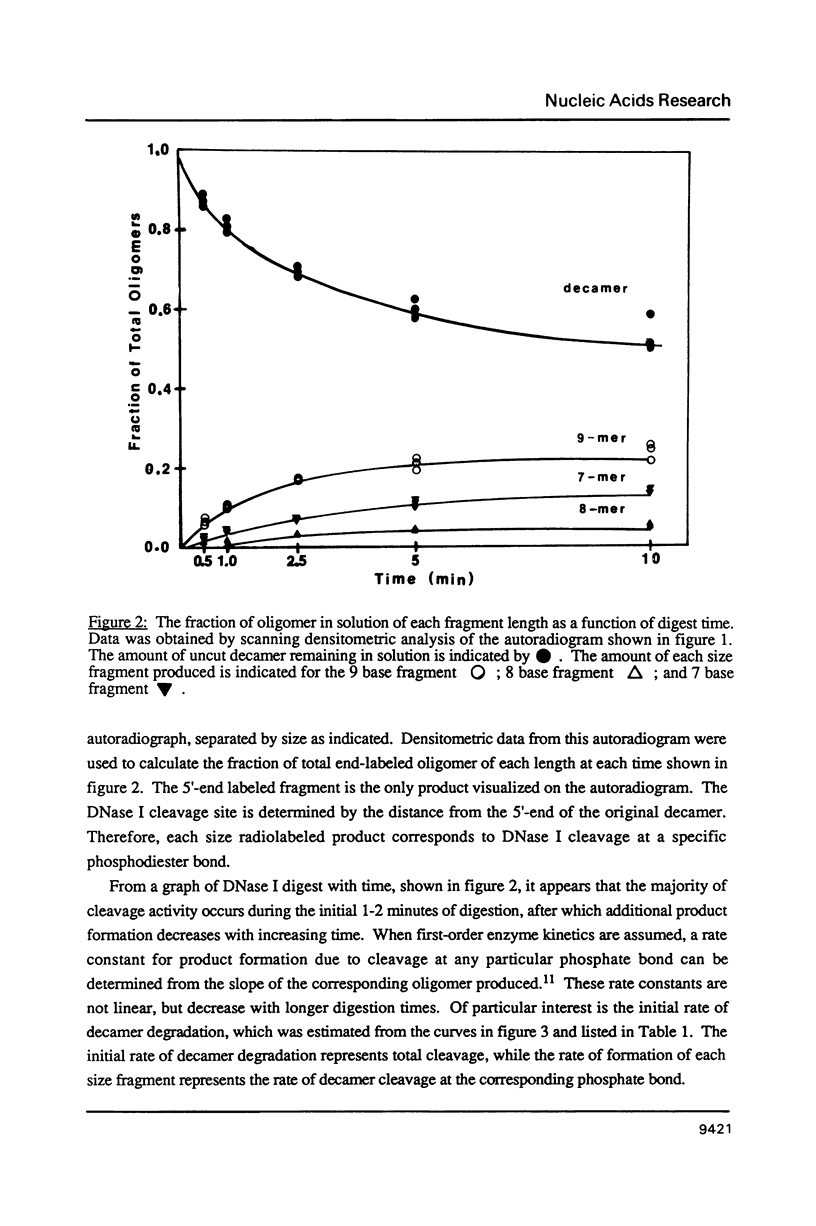
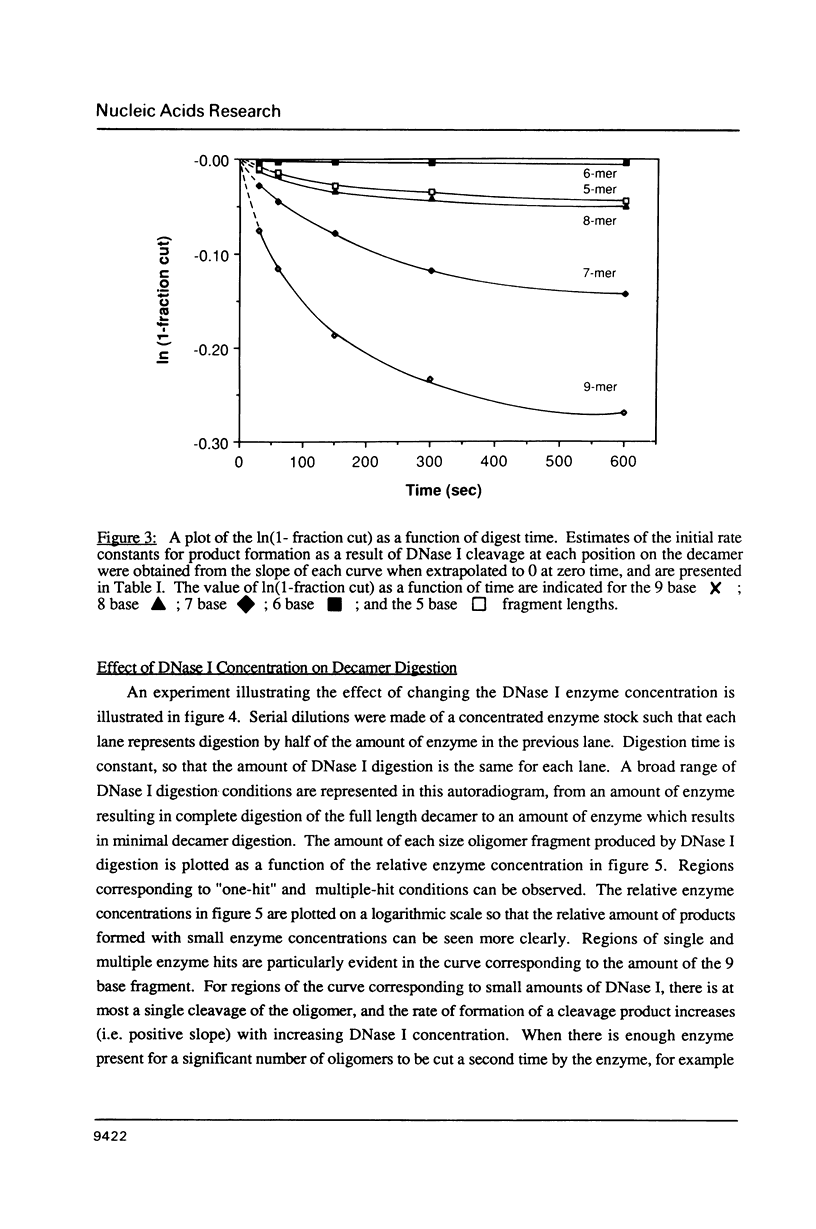

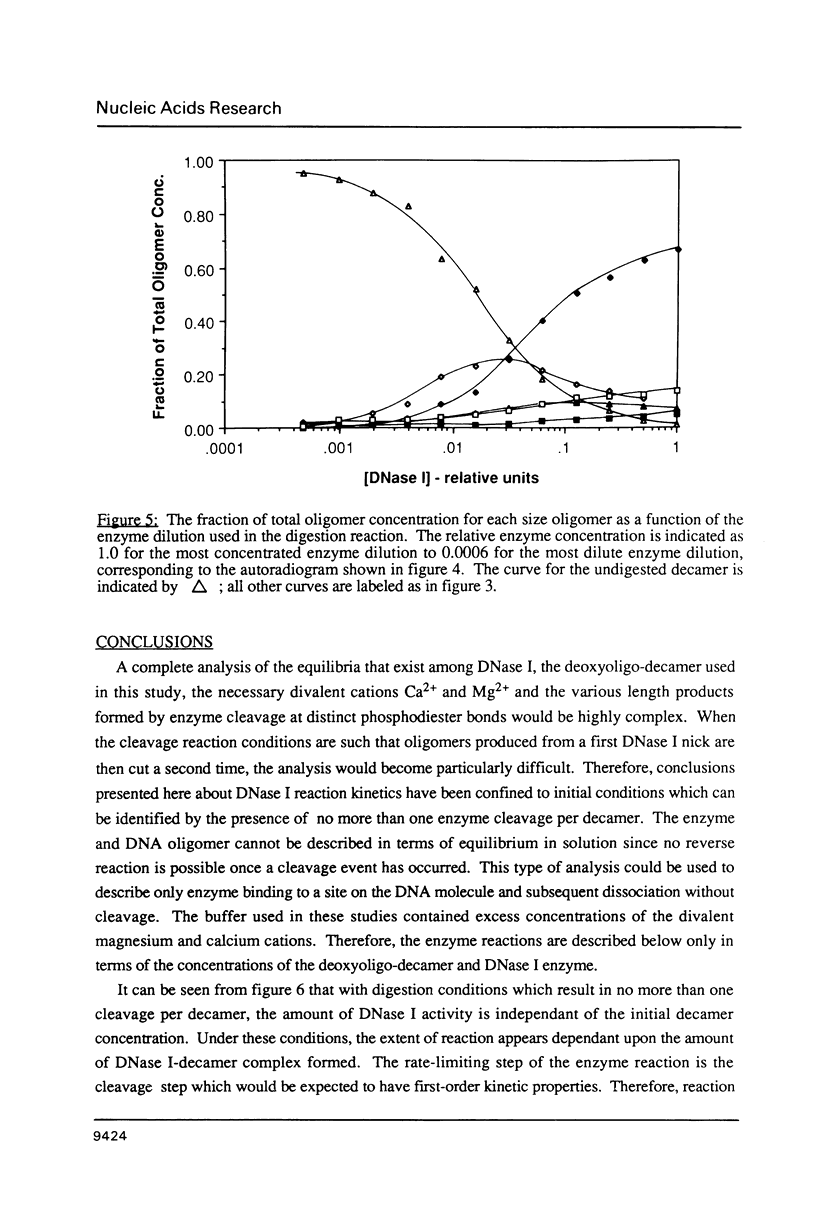
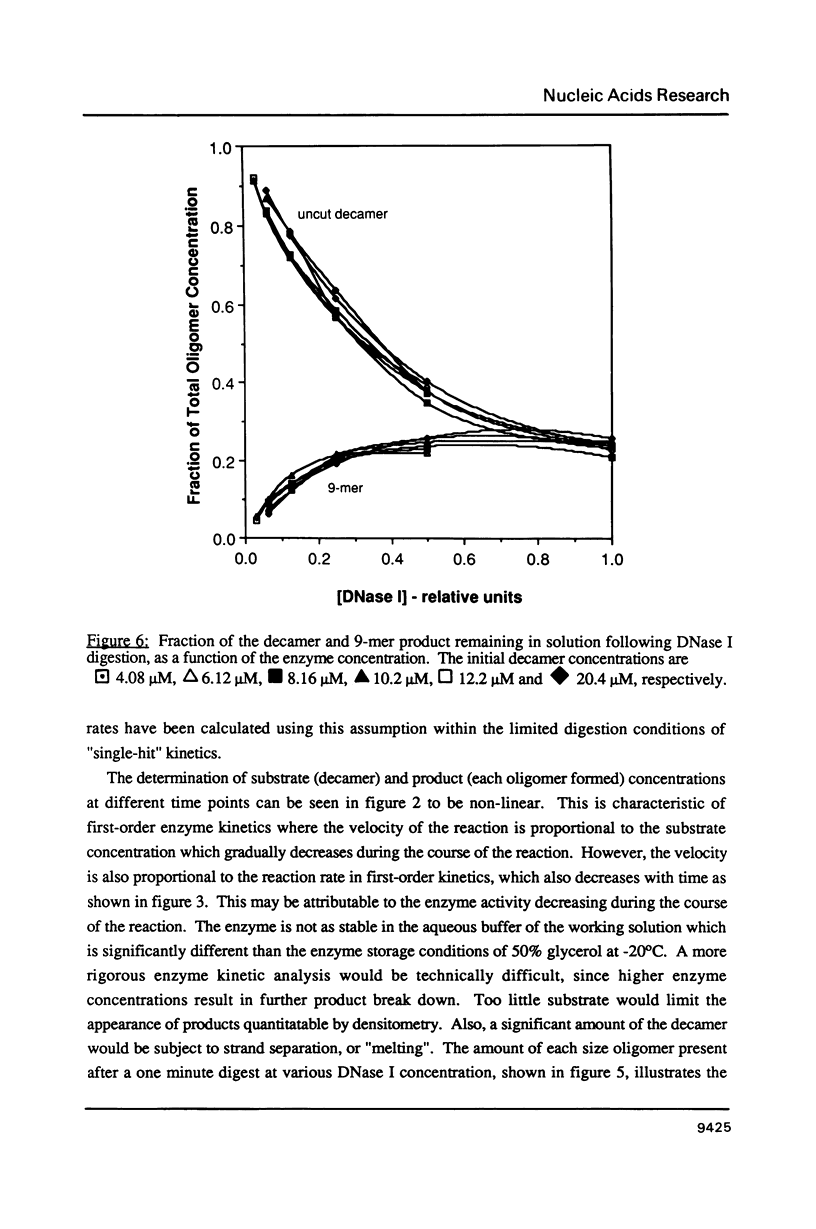


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dickerson R. E. Base sequence and helix structure variation in B and A DNA. J Mol Biol. 1983 May 25;166(3):419–441. doi: 10.1016/s0022-2836(83)80093-x. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R. Structure of a B-DNA dodecamer. II. Influence of base sequence on helix structure. J Mol Biol. 1981 Jul 15;149(4):761–786. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- Drew H. R. Structural specificities of five commonly used DNA nucleases. J Mol Biol. 1984 Jul 15;176(4):535–557. doi: 10.1016/0022-2836(84)90176-1. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA structural variations in the E. coli tyrT promoter. Cell. 1984 Jun;37(2):491–502. doi: 10.1016/0092-8674(84)90379-9. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane M. J., Dabrowiak J. C., Vournakis J. N. Sequence specificity of actinomycin D and Netropsin binding to pBR322 DNA analyzed by protection from DNase I. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3260–3264. doi: 10.1073/pnas.80.11.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonossoff G. P., Butler P. J., Klug A. Sequence-dependent variation in the conformation of DNA. J Mol Biol. 1981 Jul 15;149(4):745–760. doi: 10.1016/0022-2836(81)90356-9. [DOI] [PubMed] [Google Scholar]
- Lutter L. C. Kinetic analysis of deoxyribonuclease I cleavages in the nucleosome core: evidence for a DNA superhelix. J Mol Biol. 1978 Sep 15;124(2):391–420. doi: 10.1016/0022-2836(78)90306-6. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mehdi S., Gerlt J. A. Syntheses and configurational analyses of thymidine 4-nitrophenyl [17O,18O]phosphates and the stereochemical course of a reaction catalyzed by bovine pancreatic deoxyribonuclease I. Biochemistry. 1984 Oct 9;23(21):4844–4852. doi: 10.1021/bi00316a005. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Brown D. D., Engelke D., Ng S. Y., Shastry B. S., Roeder R. G. The binding of a transcription factor to deletion mutants of a 5S ribosomal RNA gene. Cell. 1981 Mar;23(3):665–669. doi: 10.1016/0092-8674(81)90429-3. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pescador R., Urdea M. S. Use of unpurified synthetic deoxynucleotide primers for rapid dideoxynucleotide chain termination sequencing. DNA. 1984 Aug;3(4):339–343. doi: 10.1089/dna.1.1984.3.339. [DOI] [PubMed] [Google Scholar]
- Schmitz A., Galas D. J. The interaction of RNA polymerase and lac repressor with the lac control region. Nucleic Acids Res. 1979 Jan;6(1):111–137. doi: 10.1093/nar/6.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suck D., Oefner C., Kabsch W. Three-dimensional structure of bovine pancreatic DNase I at 2.5 A resolution. EMBO J. 1984 Oct;3(10):2423–2430. doi: 10.1002/j.1460-2075.1984.tb02149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suck D., Oefner C. Structure of DNase I at 2.0 A resolution suggests a mechanism for binding to and cutting DNA. Nature. 1986 Jun 5;321(6070):620–625. doi: 10.1038/321620a0. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Hertzberg R. P., Dervan P. B. Map of distamycin, netropsin, and actinomycin binding sites on heterogeneous DNA: DNA cleavage-inhibition patterns with methidiumpropyl-EDTA.Fe(II). Proc Natl Acad Sci U S A. 1982 Sep;79(18):5470–5474. doi: 10.1073/pnas.79.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. H., Ughetto G., Quigley G. J., Hakoshima T., van der Marel G. A., van Boom J. H., Rich A. The molecular structure of a DNA-triostin A complex. Science. 1984 Sep 14;225(4667):1115–1121. doi: 10.1126/science.6474168. [DOI] [PubMed] [Google Scholar]




