Abstract
Riboswitches are non-coding RNAs that control gene expression by sensing small molecules through changes in secondary structure. While secondary structure and ligand interactions are thought to control switching, the exact mechanism of control is unknown. Using a novel two-piece assay that competes the anti-terminator against the aptamer, we directly monitor the process of switching. We find that the stabilization of key tertiary contacts controls both aptamer domain collapse and the switching of the SAM-I riboswitch from the aptamer to the expression platform conformation. Our experiments demonstrate that SAM binding induces structural alterations that indirectly stabilize the aptamer domain, preventing switching toward the expression platform conformer. These results, combined with a variety of structural probing experiments performed in this study, show that the collapse and stabilization of the aptamer domain are cooperative, relying on the sum of key tertiary contacts and the bimodal stability of the kink-turn motif for function. Here, ligand binding serves to shift the equilibrium of aptamer domain structures from a more open toward a more stable collapsed form by stabilizing tertiary interactions. Our data show that the thermodynamic landscape for riboswitch operation is finely balanced to allow large conformational rearrangements to be controlled by small molecule interactions.
INTRODUCTION
Riboswitches are a class of small non-coding RNAs that exist predominantly in the 5′-untranslated region of bacterial mRNA (1). These elements have the remarkable ability to detect a variety of fundamental metabolites (ranging from glycine to adocobalamin) and control the expression of genes involved in transport or metabolism of the ligand (Figure 1). Riboswitch-controlled genes are often critical for important biochemical pathways, such as sulphur metabolism, which is regulated by the various classes of S-adenosylmethionine (SAM) riboswitches in bacteria (2–6). Riboswitches are promising targets for antibiotics because they (i) have been found in bacteria and not humans and (ii) they are highly specific, discriminating between closely related small molecules with high accuracy. While high-resolution structures of many aptamer domain elements from various riboswitch classes have provided insight into the mechanism of ligand binding and specificity, the mechanism of switching toward the alternative expression platform conformer has not been studied in detail. In this study we are able to correlate ligand binding with aptamer domain rearrangements that lead to a closed and ‘collapsed’ form of the domain. We show that specific tertiary interactions in the bound structure are necessary for collapse and riboswitch function. We also show that in absence of the ligand, the riboswitch is able to freely switch from a more open aptamer conformation toward the anti-terminator (AT) conformation of the expression platform. In the presence of ligand, the RNA is more resistant to changing conformations and remains in a collapsed stable aptamer conformation. It is this resistance that is the key to proper riboswitch function. Understanding of the switching mechanism above and beyond metabolite binding may aid in the design of antibiotics that target bacterial riboswitches.
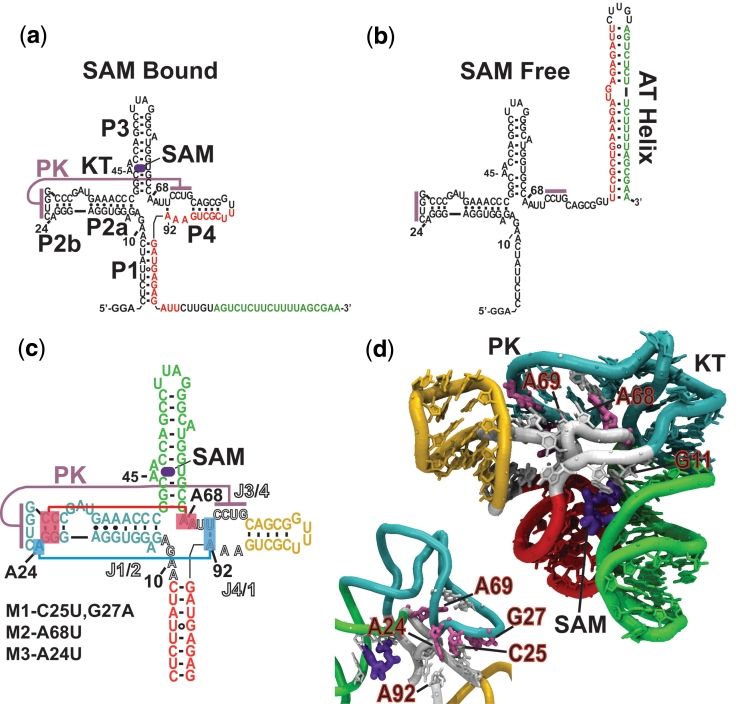
Figure 1.
(a) The T. tencongensis metF SAM I riboswitch. Upon binding SAM the aptamer domain is stabilized sequestering a shared sequence (red) from its complementary expression platform sequence (green). (b) In the absence of SAM there is an irreversible change in the secondary structure when the AT (AT) forms at the expense of the aptamer domain. (c) Secondary structure diagram with lines representing the pseudoknot and flanking base-triple tertiary contacts [helices colour coded to (d)]. Joining regions J1/2, J3/4 and J4/1, are indicated by lettering in outline. Mutant forms of the riboswitch aptamer (M1, M2 and M3) used in this study are indicated. (d) Crystallographic structure of the SAM-I riboswitch aptamer domain of Batey and co-workers (20). Tertiary contacts associated with the pseudoknot (PK) are labelled. Sites mutated are colored salmon and the KT motif facilitating the PK interaction is labelled. Inset is the reverse view showing the PK region. Construct based on the metF riboswitch but containing a truncated P3.
Riboswitches have two domains: the aptamer domain, responsible for binding, and the expression platform, responsible for gene regulation (Figure 1a). During transcription/folding, bases in the aptamer domain and the expression platform compete with each other for the same base pairing partners. As such, the domains are mutually exclusive. Thus, the switching of the riboswitch is based on sequestering a shared portion of primary sequence between these two secondary structures. This has implications for the kinetic interplay of co-transcriptional folding, ligand binding and aptamer stabilization. This is especially true in riboswitches that act through the formation of Rho-independent terminators, where a decision must be made during the transcriptional window between nascent aptamer and terminator (7,8).
In the context of an ‘off-switch’, such as the SAM-I riboswitch, when the ligand is present, the aptamer sequesters an ‘AT’ sequence, allowing a transcriptional terminator helix to form. This helix interacts with RNA polymerase, repressing gene expression by terminating transcription (Figure 1a). When the ligand is not present, the AT helix forms at the expense of both the aptamer domain and the terminator (Figure 1b). This helix prevents the terminator from forming and allows gene expression to occur.
Several mechanistic studies on the parameters of transcription, folding, ligand binding and subsequent conformational rearrangements of various classes of isolated aptamer domains have been performed. Since many of these parameters are of a similar time scale they are likely to be governed by kinetic rather than thermodynamic regimes. Studies by Wickiser et al. (9) have found that the FMN riboswitches’ function can be dictated purely by the kinetics of ligand binding, whereas the adenine riboswitch has the potential to be either kinetically or thermodynamically driven (10). In many cases, ligand binding to aptamer domains has been shown to promote a more compact, collapsed structure (11). In the case of the purine and lysine riboswitches, collapse that is mediated by ligand interactions has been shown to involve the formation of complex loop–loop tertiary contacts (8,12,13). In the SAM I riboswitch, ligand has been shown to generally decreases the mobility of flexible joining regions (5) resulting in a more compact structure (14). These transitions are analogous functionally important tertiary interactions identified in catalytic RNAs. In the hammerhead ribozyme it is now clear that catalytic activity is dependent to a large degree on stabilization of the core via distal loop–loop interactions (15,16). Also, in many ways the transition of the folded intermediate to the collapsed stable aptamer has the hallmarks of the transition of group I intron folding intermediates to their collapsed native fold. Specifically, transitions in an Azoarcus group I ribozyme have been shown to be highly cooperative (17). In another instance, the transitions were found to occur as cation concentration increased from a compact state which possessed few of the native tertiary contacts (18). Similarly, we show that the mechanism of collapse in the SAM I riboswitch is cooperative. We also find that SAM interactions act to stabilize tertiary contacts found in the collapsed conformation. The contacts are remote from the binding site and highly cooperative. Their engagement allows the aptamer to compete effectively with the expression platform for shared sequence. We also show that interactions between the expression platform sequence and terminal helix, P1 increase the dissociation constant for SAM.
MATERIALS AND METHODS
Preparation of SAM I riboswitch constructs
Synthetic DNA oligonucleotide (IDT, Coralville, IA) templates were PCR amplified for use as in vitro transcription templates. Transcription reactions were performed using Ampliscribe (EPICENTRE Biotechnologies, Madison, WI, USA) high yield T7 RNA polymerase transcription kits as per instructions.
Nucleotide Analog Interference Mapping
Transcription reactions were performed with T7 RNAP (Epicentre Biotechnologies) as per Ryder et al. (19). The construct used in Nucleotide Analog Interference Mapping (NAIM) was based on metF-H2 element from Thermoanaerobacter tengcongensis and contained extended P3 and P4 helices. Phosphorothioate nucleotide analogues were purchased from Glen research (Sterling, VA). Selection of SAM bound and free conformations were performed via native polyacrylamide gel electrophoresis (8% PA, 0.5× TBE, 1.1 mM MgCl2) at 18°C, 350 V for 3 h. Samples of RNA, 100 pmol in 16 µl, were heated to 90°C for 1.5 min. in H2O and placed on ice for 2 min followed by the addition of 4 µl of 5× binding buffer (50 mM Tris–HCl pH 7.4, 100 mM KCl, 2 mM MgCl2). SAM bound samples were made 75 µM in SAM (Sigma-Aldrich). Both SAM free and SAM bound samples were allowed to stand for 5 min at room temperature and made 10% in glycerol prior to loading. The RNA was resolved by staining with 0.01% methylene blue for 30 s. Bands representing the free and bound conformations were excised from the gel and eluted in extraction buffer (50 mM Tris–HCl pH 7.5, 250 mM NH4•acetate, 0.1% SDS, 3 mM EDTA, 5 mM DTT) for 2 h at 55°C. After phenol/chloroform extraction and ethanol precipitation, eluted RNA was 3′-end fluorescently labelled using T4 DNA ligase (New England Biolabs). A 7-nt DNA oligomer with a 3′ Alexafluor 488 (Molecular Probes) label was annealed to a bridging DNA oligomer that was also complementary to the 3′-end of the SAM-31 construct. The label/bridge was then incubated in a 1:1 molar ratio (1.5 µM in 20 µl) to selection construct RNA at room temperature for 1 h with 80 U of T4 DNA ligase in the supplied buffer. Analysis was performed on an ABI Prism 310 Genetic Analyzer capillary electrophoresis system (Applied Biosystems International, courtesy of the National Stable Isotope Resource facility, Los Alamos National Laboratory). Samples were desalted prior to loading with Dyex spin columns (Qiagen), lyophilized and resuspended in 5 µl of 10 mM I2 in deionized formamide and heated to 90°C for 2 min. then diluted to 16 µl with formamide and loaded on the sequencer (60 min at 12.1 kV, 70°C, 61 cm capillary, ABI POP-6 polymer). Control samples for non-specific cleavage were performed for each without iodine cleavage. Fluorescence electropherograms were converted to ASCII files using Data File Converter (ABI) and plotted with Origin 7. After normalizing for variability in electrokinetic injection efficiency, sites of interference were identified by the lack or significant reduction of a peak at the residue in the SAM bound sample versus the SAM free sample.
Chemical probing
All chemical probing reactions were performed a minimum of three times with and without SAM. SAM (NEB, Ipswich, MA, USA) was added to a final concentration of 10 µM. In-line probing was performed as previously described (5) in in-line probing buffer (50 mM Tris–HCl pH 8.3, 20 mM MgCl2, 100 mM KCl) with the following modifications. RNAs were folded by heating to 90°C in water for 2 min. and then crash cooled on ice for 2 min followed by addition of buffer with or without SAM. Inline probing reactions were performed at RNA concentrations of 0.3 µM for 40 h at 25°C. Reactions were purified by precipitation with three volumes EtOH and 50 µg RNase free glycogen (Ambion) and then analyzed by capillary electrophoresis after 3′ end labelling as described above.
Hydroxyl radical probing reactions were carried in 1× HMK buffer (25 mM HEPES–NaOH pH 7.4, 2 mM MgCl2, 100 mM KCl). Fe:EDTA reactions were designed to produce a burst of hydroxyl radicals by limiting the concentration of H2O2. Reactions of 0.3 µM in RNA were folded and incubated in buffer with or without ligand for 10 min at 37°C followed by addition of 3% (v/v) H2O2 in 1× HMK to a final concentration of 0.03%. The mixture was then transferred to a tube containing 40.5 mM Fe:EDTA and 208 mM ascorbate in 1 × HMK. After mixing the final concentration of Fe:EDTA was 400 µM and the reactions were incubated at 25°C for 2 min and stopped by the addition of three volumes EtOH and 50 µg glycogen and precipitated before labelling and capillary electrophoresis.
Chemical modification reactions with dimethylsulphate (Sigma-Aldrich) were performed in 1× HMK. The RNA was first folded and incubated in buffer with or without SAM for 10 min at 37°C. The reaction was initiated by the addition at a 1:100 ratio of a 10% solution of DMS in EtOH. After incubation at 25°C for 10 min the alkylation was stopped by the addition of one volume of stop buffer (1 M Tris–HCl pH 7.5, 1 M β-mercaptoetanol and 1 M sodium acetate). The mixture was then precipitated with 2.5 volumes EtOH in preparation for reverse transcription and capillary electrophoresis.
Analysis of chemical probing reactions
In-line probing and hydroxyl radical probing reactions were analyzed by fluorescently labelling of the RNA on the 3′-terminus as outlined above. The labelled RNA was then diluted 1–5 µl into 20 µl of formamide depending on recovery and labelling efficiency and then heated to 90°C for 2 min prior to loading on an ABI Prism 310 genetic analyzer (Applied Biosystems) capillary electrophoresis system equipped with a laser induced fluorescence detector. The reactions were electrokinetically injected at 12.5 kV for 30 s and run in POP-6 polymer (ABI) at 70°C for 1 h. Sequencing reactions were performed by transcribing the RNA constructs in the presences of a low level of α-phosphorothioate nucleotides followed by iodine cleavage and capillary electrophoresis.
Analysis of differential alkylation patterns with DMS was also performed using capillary electrophoresis, after reverse transcription to detect modified nucleotides. RNA constructs used in the DMS probing contained a primer-binding site at the 3′-terminus. A 15-nt primer was synthesized containing a 5′ Alexafluor 488 modification for use in reverse transcription reactions. Reverse transcription reactions contained 1:1 stoichiometry of primer to RNA at 0.5 µM in 30 µl, 0.3 mM dNTPs and 10 U of AMV-RT (Seikagaku America, Associates of Capecod) in the supplied buffer. Reactions were incubated for 1 h at 45°C. Separate sequencing reactions for G and A were performed with the addition to 100 µM of the appropriate dideoxy nucleotide triphosphate (GE lifesciences). Following reverse transcription, reactions were desalted and run on the ABI prism as outlined above. Data from capillary electrophoresis was converted to ASCII files using Data File Converter (ABI) and plotted using ORIGIN (Originlab). Runs with and without SAM were normalized using the reactivity of bases that do not change in response to ligand and overlayed for comparison.
2-aminopurine switching assay and ligand titration
Wild-type and mutant MetF-H2 SAM I riboswitch elements were folded as described above in 1× TMK (25 mM Tris–HCl pH 7.4, 2 mM MgCl2, 100 mM KCl). The synthetic RNA AT strand was purchased from Dharmacon (Thermo Fisher) containing a 2-amino purine analogue with the following sequence 5′-GA-2AP-UCUCUCAUCUUUCAGCGAA-3′. 2AP fluorescence in the association reactions was followed using a Fluoromax 4 (Horiba Jobin Yvon) with a 310-nm excitation and 372-nm emission wavelengths and 5-nm and 10-nm slit widths respectively. Solutions of folded RNA at twice the specified concentration of SAM were equilibrated to the working temperature for 2 min. These were then rapidly mixed with an equal volume and concentration of the AT piece in a 100 µl cuvette while data was being collected. The time points prior to injection were then subtracted after the run. The level of fluorescence in mock reactions (reactions mixed with buffer containing no aptamer RNA) was equivalent to that at t = 0, insuring the dead time was insignificant.
Association rate constants were determined for the different aptamer constructs in the absence of SAM. Self-association of the 2-AP labelled AT at high concentrations led to aberrant increases in fluorescence upon dilution. This placed limitations on the maximum concentration of AT possible and excluded pseudo first order conditions. Instead, initial rates were used to determine kobs. The time required for a 10% decrease in the 2AP fluorescence relative to a completely associated control at that concentration of AT were measured. The value was then used to solve for the observed rate constant in a single exponential decay function. The association rate constant, kon was then determined using the following equation:
Plots of the kobs versus RNA concentration were then analysed by linear regression and the slope taken as kon.
Determination of SAM-binding affinity by SHAPE probing
The aptamer domain RNA was folded at a concentration of 5 nM in 1× HMK buffer (50 mM HEPES–KOH pH 8.0, 2 mM MgCl2, 100 mM KCl) and various concentrations of SAM. 30 µl of 60 mM 1-methyl-7-nitroisatoic anhydride (1 M7) in DMSO was added to a 300 µl volume of RNA. The reaction proceeded for 5 min at 25°C and was then precipitated by the addition of 0.1 volumes 3 M Na:Acetate pH 6.5, 75 µg glycogen and three volumes EtOH. The recovered RNA was then subjected to reverse transcription analysis as described above. Capillary electrophoresis data was integrated by simultaneously fitting Gaussian curves to the whole trace using an in-house developed algorithm. The traces were normalized using residues whose reactivity does not change in response to SAM (Supplementary Data). In both the wild-type and mutants aptamer, nucleotides were chosen that changed in reactivity as a function of SAM concentration. The areas were plotted versus SAM concentration and fit to a two state-binding model. The reactivity change was expressed relative to the peak area in the absence of SAM. A minimum of three titration experiments were conducted on the aptamer mutants. The KD for the SAM-aptamer interaction was determined by fitting the expression:
where the left hand side of the equation is the relative SHAPE reactivity (fraction of aptamer bound) and f is the maximum decrease in reactivity. Fits were performed using the non-linear curve-fitting algorithm of Gnuplot.
Native gel switching analysis
Samples of aptamer RNA were folded as described above at twice the final concentration in the presence of the specified concentration of SAM. The folded aptamer was then mixed with an equal volume of AT RNA. Equilibrium experiments were then incubated for 3 h at 25°C and then loaded onto a 12% polyacrylamide gel with a native buffer (0.5× TBE, 1.1 mM MgCl2, 10 µM SAM). Gels were then run at 150 V for 1.5 h then stained with ethidium bromide and scanned using a Hitachi FMBIO III fluorescence imager (532-nm laser excitation, 605-nm bandpass emission filter) at 50-µm resolution. Lane traces from the fluorescence scans were generated using Hitachi analysis software. Traces were integrated using ORIGIN 7.4 (OriginLab) and areas assigned to the aptamer or aptamer/AT complex. The degree of association was relative to the associated form in the example containing no SAM.
FRET based switching assay
The ability of SAM interactions to reverse AT formation was assayed. The aptamer domain sequence was modified to contain 5′ and 3′-binding sites. DNA oligomers complementary to these extensions were made with fluorescent labels. The 5′ oligo was purchased (Integrated DNA technologies) with Cy-5 (GE lifesciences) modification on it 5′-terminus. Likewise the 3′ oligo was purchased with a 3′-Cy-3 modification. The oligos anneal to the 5′ and 3′ leaders with a 3-nt and 5-nt gap, respectively between P1 and their complementary sequence. When P1 is formed the Cy-3 Cy-5 Dyes are in a high FRET state (excitation at 535 nm 5-nM slit width, emission at 670 nm 10-nm slit width). When the anti terminator sequence (same as outlined above except with adenosine in place of 2-AP) interacts with the aptamer domain the formation of the AT helix forces the system into a low FRET state.
The AT is rapidly mixed with folded aptamer to final concentrations of 5 nM and 1 µM, respectively, in 1× HMK(50 mM HEPES–KOH pH 7.4, 2 mM MgCl2, 100 mM KCl) in the absence of SAM. The association of the two RNAs is allowed to proceed until ∼80% of the aptamer is converted to AT helix (based on the minimum FRET values of the system at equilibrium). Then 32-mM SAM in 1× HMK buffer is added to a final concentration of 1 mM. The reaction is allowed to continue and the fluorescence of the acceptor fluorophore monitored for an increase in FRET.
RESULTS
NAIM shows SAM-mediated aptamer domain collapse is dependent on a network of tertiary contacts
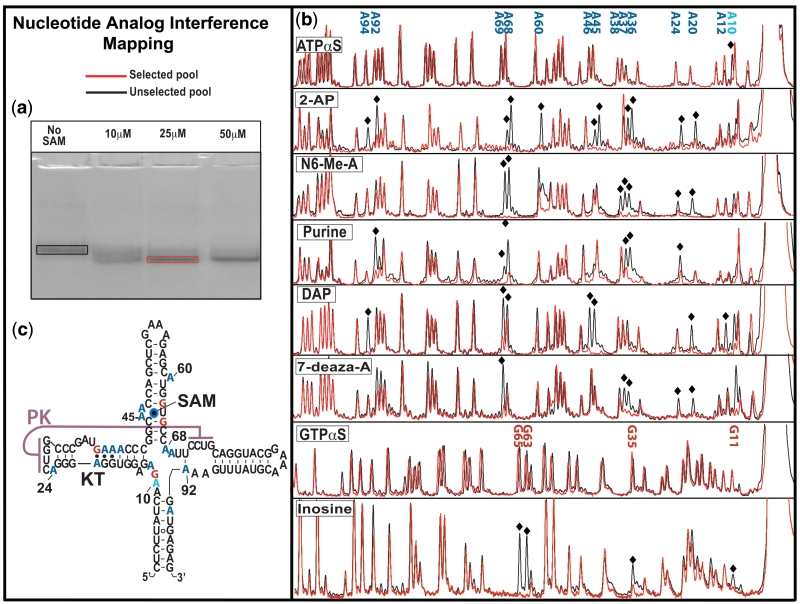
NAIM is a powerful technique for simultaneously assessing the importance of all nucleotide functional groups in a given selectable process. In response to metabolite interactions, many riboswitch aptamers have been shown to undergo a conformational change (11). We relied on the ability of the SAM I aptamer domain to adopt a more compact structure and migrate more rapidly during native polyacrylamide gel electrophoresis as a means of NAIM selection (Figure 2a). As a result, we refer to the process being studied as collapse. Sites that interfere with collapse were found by comparing the relative populations of analogue at a position between the selected pool and the unselected pool after gel purification (Figure 2c).
Figure 2.
NAIM. (a) Native polyacrylamide gel showing the shift in electrophoretic mobility in response to increasing concentrations of SAM in the loaded sample. This ‘collapse’ of the aptamer domain was used as the NAIM selection parameter. Selected or unselected pools were excised from the gels and fluorescently labelled. (b) Selected and unselected pools were analysed by capillary electrophoresis after cleavage at sites of analogue incorporation with molecular iodine. Sites of analogue interference with the process of aptamer domain collapse were chosen by visual comparison of the selected and unselected pools. (c) Secondary structure of the SAM I NAIM construct showing the locations of analogue interference; adenosine analogues in blue, guanosine in red and phosphorothioate interference at A10.
The structure of the SAM I riboswitch with ligand has been solved by the Batey group using X-ray crystallography (20). The riboswitch aptamer consists of two sets of coaxially stacked helices (P1/P4 and P2/P3) that pack together at an angle. This arrangement allows a pseudoknot interaction between the loop of helix P2 and the junction region between helices P3 and P4 (J3/4). A ligand-binding pocket forms at the interface of P1 and P3 distal to the pseudoknot. Within the riboswitch’s ligand-binding pocket, SAM forms specific hydrogen bonding interactions with helix P3 and the junction region between helices P1 and P2 (J1/2) and complementary van der Waals contacts with the minor groove of P1.
All three regions that make direct contact with ligand contained sites of analogue interference. The mechanisms of interference in the binding pocket were, for the most part, easily explained via the crystal structure. The mechanisms of interference include steric hindrance, loss of hydrogen bonding with SAM, and the weakening of base-pairing interactions flanking the P3 bulge.
The most interesting sites of interferences from a mechanistic viewpoint occurred outside the binding pocket. These can be further delineated into two classes. The first class consists of those directly disruptive to the kink-turn (KT) configuration found in P2 that is required for the formation of the pseudoknot interaction between the loop of helix P2 (L2) and the junction of helices P3 and P4 (J3/4). The second class consists of ligand-induced tertiary interactions (A68, A69, A92 and A24). These tertiary contacts flank the pseudoknot. We note that all of these interactions are involved in base triples.
The importance of two of these base triple configurations (those involving A68 and A69, respectively) in stabilizing the bound conformation is made clear by their sensitivity to all tested adenosine base analogues. Both A68 and A69 form base triples with the sugar edge of adjacent G:C pairs in P2. Loss of hydrogen bonding explains their respective interference. Similar results from analogues are found at A69. The role of these two base triple interactions can be inferred from the crystal structure as the stabilization of the pseudoknot interaction between L2 and the joining region, J3/4.
A third base triple also displayed interference. This base triple connects L2 with joining regions J4/1 and J3/4 at nucleotides A24, A92 and U71 (14). Both A24 and A92 displayed interference patterns with 2AP, N6-Me-A, purine and 7DA. The A24 to A92 hydrogen bonding pattern gleaned from the X-ray structure is comprised of a hydrogen bond between the N1 group of A24 and the 2′-hydroxyl group of A92, as well as a possible hydrogen bond between N6 of A24 and N3 of A92. This pattern explains the interference arising from the weakening or removal of a base-pairing hydrogen bond involving A24. The same holds true for analogue effects at A92 where the interference appears to arise from loss of canonical base pairing interactions with U71.
The NAIM results show that disruption of the KT’s canonical architecture inhibits the formation of a compact aptamer. In a previous study, the KT motif in the SAM I aptamer was shown to play an important role in the formation of a compact aptamer domain (14). However, given the connection between the pseudoknot and the KT, any change within the structure of one motif would require a compensatory change in the other. In a separate study of an isolated KT element, the element was found to exist in an equilibrium between two distinct conformations: a fully kinked helix and a helix with a simple three base bulge (21). The NAIM sites within the KT are consistent with a loss of interactions stabilizing the kinked conformation. In particular, A20, A36 and A37 display a pattern of interference consistent with a loss of capacity to form the Hoogsteen contacts necessary for stabilizing the KT. These results were in good agreement with a previous study of base substitutions at sheared nucleotides in an isolated KT construct (22).
The picture that emerges from the NAIM experiments is one of a network of highly cooperative tertiary interactions. These interactions are centred on the four-way junction and include base-triple contacts flanking the pseudoknot interaction. The stability of the collapsed aptamer is very sensitive at these sites of tertiary contact as well as those sites supporting a canonical KT. Disruption of any single functional group interaction severely compromises the stability of the ligand-bound conformation.
Key tertiary interactions control switching
Ligand binding by the SAM I riboswitch leads to domain collapse. Previous studies have shown that mutations disrupting the pseudoknot interaction increased the KD of the Bacillus subtilusyitJ SAM riboswitch from 5 to ∼50 nM while precluding the collapsed form of the bound aptamer domain (14) and decreasing termination in vitro (23) and in vivo (24). These observations point to the central role of the pseudoknot in collapse and allude to the possibility that the interaction is wholly or partially dependent upon ligand interactions. They also suggest collapse is the mediator of resistance to strand invasion by the AT. We wished to explore the mechanism of SAM mediated stabilization of the aptamer by looking at the relationship between ligand binding, collapse and resistance toward expression platform sequence.
The NAIM study identified sites remote from the binding pocket that are sensitive to analogue interference toward ligand mediated collapse. As such, they are targets for dissecting the connection between ligand interactions and domain collapse/stabilization. NAIM reveals the base-triple contacts flanking the pseudoknot to be more sensitive to interference than the pseudoknot base pairs. These interactions may also be responsible for modulating the ability of the aptamer domain to (i) stabilize the pseudoknot interaction observed in the X-ray structure, (ii) facilitate domain collapse and (iii) resist expression platform invasion. We created three mutant systems; M1, M2 and M3. The M1 mutant disrupted the pseudoknot directly by introducing two mutations C25U and G27A (Figure 1c) (numbering starts at the first base pair in P1). Two mutants were also made independently at the base triple contacts A68U (M2 mutant) in J3/4 and A24U (M3 mutant) in the loop region of the helix P2. We used these constructs in an assay that allows us to assess the ability of the aptamer to resist strand invasion by AT sequence found in the expression platform. We refer to this process as ‘switching’. The technique allows us to evaluate the functional implications of the NAIM inspired mutations that perturb the stability of collapse and/or ligand interactions in the aptamer. These results will provide a functional benchmark that can then be correlated with structural data from NAIM and chemical probing. In doing so we hope to gain insight into the mechanism of ligand induced aptamer stability that gives rise to resistance toward the adoption of the expression platform conformation.
Our study was performed on the SAM I riboswitch element from the metF gene of T. tengcongensis. This aptamer possesses all of the highly conserved sequence and structural elements for the SAM I class of riboswitches (25) (Figure 1a). The SAM I X-ray structure (accession code, 2GIS) was also based on this element, although its sequence differs substantially from the wild-type (20) (Figure 1d). Our assay utilizes an expression platform that is presented in trans (Figure 3a). The expression platform consists of the portion of the AT helix that competes for a complementary sequence within the aptamer domain. The AT piece was chemically synthesized with the fluorescent adenosine analogue 2-aminopurine (2-AP) at a position where it will be stacked in the AT helix. Since hydrogen bonding and base stacking interactions effectively quench the fluorescence of 2-AP, switching can be followed as the decay of 2-AP fluorescence (26).
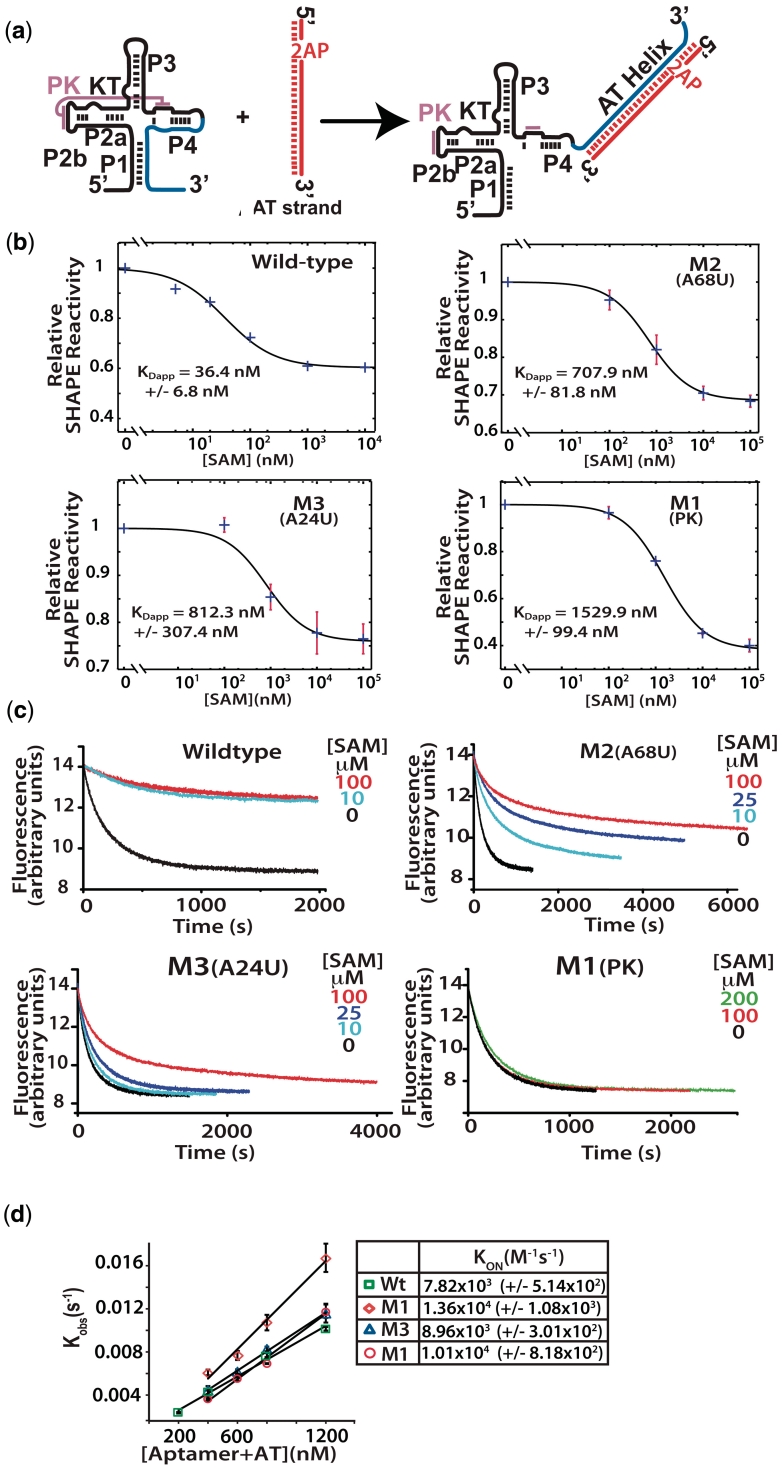
Figure 3.
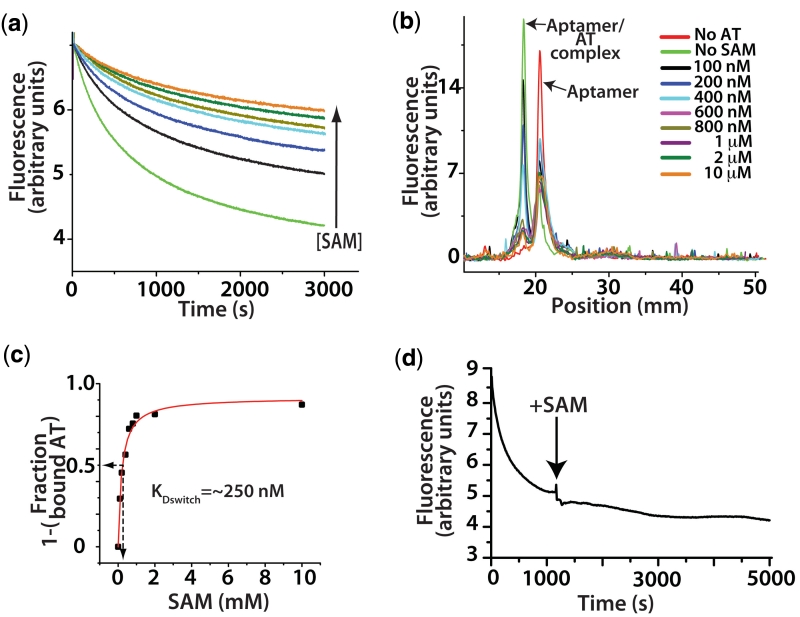
(a) Analytical switching construct. 2-Aminopurine incorporated AT strand associates with the aptamer domain forming the AT helix. (b) Fluorescence trace following the quenching of 2-AP by interaction with the wild-type aptamer and mutant aptamers. Association was followed with various concentrations of SAM as indicated. Samples were rapidly mixed at 25°C and 1:1 ratio of aptamer to AT to a final aptamer concentration of 300 nM, excitation was at 310 nm (5 nm slit width) and fluorescence detected at 372 with a 10-nm slit width. (c) Apparent dissociation constants (KDapp) were determined for the wild-type and mutant constructs. SHAPE probing reactions were performed on samples equilibrated in the presence of varying concentrations of SAM. The decreases in reactivity of a selected nucleotide relative to its reactivity in the absence of SAM were plotted versus SAM concentration and fit to a two state binding model (see ‘Materials and Methods’ section). Standard asymptotic errors for the fit are shown. Error bars represent the standard deviation of a minimum of three replica experiments. (d) Initial rates were fit to a single exponential and the observed rate constants plotted as a function of RNA concentration ([aptamer] + [AT strand]) and the second order rate constants (kon) were determined for each construct.
We also measured the apparent dissociation constants (KDapp) for SAM binding to the wild-type and mutant aptamer domains. Individual KDapp values were determined using the selective 2′-hydroxyl acylation analysed by primer extension (SHAPE) probing technique (27). SHAPE probing reports on the backbone dynamics at nucleotide resolution. Changes in the reactivity of nucleotides toward the 2′-OH acylating reagent 1-methyl-7nitroisatoic anhydride (1M7) were measured as a function of SAM. Reactivity was measured using fluorescence based capillary electrophoresis of primer extension reactions. A whole trace multiple Gaussian fit was performed to establish accurate peak areas (Supplementary Data). The areas of individual residues relative to that of a sample without SAM were plotted and a binding isotherm for a simple two state model was fit (Figure 3b).
The affinity of the mutant constructs for SAM was found to be considerably lower than wild-type, with corresponding KDapp values ranging from ∼700 to ∼1500 nM compared to ∼36 nM for wild-type (Figure 3b). The dissociation constants were also considerably higher than those reported by others on a similar pseudoknot mutant (14). The results likely reflect the decreased magnesium concentration [2 versus 10 (14) or 20 mM (5)] or increased temperature used during SHAPE probing.
The disruption of the collapsed/stabilized form in the mutant aptamer is apparent in a comparison of switching with increasing concentrations of SAM (Figure 3c). Samples of folded aptamer were rapidly mixed with an equal volume and equal concentration of AT. The association of the AT was followed by 2-AP fluorescence. In the wild-type, we observe the expected behavior. That is, in the absence of SAM, the AT forms very easily, as it should for correct riboswitch operation. In the presence of SAM, the AT does not readily form. Here, the compact aptamer domain resists invasion by the AT strand. The mutant aptamers, on the other hand, were severely impaired in their ability to resist AT association. Titrations of ligand with the M1 pseudoknot mutant failed to alter the rate or magnitude of AT strand invasion even at the highest concentration of ligand tested (200 µM). The M2 and M3 mutants were able to modulate the association rate to different extents, however, neither manifested the stability seen with the native sequence.
The AT formation seen in the presence of SAM may reflect the fluctuations in the aptamer structure that exist in the mutants even when SAM is bound. This seems likely for the M1 pseudoknot mutant that lacks switching rate response even at SAM concentrations >100-fold over KDapp. Another explanation for these results is that the mutations increased the off-rate for SAM to levels of the same order as the switching reaction. The AT kon measured with the different aptamer constructs is on the order of ∼104 M−1 × s−1 (Figure 3d). The rate constant for AT association at these concentrations in the absence of ligand is ∼0.006 s−1. As such, an off-rate for SAM on the order of that measured for ligand binding to the adenine riboswitch aptamer (∼0.022 s−1) (10) could produce the observed results. In this scenario AT strand associates with free aptamer as SAM dissociates. However, the amount of bound aptamer is also a product of the concentration of ligand and its on-rate. It is possible that the ligand-bound mutants are able to resist AT strand invasion. If this is true, then the rates of switching should reflect this at higher ligand concentrations. This is likely the case with the M2 and M3 mutants.
We have identified mutations based on NAIM data that effect both ligand affinity and riboswitch function. This suggests that the process of ligand-mediated collapse requires a complete set of tertiary interactions. This data also shows that the stable, collapsed aptamer is required for aptamer resistance to AT strand invasion. These tertiary interactions also lead to increased ligand affinity. In the case of the M1 mutant, ligand binding is not associated with collapse. We refer to this as the open-bound configuration. The M2 and M3 likely fluctuate between the open-bound and closed-bound (collapsed) configurations. The closed-bound configuration is responsible for resistance to switching.
Ligand interactions stabilize distant tertiary contacts, leading to collapse of the aptamer domain
The NAIM and switching assay results suggest that both the pseudoknot and the base triple contacts are necessary for both structural collapse and switching resistance. However, it is unclear how the bound/collapsed structure propagates and leads to expression platform resistance in the aptamer domain. To affect conformational switching, the mutations must somehow preclude stabilization of helices P1 and P4, making them susceptible to hybridization with the AT. To understand how helices P1, P2 and P4 are related in the wild-type and mutant systems, we performed an extensive structural probing study of the aptamer domain using in-line probing, DMS chemical modification and Fe:EDTA hydroxyl radical probing. The results are in good agreement with previous SAM I riboswitch in-line and ribonuclease probing studies (4,5,23). Our results provide local and global information regarding structural alterations in the aptamer in response to SAM binding for both the mutant and wild-type constructs. The data of our three probing studies indicate aptamer domain collapse (including helices P1 and P4) involves the stabilization of the pseudoknot and associated tertiary contacts remote from the SAM-binding site.
Fe:EDTA probing measures the solvent exposure of ribose moieties in the phosphate backbone and reveals changes in the global architecture of the RNA (including changes in tertiary contacts). Abstraction of sugar hydrogens by Fe produced hydroxyl radicals is the major source of strand cleavage (28). Probing of aptamer domain with and without ligand shows that the SAM-free aptamer is less compact than the SAM-bound aptamer. SAM binding promotes a structural collapse resulting in the protection of nucleotides in P1 and P3 (green bases in secondary structure diagram, Figure 4c). Bases on the 3′ strand of P1 become much less susceptible to hydroxyl radical cleavage while changes to the 5′ portion of P1 are minimal. The protected bases in P3 extend from A46 of the binding pocket toward the loop, spanning the nucleotides that make closest contact with the 3′ strand of P1 in the crystal structure of the SAM I riboswitch. This is consistent with a closure of the P1 and P3 helices in response to binding.
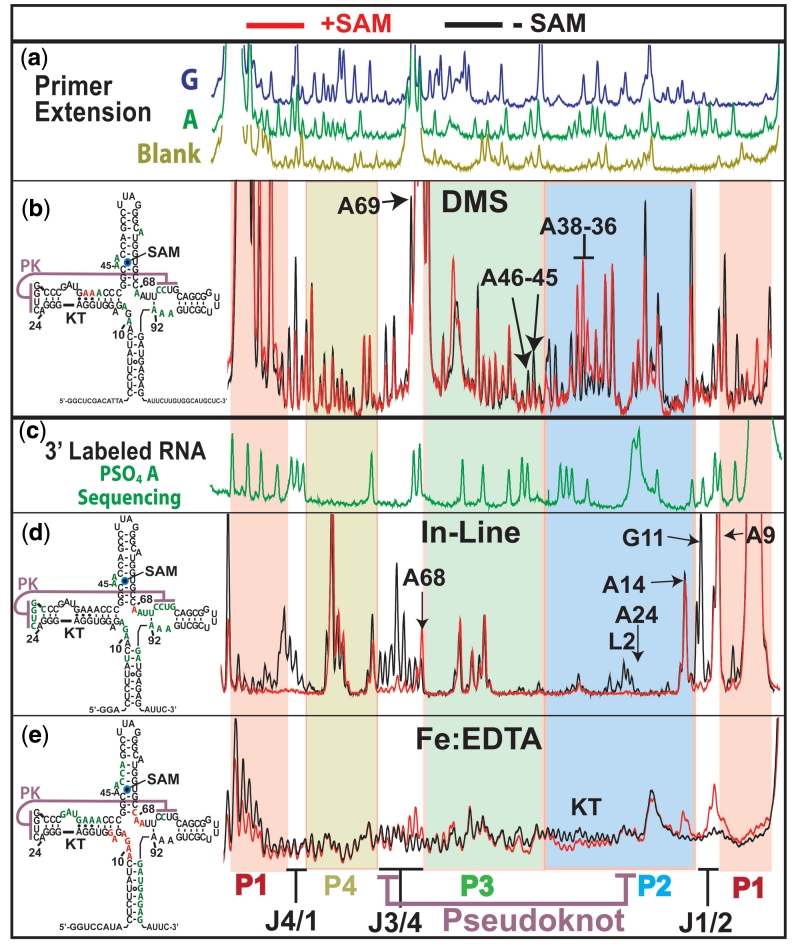
Figure 4.
Structural probing results for the wild-type aptamer domain. Red capillary electrophoresis traces are probing results in the presence of 10 µM SAM, black are without. Left, 2D structures summarize SAM dependent sites of decreased (green) and increased (red) reactivity. (a) Primer extension dideoxy sequencing reactions, for guanosine (green) and adenosine (blue) and primer extension control in brown. (b) DMS probing results. Capillary electrophoresis traces of primer extension reactions using 5′ fluorescently labelled primers. (c) Green trace shows iodine cleavage pattern of α-phosphorothioate adenosine incorporated RNA. RNA was 3′-labelled with a 3′ fluorescently labelled DNA oligo. (d) In-line probing results using 3′ fluorescently labelled RNA. (e) Fe:EDTA generated hydroxyl radical cleavage pattern from 3′ fluorescently labelled RNA. Bottom, coloured underlay coded for helices and indicating joining regions and pseudoknot interaction and KT.
In-line probing relies on the inherent capacity of RNA to undergo cleavage via a transesterification reaction. The measured reactivity is proportional to the backbone mobility and reflect the secondary or tertiary structural environment (29). Results from in-line probing data indicate that bases in helix P1 are more mobile in the SAM-free state relative to the SAM-bound state (Figure 4b). Upon SAM binding, a stabilization of the P1 helix occurs (Figure 4b, P1 red background at 5′ and 3′). There are also decreases in mobility upon SAM binding at the joining regions associated with P1, indicating an induced organization of the four-way junction(Figure 4b, joining regions have no background colour).
In our chemical probing experiments, all three helix-joining regions exhibited decreased reactivity upon SAM binding, suggesting that a rearrangement of the core nucleotides occurs upon SAM binding. DMS, in-line and hydroxyl-radical probing each show significant changes in reactivity for junctions J1/2 and J3/4 (Figure 4a–c). The large increase in reactivity for junction J4/1 in the in-line probing and the DMS studies suggests that helices P1 and P4 may not necessarily be stacked in the SAM free state (Figure 4a and b). Residues in junction J1/2 became more solvent exposed to hydroxyl radicals in response SAM. These same residues are far less mobile and protected from alkylation by DMS upon SAM binding (Figure 4a).
By in-line probing, a large reduction in mobility can also be seen in the other joining region closing the top of P1 (mainly J4/1) (Figure 4b). The alterations in mobility and solvent accessibility associated with P1, P3 and J1/2 are significant as these regions contain the nucleotides that make direct contacts with ligand. Also, since P1 contains the sequence complementary with both the aptamer and expression platform, riboswitch function is rooted in its stabilization. The local changes proximal to P1 and the ligand-binding site coincide with more remote alterations to the structure of the aptamer. Junctions J4/1 and J3/4 do not interact directly with the ligand. However, ligand binding clearly causes dramatic changes in the configurations of these distant regions (Figure 4b).
The global collapse triggered by ligand appears to involve the stabilization of the same set of tertiary contacts found to be very sensitive to perturbation in the NAIM study. In turn, the formation of a stable pseudoknot interaction requires the bimodal KT motif in P2 to be in the form of three sheared G•A pairs and a 3-nt bulge (30).
One of the two potential conformations for the SAM-I riboswitch KT can be seen in the crystal structure where the tight kink facilitates the pseudoknot interaction (23). The other conformer is apparent in our structural probing results. The exposure to hydroxyl radicals of the KT was found to decrease in the presence of SAM (Figure 4c). However, the most dramatic evidence for an alternative KT conformation comes from our DMS probing data where the sheared Adenine residues (A36–A37) increase in reactivity when SAM is present (Figure 4a). This indicates that the canonical ‘kinked’ architecture of three sheared pairs with solvent exposed base-pairing faces seen in the SAM bound X-ray structure is supplanted by a conformation where the base pairing faces of these adenosine residues are partially occluded and therefore protected from alkylation by DMS in the absence of ligand.
Concomitant with the ligand-induced change in the KT is a large change on both sides of the pseudoknot interaction. This can be seen most clearly in the in-line probing data where the P2 loop (L2) and the J3/4 half of the interaction become vastly more mobile in the absence of ligand (Figure 4b). These changes in reactivity suggest strongly that the pseudoknot interaction is significantly weaker in the absence of ligand.
Like the pseudoknot interaction, the other tertiary contacts shown to be required for riboswitch function appear to be SAM dependent. For example, A68 and A69, both increase in solvent exposure upon aptamer binding to SAM. This suggests they are displaced from a solvent inaccessible region by aptamer domain collapse to occupy the solvent exposed position seen in the X-ray structure (Figure 4c). Their susceptibility to in-line probing also changes in response to ligand with A68 becoming more reactive and A69 becoming less reactive (Figure 4b).
It has been previously shown that several factors may contribute to in-line probing reactivity, including mobility and geometric positioning (29). In certain situations, linkages show greater reactivity without greater mobility due to geometric positioning. The greater reactivity of A68 may be a result of being locked in a reactive conformation whose mechanism of transesterification is something other than nucleophilic attack by the 2′ hydroxyl group. We note that in the X-ray structure, the backbone is tightly kinked at the interface between J3/4 and P3. As such, the formation of the collapsed aptamer may induce a linkage geometry favourable to cleavage.
DMS probing cannot resolve A68 due to a strong reverse transcription stop at the base of P3. However, A69 is observed to be engaged in a protective interaction only in the SAM bound form (Figure 4a). The third base triple associated with the pseudoknot forms between A24 and a base pair between U71 of J3/4 and A92 of J4/1. The large increase in in-line probing reactivity in J3/4 and J4/1 suggest that the U71:A92 base pair is also ligand-dependent (Figure 4b). This is consistent with previous work showing a ligand induced change in the fluorescence of a 2-AP analogue inserted at A92 (14).
Mutant structures reveal ligand interactions shift the equilibrium of tertiary contacts toward engagement
The wild-type probing results suggest that a collapse of the aptamer domain involves a shift in structure toward the full engagement of the pseudoknot and its associated tertiary interactions. The conformational change is supported by the bimodal stability of a KT element and results in the reorganization of the helical junctions with concomitant formation of supporting base triple contacts.
In order to assess the relative contributions of these positions to the collapsed structure, we performed a structural probing study on the mutant aptamer domains that we have shown possess lower ligand affinity and little or no capacity to modulate switching (above). The mutant structures were analysed using in-line and hydroxyl radical structural probing (Figure 5). The results show that the mutant aptamers’ inability to resist association with the AT strand coincides with decreased stabilization of the collapsed aptamer structure. Furthermore, the data support a mechanism of collapse where the collapsed form is in equilibrium with the more open, switching competent form. Here, SAM interactions unlock tertiary contacts that act synergistically to shift this equilibrium toward the more compact and switching-resistant form.
Figure 5.
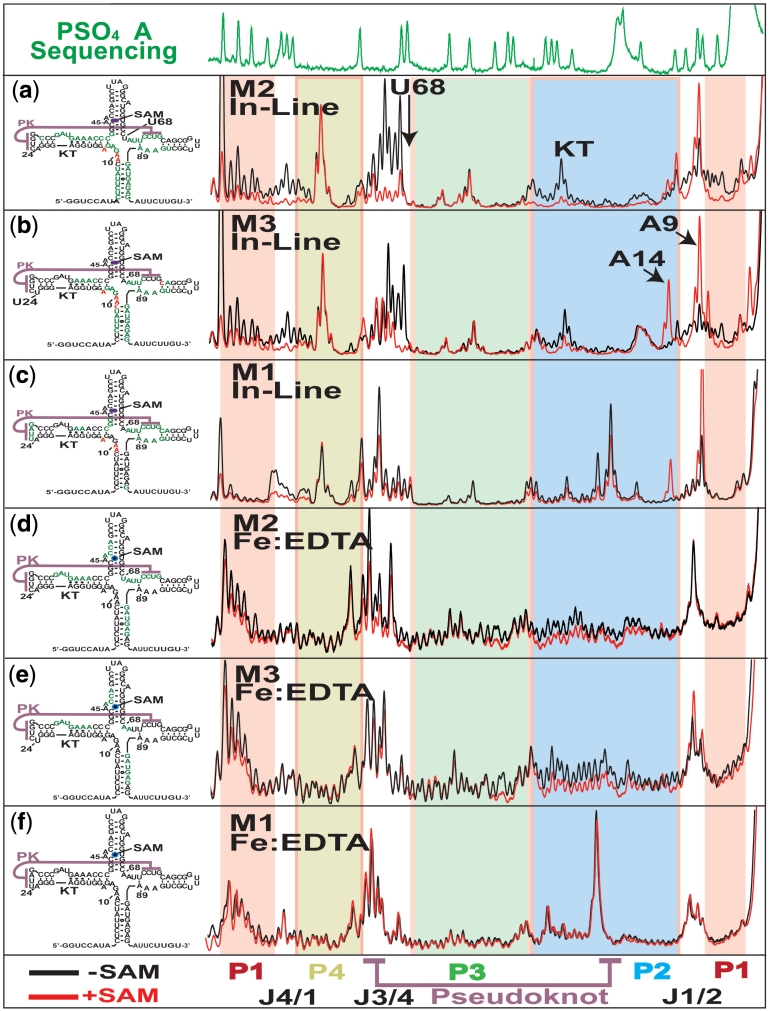
Capillary electrophoresis analysis of structure probing experiments using 3′ fluorescently labelled mutant aptamers. Top, α-phosphorothioate adenosine sequencing same as Figure 4. (a–c) Left, Fluorescence electropherograms for in-line probing of M2, M3 and M1 pseudoknot mutants showing protections patterns with (red) and without (black) 10 µM SAM. Right, secondary structure representation summarizing the probing results for each experiment indicating increasing (red) and decreasing (green) reactivity in response to ligand. (d and e) Traces for Fe:EDTA catalysed hydroxyl radical probing experiments displayed same as above.
In-line probing of the mutant aptamer domains show that the ligand induces some of the structural alterations observed for the wild-type sequence (Figure 5a–c). These included a decrease in reactivity at residues within junctions J4/1, J3/4 and the loop region of P2. However, defects were apparent in all three mutants, with the M2 mutant closest and M1 furthest from wild-type, consistent with the switching assay results. The differences between the individual mutants and wild-type were strongest in the J3/4 region containing the 3′ portion of the pseudoknot. The M2 mutant was capable of modulating the reactivity of bases in this region to a limited extent (Figure 5a).
Both M3 and M1 mutants were incapable of significantly reducing the mobility of the bases directly involved in the pseudoknot interaction from J3/4 (Figure 5a–c). However, M3 still displayed a strong stabilization of the 5′ part of J3/4 from where A68/69 make tertiary contacts (Figure 5a). In the wild-type molecule, the addition of ligand produces an aptamer with very low mobility in the two pseudoknot associated regions (Figure 4b). The mutants, on the other hand, show increased reactivity in the absence and lowered but still high reactivity in the presence of SAM in comparison to wild-type. This result is most easily explained by the formation of an equilibrium between engaged and disengaged pseudoknot conformations in the absence of ligand. In the presence of mutations, the equilibrium is shifted toward the more disengaged form in the absence of ligand. Here, the binding interactions are not sufficient to access the collapsed form. In the wild-type, the tertiary contacts maintain the equilibrium such that the ligand contacts can shift the population firmly toward the collapsed aptamer.
Hydroxyl radical probing results recapitulate the pattern observed in our in-line probing studies (Figure 5d–f). Both the M2 and M3 mutants displayed many wild-type like changes in solvent accessibility in response to SAM (Figure 5d and e). These included (i) the distinct protection pattern around the ligand-binding pocket of P3, (ii) the protection pattern of the 3′ side of helix P4 and (iii) a gross change in the exposure of helix P2. In addition, there were several features distinct to the mutant forms. These included a large increase in the solvent exposure of the J3/4 region in the bound and free forms of M2 and M3. Neither mutant displayed the pattern of increased solvent exposure for J1/2 and A68/A69. As with the in-line probing results, these results show a diminished capacity of the mutant aptamers to adopt a collapsed conformation in both the presence and absence of bound ligand. The situation is different for the M1 mutant, where alterations in residue mobility (observed via in-line probing) produce no measurable change in global solvent exposure (Figure 5f).
In the wild-type aptamer the interaction between P1 and P3 effectively encapsulates the ligand (Figure 1d). This increases affinity by decreasing the off-rate. In the M1 mutant we observe a lack of collapse related changes in solvent accessibility. Specifically, when SAM is bound, we see greater solvent accessibility in P1 and P3 in the mutant relative to wild-type. This would explain the decrease in affinity for SAM found in the mutant aptamer constructs. This can be seen in the wild-type hydroxyl radical probing results (Figure 4c). We observe differences between mutant and wild-type solvent accessibilities and backbone mobilities. We also observe differences between mutant and wild-type ligand affinities and switching rates. The degree of difference in the results between mutant and wild-type is consistent between all three techniques. Here, our data is consistent with a correlation between collapse and ligand affinity. This is best explained by the existence of a state in the mutants that is both open and bound. Ligand interactions trigger collapse of the aptamer domain by stabilizing pseudoknot-associated interactions. In this conformation ligand is encapsulated. The increased inter-aptamer contacts stabilize the domain at the expense of alternative expression platform conformations.
On the whole, these results indicate that the pseudoknot and its associated tertiary contacts are important for both the stability of the collapsed domain and also the conformation of the aptamer in absence of ligand. Their stabilizing influence is synergistic, emphasizing the fine balance of thermodynamic stabilities between the bound and unbound forms. This reinforces the proposition that these tertiary contacts exist in equilibrium without ligand and that SAM-binding acts to shift this equilibrium toward engagement of the tertiary contacts.
Ligand affinity is tuned for switching
Previous studies of the SAM I riboswitch have noted a discrepancy between the KD measured for isolated aptamer domains and the concentration of ligand required to affect half maximal termination efficiency. Analysis of the differential regulation between SAM I riboswitches observed the effect with all 11 B. subtilis SAM I riboswitches (31). The study found the KD values for individual SAM I aptamers spanned a 250-fold range. The affinities were also a poor predictor for the concentration of SAM required for regulation. These results indicate a more indirect strategy than ligand affinity may be employed for differential regulation across a class of riboswitches. The multiple steps involved in riboswitch regulation provide many possibilities for tuning a riboswitch toward differential regulation. This is especially true for the more complex process utilized by transcriptional terminating riboswitches, where the interplay between aptamer folding, ligand binding, transcription rate, pausing and competition between the alternative conformations can all conspire to produce a specific regulatory outcome. We utilized the switching assay to investigate the relationship between expression platform interactions with aptamer and ligand concentration.
A titration of SAM was performed using the fluorescence based switching assay described above. The association reaction between aptamer (100 nM) and AT strand (100 nM) was followed with various concentrations of SAM. The results show the rate and magnitude of AT association to be proportional to the concentration of ligand (Figure 6a). Importantly, the reactions tended toward equilibrium between the aptamer and AT piece that varied with SAM concentration.
Figure 6.
The effect of ligand interactions on switching. (a) A titration of SAM in the fluorescence based switching assay at a concentration of 100 nM of both aptamer and AT. Concentrations of SAM follow the legend in (b) with the exception of brown which is 500 nM. (b) Overlay of fluorescence based lane traces from native polyacrylamide gel electrophoresis analysis of equilibrium switching study. (c) Traces from (b) were integrated and the fraction of aptamer associated AT relative to the sample with no SAM was calculated and plotted versus SAM concentration and rectangular hyperbola fit. Arrows indicate the concentration at which 50% of the aptamer was associated, ∼250 nM. (d) Trace showing that the full formation of the AT helix is essentially irreversible by SAM interactions alone. Switching assay performed with the aptamer domain (5 nM) labelled on the 5′ with Cy-5 and the 3′-terminus with Cy-3. In the aptamer conformation the fluorescent dyes FRET strongly (excitation 535 nm, emission 670 nM). As the unlabelled AT (1 µM) associates the dyes are separated and go to a low FRET state. After the association reaction was allowed to proceed to ∼80% completion, SAM was added to a concentration of 1 mM (indicated by arrow). The failure to recover any FRET intensity indicates the aptamer is not reforming at the expense of the AT helix.
We next investigated the concentration of SAM required to produce a 50% decrease in AT formation at equilibrium. This was accomplished using the two-piece system and a native polyacrylamide gel for analysis. The aptamer was folded at various concentrations of ligand, after which, excess AT was mixed and the mixture was allowed to equilibrate. After incubation the samples were loaded onto a native gel to separate the switched from unswitched molecules (Figure 6b). The gel was stained with ethidium bromide and scanned with a fluorescence imager. The fluorescence lane traces were then integrated and the fraction of unassociated aptamer relative to the reaction containing no SAM was calculated. The amount of AT associated aptamer in the absence of ligand is taken as the maximum. The amount of associated aptamer in the presence of SAM is relative to the maximum. This data, when plotted versus SAM concentration, yields a concentration necessary for a 50% reduction in aptamer/AT association of ∼25 0nM (Figure 6c). This value represents a SAM dissociation constant for switching (KDswitch).
The KDswitch value is significantly higher than the binding affinity measured for the aptamer alone (∼36 nM, Figure 3b). We also show that the formation of the full AT helix is essentially irreversible by SAM interactions alone (Figure 6d). Here we attempt to recover the collapsed aptamer from the expression platform confomation (AT helix) using high concentrations of SAM. This is monitored by FRET from a 3′ Cy-3 to 5′ Cy-5 dye proximal to helix P1. We find that even after extended incubation times the loss of FRET and therefore, the P1 helix and aptamer domain is irreversible. This is not surprising considering the AT helix disrupts a considerable portion of the ligand-binding pocket specifically and aptamer domain generally (Figure 3a). As such, an equilibrium position between ligand-bound aptamer and AT requires a reversible initial interaction of AT. Since the equilibrium position (KDswitch ∼250 nM) is different from that of the aptamer and SAM (KDapp = 36 nM) the reversible interaction may increases the off-rate for ligand. This likely takes the form of a minimal set of base pairing interactions between AT complementary sequence at the base of P1. Since P1 creates a portion of the SAM-binding pocket, disruption of the helix may alter the affinity of the aptamer at large. These reversible attempts by the AT strand to nucleate an AT helix may, in turn, play a role in tuning the level of SAM required for the riboswitch to resist AT formation. Tuning would be accomplished by varying the strength of the first few base pairs and whose stability would be inversely proportional to the concentration of SAM required to resist AT strand invasion.
DISCUSSION
A role for thermodynamics
The functional outcome of riboswitch action is dependent on a number of factors during transcription. Efficient termination in the presence of ligand requires the co-transcriptional folding of the aptamer domain to produce an anti-termination resistant conformation prior to elongation of the expression platform. In the case of the FMN riboswitch it has been shown that a pause between the aptamer domain and AT sequence increases termination efficiency presumably by allowing time for the formation of the bound aptamer conformer (9). As transcription proceeds, a second pause occurs between the AT helix and the coding sequence. At this point, the aptamer and the AT conformations are allowed to compete. While the requirement for a pausing event has not been shown in the case of the SAM I class of riboswitches, the functional outcome is still dependent upon this competition. Our experimental system examines precisely this situation, competing the aptamer and AT in the presence and absence of SAM.
In the absence of SAM, the AT conformation dominates at the expense of both the aptamer domain and a termination hairpin. This is the default state for the riboswitch element in the absence of ligand. This allows gene expression as a result of the increased thermodynamic stability of this expression platform conformation. Many riboswitch elements have been shown to bind effectors with affinities that decrease with the addition of sequence from the expression domain (1,5,8,9,32,33). The decreased affinity likely results from the reversible interactions of expression platform sequence with the aptamer. This is easily explained in the SAM I aptamer, as the formation of the first few base pairs of the expression platform would utilize sequence at the expense of the P1 portion of the binding pocket.
Using the two piece switching construct we have shown that, in the case of a SAM I riboswitch aptamer, the competition for the aptamer sequence leads to the establishment of a stable equilibrium between SAM, aptamer and AT, requiring SAM concentrations similar to those required for riboswitch-mediated termination in vivo (31). We have also noted that in the SAM I riboswitch, the full formation of the AT is essentially irreversible by ligand alone without refolding. The degree to which ligand/aptamer equilibrium is affected by interactions with the AT is unknown. However, given the location of the binding pocket, which utilizes conserved contacts with helix P1 (34), the ligand/aptamer equilibrium is likely to be modified by interactions with the AT region. This may be true for many classes of riboswitch elements and provides further insight into the means by which natural selection has tuned riboswitch elements to respond at different concentrations of ligand.
SAM drives the collapse of the aptamer by stabilizing tertiary interactions
Our probing studies of the isolated aptamer reveal a compact conformation in the presence of SAM and a more open conformation in absence of SAM. The studies of aptamer collapse provide a mechanistic context to interpret the switching experiments. Overall, the results suggest a model for the collapse of the aptamer via ligand binding. Mainly, a series of mutually interdependent interactions are formed in response to binding. Aptamer stability is the product of (i) stabilization of helix P1 through the closure of P1 and P3 and (ii) reductions in mobility of the junction regions adjacent to helix P1 induced by the organization of the core junction region. A reduction in junction region fluctuations within J1/2 and J4/1, as well as P1/P3 contacts, resulting from the collapsed architecture, is undoubtedly responsible for the resistance of the aptamer domain to the expression platform sequence.
Upon examination of the X-ray structure, we can see how interactions distant from the binding site are promoted by direct ligand organization of the junction region between helices P1 and P2 (J1/2), especially at nucleotides A10 and G11. This region positions residues in the tightly packed four-way junction. We also note that the junction region J1/2 may help coordinate the pseudoknot associated contacts through a magnesium-mediated interaction. A magnesium ion bridges J1/2 (pos. A10) and J3/4 (pos. U71). This is supported by a recently published computational study showing this magnesium-binding site as being destabilized by the removal of SAM interactions with J1/2 (35).
Key tertiary contacts play an important role in secondary structure transitions
The switch assay shows that specific tertiary contacts in the aptamer domain found to be necessary for collapse are also necessary for resisting strand invasion by AT sequence in the expression platform. While many interactions within the aptamer were found to contribute to the SAM-bound fold via NAIM, a certain subset of these interactions is critical in controlling changes in the secondary structure. This subset forms a network of interactions flanking the pseudoknot. These positions do not make direct contact with SAM but interact through the four-way junction with residues that do make interactions with ligand. We find that this network is cooperative in promoting collapse and resistance. In the case of mutation M3, the removal of a single hydrogen bond in a base triple contact increases the apparent KD for ligand and destabilizes the collapsed form of the domain abolishing resistance to the expression domain. As such, the energy landscape for switching is finely balanced. In particular, the stable, bound form of the aptamer domain is structurally accessible to the apo-aptamer, yet energetically unfavourable in the absence of SAM.
Structural interpretation
During transcription, the P1/P4 coaxial stack may compete with the AT for the same set of bases (Figure 7). Our data are consistent with a dynamic equilibrium existing between a closed and open state of the aptamer. In the open state, nucleotides in P1 and P4 are more susceptible to hybridization with the AT. The key tertiary contacts (probed with mutants M1, M2 and M3) exist in equilibrium (between formed and unformed) in the wild-type system in the absence of SAM. Mutations to these tertiary contacts move the equilibrium position such that ligand interactions are no longer sufficient to stabilize the collapsed form. In this sense, the tertiary contacts control the equilibrium and are essential for correct riboswitch operation.
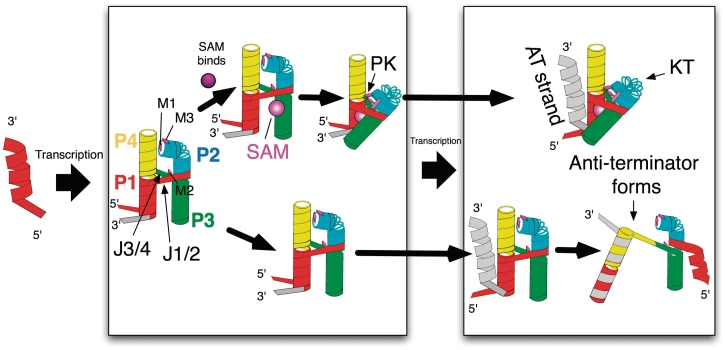
Figure 7.
The outcome for genetic regulation is determined following the synthesis of the aptamer domain. Here, SAM binding shifts the equilibrium of pseudoknot associated contacts toward engagement (top pathway). The contacts provide aptamer stability leading to the continued exclusion of the AT strand following its elongation. In the absence of SAM binding the AT strand is free to compete with an unstablized open conformation of the aptamer leading to the formation of the AT helix (lower pathway).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Laboratory Directed Research and Development; Exploratory Research program at Los Alamos National Laboratory. Funding for open access charge: Los Alamos National Laboratory LDRD-ER program funds.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Dr Cliff Unkefer and staff of the National Stable Isotope Resource for the use of space and resources necessary to perform this research. The authors would also like to thank Dr Kathleen Hall for the generous gift of 1M7. The authors are grateful to Dr Robert Batey for useful discussions.
REFERENCES
- 1.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 2.Grundy FJ, Henkin TM. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 1998;30:737–749. doi: 10.1046/j.1365-2958.1998.01105.x. [DOI] [PubMed] [Google Scholar]
- 3.McDaniel BA, Grundy FJ, Artsimovitch I, Henkin TM. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc. Natl Acad. Sci. USA. 2003;100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Epshtein V, Mironov AS, Nudler E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc. Natl Acad. Sci. USA. 2003;100:5052–5056. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 6.Wang JX, Lee ER, Morales DR, Lim J, Breaker RR. Riboswitches that sense S-adenosylhomocysteine and activate genes involved in coenzyme recycling. Mol. Cell. 2008;29:691–702. doi: 10.1016/j.molcel.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rieder R, Lang K, Graber D, Micura R. Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. Chembiochem. 2007;8:896–902. doi: 10.1002/cbic.200700057. [DOI] [PubMed] [Google Scholar]
- 8.Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chem. Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 9.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 10.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 11.Baird NJ, Ferre-D’Amare AR. Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis. RNA. 2010;16:598–609. doi: 10.1261/rna.1852310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blouin S, Lafontaine DA. A loop loop interaction and a K-turn motif located in the lysine aptamer domain are important for the riboswitch gene regulation control. RNA. 2007;13:1256–1267. doi: 10.1261/rna.560307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ottink OM, Rampersad SM, Tessari M, Zaman GJ, Heus HA, Wijmenga SS. Ligand-induced folding of the guanine-sensing riboswitch is controlled by a combined predetermined induced fit mechanism. RNA. 2007;13:2202–2212. doi: 10.1261/rna.635307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heppell B, Lafontaine DA. Folding of the SAM aptamer is determined by the formation of a K-turn-dependent pseudoknot. Biochemistry. 2008;47:1490–1499. doi: 10.1021/bi701164y. [DOI] [PubMed] [Google Scholar]
- 15.Uhlenbeck OC. Less isn’t always more. RNA. 2003;9:1415–1417. doi: 10.1261/rna.5155903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoogstraten CG, Sumita M. Structure-function relationships in RNA and RNP enzymes: recent advances. Biopolymers. 2007;87:317–328. doi: 10.1002/bip.20836. [DOI] [PubMed] [Google Scholar]
- 17.Rangan P, Masquida B, Westhof E, Woodson SA. Assembly of core helices and rapid tertiary folding of a small bacterial group I ribozyme. Proc. Natl Acad. Sci. USA. 2003;100:1574–1579. doi: 10.1073/pnas.0337743100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez-Salas UA, Rangan P, Krueger S, Briber RM, Thirumalai D, Woodson SA. Compaction of a bacterial group I ribozyme coincides with the assembly of core helices. Biochemistry. 2004;43:1746–1753. doi: 10.1021/bi035642o. [DOI] [PubMed] [Google Scholar]
- 19.Ryder SP, Ortoleva-Donnelly L, Kosek AB, Strobel SA. Chemical probing of RNA by nucleotide analog interference mapping. Methods Enzymol. 2000;317:92–109. doi: 10.1016/s0076-6879(00)17008-9. [DOI] [PubMed] [Google Scholar]
- 20.Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 21.Goody TA, Melcher SE, Norman DG, Lilley DM. The kink-turn motif in RNA is dimorphic, and metal ion-dependent. RNA. 2004;10:254–264. doi: 10.1261/rna.5176604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turner B, Lilley DM. The importance of G.A hydrogen bonding in the metal ion- and protein-induced folding of a kink turn RNA. J. Mol. Biol. 2008;381:431–442. doi: 10.1016/j.jmb.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 23.McDaniel BA, Grundy FJ, Henkin TM. A tertiary structural element in S box leader RNAs is required for S-adenosylmethionine-directed transcription termination. Mol. Microbiol. 2005;57:1008–1021. doi: 10.1111/j.1365-2958.2005.04740.x. [DOI] [PubMed] [Google Scholar]
- 24.McDaniel BA, Grundy FJ, Kurlekar VP, Tomsic J, Henkin TM. Identification of a mutation in the Bacillus subtilis S-adenosylmethionine synthetase gene that results in derepression of S-box gene expression. J. Bacteriol. 2006;188:3674–3681. doi: 10.1128/JB.188.10.3674-3681.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang JX, Breaker RR. Riboswitches that sense S-adenosylmethionine and S-adenosylhomocysteine. Biochem. Cell Biol. 2008;86:157–168. doi: 10.1139/O08-008. [DOI] [PubMed] [Google Scholar]
- 26.Xu D, Evans KO, Nordlund TM. Melting and premelting transitions of an oligomer measured by DNA base fluorescence and absorption. Biochemistry. 1994;33:9592–9599. doi: 10.1021/bi00198a027. [DOI] [PubMed] [Google Scholar]
- 27.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 28.Tullius TD, Greenbaum JA. Mapping nucleic acid structure by hydroxyl radical cleavage. Curr. Opin. Chem. Biol. 2005;9:127–134. doi: 10.1016/j.cbpa.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 29.Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winkler WC, Grundy FJ, Murphy BA, Henkin TM. The GA motif: an RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs. RNA. 2001;7:1165–1172. doi: 10.1017/s1355838201002370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomsic J, McDaniel BA, Grundy FJ, Henkin TM. Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-box elements in bacillus subtilis exhibit differential sensitivity to SAM In vivo and in vitro. J. Bacteriol. 2008;190:823–833. doi: 10.1128/JB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winkler WC, Breaker RR. Genetic control by metabolite-binding riboswitches. Chembiochem. 2003;4:1024–1032. doi: 10.1002/cbic.200300685. [DOI] [PubMed] [Google Scholar]
- 34.Montange RK, Mondragon E, van Tyne D, Garst AD, Ceres P, Batey RT. Discrimination between closely related cellular metabolites by the SAM-I riboswitch. J. Mol. Biol. 396:761–772. doi: 10.1016/j.jmb.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang W, Kim J, Jha S, Aboul-ela F. A mechanism for S-adenosyl methionine assisted formation of a riboswitch conformation: a small molecule with a strong arm. Nucleic Acids Res. 2009;37:6528–6539. doi: 10.1093/nar/gkp664. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.