Abstract
Cell growth and proliferation are tightly controlled via the regulation of the p53–MDM2 feedback loop in response to various cellular stresses. In this study, we identified a nucleolar protein called PAK1IP1 as another regulator of this loop. PAK1IP1 was induced when cells were treated with chemicals that disturb ribosome biogenesis. Overexpression of PAK1IP1 inhibited cell proliferation by inducing p53-dependent G1 cell-cycle arrest. PAK1IP1 bound to MDM2 and inhibited its ability to ubiquitinate and to degrade p53, consequently leading to the accumulation of p53 levels. Interestingly, knockdown of PAK1IP1 in cells also inhibited cell proliferation and induced p53-dependent G1 arrest. Deficiency of PAK1IP1 increased free ribosomal protein L5 and L11 which were required for PAK1IP1 depletion-induced p53 activation. Taken together, our results reveal that PAK1IP1 is a new nucleolar protein that is crucial for rRNA processing and plays a regulatory role in cell proliferation via the p53–MDM2 loop.
INTRODUCTION
Cell growth and proliferation are two precisely controlled and integrated processes throughout the whole life of an organism. The maintenance of an appropriate cell number in a well-functioning organ is dependent on the balance of the rate at which cells are generated via proliferation and eliminated by apoptosis (1). Cell proliferation is highly regulated at each phase of the cell cycle, and the G1/S checkpoint is one of the major cell-cycle transition points. At the G1/S transition, a cell can decide its fate, such as cell division, G1 arrest, quiescence, or differentiation, in response to diverse signals (2,3). Any malfunctions at this checkpoint, such as activation of an oncogene or inactivation of a tumor suppressor gene, may lead to abnormal cell proliferation and transformation, consequently cancer development (4,5). One of the most important tumor suppressor genes is TP53, which is mutated in >50% of all types of human cancers while the rest of cancers are often associated with alterations of p53 modulators that regulate p53 stability and activity. For example, the mdm2 gene, which encodes a p53 suppressor called MDM2, is upregulated in 7% of human cancers which lack the mutation of p53 (6–8). In normal unstressed cells, p53 level is low mainly due to the regulation by MDM2, which is transcriptionally stimulated by p53 and functions as an E3 ubiquitin ligase to mediate p53 ubiquitination and degradation through a feedback mechanism (8–12).
Upon stress conditions, such as genotoxic, oncogenic and nucleolar stresses, p53 is stabilized and activated largely through the inhibition of MDM2, leading to cell-cycle arrest, apoptosis, DNA repair or senescence (13). In response to nucleolar stress, several nucleolar proteins including nucleophosmin, nucleostemin, L5, L11, L23 and S7 interact with MDM2 and inhibit its activity, leading to p53 stabilization (14–21). Nucleolar/ribosomal stress is induced by the perturbation of ribosomal biogenesis, such as the interference in rRNA synthesis, processing and ribosome assembly (22,23). It has been reported that the treatment of human cells with fluorouracil (5-FU) or actinomycin D (low doses), serum starvation, disruption of nucleolar or ribosomal proteins leads to ribosomal stress, and consequently, p53 activation (14–21,24,25). Therefore, nucleolar proteins play a critical role in transmitting nucleolar stress signals to the p53 pathway, coupling this pathway with ribosomal biogenesis (25).
PAK1IP1, a PAK1-interacting protein (also called hPIP1), was previously identified as a component in highly purified nucleoli of human cells (26,27). This protein contains five G protein β-like WD40 repeats and shares high sequence homology with Mak11 in budding yeast (28) and Skb15 in fission yeast (29). The two yeast orthologs of PAK1IP1 are involved in 60 S rRNA biogenesis and essential for cell viability (30,31). Our previous study showed that PAK1IP1 is abundantly expressed in most human tissues, and negatively regulates the activity of PAK1 (27) which has been shown to regulate various cellular activities, including cell proliferation, cell survival, mitosis and transcription (32–35). Although we have demonstrated that mouse Pak1ip1 can substitute Skb15 function in fission yeast, the cellular and biochemical functions of PAK1IP1 in mammalian cells is poorly understood.
In an attempt to address this issue, we identified new functions of PAK1IP1 in regulating the cell cycle via the p53–MDM2 pathway as described in this study. With a functional nuclear/nucleolar localization signal sequence, PAK1IP1 could localize in both the nucleoplasm and nucleolus. Overexpression of PAK1IP1 stabilized p53 protein level and inhibited cell proliferation by inducing G1 arrest. PAK1IP1 executed this cellular function by interacting with MDM2 and inhibiting its activity toward p53 ubiquitination and degradation. Knockdown of PAK1IP1 by shRNA induced p53 activation through enhancement of the binding of the ribosomal proteins L5 and L11 to MDM2 in human cancer cells. Therefore, these results reveal a novel and critical role of PAK1IP1 in cell-cycle regulation through the modulation of the p53–MDM2 feedback loop.
MATERIALS AND METHODS
Cell cultures and transfection
Human HeLa, HEK293T, U2OS, Saos2, A549, H1299 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, USA), supplemented with 10% fetal bovine serum (FBS), 100 U each of penicillin and streptomycin, in an air—5% CO2 atmosphere at constant humidity. For transfection, plasmids or siRNA were mixed with Lipofectamine 2000 reagent (Invitrogen Inc.) in Opti-MEM (Gibco, USA) and incubated at room temperature for 20 min. The Lipofectamine 2000–DNA complex was added to cells and mixed by gentle agitation. Growth media containing 10% FBS were exchanged to the cells 6 h after transfection of the plasmids.
DNA construction
To generate a fusion protein of PAK1IP1 with enhanced green fluorescent protein (EGFP), the PAK1IP1 ORF was subcloned into the XhoI and EcoRI (NEB) sites of the pEGFP-C3, each mutant of PAK1IP1 sequence was subcloned into pEGFP-vector, or site-directed mutagenesis PCR according to the QuikChange® XL Site-Directed Mutagenesis Kit (stratagene). The direct PCR Primers used for constructs mentioned above are as follows: pEGFP-C3-PAK1IP1: F-5′ GGCTCGAGATGGAGCTGGTCGCTGGTTGCTAC 3′ and R-5′ CAGAATTCCTGCAGCCCTCAGGAGAC 3′; pEGFP-N2-PAK1IP1-WD40: F-5′ TAAGAATTCATATGGAGCTGGTCGCTGGTT 3′ and R-5′ GTAGTCGACCAGGCTCCTTTTTGCCAA 3′; pEGFP-N2-PAK1IP1-dWD40: F-5′ GGGGAATTCACGTGTCTTGGAGTGTGG 3′ and R-5′ GGGGTCGACACTGCATTGTTTTTATTTTC 3′.
The site-directed mutagenesis PCR primers used for constructs mentioned above as follow: pEGFP-C3-PAK1IP1-mNoLS1: F-5′ GGCCTGATATCAACCGCAGCGGCAGCAATGGTAGAAATGTTGGAAAAG 3′ and R-5′ CAACATTTCTACCATTGCTGCCGCTGCGGTTGATATCAGGCCACTTTC 3′; pEGFP-C3-PAK1IP1-mNoLS2 : F-5′ GGTAGAAATGTTGGAAGCGGCGGCAGCAAAGAAGAAAATAAAAACAATGC 3′ and R-5′ TTATTTTCTTCTTTGCTGCCGCCGCTTCCAACATTTCTACCATTTTCCTC 3′; This construct used for all experiments except where indicated, and pEGFP-C3-PAK1IP1-mNoLS3 : F-5′ AGAAGAGGAAAGCGGCGGCAATAGCAACAATGCAGTGAATCACAG 3′ and R-5′ CACTGCATTGTTGCTATTGCCGCCGCTTTCCTCTTCTTTTCCAAC 3′.
Immunoprecipitation and western blot
Binding of PAK1IP1 with human MDM2 in five-dish (10 cm) mammalian cells was examined by immunoprecipitaion and by western blot analysis, which were performed as previously described (36). Antibodies recognizing PAK1IP1 (#sc-101866, Santa Cruz, USA and #ab67348, Abcam, UK), M2 Flag Antibody and anti-Flag M2 affinity beads (F1804 and A2220, Sigma, Germany) p53 (DO1) and MDM2 (N-20) (#sc-126 and #sc-813, Santa Cruz, CA, USA), p21 (Cell Signaling Technology, MA, USA) were used in IP and WB analysis. Anti-L5 and anti-L11 have been previously described previously (21). The relative ratios of western blot signals were estimated with the Image-Pro plus 6.0 software (Media Cybernetics).
Lentivirus-based PAK1IP1 overexpression and knockdown
For the overexpression of lentiviral constructs, the lentivirus-based vector pLL3.7 was employed (37). The fragments of GFP-PAK1IP1, GFP-PAK1IP1-dNLS and GFP-PAK1IP1-mNoLS derived from pEGFP-constructs and substituted the GFP sequence of pLL3.7 vector using the NheI and EcoRI. For the knockdown constructs, we designed two shRNA oligos used the promega shRNA Design software (http://www.promega.com/siRNADesigner/program/default.asp), two shRNAs (237–255, 884–902) were designed corresponding to nuclear acids of ORF (open reading frame), and the sequences are below: shRNA1(884–902), Forward- 5′ TGGCTGACGTGTCTTGGAGTTTCAAGAGTGAGGTTCTGTGCAGTCGGTTTTTTC 3′ and Reverse- 5′ ACCGACTGCACAGAACCTCAAAGTTCTCACTCCAAGACACGTCAGCCAAAAAAGAGCT 3′; shRNA2(237-255), Farward-5′ TGCATCACAGTGGTACAATATTCAAGAGATATTGTACCACTGTGATGCTTTTTTC 3′, this construct used for all experiments except where indicated, and Reverse- 5′ ACGTAGTGTCACCATGTTATAAGTTCTCTATAACATGGTGACACTACGAAAAAAGAGCT 3′; These oliognucleotides were annealed and ligated into pLL3.7. Following DNA sequencing confirmation, the overexpression, knockdown or control construct, was transiently co-transfected into 293T cells along with the gag, env-expressing plasmids, Virus-containing supernatants were collected and used to infect target cells. Expression of wild-type and mutant PAK1IP1 was determined by Inverted Fluorescence Microscope and western blotting using antibody against PAK1IP1. The target sequence of TP53 is obtained from Thermo Scientific Dharmacon, and the target sequences for L5, L11 and the control scrambled siRNA have been described previously (17,38).
Cell proliferation and colony formation assays
For the cell growth curve assay, the infected cells were seeded at 2 × 104 cells/well and grown in 24-well plates for continual 1–7 days in triplicate. The cells were harvested and counted on the time point as indicated. For cell viability assay, a modified MTT assay (Promega, Madison, WI, USA) was used. 5000 infected cells were plated into 96-well plates in triplicate for 3 days. The 20 μl MTS was added according to manufacturer instruction. Absorbance was measured at 490 nm using a microplate reader and is directly proportional to the number of viable cells in the cultures. For colony formation assay, the infected cells were sorted by FACS, and 1000 cells were plated into 6cm dish in triplicate. After cells were cultured for 2–4 weeks, the cells were stained with 0.4% crystal violet (dissolved in 95% ethanol). The number of colonies, in triplicate wells were photographed and counted to generate the histograms.
Cell-cycle analysis
To determination of cell-cycle distribution, the transfected or infected cells were let grow to ∼30–40% conflucence. The cells were treated with 2 mM thymidine (Sigma) for 12 h, and released into DMEM growth media for 10 h, then secondary thymidine block for 12 h, finally release cells into DMEM growth medium. The cells were harvested after release in the indicated time, then fixed with 70% cold ethanol and stained with 50 μg/ml propidium iodide (Sigma, St Louis, MO, USA) followed with RNase A (Sigma) treatment for 30 min at room temperature. DNA content was analyzed by FACscan cell analyzer (BD Biosciences, San Jose, CA, USA) equipped with Cellquest software (BD Biosciences). The population of cells in each phase was determined using ModFit LT software (BD Biosciences).
Indirect immunofluorescence
The transfected or infected cells were seeded onto glass cover slips. Cells were washed with phosphate-buffered saline, fixed at room temperature using 4% paraformaldehyde, followed by 0.5% Triton-X 100 in phosphate-buffered saline for 15 min at room temperature. Cells were stained with indicated antibodies, followed by FITC or TRITC-conjugated immunoglobulins. Nuclei were counterstained with DAPI. The images were taken by confocal laser scanning microscopy (CLSM; Leica TCS SP5).
BrdU incorporation assays
Bromodeoxyuridine (BrdU) incorporation assays were conducted as described previously (21). Tranfected cells were incubated in the presence of 10 μM BrdU (Sigma) for 10 h after starvation overnight. Cells were then fixed with 4% paraformaldehyde and treated with 2M HCl containing 1% Triton X-100. The cells were stained with monoclonal anti-BrdU (Sigma) antibody and then stained with TRITC-conjugated goat anti-mouse antibodies and DAPI. The red immunofluorescence, indicating BrdU staining, The total numbers of GFP-positive cells, as well as BrdU-labeled cells, were counted in 5–10 different fields of each well.
In vivo ubiquitylation assay
Human p53-null H1299 cells were transfected with indicated plasmids. The cells were treated with 20 μM of MG132 for 6 h before they were harvested at 36 h post-transfection. In vivo ubiquitinlation assays were conducted as described previously (21). In brief, 10% of the total suspension was retained for total protein analysis. The remaining cell fraction was lysed under denaturing conditions with the 6 M guanidinium–HCl buffer. Total His-ubiquitin protein conjugates were purified by affinity chromatography using Ni2+ NTA agarose beads and the levels of ubiquitination were determined using anti-p53 antibody or anti-MDM2 antibody.
Sucrose gradient centrifugation
Sucrose gradient sedimentation of polysomes, and analysis of the polysome and mRNP distribution of proteins were carried out as previously described with minor modifications (17). Briefly, cells were incubated with 50 μg of cycloheximide/ml for 5 min. The cells were sonicated on ice using an ultrasonic processor equipped with a microtip and sonication buffer containing 25 mM Tris–Cl, pH 7.5, 100 mM KCl, 1 mM NaF, 2 mM EDTA, 0.05% (v/v) NP-40, 1 mM DTT, 10 μl/ml protease inhibitor cocktail (Sigma), 50–100 U/ml RNase inhibitor (TaKaRa). Each supernatant was subjected to sedimentation centrifugation in a 10–50% sucrose gradient solution containing 25 mM Tris–HCl (pH 7.5), 2 mM EDTA and 100 mM KCl in a Beckman SW41 rotor at 37 000 rpm for 3 h. Fourteen fractions were collected and the proteins were concentrated using 20% trichloroacetic acid (TCA) for western blotting.
RESULTS
PAK1IP1 is a nucleolar stress induced nucleolar protein with nuclearoplasmic localization
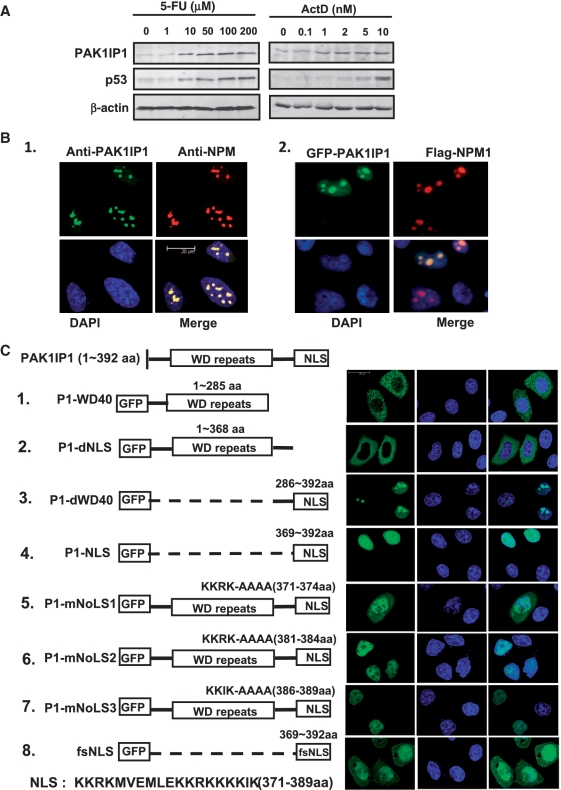
It was recently reported that Mak11 and Skb15, the yeast orthologs of PAK1IP1, play important roles in 60 S ribosome biogenesis (26,31), but the cellular and biochemical functions of PAK1IP1 in mammalian cells remain largely elusive. To understand the potential role of PAK1IP1 in ribosomal stress response, we first examined the protein expression level of PAK1IP1 in response to ribosomal stress using fluorouracil (5-FU) or actinomycin D (ActD). These two chemotherapeutic agents inhibit ribosome biogenesis and trigger nucleolar stress, and subsequently, lead to p53 stabilization and cell growth arrest (15,39). As shown in Figure 1A, when HeLa cells were treated with 5-FU for 18 h, the protein level of PAK1IP1 was significantly upregulated with the increasing amount of 5-FU in a dose-dependent fashion (Figure 1A, left panel). As a positive control, the expression of p53 is also increased with the treatment of 5-FU (Figure 1A, middle). Similarly, ActD also induced the expression of PAK1IP1 protein in a dose-dependent manner (Figure 1A, right panel). These results suggest that PAK1IP1 is a ribosomal stress-induced protein and could play an important role in ribosomal stress responses.
Figure 1.
PAK1IP1 is a nucleolar stress induced protein with nuclear localization. (A) Nucleolar stress induces PAK1IP1 expression. HeLa cells were treated with increasing doses of 5-FU and ActD as indicated for 18 h. Cell lysates were assayed for the expression of PAK1IP1 and p53 by western blot. (B) Subcellular localization analysis of PAK1IP1. (B-1) Endogenous PAK1IP1 colocalizes with NPM in the nucleus and accumulates in the nucleolus. Immunostaining of HeLa cells was performed with anti-PAK1IP1 (green) and anti-NPM1 (red) antibodies. The nuclei were stained with DAPI. (B-2) GFP-PAK1IP1 recombinant protein colocalizes with NPM in the nucleus and accumulates in the nucleolus. HeLa cells were co-transfected with GFP-PAK1IP1 and flag-NPM. Twenty-four hours post-transfection, the cells were immunostained with anti-flag antibody and DAPI. (C) Subcellular localization analysis of PAK1IP1 mutants. HeLa cells were transfected with different PAK1IP1 mutants, respectively. After 24 h, cells were fixed and examined under confocal laser scanning microscopy. 1, GFP-PAK1IP1-WD40; 2, GFP-PAK1IP1-dNLS; 3, GFP-PAK1IP1-dWD40; 4, GFP-PAK1IP1-NLS; 5, GFP-PAK1IP1-mNoLS1; 6 GFP-PAK1IP1-mNoLS2; 7, GFP-PAK1IP1-mNoLS3; 8, GFP-fsNLS. P1, PAK1IP1; d, delta; m, mutant; fs, frame-shift. Diagram of each transfected construct was presented on the left panel. Bars, 20 μm.
The nucleolus plays an important role in ribosomal stress response (23). We then examined the localization of PAK1IP1 using immunofluorescence staining. As shown in Figure 1B, both endogenous and exogeneous PAK1IP1 proteins exhibited diffuse nucleoplasmic distribution with apparently nucleolar accumulation, colocalizing with nucleophosmin (NPM1), the granular component marker, and the high quality image was shown in Supplementary Figure S1. Similar results were also observed in U2OS and 293T cells (data not shown). Thus, PAK1IP1 mainly resides in the nucleolus with minor nucleoplasmic distribution.
To identify the nuclear and nucleolar localization signal sequence(s) (NLS/NoLS) of PAK1IP1, we used the LOCtree software (40) and identified three consensus (R/K) (R/K) X (R/K) sequences in the C-terminus [from 371 to 389 amino-acid residues (KKRKMVEMLEKKRKKKKIK)] of PAK1IP1 as potential NLS/NoLSs (41). To validate these sites, we generated a set of deletion mutations of PAK1IP1 (abbreviated to P1) using the GFP fusion construct as a template (Figure 1C, left panel), and the subcellular localization was analyzed using confocal laser scanning microscopy as shown in the right panel of Figure 1C. With the deletion of the NLS, P1-WD40 (1–285aa) and P1-dNLS (deltaNLS, 1–368 aa), the GFP fusion proteins were excluded from the nuclei (Figure 1C-1 and 2), suggesting that the predicted NLS is essential for the nuclear localization of PAK1IP1. Consistently, the GFP-putative NLS fusion proteins were only detected in the nucleus and nucleolus (Figure 1C-3 and 4). The flanking sequence (286–368aa) did not affect the nuclear localization of the fusion protein, but it might play a regulatory role in nucleolar localization of this protein, as deletion of the flanking sequence of NLS increased the fluorescence intensity of the GFP-fusion protein in nucleoplasm (comparing Figure 1C-3 and 4). Further mutation analysis of each (R/K) (R/K) X (R/K) motif indicated that each of the motifs is important for the nucleolar localization of the protein (Figure 1C-5–7). Mutation of each motif excluded the protein from the nucleolus (Figure 1C-5–7). When the frame-shifted NLS (369–392 aa) was fused with GFP, the chimera protein was expressed in both nucleus and cytoplasm in all transfected cells (Figure 1B-8). These results demonstrate that three (R/K) (R/K) X (R/K) motifs within the C-terminal NLS/NoLS region of PAK1IP1 are essential for nucleolar and nuclear localization of the protein and suggest that they may play a regulatory role in PAK1IP1 cellular function.
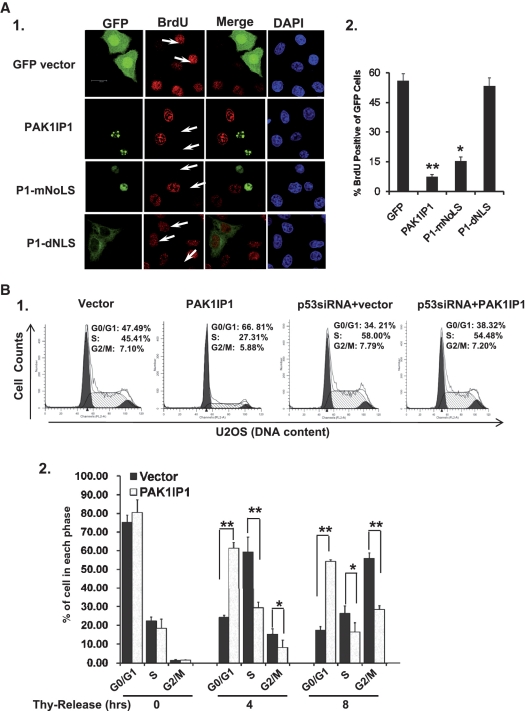
Figure 3.
Overexpression of PAK1IP1 inhibits BrdU incorporation and induces G1/S cell-cycle delay. (A) Overexpression of PAK1IP1 inhibits BrdU incorporation. HeLa cells were transfected with control, PAK1IP1, P1-mNoLS or P1-dNLS, respectively. The cells were immunostained with BrdU specific antibody (A-1). The percentages of BrdU positive cells in GFP labeled population (A-2) were determined as described in ‘Materials and Methods’ section. The arrows showed the GFP-positive cells with much less BrdU incorporation, and the data were from three independent experiments. (B) Overexpression of PAK1IP1 induces G1/S cell-cycle delay. (B-1), U2OS cells were transfected with or without p53 siRNA respectively, after 12 h, the cells were infected with lentivirus of PAK1IP1 and control vector, then the cells were analyzed by FACS analysis. (B-2), overexpression of PAK1IP1 regulates G1/S phase transition in synchronized cells. The post-infected cells were synchronized with double-thymidine block, and then the cells were collected at the indicated time points after release. Cell-cycle distributions of the cells were analyzed by FACS analysis. The error bars indicate standard deviations from three independent experiments, and the differences in both variables were analyzed for significance by Student’s t-test. P1, PAK1IP1. Thy, thymidine. **P < 0.01 and *P < 0.05. Bars, 20 μm.
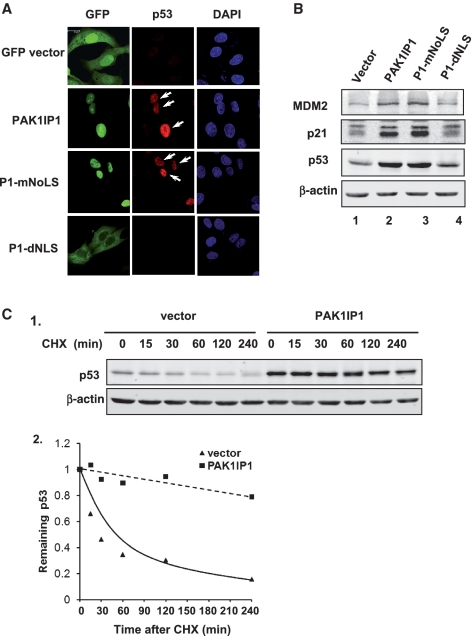
Figure 4.
PAK1IP1 induces the accumulation of tumor suppressor p53. (A) PAK1IP1 upregulates p53. U2OS cells were transfected with PAK1IP1, P1-mNoLS or P1-dNLS GFP fusion constructs respectivly. The arrows showed GFP-positive cells that have more p53 protein. (B) Overexpression of PAK1IP1 regulates p53 downstream target proteins. Different PAK1IP1 constructs as indicated were transfected into U2OS cells. Whole cell lysates were harvested and separated by 10% SDS–PAGE for western blot analysis using antibodies specific for p53, p21, MDM2 and β-actin. (C) PAK1IP1 stabilizes p53. U2OS cells were infected with control vector or PAK1IP1 lentivirus for 48 h and then cycloheximide (CHX) was supplemented into the media (concentration of 50 μg/ml). (C-1), the cells were harvested at indicated time points and subjected to western blot analysis with antibodies specific for p53 and β-actin (C-1). (C-2), the bands were quantified and normalized with loading controls determined by β-actin expression in U2OS (C-2). Bars, 20 μm.
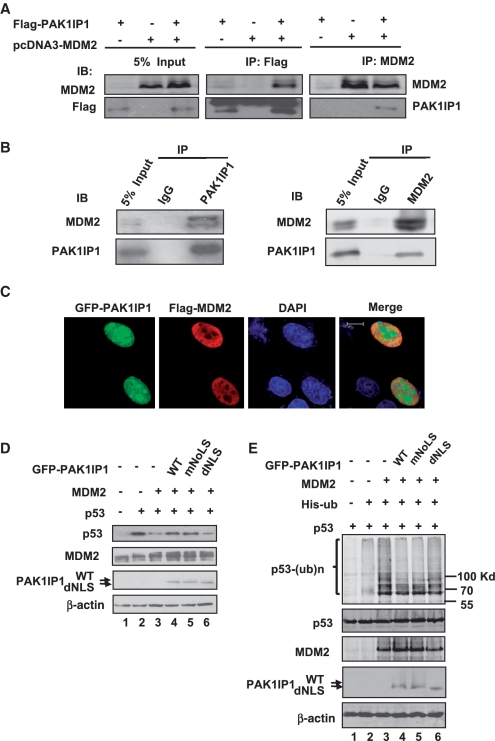
Figure 5.
PAK1IP1 associates with MDM2 and inhibits MDM2-mediated p53 ubiquitylation and degradation. (A) Ectopically expressed PAK1IP1 interacts with ectopically expressed MDM2 in 293T cells. Cells were transfected with Flag-PAK1IP1 and pcDNA3-MDM2 individually or together. Cell lysates were immunoprecipitated with anti-Flag or anti-MDM2 antibodies, followed by IB with anti-Flag or anti-MDM2 antibodies. (B) Endogenous PAK1IP1 interacts with endogenous MDM2 in U2OS cells. Cell lysates were immunoprecipitated with anti-PAK1IP1 polyclonal antibodies or rabbit immunoglobulin G (IgG), anti-MDM2 antibodies, or IgG, followed by anti-PAK1IP1 and anti-MDM2 antibodies, respectively. (C) PAK1IP1 colocalizes with MDM2 in the nucleus. U2OS cells were co-transfected with GFP-PAK1IP1 and Flag-MDM2. The cells were observed with a confocal laser scanning microscopy for GFP (green) or were immunostained with anti-Flag antibody, followed by staining with goat anti-rabbit secondary antibody (red) and DAPI. (D) PAK1IP1 inhibits MDM2-mediated p53 degradation. H1299 cells were transfected with the combinations of plasmids as indicated. Cell lysates were subjected to immunoblot using antibodies as indicated. (E) PAK1IP1 inhibits MDM2-mediated p53 ubiquitylation. H1299 cells were transfected with the combinations of indicated plasmids. The transfected cells were treated with MG132 (20 μM) for 6 h before harvest. Total p53 and ubiquitylated proteins p53 [p53-(ub)n] were detected by immunobloting with specific anti-p53 antibody (upper panel). Ubiquitylated was indicated. The expression of total p53, MDM2, and PAK1IP1 proteins were shown in the lower panels. Bars, 10 μm.
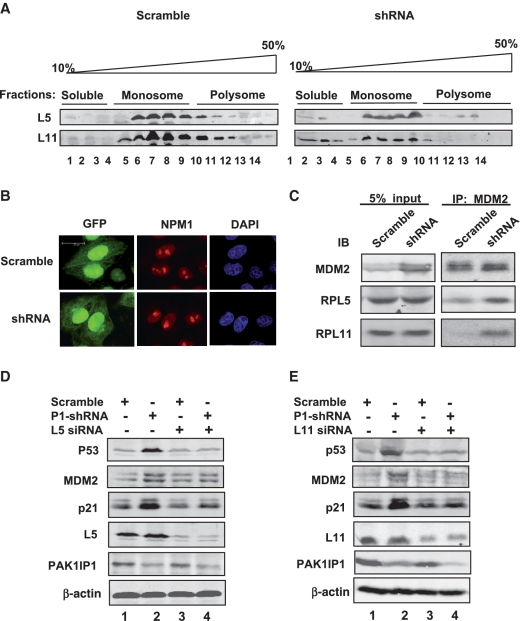
Figure 7.
Knockdown of PAK1IP1 increases the free ribosomal proteins and enhances the interaction of MDM2 with RPL5 and RPL11. (A) Polysome profiles from scramble (left panel) or PAK1IP1 shRNA (right panel) infected HeLa cells are shown. The fractions were precipitated with 20% TCA, and the precipitated proteins were analyzed by western blotting. (B) Knockdown of endogenous PAK1IP1 does not disrupt the nucleolus structure. HeLa cells were transfected with scramble or PAK1IP1 shRNA as indicated. The cells were then stained with anti-NPM1 (red) antibodies and nucleus was stained with DAPI. (C) Knockdown of endogenous PAK1IP1 enhances the interaction of MDM2 with ribosomal proteins, RPL5 and RPL11. HeLa cells were infected with the indicated lentiviruses. The same number of cells (scramble and shRNA) was lysated and 5% were loaded as input control. To equal the total amount of MDM2 protein, all of the scramble cell lysates and 30% shRNA treated cell lysates were subjected to IP using anti-MDM2 antibody, followed by IB to detect the protein levels of L11, L5 and MDM2, respectively. (D and E) The knockdown of L5 or L11 abolishes the induction of p53 by the knockdown of PAK1IP1. HeLa cells were infected with PAK1IP1-shRNA lentiviruses for 24 h, and then transfected with scrambled siRNA, L5 siRNA (D), or L11 siRNA (E) as indicated. Cell lysates were subjected to IB to detect the expression of p53, MDM2, p21, L5, or L11, as indicated. The knockdown efficiency of L5 and L11 protein compared to actin was: L5-1(1.0), L5-2(1.2), L5-3(0.19), L5-4(0.15), and L11-1(1.0), L11-2 (1.05), L11-3(0.15), L11-4 (0.13). P1, PAK1IP1. Bars, 20 μm.
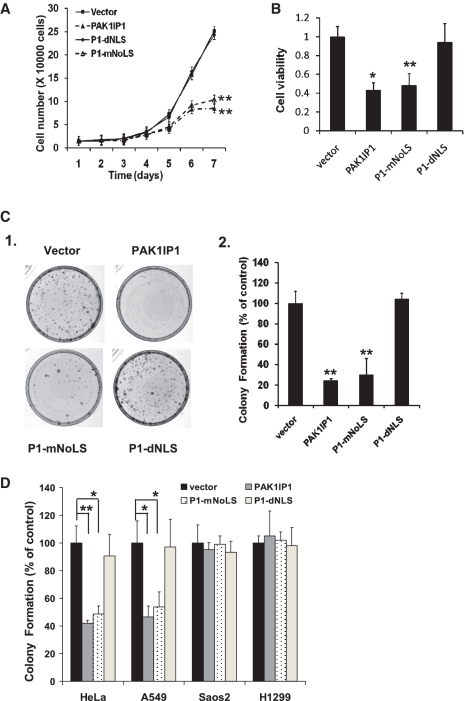
Ectopic expression of PAK1IP1 inhibits cell proliferation and colony formation in p53-containing, but not p53-deficient, cells
Since PAK1IP1 is induced by ribosomal stress, we initiated our analysis by asking what the subsequent consequence is when PAK1IP1 is overexpressed. A modified lentiviral-based pLL3.7 vector system (37) was employed for the overexpression of GFP-PAK1IP1 or its mutant proteins. U2OS cells were infected with control, wild-type and mutant PAK1IP1 lentiviruses, respectively. Cell number was counted every day for 7 days post-infection for analysis of cell growth (Figure 2A). The growth of the cells infected with wild-type PAK1IP1 or P1-mNoLS (nuclear localization excluded in nucleolus) was significantly inhibited in comparison with that of control and P1-dNLS (cytoplasmic localization) infected cells. These results suggest that high levels of PAK1IP1 suppress cell proliferation. To confirm the inhibitory effect of PAK1IP1 on cell proliferation, we carried out MTS and colony formation assays in U2OS cells. As shown in Figure 2B and C, wild-type PAK1IP1 and P1-mNoLS mutant dramatically inhibited cell viability and colony formation, whereas vector-control and P1-dNLS mutant did not show any effect. Of note, the inhibitory effect of wild-type PAK1IP1 was stronger than that of the nuclear-localized P1-mNoLS mutant. These results demonstrate that ectopic overexpression of PAK1IP1 can inhibit cell proliferation and that PAK1IP1-mediated inhibition relies on its subcellular localization.
Figure 2.
Overexpression of PAK1IP1 inhibits cell proliferation. (A) PAK1IP1 inhibits the U2OS cell proliferation. A total of 1.5 × 104 infected cells were seeded in 24-well plate, and grown for 1–7 days and the cell number was counted every day. (B) PAK1IP1 reduces the number of viable U2OS cells. Equal numbers of cells infected with different constructs were seeded in multiples in 96-well plate. After 72 h, the plate was subjected to the MTS assay. (C) Overexpression of PAK1IP1 inhibits colony formation. Equal numbers of infected U2OS cells were seeded into 33-mm dishes. After a period of 14 days, the cells were stained with crystal violet. The average numbers of colonies were shown in (C-2). (D) Overexpression of PAK1IP1 inhibits colony formation selectively in p53 positive cells. HeLa (p53 positive), Saos2 (p53 null), A549 (p53 positive) and H1299 (p53 null) were infected with lentivirus and seeded into 33-mm dishes as described in ‘Materials and Methods’ section. The average numbers of colonies were shown compared to vector control. P1: PAK1IP1. n = 3, **P < 0.01 and *P < 0.05 compared to control.
To determine whether the inhibition of cell proliferation by PAK1IP1 has any relationship with p53, we examined the effects of PAK1IP1 on p53-positive cells (HeLa, A549) and p53-deficient cells (Saos2 and H1299) using cell proliferation and colony formation assays. As shown in Figure 2D, overexpression of PAK1IP1 and P1-mNoLS inhibited colony formation in p53-containing, but not p53-deficient, tumor cells. Similar data were obtained with cell proliferation assays (data not shown). Taken together, these results suggest that the inhibitory effect of PAK1IP1 on cell proliferation and colony formation is associated with the expression of p53.
Overexpression of PAK1IP1 leads to inhibition of DNA synthesis and G1 phase arrest
To gain more insights into the PAK1IP1 inhibition of cell proliferation, we conducted 5-bromodeoxyuridine (BrdU) incorporation assays. Overexpression of PAK1IP1 and P1-mNoLS, but not PAK1IP1-dNLS, significantly reduced the number of BrdU positive cells (Figure 3A), suggesting that PAK1IP1 indeed inhibits cell proliferation via reduction of DNA synthesis.
To determine the effect of PAK1IP1 on cell-cycle progression, we performed cell-cycle analysis of cells infected with vector control or PAK1IP1 using FACS analysis in U2OS cells. As shown in Figure 3B, significantly more cells that expressed GFP-PAK1IP1 were accumulated at G1 phase than that expressing GFP vector control (∼67% over ∼47%) (Figure 3B-1, left panel). Correspondingly, the percentage of cells in S phase was reduced in PAK1IP1 infected cells (27%) compared with that in the control cells (45%). To determine whether the cell-cycle delay by PAK1IP1 is dependent on p53, U2OS cells were infected with vector or PAK1IP1 after knocking down of p53 by siRNA (Supplementary Figure S2D). Through FACS analysis, the siRNA-mediated silencing of p53 significantly reduced PAK1IP1 induced G1 arrest (Figure 3B-1, right panel). Consistent with the result in Figure 2D, overexpression of PAK1IP1 did not affect cell-cycle progression in p53-deficient H1299 cells (data not shown) either, indicating that these effects of PAK1IP1 are p53-dependent.
To further analyze cell-cycle progression in PAK1IP1 overexpressed cells, the infected U2OS cells were synchronized with double thymidine treatment, and harvested at different time points after withdrawing thymidine in the culture medium. PAK1IP1 infected and control cells exhibited similar cell-cycle distribution after thymidine treatment, indicating equally synchronized efficiency of the cells (Figure 3B-2, time 0). However, cells transfected with PAK1IP1 were accumulated at G1 phase more significantly, particularly at the 4-h time point, in comparison with the control cells (Figure 3B-2, 4 h). Similar results were also obtained in HeLa cells but not in p53-deficient H1299 cells (data not shown). These results demonstrate that ectopic overexpression of PAK1IP1 induces p53-dependent G1 delay.
Overexpression of PAK1IP1 leads to the increase of p53 level and activity
To determine whether PAK1IP1 induces p53-dependent G1 arrest by affecting p53 level and activity, we analyzed p53 levels after overexpression of PAK1IP1 by immunofluorescence staining and western blot analysis in U2OS cells. Overexpression of PAK1IP1 or P1-mNoLS, but not the P1-dNLS, led to p53 accumulation in the nuclei using GFP fluorescence and specific anti-p53 antibody (Figure 4A, arrows). Furthermore, PAK1IP1 increased the protein expression levels of p53, MDM2, and p21 using western blot analysis (Figure 4B). By contrast, the cytoplasmic mutant of PAK1IP1 failed to induce p53 and its target proteins (Figure 4A and B). To investigate how PAK1IP1 induces p53 levels, we further examined the half-life of p53 in PAK1IP1 or vector infected U2OS cells by western blot analysis. As shown in Figure 4C, the half-life of the p53 protein was significantly extended to >4 h in PAK1IP1 infected cells from half an hour in control cells (Figure 4C), suggesting that overexpression of PAK1IP1 can lead to p53 stabilization.
PAK1IP1 associates with MDM2 and inhibits MDM2-mediated p53 ubiquitination and degradation
To gain further insight into the mechanisms underlying the stabilization of p53, we sought to test whether PAK1IP1 interacts with MDM2, because the stability of p53 is tightly controlled by MDM2 (8,9,42). To determine whether exogenous PAK1IP1 and MDM2 can interact with each other, we co-transfected 293T cells with Flag-PAK1IP1 and pcDNA3-MDM2, followed by co-IP-IB assays. As shown in Figure 5A, MDM2 was specifically co-immunoprecipitated with PAK1IP1 by the anti-Flag antibody in cells co-transfected with both pcDNA3-MDM2 and Flag-PAK1IP1. Conversely, PAK1IP1 was specifically co-immunoprecipitated with MDM2 by the anti-MDM2 antibody in cells co-transfected with pcDNA3-MDM2. These results suggest that overexpressed PAK1IP1 associates with MDM2. To confirm whether endogenous PAK1IP1 and MDM2 also associate with each other, we conducted immunoprecipiation assays in U2OS cells. As shown in the representative Figure 5B, endogenous PAK1IP1 and MDM2 were co-immunoprecipitated by anti-MDM2 or anti-PAK1IP1 antibodies respectively, but not IgG (Figure 5B). To further determine the association of PAK1IP1 with MDM2, U2OS cells were co-transfected with GFP-PAK1IP1 and Flag-MDM2, followed by immunofluorescence staining using anti-Flag antibodies. As shown in Figure 5C, ectopic MDM2 was expressed primarily in the nucleoplasm, while GFP-PAK1IP1 was expressed in the whole nuclear but predominantly in the nucleolus. Confocal microscopy indicates that the subcellular distribution patterns of PAK1IP1 and MDM2 are compatible with their interaction in the nucleoplasm. To investigate if the association affects MDM2-mediated p53 ubiquitylation and degradation, co-transfection and ubiquitination assays were performed in p53-deficient H1299 cells to avoid the interference of endogenous p53 protein. As shown in Figure 5D, when exogenous proteins were introduced into H1299 cells, nuclear PAK1IP1, but not the cytoplasmic mutant (dNLS), inhibited MDM2 mediated p53 degradation. Overexpression of PAK1IP1 as well as its nucleus-localized mutant, but not cytoplasm-localized mutant, also inhibited MDM2-mediated p53 ubiquitination (Figure 5E). These results indicate that PAK1IP1 can stabilize p53 by inhibiting MDM2-mediated p53 ubiquitination and degradation, and the nuclear localization of PAK1IP1 is critical for this inhibition.
Knockdown of PAK1IP1 inhibits cell proliferation and induces cell-cycle delay
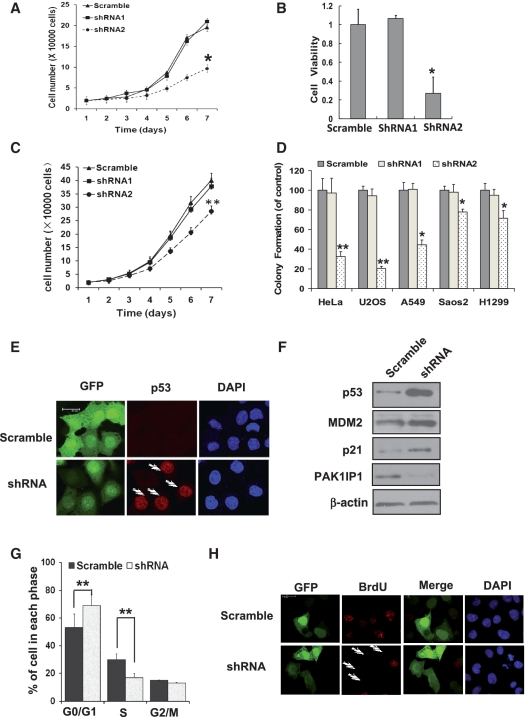
To determine the physiological relevance of PAK1IP1 in the regulation of the cell proliferation, we employed a lentiviral based RNA interference system using two different shRNA for PAK1IP1. Both the mRNA and protein levels of endogenous PAK1IP1 were efficiently ablated when shRNA2 was introduced into the cells while shRNA1 has no effect on the level of PAK1IP1 (Supplementary Figure S2A–C). Surprisingly, knockdown of endogenous PAK1IP1 also inhibited cell proliferation by cell growth and MTS assays in HeLa cells (Figure 6A and B). Since overexpression of PAK1IP1 inhibited cell proliferation in a p53-dependent manner, we next tested whether the inhibition of cell proliferation by knocking down PAK1IP1 is also p53 dependent. As shown in Figure 6C, knockdown of PAK1IP1 inhibited proliferation of p53-deficient H1299 cells by ∼30%. To further confirm the data, colony formation assays were performed in p53-proficient (HeLa, U2OS, A549) and p53–deficient (Saos2 and H1299) cells after knockdown of endogenous PAK1IP1 by RNA interference. As shown in Figure 6D, knockdown of PAK1IP1 in all five cell lines led to inhibition of cell proliferation and colony formation. However, this inhibition was more apparent in p53-proficient cells than in p53-deficient cells, suggesting that knockdown of PAK1IP1 may also induce p53-dependent cell growth arrest. These results indicate that PAK1IP1 is essential for cell proliferation while also plays a role in p53 response to nucleolar stress since both the up- or down-regulation of PAK1IP1 leads to p53-dependent cell growth arrest.
Figure 6.
Knockdown of PAK1IP1 inhibits cell proliferation with p53 activation. (A, B and C) Knockdown of PAK1IP1 inhibits cell growth and viability. Cell growth curves of HeLa (A) and H1299 (C) cells, infected with the indicated lentivirus, were determined from Day 1 to Day 7 after infections. The same number of infected cells was seeded in 24-well plates to grow for 1–7 days and the cell number was counted every day. The number of viable cells in HeLa cells was measured by MTS (B). (D) Knockdown of PAK1IP1 reduces the efficiency of colony formation in p53 positive and negative cells. Indicated cells were infected with scramble or PAK1IP1 shRNA lentiviruses (shRNA1 or shRNA2) and seeded into 33-mm dish with 1000 cells. After a 14-day growth, the numbers of colonies were counted as described in ‘Materials and Methods’ section. Data was representative of three independent experiments (n = 3), *P < 0.05, **P < 0.01 compared to scramble RNA-infected cells. (E) Knockdown of PAK1IP1 upregulates p53 protein level. HeLa cells were infected with indicated lentiviruses for 48 h. GFP and p53 were detected by immunostaining with confocal laser scanning microscopy. The arrows showed that the p53 protein was accumulated in GFP-positive cells indicating the knockdown of PAK1IP1. (F) Down-regulation of PAK1IP1 increased protein level of p53 and its target proteins, MDM2 and p21. (G) Knockdown of PAK1IP1 induces G1/S cell-cycle delay. HeLa cells were infected with the indicated lentiviruses for 48 h. Cell-cycle distributions were analyzed by FACS analysis. (H) Knockdown of PAK1IP1 inhibits BrdU incorporation. HeLa cells were transfected with the indicated plasmids for 48 h and then BrdU was supplemented. After 10 h, the cells were immunostained with BrdU specific antibody (red) and the nucleus was stained with DAPI. The arrows showed the GFP-positive cells with undetectable BrdU incorporation. Bars, 20 μm.
To determine whether knockdown of PAK1IP1 is associated with the regulation of the MDM2-p53 pathway, we examined p53 levels using immunofluorescence staining and western blot analysis. As shown in Figure 6E and F, knockdown of endogenous PAK1IP1 led to p53 accumulation in the nuclei (Figure 6E) as well as the increase of p21 and MDM2 levels (Figure 6F). Furthermore, knockdown of PAK1IP1 drastically induced cell-cycle arrest in the G1 phase (Figure 6G), and inhibited DNA synthesis in HeLa cells as determined by BrdU incorporation assays (Figure 6H). These results indicate that the depletion of PAK1IP1 also induces p53 activation and G1 arrest.
Knockdown of PAK1IP1 increases the free ribosomal proteins and enhances the interaction of MDM2 with RPL5 and RPL11
The functional inactivation of yeast PAK1IP1 homolog Mak11 and Skb15 impaired the rRNA synthesis, and led to a cell-cycle delay in G1 (31). As we know that inhibition of rRNA synthesis leads to disassembly of ribosomal precursors and release of ribosome-unbound ribosomal proteins from the nucleolus (43). Since PAK1IP1 is a evolutionarily conserved protein, we hypothesize that knockdown of PAK1IP1 may lead to the release of ribosomal proteins which then activates p53. To confirm the role of PAK1IP1 in ribosomal protein release, we carried out a sucrose gradient fractionation assay to separate soluble proteins from ribosomal monosomes/polysomes in scramble- and PAK1IP1- shRNA infected HeLa cells. The collected fractions were analyzed for L5 and L11 proteins by western blotting. Notably, the levels of L5 and L11 in the soluble, ribosome-unbound fractions were evidently increased in PAK1IP1-depleted cells, indicating that L5 and L11 were released from the ribosomal subunits as ribosome free, proteins in the cell lysates (Figure 7A), but the total level of these two ribosomal proteins was not affected (Figure 7C, lysate).
Since the protein level of PAK1IP1 was induced when the cells were treated with either actinomycin D or 5-FU (Figure 1A), we suspect that PAK1IP1 is an integrated component of the nucleolus and that depletion of PAK1IP1 could trigger a nucleolar stress that activates p53. Nucleolar stress usually leads to the disruption of the nucleolus (44), so we first examined the nucleolar structure upon the PAK1IP1 knockdown by using immune-fluorescence staining. Surprisingly, as determined by the nucleolar marker NPM1, knockdown of PAK1IP1 did not significantly alter the nucleolar structure (Figure 7B). Since the free L5 and L11 were increased in PAK1IP1 knockdown cells, we determined whether knockdown of PAK1IP1 by shRNA could activate the ribosomal protein–MDM2–p53 pathway by examining the interaction of L5 and L11 with MDM2, respectively. HeLa cells infected with scrambled or PAK1IP1 shRNA were analyzed by co-IP assays with anti-MDM2 antibodies. As shown in Figure 7C, knockdown of PAK1IP1 indeed enhanced the interaction between MDM2 and L5 or L11 but did not change the protein levels of these two ribosomal proteins. These results suggest that the interaction between MDM2 and ribosomal proteins (L5 or L11) is also responsive to the depletion of PAK1IP1 by RNA interference.
Knockdown of L5 or L11 attenuates PAK1IP1 depletion-induced p53 activation
The enhanced interaction of MDM2 with L5 and L11 suggests that these ribosomal proteins may play a role in PAK1IP1 knockdown induced p53 activation. To confirm the role of ribosomal proteins in PAK1IP1 depletion-induced p53 activation, the expression of L5 or L11 was depleted by siRNAs in PAK1IP1-deficit or control cells. As shown in Figure 7, reduction of either L5 (Figure 7D) or L11 (Figure 7E) protein by siRNAs markedly inhibited the PAK1IP1 knockdown-induced p53 levels in comparison with that of the scrambled RNA-transfected cells. Consistently, the induction of MDM2 and p21 protein levels by the PAK1IP1 knockdown was drastically reduced by shRNAs against L5 or L11 but not by the scrambled sequences (Figure 7D and E). This reduction of MDM2 and p21 was also observed in mRNA levels as measured by real-time-PCR assays (Supplementary Figure S3). Together, these results demonstrate that the ribosomal protein–MDM2–p53 pathway play a critical role in PAK1IP1 knockdown-caused p53 activation.
DISCUSSION
We previously showed that PAK1IP1 is a WD-repeat containing protein, which interacts with PAK1 and inhibits its kinase activity. A null mutation of the fission yeast ortholog, Skb15, is lethal and results in the deregulation of actin polymerization, microtubule biogenesis, and cell-cycle arrest (29). However, the physiological function of this evolutionarily conserved protein remains obscure. In the current study, we show that PAK1IP1 is mainly localized in the nucleolus with some in the nucleus. In addition, we identified the NLS and NoLs sequences in the C-terminus of PAK1IP1, which are essential for the subcellular localization of this protein. The protein level of PAK1IP1 was induced when cells were treated with nucleolar stress-inducing agents. Interestingly, PAK1IP1 was associated with MDM2 and reduced MDM2-mediated p53 ubiquitination and degradation. Knockdown of endogenous PAK1IP1 induces free ribosomal proteins (L5 and L11) which interact with MDM2 and activate p53 pathway. Together, either overexpression or knockdown of PAK1IP1 in human cancer cells induced G1 arrest and inhibited cell proliferation via a p53-dependent pathway.
The subcellular localization of PAK1IP1 is dependent on three essential tandem KKXK motifs. The nuclear or nucleolar localization of this protein appears to be crucial for the function to block cell proliferation, as its cytoplasm-localized mutant had no such function. Furthermore, since the inhibitory effect of the nucleus-localized mutant on cell proliferation was not as severe as that of wild-type, this suggests that both nucleoplasmic and nucleolar fractions of PAK1IP1 play an important role in regulation of cell proliferation. It has been reported that the Mak11 protein, the budding yeast ortholog of PAK1IP1, is necessary for the maintenance of killer M1 double-stranded RNA and the C-terminal lysine-rich region is essential for Mak11-complementing activity (28). The identification of the NLS/NoLS in PAK1IP1 in the current study explains why the C-terminus is essential for Mak11 function. Since the subcellular localization of PAK1IP1 is very important for its function, it is very interesting to further study whether the subcellular localization of PAK1IP1 is regulated by extracellular stimuli and the mechanism through which the translocation of PAK1IP1 is regulated.
Malfunction of the rRNA biogenesis machinery causes ribosomal stress and induces cell-cycle arrest, apoptosis or senescence (25). Over the past ten years, numerous nucleolar proteins (ARF, NS, Bop1) or ribosomal proteins (L5, L11, L23) have been identified to regulate cell proliferation through the MDM2-p53 feedback loop in response to nucleolar stress (16,17,21,38,45,46). Although there is no evidence for the function of PAK1IP1 in nucleolus, it was reported that its orthologs, Mak11 and Skb15, act as a cofactor of ribosomal protein Rlp24 and regulate 60S rRNA maturation (31). We observed that the protein level of PAK1IP1 was increased by ribosomal stress caused by 5-FU and ActD treatment (Figure 1A). This observation led us to illustrate how PAK1IP1 responded to ribosomal stress and regulated cell proliferation. It is possible that ribosomal malfunction induced PAK1IP1 accumulation and the superfluous of PAK1IP1 associates with MDM2 to induce p53 level and activity by inhibiting MDM2-mediated p53 ubiquitination and degradation. PAK1IP1 inhibits cell proliferation is dependent on p53 which is observed in several cell lines including HeLa cells. However, the main p53 regulator in HeLa cell is HPV E6, which causes its ubiquitylation irrespective of MDM2 (47,48). It remains unclear whether PAK1IP1 also modulates HPV E6 activity.
In addition, we found that not only abnormally high level, but also aberrant low level, of PAK1IP1 can induce G1 arrest and inhibit cell proliferation. Our results support the idea that aberrantly high levels of PAK1IP1 can activate p53 by inhibiting MDM2 function. Furthermore, we also found that aberrantly low levels of PAK1IP1 could also induce p53 response probably by causing nucleolar or ribosomal stress. In past years, the nucleolar proteins have been shown to be critical for 60S ribosome maturation (49–52). Such as the case for nucleostemin, whose level is critically balanced and any off-balance would trigger p53 response (21). In cells, the pre-ribosomal RNA transcripts are processed and modified by small nucleolar ribonucleoproteins (snoRNPs). The modified rRNAs associate with ribonucleoprotein and assembly with ribosomal proteins to form 40S and 60S ribosome subunits which are both exported to the cytoplasm where they bind to mRNA to form functional 80S ribosome for protein translation (22). Our unpublished data reveals that PAK1IP1 depletion by shRNA impaired 28S rRNA processing and leads to reduction of 60S ribosome subunits (data not shown). Furthermore, the depletion of endogenous PAK1IP1 increases the ribosome-unbound form of L5 and L11, and enhances their interactions with MDM2 (Figure 7C). These ribosomal proteins are associated with ribosomal biogenesis and required for nucleolar stress-induced p53 activation and cell-cycle arrest (21,53,54). The reason why knockdown of PAK1IP1 mildly inhibits cell proliferation of p53-deficient cells remains unclear. It is likely that the ribosomal stress induced by disruption of PAK1IP1 expression causes a p53-independent pathway which inhibits cell proliferation. In summary, our study not only identifies PAK1IP1 as a new regulator of the p53–MDM2 pathway potentially in response to nucleolar stress, but also reveals that the adequate level of this protein in cells is crucially important for maintaining cell growth and proliferation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research Platform of Cell Signaling Networks from the Science and Technology Commission of Shanghai Municipality (06DZ22923, partial); National Natural Science Foundation of China (no. 30800627 and 30930055); State Key Development Programs of China (2010CB945403, 2009CB918402); National Institutes of Health/NCI grants (CA127724, CA095441 and CA129828 to H.L.). Funding for open access charge: State Key Development Programs of China (2010CB945403).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank all members in their institute for active discussion.
REFERENCES
- 1.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Norbury C, Nurse P. Animal cell cycles and their control. Annu. Rev. Biochem. 1992;61:441–470. doi: 10.1146/annurev.bi.61.070192.002301. [DOI] [PubMed] [Google Scholar]
- 3.Du YC, Stillman B. Yph1p, an ORC-interacting protein: potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell. 2002;109:835–848. doi: 10.1016/s0092-8674(02)00773-0. [DOI] [PubMed] [Google Scholar]
- 4.Vousden KH. Activation of the p53 tumor suppressor protein. Biochim. Biophys. Acta. 2002;1602:47–59. doi: 10.1016/s0304-419x(02)00035-5. [DOI] [PubMed] [Google Scholar]
- 5.Green DR, Evan GI. A matter of life and death. Cancer Cell. 2002;1:19–30. doi: 10.1016/s1535-6108(02)00024-7. [DOI] [PubMed] [Google Scholar]
- 6.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat. Rev. Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 7.Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Res. 1998;26:3453–3459. doi: 10.1093/nar/26.15.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 9.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 10.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 11.Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 12.Michael D, Oren M. The p53-Mdm2 module and the ubiquitin system. Semin. Cancer Biol. 2003;13:49–58. doi: 10.1016/s1044-579x(02)00099-8. [DOI] [PubMed] [Google Scholar]
- 13.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 14.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell. Biol. 2003;23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 2004;279:44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 17.Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol. Cell. Biol. 2004;24:7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurki S, Peltonen K, Latonen L, Kiviharju TM, Ojala PM, Meek D, Laiho M. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell. 2004;5:465–475. doi: 10.1016/s1535-6108(04)00110-2. [DOI] [PubMed] [Google Scholar]
- 19.Chen D, Zhang Z, Li M, Wang W, Li Y, Rayburn ER, Hill DL, Wang H, Zhang R. Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene. 2007;26:5029–5037. doi: 10.1038/sj.onc.1210327. [DOI] [PubMed] [Google Scholar]
- 20.Sun XX, Dai MS, Lu H. 5-fluorouracil activation of p53 involves an MDM2-ribosomal protein interaction. J. Biol. Chem. 2007;282:8052–8059. doi: 10.1074/jbc.M610621200. [DOI] [PubMed] [Google Scholar]
- 21.Dai MS, Sun XX, Lu H. Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol. Cell. Biol. 2008;28:4365–4376. doi: 10.1128/MCB.01662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 23.Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 24.Zhu Y, Poyurovsky MV, Li Y, Biderman L, Stahl J, Jacq X, Prives C. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol. Cell. 2009;35:316–326. doi: 10.1016/j.molcel.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scherl A, Coute Y, Deon C, Calle A, Kindbeiter K, Sanchez JC, Greco A, Hochstrasser D, Diaz JJ. Functional proteomic analysis of human nucleolus. Mol. Biol. Cell. 2002;13:4100–4109. doi: 10.1091/mbc.E02-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia C, Ma W, Stafford LJ, Marcus S, Xiong WC, Liu M. Regulation of the p21-activated kinase (PAK) by a human Gbeta -like WD-repeat protein, hPIP1. Proc. Natl Acad. Sci. USA. 2001;98:6174–6179. doi: 10.1073/pnas.101137298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Icho T, Wickner R. The MAKll protein is essential for cell growth and replication of M Double-stranded RNA and is apparently a membrane-associated protein. J. Biol. Chem. 1988;263:1467–1475. [PubMed] [Google Scholar]
- 29.Kim HW, Yang P, Qyang Y, Lai H, Du H, Henkel JS, Kumar K, Bao S, Liu M, Marcus S. Genetic and molecular characterization of Skb15, a highly conserved inhibitor of the fission yeast PAK, Shk1. Mol. Cell. 2001;7:1095–1101. doi: 10.1016/s1097-2765(01)00248-9. [DOI] [PubMed] [Google Scholar]
- 30.Saveanu C, Namane A, Gleizes PE, Lebreton A, Rousselle JC, Noaillac-Depeyre J, Gas N, Jacquier A, Fromont-Racine M. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 2003;23:4449–4460. doi: 10.1128/MCB.23.13.4449-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saveanu C, Rousselle JC, Lenormand P, Namane A, Jacquier A, Fromont-Racine M. The p21-activated protein kinase inhibitor Skb15 and its budding yeast homologue are 60S ribosome assembly factors. Mol. Cell. Biol. 2007;27:2897–2909. doi: 10.1128/MCB.00064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maroto B, Ye MB, von Lohneysen K, Schnelzer A, Knaus UG. P21-activated kinase is required for mitotic progression and regulates Plk1. Oncogene. 2008;27:4900–4908. doi: 10.1038/onc.2008.131. [DOI] [PubMed] [Google Scholar]
- 33.Wang RA, Zhang H, Balasenthil S, Medina D, Kumar R. PAK1 hyperactivation is sufficient for mammary gland tumor formation. Oncogene. 2006;25:2931–2936. doi: 10.1038/sj.onc.1209309. [DOI] [PubMed] [Google Scholar]
- 34.Vadlamudi RK, Bagheri-Yarmand R, Yang Z, Balasenthil S, Nguyen D, Sahin AA, den Hollander P, Kumar R. Dynein light chain 1, a p21-activated kinase 1-interacting substrate, promotes cancerous phenotypes. Cancer Cell. 2004;5:575–585. doi: 10.1016/j.ccr.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 35.Balasenthil S, Sahin AA, Barnes CJ, Wang RA, Pestell RG, Vadlamudi RK, Kumar R. p21-activated kinase-1 signaling mediates cyclin D1 expression in mammary epithelial and cancer cells. J. Biol. Chem. 2004;279:1422–1428. doi: 10.1074/jbc.M309937200. [DOI] [PubMed] [Google Scholar]
- 36.Cai Y, Stafford LJ, Bryan BA, Mitchell D, Liu M. G-protein-activated phospholipase C-beta, new partners for cell polarity proteins Par3 and Par6. Oncogene. 2005;24:4293–4300. doi: 10.1038/sj.onc.1208593. [DOI] [PubMed] [Google Scholar]
- 37.Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Zhang M, Ihrig MM, McManus MT, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 38.Dai MS, Shi D, Jin Y, Sun XX, Zhang Y, Grossman SR, Lu H. Regulation of the MDM2-p53 pathway by ribosomal protein L11 involves a post-ubiquitination mechanism. J. Biol. Chem. 2006;281:24304–24313. doi: 10.1074/jbc.M602596200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghoshal K, Jacob ST. Specific inhibition of pre-ribosomal RNA processing in extracts from the lymphosarcoma cells treated with 5-fluorouracil. Cancer Res. 1994;54:632–636. [PubMed] [Google Scholar]
- 40.Nair R, Rost B. Mimicking cellular sorting improves prediction of subcellular localization. J. Mol. Biol. 2005;348:85–100. doi: 10.1016/j.jmb.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 41.Horke S, Reumann K, Schweizer M, Will H, Heise T. Nuclear trafficking of La protein depends on a newly identified nucleolar localization signal and the ability to bind RNA. J. Biol. Chem. 2004;279:26563–26570. doi: 10.1074/jbc.M401017200. [DOI] [PubMed] [Google Scholar]
- 42.Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 43.Bhat KP, Itahana K, Jin A, Zhang Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 2004;23:2402–2412. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pestov DG, Strezoska Z, Lau LF. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol. Cell. Biol. 2001;21:4246–4255. doi: 10.1128/MCB.21.13.4246-4255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Itahana K, Bhat KP, Jin A, Itahana Y, Hawke D, Kobayashi R, Zhang Y. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol. Cell. 2003;12:1151–1164. doi: 10.1016/s1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- 47.Hengstermann A, Linares LK, Ciechanover A, Whitaker NJ, Scheffner M. Complete switch from Mdm2 to human papillomavirus E6-mediated degradation of p53 in cervical cancer cells. Proc. Natl Acad. Sci. USA. 2001;98:1218–1223. doi: 10.1073/pnas.031470698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wesierska-Gadek J, Schloffer D, Kotala V, Horky M. Escape of p53 protein from E6-mediated degradation in HeLa cells after cisplatin therapy. Int. J. Cancer. 2002;101:128–136. doi: 10.1002/ijc.10580. [DOI] [PubMed] [Google Scholar]
- 49.Sun C, Woolford JL., Jr The yeast NOP4 gene product is an essential nucleolar protein required for pre-rRNA processing and accumulation of 60S ribosomal subunits. EMBO J. 1994;13:3127–3135. doi: 10.1002/j.1460-2075.1994.tb06611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zanchin NI, Goldfarb DS. Nip7p interacts with Nop8p, an essential nucleolar protein required for 60S ribosome biogenesis, and the exosome subunit Rrp43p. Mol. Cell. Biol. 1999;19:1518–1525. doi: 10.1128/mcb.19.2.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strezoska Z, Pestov DG, Lau LF. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5. 8S RRNA processing and 60S ribosome biogenesis. Mol. Cell. Biol. 2000;20:5516–5528. doi: 10.1128/mcb.20.15.5516-5528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sydorskyy Y, Dilworth DJ, Halloran B, Yi EC, Makhnevych T, Wozniak RW, Aitchison JD. Nop53p is a novel nucleolar 60S ribosomal subunit biogenesis protein. Biochem. J. 2005;388:819–826. doi: 10.1042/BJ20041297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steitz JA, Berg C, Hendrick JP, La Branche-Chabot H, Metspalu A, Rinke J, Yario T. A 5S rRNA/L5 complex is a precursor to ribosome assembly in mammalian cells. J. Cell Biol. 1988;106:545–556. doi: 10.1083/jcb.106.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nariai M, Tanaka T, Okada T, Shirai C, Horigome C, Mizuta K. Synergistic defect in 60S ribosomal subunit assembly caused by a mutation of Rrs1p, a ribosomal protein L11-binding protein, and 3'-extension of 5S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2005;33:4553–4562. doi: 10.1093/nar/gki772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.