Abstract
Trans-cleaving hammerheads with discontinuous or extended stem I and with tertiary stabilizing motifs (TSMs) have been tested previously against short RNA substrates in vitro at low Mg2+ concentration. However, the potential of these ribozymes for targeting longer and structured RNAs in vitro and in vivo has not been examined. Here, we report the in vitro cleavage of short RNAs and of a 464-nt highly structured RNA from potato spindle tuber viroid (PSTVd) by hammerheads with discontinuous and extended formats at submillimolar Mg2+. Under these conditions, hammerheads derived from eggplant latent viroid and peach latent mosaic viroid (PLMVd) with discontinuous and extended formats, respectively, where the most active. Furthermore, a PLMVd-derived hammerhead with natural TSMs showed activity in vivo against the same long substrate and interfered with systemic PSTVd infection, thus reinforcing the idea that this class of ribozymes has potential to control pathogenic RNA replicons.
INTRODUCTION
The hammerhead ribozyme is a catalytic RNA motif that in viroids and viroid-like satellite RNAs, wherein it was initially discovered, mediates cis-cleavage of the multimeric strands resulting from a rolling-circle replication (1,2). Most of the natural hammerheads are formed by a central conserved core flanked by three double-stranded regions with relaxed sequence requirements (helices I, II and III), two of which (I and II) are capped by short loops (1 and 2, respectively) (3). Minimal trans-cleaving hammerheads including the central conserved core and two hybridizing arms have been generated by removing the peripheral loop 1, which initially was thought nonessential for catalytic activity, and extending helix I to specifically target for cleavage an RNA after a GUH sequence (where H is any nucleotide except G) (4,5). These and other ribozymes have received considerable attention because of their potential for the specific inactivation of cellular or viral RNAs. However, the target accessibility, the subcellular co-localization of ribozyme and substrate, and the catalytic activity at the low physiological concentration of Mg2+, are still barriers that limit the use of hammerheads in vivo (6).
A re-examination of natural hammerheads—in which the helix-loop motifs flanking the central conserved core are preserved—has shown that these ribozymes display significantly higher self-cleavage rates, suggesting the existence of tertiary interactions between loops 1 and 2 critical for catalysis (7,8). The tertiary interactions between loops indeed exist and stabilize the catalytically active structure at the submillimolar concentrations of Mg2+ present in vivo (9), thus explaining why minimal trans-cleaving hammerheads require higher concentrations of this cation for adopting the active folding (10–12). Recent studies of these tertiary interactions in the hammerhead of tobacco ring spot virus satellite RNA (sTRSV) (2) have revealed a Hoogsteen pair between an A in stem–loop II and a U in a nonhelical region of stem I that is apparently conserved in most natural hammerheads possibly due to its functional relevance (13). Moreover, analysis of loops 1 and 2 of the hammerheads of chrysanthemum chlorotic mottle viroid (CChMVd) (14) by nuclear magnetic resonance (NMR) spectroscopy, site-directed mutagenesis, self-cleavage kinetics and infectivity bioassays has shown that loop 1 contains an exposed 5′-U and an extra-helical 3′-U, and that loop 2 has an opened 3′-A (15). Contacts between loops 1 and 2 of most natural hammerheads may take place across the major grove of the RNA, and as a consequence of the resulting conformational changes, the 3′-A of loop 2 can form a base-pair with the 5′-U of loop 1 and the extra-helical pyrimidine of loop 1 can interact with the 3′ portion of loop 2 (15). The relevance of these residues is evidenced by their conservation in most natural hammerheads (15).
Tertiary stabilizing motifs (TSMs), specifically interactions between peripheral loops, have been incorporated into a new generation of more efficient trans-cleaving hammerheads in two different manners: (i) by extending stem I and including loop 1 as a bulge in the hybridizing arm of this stem (extended format) (16,17) and (ii) by embedding within stem I the 5′ and 3′ termini of the ribozyme and substrate, respectively (discontinuous format) (18). Some of these hammerheads are active in vitro at low Mg2+ concentration against short RNA substrates. In particular, discontinuous hammerheads derived from sTRSV and extended hammerheads derived from peach latent mosaic viroid (PLMVd) (19) are the most efficient when compared to other trans-cleaving hammerheads (16–18).
However, the recently characterized hammerheads of eggplant latent viroid (ELVd) (20) have not been yet adapted into a trans design, despite displaying higher self-cleavage rates than other natural hammerheads at very low Mg2+ concentrations (21). Moreover, these hammerheads seem particularly appropriate for the discontinuous format because their long stem I (of 7 bp) should facilitate substrate binding and folding of loop 1.
Here, we report a comparative trans-cleavage analysis in vitro of some discontinuous and extended hammerheads, derived from ELVd, PLMVd and sTRSV, against RNAs of the pathogen potato spindle tuber viroid (PSTVd) (22). We have examined the ability of the hammerheads to catalyze cleavage of short RNA substrates and of a long and highly structured RNA containing the complete sequence of PSTVd (23,24). Our results, particularly at submillimolar Mg2+, show that the ELVd-derived hammerheads are functional, with the discontinuous variants being more efficient against the full-length PSTVd-RNA than their sTRSV counterparts, and that an extended PLMVd-derived hammerhead with natural TSMs displays the highest cleavage rate. Further analyses with transient expression bioassays in Nicotiana benthamiana plants have revealed that this latter hammerhead is also active in vivo and interferes with systemic PSTVd infection.
MATERIALS AND METHODS
Synthesis, purification and labeling of short RNA substrates
The oligonucleotides RF-979 (5′-GCUCAGGAGGUCAGGU-3′), RF-992 (5′-GCUCAGGAGGUCAGG-3′), RF-993 (5′-GCUCAGGAGGUCAGGUGU-3′) and RF-994 (5′-GCUCAGGAGGUCAGGUGUGAACCAC-3′), were chemically synthesized by Sigma-Aldrich, purified by PAGE in 20% denaturing gels, eluted by extracting the crushed gel pieces with buffer-saturated phenol (Tris–HCl 10 mM pH 7.5, EDTA 1 mM and SDS 0.1%), and recovered by ethanol precipitation and resuspended in deionized sterile water. The integrity and concentration of the purified RNA substrates were confirmed by denaturing gel electrophoresis. The RNAs were 5′-labeled with [γ-32P]ATP (3000 Ci/mmol; Perkin Elmer) and T4 polynucleotide kinase (25).
Preparation of cDNAs for expressing the trans-cleaving hammerheads and the long PSTVd (–) RNAs
Ribozymes were designed to target the GUC trinucleotide located at positions 322–324 in the PSTVd (–) RNA. This site was previously shown to be a suitable target for hammerhead-mediated cleavage in vitro and in vivo (26). The hammerhead cDNA constructs were prepared by extension and amplification of partially overlapping sense and antisense primers (five cycles at 94°C for 30 s, 50°C for 30 s and 72°C for 10 s, with a final extension at 72°C for 2 min), with sense primers including the T7 promoter. PCR products were separated by PAGE in 5% non-denaturing gels and those with the expected length were eluted and cloned into pUC18 digested with SmaI. Monomeric and dimeric head-to-tail PSTVd cDNAs (intermediate strain, M16826) were cloned into pBluescript II KS (+) digested with EcoRI/HindIII.
Synthesis and purification of the ribozymes and the long RNA substrate
Hammerheads were generated by in vitro transcription of the corresponding recombinant plasmids digested with BamHI. Transcription reactions (100 µl) contained 40 mM Tris–HCl pH 8, 6 mM MgCl2, 2 mM spermidine, 10 mM dithiothreitol, 10 mM NaCl, 0.4 mM each of ATP, CTP, GTP and UTP, 0.8 U/µl of ribonuclease inhibitor (rRNasin, Promega), 100 ng/µl of plasmid and 1.2 U/µl of T7 RNA polymerase (Roche). Radiolabeled full-length monomeric PSTVd (–) RNA (464 nt) was obtained by in vitro transcription as described above, but the UTP concentration was 0.08 mM and 0.25 µCi/µl [α-32P]UTP was added to the transcription mixture. After incubation at 37°C for 1 h the transcription products were fractionated by PAGE in 5% gels with 8 M urea. The primary transcripts were eluted as described above, recovered by ethanol precipitation and resuspended in deionized sterile water. The integrity and concentration of the purified ribozyme and substrate were confirmed by denaturing PAGE in 20% and 5% gels, respectively, with appropriate size markers.
Kinetic analysis
Trans-cleaving rate constants were determined under single-turnover conditions using an excess of the ribozyme (200 nM) and traces of the 32P-labeled substrate (2 nM). Ribozyme and substrate were first annealed in Tris–HCl 50 mM pH 7.5, by heating at 95°C for 1 min and slowly cooling to 25°C (ramping decrease 1°C/10 s), and then incubated at this temperature for 5 min. After taking a zero-time aliquot, cleavage reactions were triggered by adding MgCl2 to the desired final concentration (10, 1 or 0.1 mM). Aliquots were removed at different time intervals and quenched at 0°C after adding a 10- or 5-fold excess of stop solution (8 M urea, 50% formamide, 50 mM EDTA, 0.1% xylene cyanol and bromophenol blue) for short and long substrates, respectively. Substrate and cleavage products were separated by PAGE in 20% (short RNAs) or 5% (long RNA) denaturing gels. The product fraction at different times, Ft, was determined by quantitative scanning of the corresponding gel bands and data adjustment to a single-exponential equation Ft = F0 + F∞(1 – e-kt), where F0 and F∞ are the product fractions at zero time and at the reaction end point, respectively, and k is the first-order rate constant of cleavage (kcat). Those data sets that could not be adequately fitted to a single exponential were adjusted to a double-exponential using the equation Ft = Fa(1 – e-kat) + Fb(1-e-kbt), where Fa and ka correspond to the product fraction and the rate constant for a rapid process ‘a’, and Fb and kb to the product fraction and the rate constant for a slow process ‘b’.
Agroinfiltration
The monomeric and dimeric PSTVd-cDNAs and the hammerhead-cDNAs were subcloned into a modified version of the pMOG180 vector between a double copy of the 35S CaMV promoter and the Nos-terminator. The expression cassettes were then subcloned into the plant binary expression vector pBIN19sGFP by replacing the sGFP cassette to obtain the recombinant plasmids pBINmPSTVd(–), pBINdPSTVd(–), pBINHHePLMVd, pBINHHePLMVdG5→U, pBINHHesTRSVΔL1, and pBINØ (the empty vector). Protocols for agroinfiltrating N. benthamiana plants were described previously (27,28). Bioassays were performed in a growth chamber at 23°C for 16 h with fluorescent light and at 19°C for 8 h in darkness. RNAs from the infiltrated and non-infiltrated upper leaves were extracted with a phenol-based protocol (29). The PSTVd (–) primary transcripts and the monomeric (+) circular and linear RNAs resulting from viroid replication were detected by denaturing PAGE in 5% gels containing 8 M urea, followed by northern-blot hybridization at 70°C in 50% formamide with strand-specific 32P-labeled riboprobes transcribed in vitro (30).
RESULTS
In vitro trans-cleavage activity of discontinuous hammerheads (HHd) against short RNA substrates
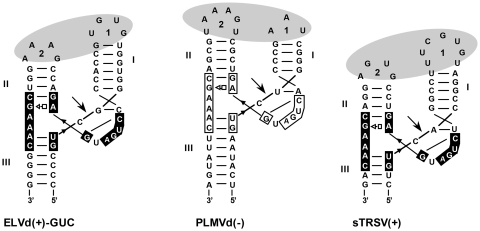
Trans-cleaving hammerheads with TSMs were designed against RNA oligonucleotides corresponding to a fragment of the minus (–) polarity strand of PSTVd that includes a GUC target site (positions 322–324). Previous studies on the self-cleavage kinetics of variants of the ELVd (+) hammerhead have shown that those with a GUC or AUC trinucleotide preceding the self-cleavage site are the most active at very low Mg2+ concentration (21). We therefore reasoned that the ELVd(+)-GUC hammerhead (Figure 1) could serve for designing discontinuous trans-cleaving hammerheads with preserved TSMs, which were named according to the base-pair of stem I adjacent to loop 1 and to the sequence of this loop (Figure 2A). First, HHd-ELVd-UA/GUGU was designed with its loop 1 closed by part of stem I (Ia, of three base-pairs), and the U-G base-pair adjacent to loop 1 substituted by the stronger U-A base-pair present in some ELVd variants (20) for increasing the stability of stem Ia. The other part of stem I (Ib, of 4 bp) served for hybridizing with the substrate. Therefore, stem I was separated into two discrete segments in HHd-ELVd-UA/GUGU.
Figure 1.
Secondary structures of the ELVd(+)-GUC, PLMVd(–) and sTRSV(+) self-cleaving hammerheads represented according to crystallographic data obtained for the Schistosoma mansoni and the sTRSV(+) hammerheads (9,13). Motifs conserved in most natural hammerheads are within boxes and self-cleavage sites are marked by arrows. Black and white backgrounds refer to (+) and (–) polarities, respectively. Dashes denote Watson–Crick (and wobble) pairs and the open square-triangle a Hoogsteen/sugar edge interaction. Nomenclature of helices and loops follows the standard criterion (43). Ovals represent the proposed tertiary interactions between loops 1 and 2.
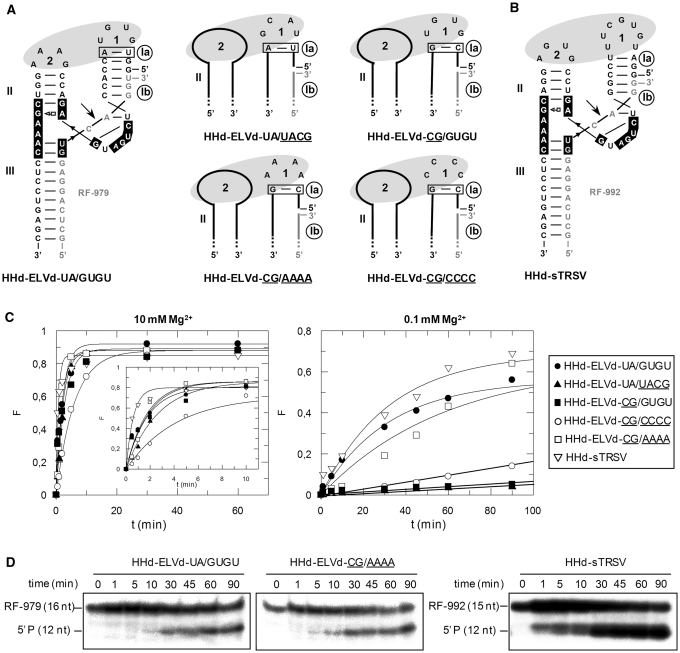
Figure 2.
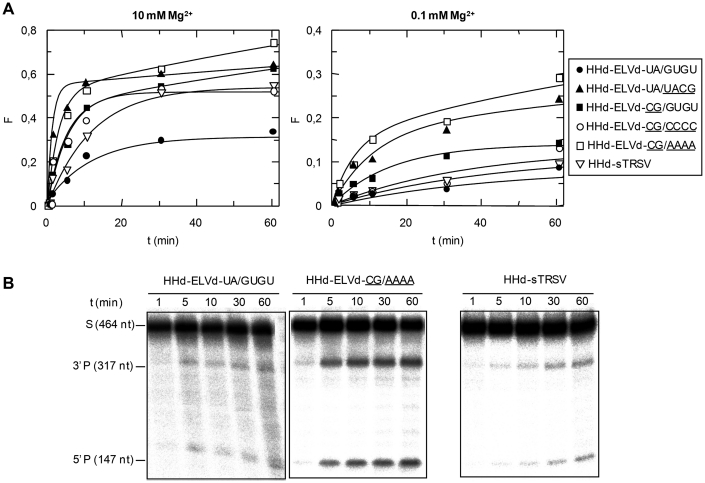
Structure and properties of discontinuous hammerheads (HHd) against short RNA substrates. (A) Schematic representation of the complex formed by the HHd-ELVd-UA/GUGU hammerhead and the substrate RF-979 (left), and schemes for complexes involving other discontinuous ELVd-derived hammerheads (right). Hammerhead and substrate nucleotides are shown with black and grey fonts, respectively, and unchanged ribozyme and substrate nucleotides are represented with continuous black and grey lines, respectively. Ia and Ib refer to the distal and proximal halves of stem I, respectively, with the base-pair of stem Ia adjacent to loop 1 being boxed. (B) Predicted secondary structure for the complex between HHd-sTRSV and the substrate RF-992. Other details as in (A). (C) Diagrams representing the product fraction (F) as a function of time at 10 and 0.1 mM Mg+2 (left and right, respectively) generated by five different hammerheads: HHd-ELVd-UA/GUGU, HHd-ELVd-UA/UACG, HHd-ELVd-CG/AAAA, HHd-ELVd-CG/CCCC and HHd-sTRSV. F-values represent the mean of two independent experiments, and the inset displays the first 10 min of each reaction. (D) Analysis by denaturing PAGE (20%) and autoradiography of reactions catalyzed by three discontinuous hammerheads at 0.1 mM Mg+2. The positions and size of the 5′-labeled substrates (RF-979 and RF-992) and of the resulting 5′-cleavage products (P) are indicated.
This hammerhead was very active at 10 and 1 mM Mg2+, catalyzing specifically and efficiently cleavage of the substrate RF-979 (92% and 90% at the end point of the reaction, respectively), and still keeping a significant activity at 0.1 mM Mg2+ (Table 1, Figure 2C and D). We also designed other ELVd-derived artificial hammerheads with TSMs (Figure 2A), in which the sequence of the loop 1 and/or the base-pair adjacent to this loop were modified to evaluate the effect of these changes on the hammerhead activity. In hammerheads HHd-ELVd-UA/UACG, HHd-ELVd-CG/AAAA and HHd-ELVd-CG/CCCC, the wild-type loop 1 (GUGU) was replaced by UACG, AAAA or CCCC, respectively. Formation of loop 1 should be facilitated by the high stability of the UACG tetraloop belonging to the UNCG family (31) in HHd-ELVd-UA/UACG, and by the stronger C-G base-pair of stem Ia in HHd-ELVd-CG/AAAA and HHd-ELVd-CG/CCCC. These three hammerheads catalyzed cleavage of a high fraction of the substrate at 10 and 1 mM Mg2+, but only HHd-ELVd-CG/AAAA remained active at 0.1 mM Mg2+ (Table 1, Figure 2C and D), suggesting that alternative tertiary interactions between artificial loops 1 and the wild-type loop 2 might promote cleavage at submillimolar Mg2+ (32). In support of this notion, RNase T1 probing was consistent with formation of the ribozyme–substrate complex by the HHd-ELVd-UA/GUGU, HHd-ELVd-UA/UACG and HHd-ELVd-CG/AAAA hammerheads (Supplementary Figures S1 and S2). The other ELVd-derived hammerhead, HHd-ELVd-CG/GUGU, in which the U-A base-pair adjacent to loop 1 was replaced by a stronger C-G pair that should facilitate formation of this loop, did not show higher efficiency (Table 1, Figure 2C).
Table 1.
Trans-cleavage activity of discontinuous (HHd) and extended (HHe) hammerheads against short RNA substrates
| Hammerhead | [MgCl2] | kcat (min−1)a | F∞b |
|---|---|---|---|
| HHd-ELVd-UA/GUGU | 10 | 0.52 ± 0.065 | 0.92 |
| 1 | 0.49 ± 0.062 | 0.90 | |
| 0.1 | 0.03 ± 0.002 | 0.47 | |
| HHd-ELVd-UA/UACG | 10 | 0.36 ± 0.002 | 0.89 |
| 1 | 0.31 ± 0.015 | 0.87 | |
| 0.1 | 0.05 ± 0.003 | 0.04 | |
| HHd-ELVd-CG/AAAA | 10 | 0.52 ± 0.058 | 0.88 |
| 1 | 0.42 ± 0.024 | 0.85 | |
| 0.1 | 0.02 ± 0.003 | 0.43 | |
| HHd-ELVd-CG/CCCC | 10 | 0.18 ± 0.008 | 0.88 |
| 1 | 0.13 ± 0.008 | 0.88 | |
| 0.1 | n.m.c | 0.09 | |
| HHd-ELVd-CG/GUGU | 10 | 0.45 ± 0.069 | 0.88 |
| 1 | 0.25 ± 0.059 | 0.89 | |
| 0.1 | n.m. | 0.05 | |
| HHd-sTRSV | 10 | 1.41 ± 0.194 | 0.85 |
| 1 | 0.53 ± 0.090 | 0.79 | |
| 0.1 | 0.03 ± 0.003 | 0.62 | |
| HHe-PLMVd | 10 | 1.11 ± 0.148 | 0.87 |
| 1 | 0.28 ± 0.051 | 0.79 | |
| 0.1 | 0.08 ± 0.008 | 0.79 | |
| HHe-ELVd | 10 | 0.41 ± 0.043 | 0.85 |
| 1 | 0.07 ± 0.003 | 0.86 | |
| 0.1 | 0.02 ± 0.003 | 0.18 | |
| HHe-ELVdΔL1 | 10 | 0.14 ± 0.010 | 0.88 |
| 1 | 0.04 ± 0.003 | 0.61 | |
| 0.1 | n.m. | 0.00 | |
| HHe-sTRSVΔL1 | 10 | 0.63 ± 0.028 | 0.97 |
| 1 | 0.45 ± 0.031 | 0.98 | |
| 0.1 | 0.02 ± 0.002 | 0.48 |
aCleavage rate constant.
bFraction of product at the end point of the reaction.
cNon-measurable.
Because previous studies have shown that discontinuous ribozymes derived from sTRSV (+) hammerhead (Figure 1), with a stem Ib of only three base-pairs, can catalyze cleavage of short RNA substrates at low Mg2+ concentration (18), a variant thereof (HHd-sTRSV) was designed against a short RNA (RF-992) derived from the PSTVd (–) strand that was identical to the previous one (RF-979) but one nucleotide shorter at the 3′ end (Figure 2B). HHd-sTRSV promoted very efficient cleavage of the substrate in vitro (Figure 2D), showing the highest values of catalytic constant (kcat) (Table 1). Therefore, the most efficient hammerheads, HHd-ELVd-UA/GUGU and HHd-sTRSV, were derived from natural hammerheads in which the sequence of the loops 1 and 2 remain unaltered. Both hammerheads with natural loops, and one ELVd-derived ribozyme with an artificial loop 1 (HHd-ELVd-CG/AAAA), still remained active at low Mg2+ (0.1 mM) promoting cleavage of 47%, 62% and 43% of the substrate, respectively (Table 1). No experiments were attempted with PLMVd-derived hammerheads, because a stem I of only 5 bp (Figure 1) lacks sufficient stability to support the discontinuous format (18).
In vitro trans-cleavage activity of extended hammerheads (HHe) against short RNA substrates
TSMs have also been incorporated into extended hammerheads by including loop 1 as a bulge in the hybridizing arm of stem I. Because these extended PLMVd-derived ribozymes against short RNAs are more efficient than those derived from other natural hammerheads (16,17), we designed a variant (HHe-PLMVd) against a short RNA (RF-994) corresponding to a segment of the (–) strand of PSTVd (Figure 3A). HHe-PLMVd has unmodified wild-type loops 1 (UAA) and 2 (UAAAGU) (Figure 1) (19) to preserve loop–loop interactions. This hammerhead displayed high kcat at 10 and 1 mM Mg2+, catalyzing cleavage of 87% and 79% of the substrate, respectively, and still keeping high activity at 0.1 mM Mg2+ (Table 1, Figure 3B and C).
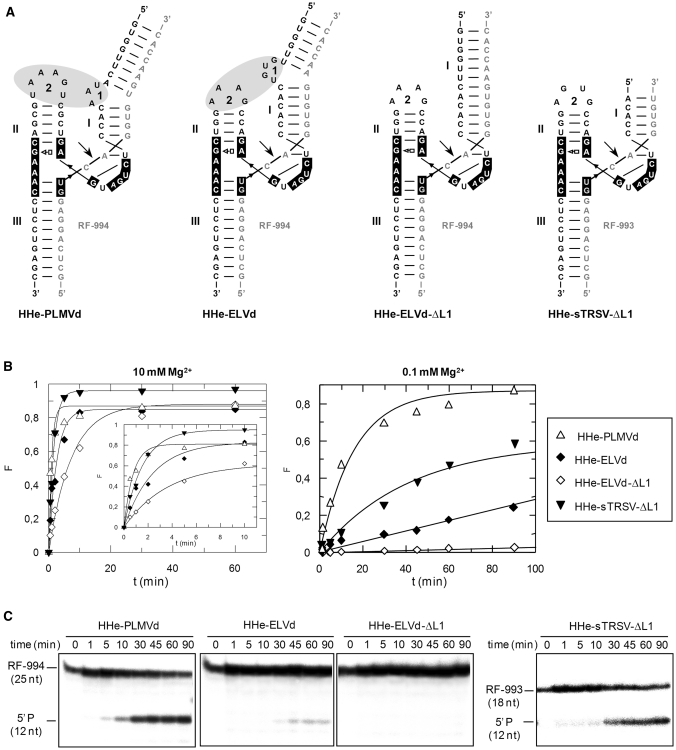
Figure 3.
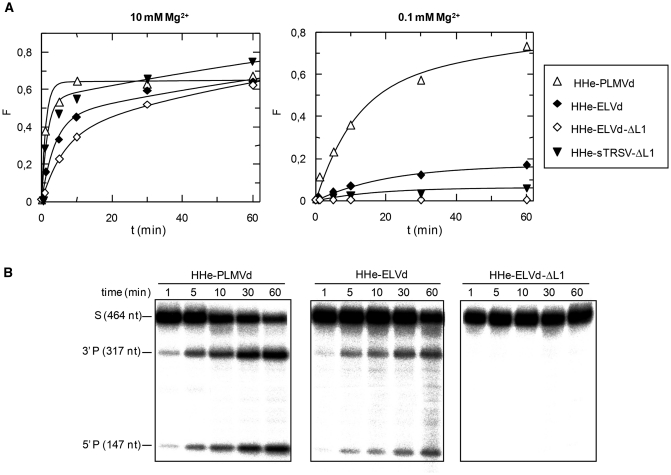
Structure and properties of extended hammerheads (HHe) against short RNA substrates. (A) Schematic representation of the different ribozyme–substrate complexes. HHe-PLMVd and HHe-ELVd hammerheads contain TSMs, while the minimal HHe-sTRSV-ΔL1 and HHe-ELVd-ΔL1 hammerheads lack these motifs. (B) Diagrams representing the product fraction (F) as a function of time at 10 and 0.1 mM Mg+2 (left and right, respectively) generated by four different hammerheads: HHe-PLMVd, HHe-ELVd, HHe-ELVd-ΔL1 and HHe-sTRSV-ΔL1. The inset displays the first 10 min of each reaction. (C) Analysis by denaturing PAGE (20%) and autoradiography of reactions catalyzed by four extended hammerheads at 0.1 mM Mg+2. The positions and size of the 5′-labeled substrates (RF-994 and RF-993) and of the resulting 5′-cleavage products (P) are indicated.
Two extended ELVd-derived hammerheads were also generated against the same substrate: (i) HHe-ELVd, wherein the wild-type loop 1 was included in the hybridizing arm of stem I as a bulging loop 7 nt apart from the catalytic core as in the natural ribozyme and (ii) HHe-ELVd-ΔL1, a minimal hammerhead wherein loop 1 was deleted (Figure 3A). At 10 mM Mg2+, HHe-ELVd displayed a significant higher catalytic constant when compared with HHe-ELVd-ΔL1, although the fraction of the substrate cleaved at the reaction end point was approximately the same (Table 1, Figure 3B). In contrast, HHe-ELVd-ΔL1 was essentially inactive at 0.1 mM Mg2+, while HHe-ELVd was able to catalyze cleavage of 18% of the substrate (Figure 3B and C). Moreover, RNase T1 probing was consistent with the formation of a ribozyme–substrate complex by the HHe-ELVd hammerhead (Supplementary Figure S3). These results indicate that extended ELVd-derived hammerheads with TSMs retain activity at submillimolar Mg+2, although they are less efficient than their discontinuous counterparts.
In addition, a minimal sTRSV-derived hammerhead lacking loop 1 (HHe-sTRSV-ΔL1), a modified version of a ribozyme without TSMs (Figure 3A) but with in vivo activity when stably expressed in transgenic potato plants (26), was included for comparative purposes. HHe-sTRSV-ΔL1 was designed with 5′ and 3′ hybridizing arms of 11 and 6 nt, respectively, instead of the 11 and 10 nt of the original ribozyme (26), because shorter hybridizing arms generally result in specific substrate binding and faster product release, thus maximizing turnover rate (33,34). The hammerhead was active against the short RNA at 10 and 1 mM Mg2+ (catalyzing cleavage of 97% and 98% of the substrate, respectively) although the kcat values were lower than those of the HHe-PLMVd (Table 1, Figure 3B). As expected, the catalytic constant and the fraction of cleaved substrate at the end point decreased when the Mg2+ concentration was reduced to 0.1 mM (Table 1, Figure 3B and C), most likely as a result of the absence of TSMs in this hammerhead.
In vitro trans-cleavage of a highly-structured RNA by discontinuous hammerheads
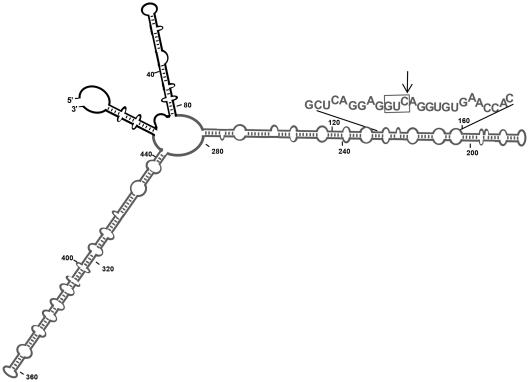
We next examined the ability of these hammerheads to catalyze cleavage of a long and highly structured RNA in vitro. To this aim, we generated a 464-nt RNA by in vitro run-off transcription of a monomeric PSTVd-cDNA clone. The resulting transcript included a GUC target site within the PSTVd (–) full-length strand (359 nt) flanked by vector sequences at both 5′ and 3′ termini (17 and 88 nt, respectively) (Figure 4). Previous analysis by temperature-gradient gel electrophoresis (35) support the rod-like conformation of PSTVd (–) RNA predicted by the Mfold program (36), and our own results obtained by RNase T1 probing (data not shown) indicate that the flanking plasmid sequences do not disturb this conformation.
Figure 4.
Schematic representation of the Mfold-predicted secondary structure of the long RNA substrate. Major features of this compact structure have been validated by temperature-gradient gel electrophoresis (35). PSTVd and flanking vector sequences are indicated in grey and black, respectively. The Mfold program (36) and our results obtained with RNase T1 probing (data not shown), indicated that the vector sequences do not disturb the rod-like conformation of the PSTVd(–) RNA. The hammerhead-binding sequence is denoted with capital fonts, with the trinucleotide GUC preceding the cleavage site boxed and the cleavage site marked with an arrow.
First, we analyzed the activity of the HHd-ELVd hammerheads. In contrast with the results obtained with the short substrate, the HHd-ELVd-UA/GUGU hammerhead showed very low catalytic activity against the PSTVd (–) RNA even at 10 mM Mg2+ (Table 2, Figure 5A). The most likely interpretation for this result is that the compact rod-like conformation of PSTVd (–) RNA disfavors hybridization with the ribozyme arms. Indeed, RNase T1 probing revealed formation of the hammerhead–substrate complex with the short RNA, but neither with the PSTVd (–) RNA nor with short and long non-substrate RNAs (Supplementary Figure S2A).
Table 2.
Trans-cleavage activity of discontinuous (HHd) and extended (HHe) hammerheads against the full-length PSTVd (–) RNA
| Hammerhead | [MgCl2] | ka (min−1)a | kb (min−1)b | F∞c | Fad |
|---|---|---|---|---|---|
| HHd-ELVd-UA/GUGU | 10 | 0.08 ± 0.006 | 0.32 | ||
| 1 | 0.03 ± 0.005 | 0.21 | |||
| 0.1 | n.m.e | 0.08 | |||
| HHd-ELVd-CG/GUGU | 10 | 0.21 ± 0.032 | 0.005 ± 0.001 | 0.62 | 0.45 |
| 1 | 0.25 ± 0.038 | 0.009 ± <0.001 | 0.58 | 0.34 | |
| 0.1 | 0.06 ± 0.004 | 0.14 | |||
| HHd-ELVd-UA/UACG | 10 | 0.76 ± 0.206 | 0.002 ± 0.001 | 0.64 | 0.55 |
| 1 | 0.18 ± 0.007 | 0.013 ± 0.002 | 0.58 | 0.23 | |
| 0.1 | 0.09 ± 0.002 | 0.005 ± 0.002 | 0.24 | 0.17 | |
| HHd-ELVd-CG/AAAA | 10 | 0.33 ± 0.048 | 0.006 ± 0.001 | 0.74 | 0.52 |
| 1 | 0.26 ± 0.026 | 0.007 ± <0.001 | 0.76 | 0.48 | |
| 0.1 | 0.16 ± 0.074 | 0.010 ± 0.003 | 0.29 | 0.15 | |
| HHd-ELVd-CG/CCCC | 10 | 0.18 ± 0.038 | 0.52 | ||
| 1 | 0.11 ± 0.024 | 0.005 ± 0.001 | 0.50 | 0.35 | |
| 0.1 | 0.06 ± 0.005 | 0.13 | |||
| HHd-sTRSV | 10 | 0.08 ± 0.013 | 0.54 | ||
| 1 | 0.06 ± 0.007 | 0.43 | |||
| 0.1 | n.m. | 0.09 | |||
| HHe-PLMVd | 10 | 0.83 ± 0.188 | < 0.001 ± 0.001 | 0.67 | 0.64 |
| 1 | 0.77 ± 0.274 | < 0.001 ± <0.001 | 0.6 | 0.58 | |
| 0.1 | 0.09 ± 0.012 | 0.004 ± 0.001 | 0.73 | 0.57 | |
| HHe-ELVd | 10 | 0.27 ± 0.045 | 0.007 ± <0.001 | 0.64 | 0.45 |
| 1 | 0.08 ± 0.007 | 0.56 | |||
| 0.1 | 0.05 ± 0.004 | 0.17 | |||
| HHe-ELVdΔL1 | 10 | 0.17 ± 0.011 | 0.010 ± <0.001 | 0.63 | 0.35 |
| 1 | 0.02 ± 0.003 | 0.015 ± 0.002 | 0.61 | 0.35 | |
| 0.1 | n.m. | 0.00 | |||
| HHe-sTRSVΔL1 | 10 | 0.61 ± 0.133 | 0.005 ± 0.001 | 0.76 | 0.55 |
| 1 | 0.13 ± 0.009 | 0.004 ± 0.002 | 0.73 | 0.53 | |
| 0.1 | n.m. | n.m. | 0.00 |
aCleavage rate constant (in biphasic cleavage reactions refers to the rapid ‘a’ process).
bCleavage rate constant for the slow ‘b’ process in biphasic cleavage reactions
cFraction of product at the end point of the reaction
dFraction of product at the end point of the rapid ‘a’ process in biphasic cleavage reactions.
eNon-measurable.
Figure 5.
Discontinuous hammerheads (HHd) against PSTVd (–) RNA. (A) Diagrams representing the product fraction (F) as a function of time at 10 and 0.1 mM Mg+2 (left and right, respectively) generated by five different hammerheads: HHd-ELVd-UA/GUGU, HHd-ELVd-UA/UACG, HHd-ELVd-CG/AAAA, HHd-ELVd-CG/CCCC and HHd-sTRSV. F-values represent the mean of two independent experiments. (B) Analysis by denaturing PAGE (5%) and autoradiography of reactions catalyzed by three of the hammerheads at 0.1 mM Mg+2. The positions and size of the substrate (S) and of the resulting 3′ and 5′ cleavage products (P) are indicated.
The HHd-ELVd-CG/GUGU variant (with the U-A base-pair adjacent to loop 1 replaced by a stronger C-G base-pair that should facilitate formation of this loop), which was moderately efficient against the short RNA substrate, was more efficient than the wild-type HHd-ELVd-UA/GUGU hammerhead against the full-length PSTVd (–) RNA (Table 2, Figure 5A and B). These results support the idea that stem Ia stability may be critical for loop 1 formation and, by extension, for active discontinuous hammerheads as proposed previously (18).
HHd-ELVd-UA/UACG, HHd-ELVd-CG/AAAA and HHd-ELVd-CG/CCCC, the three additional hammerheads designed with artificial sequences in loop 1, should not form, according to Mfold, stable interactions with the substrate alternative to the catalytically active folding. In consonance with these predictions, HHd-ELVd-UA/UACG and HHd-ELVd-CG/AAAA displayed high cleavage rates and, particularly, the latter hammerhead catalyzed end-point cleavage of 74%, 76% and 29% of the substrate at 10, 1 and 0.1 mM Mg2+, respectively (Table 2, Figure 5A and B). In vitro probing with RNase T1 supports that HHd-ELVd-CG/AAAA forms the expected complex with the substrate (Supplementary Figure S2C). These data suggest again that alternative tertiary interactions between artificial loop 1 sequences and the wild-type loop 2 might promote hammerhead stability and cleavage at submillimolar Mg2+.
On the other hand, HHd-sTRSV, which was the most efficient discontinuous ribozyme against the short RNA substrate, was less efficient than its ELVd counterparts against the PSTVd full-length substrate (Table 2). The reduction in the catalytic activity of HHd-sTRSV was especially important at 0.1 mM Mg2+, with only 9% of the substrate cleaved (Table 2, Figure 5A and B), probably as a consequence of poor substrate binding by the 1-nt-shorter stem Ib.
In vitro trans-cleavage of a highly structured RNA by extended hammerheads
The HHe-PLMVd hammerhead, which was the most efficient against the short RNA substrate (Table 1), was also very active against the highly structured full-length PSTVd (–) RNA (Table 2). This hammerhead displayed high catalytic constants at 10, 1 and 0.1 mM Mg2+ cleaving also a significant fraction of the substrate at all Mg2+ concentrations (Table 2, Figure 6A and B).
Figure 6.
Extended hammerheads (HHe) against against PSTVd (–) RNA. (A) Diagrams representing the product fraction as a function of time at 10 and 0.1 mM Mg+2 (left and right, respectively) generated by HHe-PLMVd and HHe-ELVd hammerheads, and the minimal HHe-ELVd-ΔL1 and HHe-sTRSV-ΔL1 hammerheads. (B) Analysis by denaturing PAGE (5%) and autoradiography of reactions catalyzed by three of the hammerheads at 0.1 mM Mg+2. Other details as in Figure 5.
The extended hammerhead HHe-ELVd and the minimal hammerhead HHe-ELVd-ΔL1 were also tested against the same long RNA substrate. At 10 mM Mg2+, HHe-ELVd displayed a 2-fold increase of the catalytic constant when compared with HHe-ELVd-ΔL1, although the fraction of the substrate cleaved at the end point of the reaction was approximately the same (64% and 63%, respectively) (Table 2, Figure 6A). In contrast, HHe-ELVd-ΔL1 was inactive at 0.1 mM Mg2+, while HHe-ELVd was able to catalyze cleavage of 17% of the substrate (Figure 6A and B). These results indicate that extended ELVd-derived hammerheads with TSMs are also active against a long and structured substrate at submillimolar Mg+2, although they are less efficient than their discontinuous counterparts. A plausible explanation for these results was provided by Mfold analysis, which predicted for the ribozyme–substrate complex alternative secondary structures more stable than the catalytically active folding (Supplementary Figure S4). Particularly, the four nucleotides of loop 1 and the adjacent 6 nt of the distal part of stem I can base-pair with nucleotides of the substrate, thus disrupting the TSMs and disfavoring the efficiency of HHe-ELVd at low Mg2+ concentration.
The minimal sTRSV-derived hammerhead lacking loop 1, HHe-sTRSV-ΔL1, was also active against the same long RNA substrate at 10 and 1 mM Mg2+, although the catalytic constants were moderate (Table 2, Figure 6A). As expected, the catalytic constant and the fraction of cleaved substrate dropped to essentially undetectable levels when the Mg2+ concentration was reduced to 0.1 mM (Table 2, Figure 6A), most likely as a result of the absence of TSMs in this hammerhead.
In vivo cleavage of a viroid RNA by an extended PLMVd-derived hammerhead with natural TSMs
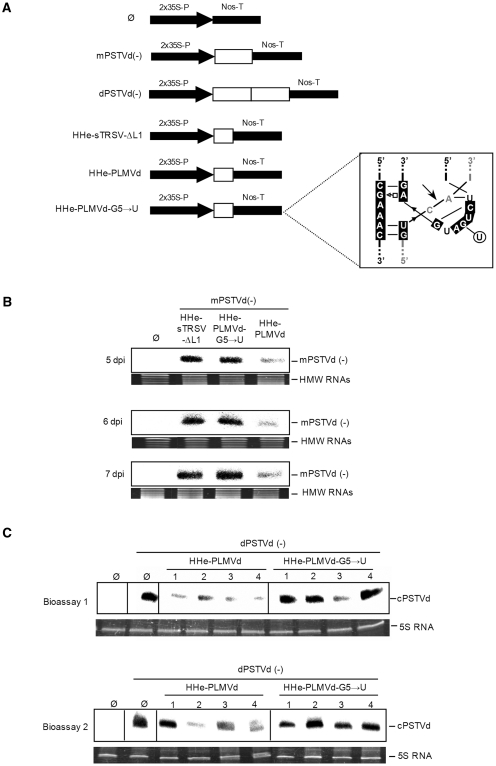
Because of its high in vitro activity at low Mg2+ concentration against short and long RNAs, the HHe-PLMVd was selected for further in vivo testing. To this aim, we used an in planta approach in which two cultures of Agrobacterium tumefaciens transformed with constructs expressing the ribozyme and the substrate were co-infiltrated in N. benthamiana leaves (Figure 7A). Two substrate constructs were used for the in planta assays: mPSTVd(–), which results in a noninfectious monomeric PSTVd RNA of minus (–) polarity, and dPSTVd (–), which generates a head-to-tail dimeric PSTVd (–) RNA that acts as template for synthesis of the complementary (+) RNA; this RNA is subsequently cleaved and ligated to produce the infectious monomeric (+) circular RNA that initiates replication through a rolling-circle mechanism (37,38). Controls for the experiment included the empty vector and the constructs for HHe-sTRSV-ΔL1 and HHe-PLMVd-G5→U, in which the CUGA box of the central core was mutated into CUUA (Figure 7A, inset) leading to a ribozyme catalytically inactive in vitro (data not shown).
Figure 7.
In vivo effects on PSTVd RNAs of three extended hammerheads co-agroinfiltrated in N. benthamiana. (A) Schematic diagrams of the constructs with the expression cassettes (boxed and with white background) between a double copy of the 35S promoter and the Nos-terminator: empty vector (Ø), monomeric PSTVd (–) RNA [mPSTVd(–)], dimeric PSTVd (–) RNA [dPSTVd(–)], the minimal hammerhead HHe-sTRSV-ΔL1, and the hammerheads HHe-PLMVd and HHe-PLMVd-G5→U (with the mutation affecting the catalytic center indicated within the inset). (B) Analysis by denaturing PAGE (5%) and northern-blot hybridization with a riboprobe for detecting PSTVd (–) strands of RNAs extracted from pools of co-infiltrated leaves from independent plants collected at 5, 6 and 7 days post-infiltration (dpi). Leaves were co-infiltrated with the mPSTVd(–) construct and with either the constructs Ø, HHe-sTRSV-ΔL1, HHe-PLMVd or HHe-PLMVd-G5→U. The position of the PSTVd primary transcript mPSTVd(–) is indicated at the right. High-molecular-weight RNAs (HMW RNAs) stained with ethidium bromide were used as loading controls. (C) Analysis by denaturing PAGE and northern-blot hybridizations with a riboprobe for detecting PSTVd (+) strands of RNAs extracted from the upper-non-infiltrated leaves of four individual plants collected at 20 and 15 dpi in bioassays 1 and 2, respectively. 5S RNA stained with ethidium bromide was used as loading control.
Each of the three ribozyme constructs was co-agroinfiltrated with the construct expressing the full-length monomeric PSTVd (–) RNA. Northern-blot hybridization of RNAs extracted 5 days-post-inoculation (dpi) from six independent plants revealed that the monomeric PSTVd transcript was significantly reduced in leaves co-agroinfiltrated with the HHe-PLMVd construct with respect to those co-agroinfiltrated with the HHe-sTRSV-ΔL1 or HHe-PLMVd-G5→U variants (Figure 7B). The differential effects of HHe-PLMVd were also observed in RNAs extracted 6 and 7 dpi (Figure 7B), thus confirming that only this hammerhead with TSMs was able to cleave efficiently the highly structured RNA substrate in vivo. However, the resulting cleavage products could not be detected by Northern-blot hybridization, most likely because of their rapid degradation by cellular RNases (39).
The observations were extended to RNA preparations from the upper non-inoculated leaves. Northern-blot hybridizations revealed that the construct expressing the HHe-PLMVd ribozyme when co-agroinfiltrated with a construct expressing an infectious dimeric PSTVd (–) RNA affected negatively the accumulation of the monomeric circular PSTVd (+) RNAs (resulting from replication and systemic invasion) with respect to parallel co-agroinfiltrations with the constructs expressing the HHe-PLMVd-G5→U or the empty vector (Figure 7C). The effects on PSTVd infection, which were reproduced in two independent bioassays (Figure 7C), could result from HHe-PLMVd mediating cleavage not only of the PSTVd (–) primary transcript but also of the PSTVd (–) oligomeric RNAs generated during viroid replication in the infiltrated leaves.
DISCUSSION
Developing ribozymes for intracellular applications requires their efficient action against long and usually structured RNAs at the low Mg+2 concentrations existing in vivo. Efforts aimed at designing minimal hammerheads against long substrates have met with limited success, with in vitro trans-cleavage constants being ∼100-fold lower than those observed with short RNA substrates, most likely due to alternative interactions with nucleotides of the ribozyme (40) or to higher-order structures of the substrate that restrict proper base-pairing with the ribozyme in the vicinity of the cleavage site (41). More recently, the study of trans-acting hammerheads at low Mg+2 concentration has received increasing attention after discovery of the TSMs in natural hammerheads (7,8) which, when incorporated into ribozymes with discontinuous or extended formats, provide enhanced activity. However, these studies have been performed only in vitro and against short substrates that entirely base-pair with the ribozyme or that leave few unpaired nucleotides (16–18). Only an extended hammerhead derived from PLMVd has been tested in vitro against a long RNA (a 258-nt fragment of the human immunodeficiency virus 1, HIV-1) (16). Here, we have examined the in vitro cleavage of short RNA substrates and of a long (464-nt) highly structured RNA by trans-acting hammerheads with discontinuous and extended formats, and selected the most efficient hammerhead variant for further analysis against PSTVd in vivo.
Some ELVd-derived hammerheads in discontinuous format catalyzed cleavage of the short RNA substrate, although they were less efficient than the sTRSV-derived hammerhead, suggesting that a hybridizing stem of only three nucleotides is enough to ensure proper substrate binding. On the other hand, extended versions of ELVd hammerhead were active against the short RNA substrate but their catalytic constant was lower than that derived from PLMVd. These results indicate that, as suggested before (17), the simple transposition of loop 1 to adapt a hammerhead to the trans format may not preserve a high catalytic activity. In agreement with previous data (16,17), HHe-PLMVd was the most efficient extended hammerhead.
In contrast with the situation observed with short RNA substrates, most of the ELVd-derived discontinuous hammerheads catalyzed cleavage of the long substrate in vitro more efficiently than their sTRSV-derived counterpart, probably because the longer stem Ib improves substrate binding and the TSMs are not disrupted with alternative interactions with nucleotides of the long substrate. Moreover, stem Ib stability appears critical for preserving the TSMs, as revealed by the higher cleavage rates of a variant in which the U-A base-pair closing loop 1 was substituted by a stronger C-G base-pair. In a previous work, the lack of activity of a discontinuous PLMVd-derived hammerhead at low Mg+2 concentrations was explained by the insufficient stability of stem Ib (18). Interestingly, two of the hammerheads with artificial loop 1 sequences (UACG and AAAA) were active at 0.1 mM Mg+2, thus suggesting that alternative TSMs between artificial sequences of loop 1 and the wild-type loop 2 might promote cleavage at submillimolar Mg2+, as reported for a discontinuous sTRSV-derived hammerhead with an artificial UUCG tetraloop (18). Pertinent to this context, non-natural hammerhead sequences forming part of the TSMs (8,16) or the catalytic core (21) can even enhance activity at low Mg+2 concentrations, probably because the sequences of natural hammerheads have been selected not only for high cleavage rates but also for mediating other functions (discussed in refs. 21 and 42).
The extended hammerhead derived from PLMVd was the most efficient in vitro against the long RNA substrate, especially at submillimolar Mg+2, in line with previous in vitro selection studies at low Mg2+ concentration in which a PLMVd-derived hammerhead with only two transitions in loop 2 with respect to the wild-type (UAGGGU) was selected for the fastest self-cleavage (16). The nucleotides of the asymmetric bulging loop of the HHe-PLMVd most likely generate TSMs resembling those existing in the natural hammerhead, because a bulging loop of only 3 nt permits less alternative interactions than in the HHe-ELVd (with a bulging loop of 4 nt). Supporting this view, extended hammerheads derived from sTRSV and CChMVd, with bulging loops of seven nucleotides, also display low catalytic efficiency (17).
Bioassays in which constructs expressing three hammerheads and a monomeric PSTVd (–) RNA substrate were co-agroinfiltrated in N. benthamiana revealed that only the HHe-PLMVd was active in vivo, whereas the minimal HHe-sTRSV-ΔL1 and the catalytically deficient HHe-PLMVd-G5→U were not. These results strongly suggest that the lower accumulation of the PSTVd transcript in plants expressing the HHe-PLMVd most likely results from ribozyme-mediated cleavage, and that TSMs are critical in this respect. Moreover, this hammerhead interfered with viroid infection when co-expressed with an infectious PSTVd (–) dimeric RNA, indicating that it may be active against the primary dimeric transcript and perhaps also against the oligomeric (–) replicative intermediates. Because a minimal hammerhead similar to HHe-sTRSV-ΔL1 only conferred resistance against PSTVd in some potato transgenic lines but not in transgenic tomato (26), we believe that HHe-PLMVd constitutively expressed in transgenic plants could serve to control PSTVd more efficiently. We also propose that agroinfiltration assays in N. benthamiana, which provide an easy and rapid test of the catalytic performance in vivo of trans-cleaving hammerheads, should be carried out before attempting stable plant genetic transformation that demands considerable more time.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministerio de Ciencia e Innovación (BFU2008-03154/BMC to R.F.); Generalidad Valenciana (ACOMP/2010/278 to R.F.); Ministerio de Educación y Ciencia (Predoctoral fellowship to A.C.); and Consejo Superior de Investigaciones Científicas (Postdoctoral I3P contract to S.G.) of Spain. Funding for open access charge: Ministerio de Ciencia e Innovación (BFU2008-03154/BMC to R.F.); Generalidad Valenciana (ACOMP/2010/278 to R.F.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to A. Ahuir for excellent technical assistance.
REFERENCES
- 1.Hutchins CJ, Rathjen PD, Forster AC, Symons RH. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 1986;14:3627–3640. doi: 10.1093/nar/14.9.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prody GA, Bakos JT, Buzayan JM, Schneider IR, Bruening G. Autolytic processing of dimeric plant virus satellite RNA. Science. 1986;231:1577–1580. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- 3.Flores R, Hernández C, De la Peña M, Vera A, Daròs JA. Hammerhead ribozyme structure and function in plant RNA replication. Methods Enzymol. 2001;341:540–552. doi: 10.1016/s0076-6879(01)41175-x. [DOI] [PubMed] [Google Scholar]
- 4.Uhlenbeck OC. A small catalytic oligoribonucleotide. Nature. 1987;328:596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- 5.Haseloff J, Gerlach WL. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature. 1988;334:585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- 6.Castanotto D, Li JR, Michienzi A, Langlois MA, Lee NS, Puymirat JY, Rossi JJ. Intracellular ribozyme applications. Biochem Soc Trans. 2002;30:1140–1145. doi: 10.1042/bst0301140. [DOI] [PubMed] [Google Scholar]
- 7.De la Peña M, Gago S, Flores R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003;22:5561–5570. doi: 10.1093/emboj/cdg530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- 9.Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:1–12. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rueda D, Wick K, McDowell SE, Walter NG. Diffusely bound Mg2+ ions slightly reorient stems I and II of the hammerhead ribozyme to increase the probability of formation of the catalytic core. Biochemistry. 2003;42:9924–9936. doi: 10.1021/bi0347757. [DOI] [PubMed] [Google Scholar]
- 11.Canny M, Jucker F, Kellogg E, Khvorova A, Jayasena S, Pardi A. Fast cleavage kinetics of a natural hammerhead ribozyme. J. Am. Chem. Soc. 2004;126:10848–10849. doi: 10.1021/ja046848v. [DOI] [PubMed] [Google Scholar]
- 12.Penedo J, Wilson T, Jayasena S, Khvorova A, Lilley D. Folding of the natural hammerhead ribozyme is enhanced by interaction of auxiliary elements. RNA. 2004;10:880–888. doi: 10.1261/rna.5268404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chi Y, Martick M, Lares M, Kim R, Scott WG, Kim S. Capturing hammerhead ribozyme structures in action by modulating general base catalysis. PLoS Biol. 2008;6:2060–2068. doi: 10.1371/journal.pbio.0060234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Navarro B, Flores R. Chrysanthemum chlorotic mottle viroid: unusual structural properties of a subgroup of self-cleaving viroids with hammerhead ribozymes. Proc. Natl Acad. Sci. USA. 1997;14:11262–11267. doi: 10.1073/pnas.94.21.11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dufour D, De la Peña M, Gago S, Flores R, Gallego J. Stucture–function analysis of the ribozymes of chrysanthemum chlorotic mottle viroid: a loop–loop interaction motif conserved in most natural hammerheads. Nucleic Acids Res. 2009;37:368–381. doi: 10.1093/nar/gkn918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saksmerprome V, Roychowdhury-Saha M, Jayasena S, Khvorova A, Burke DH. Artificial tertiary motifs stabilize trans-cleaving hammerhead ribozymes under conditions of submillimolar divalent ions and high temperatures. RNA. 2004;10:1916–1924. doi: 10.1261/rna.7159504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinberg MS, Rossi JJ. Comparative single-turnover kinetic analyses of trans-cleaving hammerhead ribozymes with naturally derived non-conserved sequence motifs. FEBS Lett. 2005;579:1619–1624. doi: 10.1016/j.febslet.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 18.Burke DH, Greathouse ST. Low-magnesium, trans-cleavage activity by type III, tertiary stabilized hammerhead ribozymes with stem 1 discontinuities. BMC Biochem. 2005;6:14. doi: 10.1186/1471-2091-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernández C, Flores R. Plus and minus RNAs of peach latent mosaic self-cleave in vitro via hammerhead structures. Proc. Natl Acad. Sci. USA. 1992;89:3711–3715. doi: 10.1073/pnas.89.9.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fadda Z, Daròs JA, Fagoaga C, Flores R, Durán-Vila N. Eggplant latent viroid, the candidate type species for a new genus within the family Avsunviroidae. J. Virol. 2003;77:6528–6532. doi: 10.1128/JVI.77.11.6528-6532.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carbonell A, De la Peña M, Flores R, Gago S. Effects of the trinucleotide preceding the self-cleavage site on eggplant latent viroid hammerheads: differences in co- and post-transcriptional self-cleavage may explain the lack of AUC in most natural hammerheads. Nucleic Acids Res. 2006;34:5613–5622. doi: 10.1093/nar/gkl717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diener TO. Potato spindle tuber virus. IV. Replicating, low molecular weight RNA. Virology. 1971;45:, 411–428. doi: 10.1016/0042-6822(71)90342-4. [DOI] [PubMed] [Google Scholar]
- 23.Gross HJ, Domdey H, Lossow C, Jank P, Raba M, Alberty H, Sänger HL. Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature. 1978;273:203–208. doi: 10.1038/273203a0. [DOI] [PubMed] [Google Scholar]
- 24.Sänger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl Acad. Sci. USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Yang X, Yie Y, Zhu F, Liu Y, Kang L, Wang X, Tien P. Ribozyme-mediated high resistance against potato spindle tuber viroid in transgenic potatoes. Proc. Natl Acad. Sci. USA. 1997;94:4861–4865. doi: 10.1073/pnas.94.10.4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenllado F, Díaz-Ruiz JR. Double-stranded RNA-mediated interference with plant virus infection. J. Virol. 2001;75:12288–12297. doi: 10.1128/JVI.75.24.12288-12297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carbonell A, Martínez de Alba AE, Flores R, Gago S. Double-stranded RNA interferes in a sequence-specific manner with infection of representative members of the two viroid families. Virology. 2008;371:44–53. doi: 10.1016/j.virol.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 29.Navarro JA, Botella F, Marhuenda A, Sastre P, Sánchez-Pina MA, Pallás V. Comparative infection progress analysis of lettuce big-vein virus and mirafiori lettuce virus in letucce crops by developed molecular diagnosis techniques. Phytopathology. 2004;94:470–477. doi: 10.1094/PHYTO.2004.94.5.470. [DOI] [PubMed] [Google Scholar]
- 30.Daròs JA, Marcos JF, Hernández C, Flores R. Replication of avocado sunblotch viroid: evidence for a symmetric pathway with two rolling circles and hammerhead ribozyme processing. Proc. Natl Acad. Sci. USA. 1994;91:12813–12817. doi: 10.1073/pnas.91.26.12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molinaro M, Tinoco Jr I. Use of ultrastable UNCG loop hairpins to fold RNA structures: thermodynamic and spectroscopic applications. Nucleic Acids Res. 1995;23:3056–3063. doi: 10.1093/nar/23.15.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stage-Zimmermann T, Uhlenbeck OC. Hammerhead ribozyme kinetics. RNA. 1998;4:875–889. doi: 10.1017/s1355838298980876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hershlag D, Cech TR. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction on an RNA substrate complementary to the active site. Biochemistry. 1990;29:10159–10171. doi: 10.1021/bi00496a003. [DOI] [PubMed] [Google Scholar]
- 34.Edwards GA, Hepher A, Clerk SP, Boulter D. Pea lectin is correctly processed, stable and active in leaves of transgenic potato plants. Plant Mol. Biol. 1991;17:89–100. doi: 10.1007/BF00036809. [DOI] [PubMed] [Google Scholar]
- 35.Hecker R, Wang ZM, Steger G, Riesner D. Analysis of RNA structures by temperature-gradient gel electrophoresis: viroid replication and processing. Gene. 1988;72:59–74. doi: 10.1016/0378-1119(88)90128-x. [DOI] [PubMed] [Google Scholar]
- 36.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Branch AD, Benenfeld BJ, Robertson HD. Evidence for a single rolling circle in the replication of potato spindle tuber viroid. Proc. Natl Acad. Sci. USA. 1988;85:9128–9132. doi: 10.1073/pnas.85.23.9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qi Y, Ding B. Differential subnuclear localization of RNA strands of opposite polarity derived from an autonomously replicating viroid. Plant Cell. 2003;15:2566–2577. doi: 10.1105/tpc.016576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sullenger BA, Cech TR. Tethering ribozymes to a retroviral packaging signal for destruction of viral RNA. Science. 1993;262:1566–1569. doi: 10.1126/science.8248806. [DOI] [PubMed] [Google Scholar]
- 40.Hormes R, Sczakiel G. The size of hammerhead ribozymes is related to cleavage kinetics: the role of substrate length. Biochimie. 2002;84:897–903. doi: 10.1016/s0300-9084(02)01461-x. [DOI] [PubMed] [Google Scholar]
- 41.Campbell TB, McDonald CK, Hagen M. The effect of structure in a long target RNA on ribozyme cleavage efficiency. Nucleic Acids Res. 1997;25:4985–4993. doi: 10.1093/nar/25.24.4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De la Peña M, Flores R. An extra nucleotide in the consensus catalytic core of a viroid hammerhead ribozyme: implications for the design of more efficient ribozymes. J. Biol. Chem. 2001;276:34586–34593. doi: 10.1074/jbc.M103867200. [DOI] [PubMed] [Google Scholar]
- 43.Hertel KJ, Pardi A, Uhlenbeck OK, Koizumi M, Ohtsuka E, Uesugi S, Cedergren R, Eckstein F, Gerlach WL, Hodgson R, et al. Numbering system for the hammerhead. Nucleic Acids Res. 1992;20:3252. doi: 10.1093/nar/20.12.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.