Abstract
Translation initiation on HIV genomic RNA relies on both cap and Internal Ribosome Entry Site (IRES) dependant mechanisms that are regulated throughout the cell cycle. During a unique phenomenon, the virus recruits initiation complexes through RNA structures located within Gag coding sequence, downstream of the initiation codon. We analyzed initiation complexes paused on the HIV-2 gag IRES and revealed that they contain all the canonical initiation factors except eIF4E and eIF1. We report that eIF3 and the small ribosomal subunit bind HIV RNA within gag open reading frame. We thus propose a novel two step model whereby the initial event is the formation of a ternary eIF3/40S/IRES complex. In a second step, dependent on most of the canonical initiation factors, the complex is rearranged to transfer the ribosome on the initiation codons. The absolute requirement of this large structure for HIV translation defines a new function for a coding region. Moreover, the level of information compaction within this viral genome reveals an additional level of evolutionary constraint on the coding sequence. The conservation of this IRES and its properties in rapidly evolving viruses suggest an important role in the virus life cycle and highlight an attractive new therapeutic target.
INTRODUCTION
Once in the host cell cytoplasm, lentiviral genomes are reverse transcribed into proviral DNA which is subsequently permanently integrated into the host cell genome. Once integrated, the provirus first follows the fate of a cellular gene. It is transcribed by the host RNA polymerase II, consequently capped, then spliced and exported to the cytoplasm where it is translated by the host protein synthesis machinery. Later, the virally encoded Rev protein allows a certain proportion of unspliced transcripts to be exported to the cytoplasm (1). This full length RNA, known as the genomic RNA (gRNA), is multifunctional. It acts both as an mRNA that is translated to yield the Gag and Gag-Pol polyproteins and as a genome that is encapsidated within the virions (2). The 5′-UTR of gRNAs is long, highly structured and thus predicted to impede the scanning step of the canonical cap-dependent initiation pathway (3–5). Indeed, structured motifs within the 5′-UTR, mainly the 5′–terminal Tar stem-loop, have been shown to inhibit the translation of reporter genes in vitro and in transiently transfected cells (6–8). Under certain conditions, this can be relieved by the tat protein, although the mechanisms involved are not clear (9). In addition, distal portions of the HIV-1 (Human Immunodeficiency virus type 1) and the SIVMAC (Simian Immunodeficiency virus Macaque) 5′-UTRs have been shown to function as Internal Ribosome Entry Site (IRES) (10–12) attracting initiation complexes independently of the 5′-terminal cap. Interestingly, the IRES present in HIV-1 5′-UTR is activated during the G2/M phase (10). While work on HIV-2 failed to identify an IRES in the 5′-UTR, it evidenced the presence of one or several ribosome entry sites within the Gag coding region (13,14). We further showed that in addition to the full length polyprotein, translation from this IRES yields two shorter Gag isoforms initiated from in frame initiation triplets (AUG2 and AUG3) (13). This IRES is unique, because it directs ribosomes to the 5′-terminal AUG (AUG1) and therefore upstream from its determinant(s). As a consequence, an artificial cognate leaderless RNA starting with the initiation codon is very efficiently translated. Also atypical, this IRES recruits up to three initiation complexes paused on each initiation triplet on a single RNA molecule (15). Originally discovered in HIV-2, this IRES was shown to be conserved among primate lentiviruses and to rely on common secondary structure determinants adopted by the Gag coding sequence itself (15). Lentivirus gRNA translation can thus be initiated through an inefficient canonical cap-dependent mechanism (16) or by internal entry of the ribosomes onto the 5′-UTR or within the coding region (1). These mechanisms are conserved among primate lentiviruses, and are therefore likely to each play a role, during one or the other of the different stages of the virus cycle (17). As a first step to understand translation regulation and the interconnection between the three pathways, we undertook the characterization of the molecular events leading to internal entry of the ribosomes within the coding region. We used RNA affinity chromatography to purify initiation complexes formed in vitro on a leaderless lentiviral gRNA, and analyzed their initiation factor content. This first set of results is compatible with picornaviral type I and II IRES which recruit the 40S ribosomal subunit through their interaction with eIF4G (18,19). To our surprise we found that the coding region actually binds the 40S subunit and the central initiation factor eIF3 but not eIF4G. The use of deletion mutants revealed that this complex is formed on a large conserved RNA structure. We characterized a primary initiation complex containing the 40S ribosomal subunit and eIF3 bound to the coding region, which likely represents the first step of the initiation mechanism. HIV gRNA is the first example of a coding sequence adopting such a large and complex structure to regulate its own translation.
MATERIALS AND METHODS
RNAs
The control globin RNA was in vitro transcribed from a plasmid containing the 5′-UTR and the coding region of the β-globin gene (YPGlo) using the mMessage mMachine and poly(A) tailing kits (Ambion) following the manufacturer instructions. The DNA sequence corresponding to the coding region of gag HIV-1 (pNL4.3 F522Y) was amplified by PCR using a 5′ oligonucleotide starting at the T7 promoter sequence followed by a 32-nt stem loop, a 15-nt spacer sequence and the 11 first nucleotides of gag open reading frame (5′-TAATACGACTCACTATAGGACAGATCTACGCGTACGTACGCGTAGATCTGTAGTAGAAGGAGAGAGaTGGGTGCGAG-3′) and the 5′-AAACATGGGTATTACTTCTGG-3′ oligonucleotide.
To create a DNA construct containing the wt HIV-2 gag IRES (545–965) and the streptomycin aptamer. HindIII and XbaI restriction sites were introduced on the 5′ and 3′ end of the HIV-2 gag IRES by PCR, and the resulting fragment was inserted into the StreptoTag vector (20). To generate the truncated gag IRES constructs, the pcDNA3 HIV-2 gag (13) was amplified with various 5′-PCR primers including the T7 polymerase promoter sequence followed by the first 24 nt of the internal starting domain, and a 3′-PCR primer starting at the end of the internal domain to be amplified (for detailed location see Figures 1 and 5). The HIV-2 gag IRES containing deletion of the P4 domain was previously described (15). For the synthesis of HIV-1 and SIVMac gag IRES, the DNA sequence of gag HIV-1 (pNL4.3) and SIVMac (mac251, 13 001) were amplified by PCR using a 3′ oligonucleotide starting at the end of the conserved core domain respectively nucleotides 572 and 812, according to our predicted secondary structure model, and a 5′ oligonucleotide starting with the T7 promoter sequence and the first 24 nt of the gag coding sequence respectively nucleotides 360 and 562. All RNAs were directly transcribed using the T7 RNA polymerase from PCR products containing the T7 polymerase promoter sequence and purified as previously described (21,22). Radiolabeled IRES RNAs were transcribed as above in the presence of α-32P-UTP (3000 mCi/mmol) and purified by size exclusion chromatography.
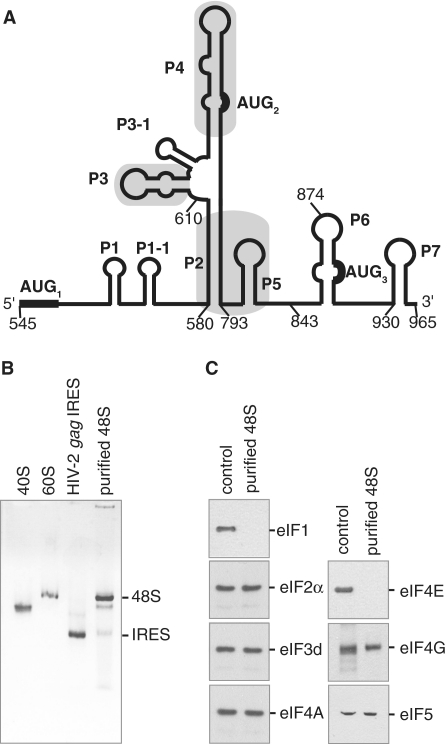
Figure 1.
HIV-2 gag IRES ribosomal 48S complexes contain most of the canonical initiation factors except for eIF4E and eIF1. (A) Schematic representation of the HIV-2 gag IRES secondary structure adapted from (15). Pairings are designated by Pn in order of appearance from 5′ to 3′. In frame AUG codons leading to the different Gag isoforms are indicated and numbered. Positions of the deletions for binding studies are indicated. (B) The mobility of 48S complexes assembled onto HIV-2 gag IRES purified on streptomycin derived sepharose beads is compared to the mobility of 40S, 60S subunits as well as HIV-2 gag IRES RNA as indicated. (C) The composition of 48S complexes assembled onto HIV-2 gag RNA was assessed by comparing western blots of purified complexes (purified 48S) to those of total Rabbit Reticulocyte Lysate (control). Initiation factor specificity of the primary antibody is indicated.
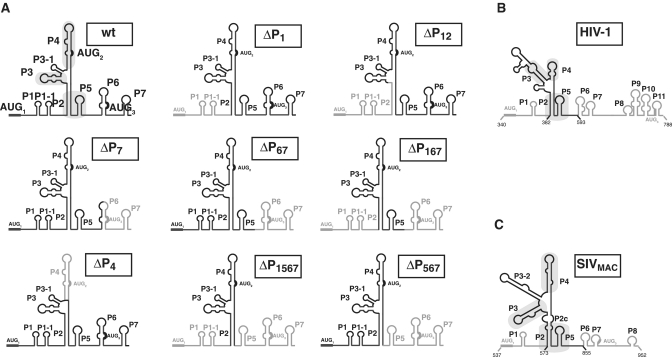
Figure 5.
eIF3 and the 40S ribosomal subunit bind to the conserved core of the HIV-2 IRES. (A) Secondary structure schematic representation of the mutants used in eIF3 and 40S subunit binding studies. Deleted domains are in light grey. Positions of the deletion are noted on the structures. Secondary structure schematic representation of the HIV-1 gag IRES (B) and SIVMac (C), conserved structural elements between primates lentivirus are boxed in grey. The stabilities of each structures as calculates by Mfold are HIV-1 = −58.38 KICal, HIV-2 = −72.37 and SIVMAC = −60.18 Kcal. Note that this portion of HIV-1 is only 60% homologous to HIV-2, while SIVMAC is 80% homologous to HIV-2.
Mobility of complexes during sucrose density gradient centrifugation
Complexes were assembled on 32P-labeled HIV-1 RNA or capped and polyadenylated globin mRNA. Rabbit reticulocyte lysate (FlexiR-RRL-Promega) was preheated with 2 mM GMP-PNP for 10 min at 30°C. An amount of 2 pmol of RNA (100 µl) was incubated at 30°C during 20 min with 100 µl of pretreated RRL in the presence of 0.5 mM magnesium acetate, 75 mM potassium acetate, 20 µM amino acid and 8 U RNAsin (Promega). Determination of complex formation with purified components was performed by incubating 2 pmol of RNA (50 nM) with 75 nM 40S subunits and/or 100 nM purified eIF3 for 10 min at 30°C in 20 mM Tris pH 7.6, 100 mM potassium acetate, 2.5 mM magnesium acetate, 2 mM DTT, 0.25 mM spermidine, 2 mM ATP and 8 U RNAsin (Promega). Reactions were stopped on ice, then layered over 10–30% sucrose gradients (25 mM Tris pH 7.6, 6 mM MgCl2, 75 mM KCl) and sedimented by ultracentrifugation at 39 000 r. p. m. in a SW40 Ti rotor for 3.5 h at 4°C. Three-hundred-microliter fractions were collected, one-third of which were vacuum blotted onto a Hybond N+ membrane (Amersham), exposed and scanned. The amount of RNA in each fraction was determined and expressed as the percentage of total counts.
In vitro assembly and affinity purification of the 48S complexes
The 48S complexes were assembled in RRL onto IRES RNAs in the presence of 2 mM GMP-PNP (GMP-PNP is a non-hydrolysable GTP analog that blocks the 60S subunit joining to the initiation complex) and then isolated following the StreptoTag-based affinity purification method (20).
Immunoblotting
To test eIF composition of 48S complexes, proteins from 5 pmol of purified particles were resolved on a 12% SDS–PAGE gel, transferred onto a nitrocellulose membrane, and detected using antibodies against eIF4E, eIF2a, eIF3-p66, eIF5, eIF4A, eIF4G (santa-cruz), then with appropriate HRP-conjugated secondary antibodies (Abcam), revealed with the supersignal West Pico Kit (Pierce), and visualized with a LAS 4000 (Fuji).
Translation assay
RNA (0.5 pmol) was incubated for 60 min at 30°C in a final volume of 10 µl containing 7 µl of RRL (Promega Flexi RRL) in the presence of 0.5 mM magnesium acetate, 75 mM potassium acetate, 20 mM of each amino acid (minus methionine) and 0.2 mCi/ml 35S methionine and 5 U RNasin (Promega). Various concentrations of Hippuristanol or 4EGI-1 were included during incubation. The reaction was stopped with 90 µl of protein loading buffer. An amount of 10 µl of the reaction were loaded onto a 12% SDS–PAGE gel. Electrophoresis were analyzed and quantified using a storm phosphorImager (GE healthcare). Each experiment was repeated independently three times using different RNA preparations.
Preparation of the factors and ribosomal subunits
The initiation factors and ribosomal subunits were prepared following previously established procedures (23).
Binding studies
Radiolabeled RNA (50 fmol) was denatured by heating to 85°C for 1 min and then cooled to room temperature containing binding buffer (20 mM Tris pH 7.6, 100 mM KCl, 2 mM DTT, 2 mM MgCl2). Serial dilution of eIF3 or 40S subunit were prepared extemporaneously, added to a 10-µl reaction, and incubated at 37°C for 30 min. Reactions were then either used for filter binding assay, or resolved on native polyacrylamide gel electrophoresis in 0.5× Tris Borate buffer (45 mM, pH 8.3). Filter binding was accomplished essentially as previously described using two filters (24). From top to bottom a nitrocellulose filter and a charged nylon filter. The filters were presoaked in 1× binding buffer, assembled in a dot blot apparatus or on glass filter funnel and the reactions were applied and directly vacuum filtered. The filters were then rinsed, removed, dried and radioactivity was quantified using a storm phosphorImager (GE healthcare). To determine the apparent Kd, the data was fit to a Langmuir isotherm described by the equation θ = (P)/(P) + Kd where θ is the fraction of RNA bound and P is either 40S subunit or eIF3 concentration.
For eIF3 competition binding experiments, increasing concentration of various unlabeled RNA were incubated 15 min with eIF3 (350 nM) prior to incubation with wt 32P-labeled RNA (5 nM).The competition constant (Kc) was determined fitting the data to a sigmoidal dose-response curve and using the Lin and Riggs equation (25).
Stoichiometric binding assay were performed as previously described (25). A concentration of 200 nM of 32P-labeled IRES RNA, 10-fold over the Kd for the 40S subunit, was incubated with increasing quantities of 40S subunits and the amount of binary complex was measured by filter binding assay.
All binding studies were performed in parallel with wild-type IRES RNA as an internal standard. Reported values are the average of three repetitions with standard errors. All calculations were performed with GraphPad Prism 5.
UV cross-linking
Five picomoles of HIV-2 gag IRES RNA synthesized in the presence of 32P UTP were incubated with purified eIF3. The mix was irradiated at 260 nm for 30 min on ice using a handheld UV lamp, treated with RNase A, the products were resolved on a 12% SDS–PAGE, and the radioactivity was quantified using a storm phosphorImager (GE healthcare).
RESULTS
Canonical factors present in 48S initiation complexes assembled onto HIV-2 gag IRES
To identify the initiation factors present in 48S complexes assembled onto the HIV-2 gag IRES, we used a previously described RNA-based affinity purification method (20,26,27). To this end, an RNA construct containing the HIV-2 gag IRES (first 420 nt of Gag coding sequence—Figure 1A and Supplementary Figure S1) followed by the streptomycin aptamer (28) was designed. This hybrid RNA was incubated in GMP-PNP treated reticulocyte lysate, and the resulting 48S initiation complexes were isolated using streptomycin derivatized sepharose beads (20,26,27). Purified HIV-2 gag IRES-bound ribosomal complexes were first analyzed on a native agarose gel. As observed in Figure 1B, the vast majority of HIV-2 RNA is engaged in a slow migrating complex running close to the 60S subunit, in agreement with the purified complexes being 48S initiation complexes. Determination of the canonical initiation factor content was subsequently performed by probing immunoblots with antibodies directed against eIF1, eIF2γ, eIF3d, eIF4E, eIF4G, eIF4A and eIF5. As shown in Figure 1C, eIF2, eIF3, eIF4A, eIF4G and eIF5 were detected in 48S complexes assembled onto the HIV-2 gag IRES. In contrast, although present in the lysate, eIF1 and eIF4E are absent from the purified complexes. Importantly, the identification of eIF4G in the purified 48S complexes could indicate that the HIV-2 gag IRES is related to the type I or II IRESes. Indeed, while type III and IV IRESes (Hepatitis C virus, HCV, IRESes and Cricket Paralysis Virus, CrPV, respectively) have no requirement for any components of the eIF4F complex, type I and II IRESes (Encephalomyocarditis Virus, EMCV and Poliovirus, PV, IRESes, respectively) strictly require eIF4G/4A (18,19).
Functional requirements for the eIF4F complex
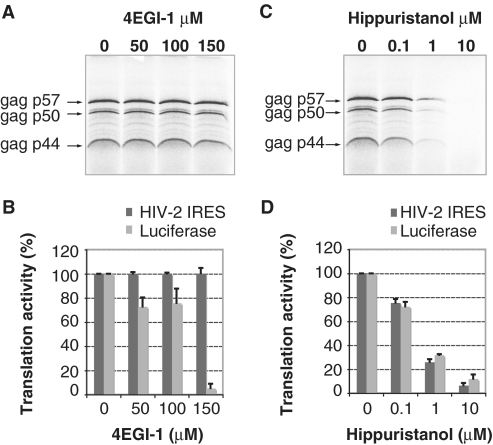
The presence or absence of a given factor in the ‘frozen’ complexes on the HIV-2 gag IRES does not directly address its functional requirement. We thus performed in vitro translation assays in the presence of specific inhibitors to validate the functional role of eIF4F components. More precisely, the necessity for the eIF4E:eIF4G interaction was assessed by using increasing concentrations of 4EGI-1, a known inhibitor of the eIF4E:eIF4G interaction (29). While 150 µM of 4EGI-1 drastically inhibits the translation of a control mRNA (Supplementary Figure S2); it does not affect either the yield or the pattern of HIV-2 gag translation (Figure 2A and B). This confirmed that the presence of eIF4E and the eIF4E-binding domain of eIF4G are dispensable for internal entry of ribosomes mediated by the HIV-2 gag IRES, even though eIF4G is present in the initiation complexes. We further evaluated the requirement for eIF4A, using hippuristanol, a natural compound that binds the C-terminal domain of eIF4A and thus inhibits RNA binding, helicase and ATPase activities of eIF4A (30). Addition of increasing amounts of hippuristanol inhibits the translation of both the control luciferase and the three Gag isoforms in a similar dose-response manner (Figure 2C and D); confirming a functional requirement for eIF4A. Taken together these results confirm our analysis of purified 48S complexes and clearly demonstrate that functional 48S complexes assembled onto the HIV-2 gag IRES require the eIF4A helicase, but not eIF4E.
Figure 2.
Translation of the three Gag isoforms in the presence of inhibitors of translation. (A, C) Autoradiogram showing products of in vitro translation of HIV-2 gag transcripts. Transcripts (10 ng/µl) were translated in RRL in the presence of increasing concentration of 4EGI-1 (0, 50, 100 and 150 µM) (A) or Hippuristanol (0, 0.1, 1 and 10 µM) (C) and their products resolved on a 20% SDS–PAGE gel. Gag isoforms from the three in frame AUG codons are indicated. (B, D) Relative intensity, expressed as a percentage of the initial product translated in the absence of 4EGI-1 (B) or in the absence of hippuristanol (D), of the three Gag isoforms (dark grey) compared to that of canonical luciferase (light grey) performed under the same experimental conditions. Data are representative of three independent experiments. Error bars are standard deviation of the mean.
Specific interaction of eIF3 with the HIV-2 gag IRES
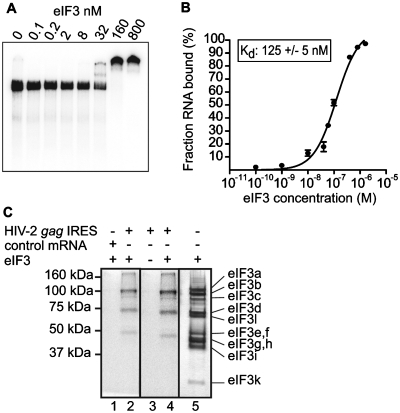
The canonical initiation factors eIF2, eIF3, eIF4A, eIF5 and eIF4G are components of the 48S complexes paused on the HIV-2 gag IRES. In order to identify the possible triggering event leading to 48S complex formation, we assayed whether individual initiation factors or 40S ribosomal subunit could directly interact with the IRES. To this end, the individual components of the translation machinery, eIFs and ribosomal subunits, were purified according to described procedures (23). Then, each individual purified component was independently incubated with 32P-labeled HIV-2 gag IRES and the putative interactions were analyzed by electrophoresis-mobility shift assay. No specific interactions could be detected between the HIV-2 gag IRES and eIF4A, eIF4F, eIF2 or eIF5 (Data not shown). In contrast, incubation of the HIV-2 gag IRES with eIF3 led to the formation of a low mobility complex, reflecting a direct interaction (Figure 3A). To analyze the affinity and specificity of purified eIF3 binding to the HIV-2 gag IRES, we conducted filter binding assays. In vitro transcribed 32P-radiolabeled RNA was incubated with purified eIF3, and the amount of resulting complexes was determined. The HIV-2 gag IRES binds eIF3 with an apparent equilibrium dissociation constant (Kd) of 125 nM (Figure 3B). Competitive binding assays were performed by incubating preformed binary eIF3:HIV-2 gag IRES complexes with a non-specific RNA competitor (globin mRNA). The proportion of complex remained unchanged, showing that eIF3 specifically binds to the HIV-2 gag IRES (data not shown). We further showed by mutating AUGs to CUCs, that eIF3 binding is independent of the presence of the initiation codons (Supplementary Figure S3).
Figure 3.
The initiation factor eIF3 directly binds to the HIV-2 gag IRES. (A) Autoradiograph of 4% acrylamide native gel with 0.5 nM of 32P labeled HIV-2 gag IRES in the absence or in the presence of increasing quantities (from 0.1 nM to 0.8 µM) of purified eIF3. (B) Binding curve of 32P-labeled HIV-2 gag IRES RNA to purified eIF3. Binary complexes were formed under the conditions used for native gels and analyzed by filter binding assay. (C) The determination of the eIF3 subunits involved in HIV-2 gag IREs binding was performed by crosslinking experiments. Binary complexes between eIF3 and 32P-labeled globin mRNA (lane 1) or 32P-labeled HIV-2 gag IRES RNA (lane 2 and 4) or HIV-2 gag IRES RNA alone (lane 3) were resolved by gel electrophoresis after UV cross-linking, and RNase digestion. Binary complexes are formed under the conditions used for native gels. The positions of molecular weight markers are indicated to the left and eIF3 subunits were visualized by silver staining as a reference (lane 5) [(note that eIF3a, a 170-kDa protein, is classically proteolysed as a 100- to 130-kDa protein in RRL (23,48)].
Identification of eIF3 subunits that mediates eIF3 binding to HIV-2 gag IRES
While the mammalian eIF3 complex consists of 10–13 subunits (eIF3a-m), only a subset of those is directly involved in type III IRES binding (31–33). Therefore, we used UV cross-linking to determine which specific eIF3 subunits are in close contact with HIV-2 gag IRES RNA. Binary eIF3-HIV IRES complexes were assembled by incubating 32P-UTP-labeled HIV-2 gag IRES and purified eIF3. Complexes were then irradiated with UV, treated with RNases and analyzed by SDS–PAGE. Only proteins that become covalently linked to the RNA upon UV irradiation are revealed by autoradiography of the gel. 32P-labeled bands were observed in the eIF3/RNA lane and not in the mock controls (Figure 3C). The bands run approximately as 100-, 70- and 50-kDa proteins; comparison with a silver stained gel of purified eIF3 allowed us to identify the subunits. The 100-kDa band can be attributed to the b and/or c subunits, the 70- and 50-kDa bands suggest it is the d subunit (66 kDa) and the e, and/or f subunits (47 and 48 kDa), respectively, that directly interacts with the HIV-2 gag IRES RNA. None of the bands described above were detected if eIF3 was omitted from the reaction mixture (Figure 3C, lane 3) or if eIF3 was incubated with a non-specific RNA (Figure 3C, lane 2). The cross-linking pattern of eIF3 subunits with the HIV-2 gag IRES suggests that central scaffold (34) subunits directly interact with the RNA.
HIV-2 Gag IRES binds 40S ribosomal subunit in the absence of initiation factors
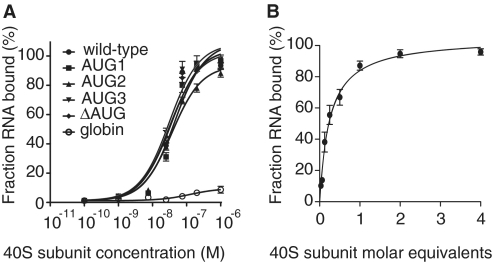
To detect a potential direct binding of the 40S subunit to the HIV-2 gag IRES, we performed filter binding assays (Figure 4A) and determined that the HIV-2 gag IRES binds the 40S subunit with an apparent equilibrium dissociation constant (Kd) of 34 nM (Figure 4A and Table 1). In contrast, the 40S subunit was unable to bind the capped globin mRNA under similar conditions (Figure 4A). The HIV-2 gag IRES contains three functional AUG start codons (13) and we previously demonstrated that up to three initiation complexes could be assembled onto the HIV-2 gag IRES (15). This prompted us to examine the stoichiometry of the observed IRES/ribosome interaction. We used stoichiometric binding assays to determine the ratio of 40S subunits bound per RNA molecule (25). A RNA concentration 20-fold over the calculated Kd was incubated with increasing concentration of ribosomes. Saturation of RNA molecules was reached after addition of 1 M equivalent of ribosomal subunits (Figure 4B). Therefore, the HIV-2 gag IRES:40S complex shows a 1:1 stoichiometry, one IRES molecule can only bind one 40S subunit molecule in the absence of initiation factors. We next asked whether any of the AUGs were essential for the recruitment of the 40S subunit. HIV-2 gag IRES mutated constructs were engineered where AUG codons were replaced by CUC codons two at a time or all together (13,15). None of those mutations impaired the binding of the 40S subunit, showing that complex formation is independent of the initiation codons (Figure 4A).
Figure 4.
HIV-2 gag IRES directly binds to the 40S subunit. (A) Binding curves of 32P-labeled wild-type, AUG mutants and globin control to purified 40S subunits. (B) Stoichiometric binding assays of 32P-labeled HIV-2 gag IRES to 40S subunit. Results are from three independent experiments.
Table 1.
40S subunit binding affinities of HIV-2 gag IRES and AUG mutants
| RNA | Kd binding to 40S subunit ± SEM (nM) |
|---|---|
| wild-type | 34.1 ± 6.8 |
| AUG1 | 44.8 ± 7.9 |
| AUG2 | 36.9 ± 4.5 |
| AUG3 | 31.2 ± 6.3 |
| ΔAUG | 31.7 ± 5.8 |
| globin | >500 |
AUG1, AUG 2 and AUG3 refer to constructs where two of the three AUGs have been mutated to CUC, the number indicating the non-mutated initiation codon. ΔAUG refers to the mutation of all three AUGs into CUCs.
IRES domain requirement for the recruitment of eIF3 and the 40S subunit by lentivirus gag IRESes
To identify minimal RNA structures that bind eIF3 and the 40S ribosomal subunit, we engineered RNA constructs in which individual domains were deleted (Figure 5A). Those truncated RNAs were used as cold competitors for binding of eIF3 to the full-length HIV-2 gag IRES. The full length IRES, or progressive deletions from the 5′ or 3′ end preserving the P2 pairing, compete out the HIV-2 gag IRES/eIF3 complexes (Table 2, ΔP1, ΔP7, ΔP67, ΔP567, ΔP1567). In contrast, deletions affecting the P2 pairing (Table 2, ΔP12), or internal deletion of the P4 domain severely impair the ability of the cold RNAs to compete for eIF3 (Table 2, ΔP4). We next evaluated the ability of the same truncated IRESes to directly interact with the 40S subunit. They appear to fall in two classes. Constructs deleted from the 5′-end and/or the 3′-end up to P2 or P5, respectively (ΔP1, ΔP7, ΔP67, ΔP167), show a 2-fold increase in Kd but retain their ability to be fully bound (Table 3). This suggests that P1, P6 and P7 are involved in the 40S binding site, although with a modest contribution. Deletions of the P2, P4 or the P5 domain (ΔP12, ΔP567, ΔP4) impair more drastically the recruitment of the 40S subunit with an increase in Kd between 2- and 6-fold, and a strongly affected maximum binding (Plateau << 100%, see Table 3). The reduced amount of maximum binding suggests that P2, P4 and P5 are not only part of the 40S binding site but that these deletions further destabilize the tertiary folding of the IRES active conformation. Taken together, these results indicate that the crucial elements of the 40S ribosomal subunit binding site lies between P2 and P5, while eIF3 binding does not need the P5 pairing. This defines the P2-P3-P4-P5 region as the central scaffold of the HIV-2 IRES. Interestingly, we had previously shown that the Gag coding regions of two other primate lentiviruses (HIV-1 and SIVMac) are not only endowed with a similar IRES activity but also present these conserved structural elements (15) (Figure 5). This prompted us to investigate the ability of the HIV-1 and SIVMAC P2-P3-P4-P5 domains to directly interact with eIF3 and the small ribosome subunit. Gel shift and filter binding assays unambiguously show that the HIV-1 and SIVMac central scaffold binds eIF3 and the 40S subunit, with affinities comparable to what has been observed for HIV-2 (Supplementary Figure S4).
Table 2.
eIF3 binding affinity of HIV-2 Gag IRES and mutants
| RNA | Kc binding to eIF3 ± SEM (nM) |
|---|---|
| wild-type | 122.8 ± 1.3 |
| ΔP1 | 128.8 ± 1.3 |
| ΔP123 | 790.4 ± 1.2 |
| ΔP7 | 107 ± 1.3 |
| ΔP67 | 132.1 ± 1.6 |
| ΔP567 | 129.7 ± 1.3 |
| ΔP1567 | 131 ± 1.3 |
| ΔP4 | 435.5 ± 1.1 |
Competition reactions were plotted as the fraction of bound labeled RNA versus the concentration of unlabeled competitor. The competition constant Kc was determined by fitting the data to a sigmoidal dose-response curve, using the Lin and Riggs equation (25).
Table 3.
40S subunit binding affinity of HIV-2 gag IRES and mutants defined by filter binding assay
| RNA | Kd binding to 40S subunit ± SEM (nM) | Bmax binding to 40S subunit ± SEM (nM) |
|---|---|---|
| wild-type | 36.1 ± 5.5 | 95.5 ± 3.3 |
| ΔP1 | 70.18 ± 14.78 | 99.76 ± 5.4 |
| ΔP12 | 134.6 ± 21.5 | 38.2 ± 1.8 |
| ΔP7 | 95.1 ± 2.72 | 101.4 ± 6.6 |
| ΔP67 | 67.8 ± 11.6 | 95.9 ± 4.2 |
| ΔP567 | 81.8 ± 13.1 | 53.1 ± 2.2 |
| ΔP4 | 233.3 ± 53.8 | 65.4 ± 5.1 |
| ΔP167 | 70.4 ± 15.2 | 99.7 ± 5.5 |
Formation of an IRES/eIF3/40S subunit ternary complex
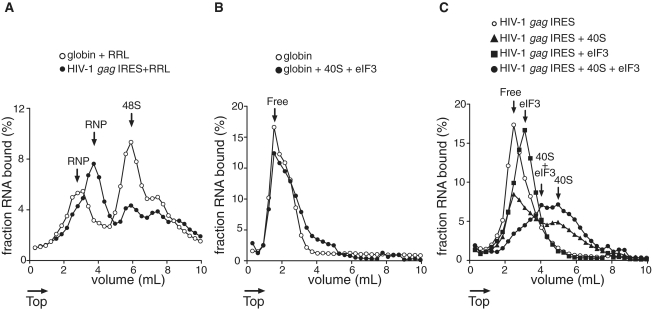
To further investigate the capability of eIF3 and/or 40S subunits to form stable complexes on the HIV gag IRES, we analyzed the mobility of the complexes during sucrose density gradient centrifugation. To define landmarks, HIV-1 or β-globin RNAs were incubated with GMP-PNP-treated Rabbit Reticulocytes Lysate (RRL). We used HIV-1 gag IRES which contains only two initiation codons, to simplify the profile analysis. Translation of the full length Gag polyprotein starts at the first AUG homologous to HIV-2 AUG1, while the second AUG allows the translation of a shorter isoform that is not homologous to any of HIV-2 shorter isoforms (11,15,16). As observed in Figure 6, both RNAs sediment as three peaks, the lighter is usually attributed to various undefined ribonucleoproteins (RNPs) (Figure 6A). Of note, RNPs assembled onto globin mRNA display a reduced mobility as compared to those assembled onto HIV-1 RNA indicating possible differences in their composition. Importantly, both HIV-1 RNA and globin mRNA can efficiently form 48S initiation complexes which migrate at 6 ml from the top of the gradient. While the sedimentation profile of globin mRNA results in only one major peak [the second minor peak can be attributed to the recruitment of an additional 43S complex as previously observed on an unstructured 5′-UTR (35)], the profile of HIV-1 RNA indicates the presence of two quantitatively equivalent complexes. The presence of this second peak in the HIV-1 profile has been attributed to the presence of 48S initiation complexes formed at both of the in frame AUGs (15). Next, we assessed whether the direct interaction of 40S subunits and/or eIF3 with HIV-1 RNA could lead to the formation of stable complexes. The control globin RNA (Figure 6B) displays the mobility of free RNA in the presence of both 40S subunits and purified eIF3, confirming the absence of direct interaction between the mRNA and the two other components. Interestingly, the profiles obtained with HIV-1 RNA in the presence of eIF3 or 40S subunits show peaks with an increased mobility in the gradient (3 and 5 ml from the top of the gradient, respectively—see Figure 6C) which is consistent with the formation of RNA/eIF3 and RNA/40S stable complexes. Furthermore, when both eIF3 and 40S subunits are allowed to assemble onto HIV-1 RNA, one can observe an additional complex with a reduced mobility (4 ml from the top) as compared to the RNA/40S complex. Importantly, the binding of eIF3 to ribosomal 40S subunits is known to reduce the mobility of ribosomal complexes in sucrose density gradients. The previously proposed explanation for this observation is that conformational changes in the 40S subunit, or changes in the shape of the complex to a less globular form, could increase its frictional coefficient (36). Therefore, this additional complex can unambiguously be attributed to the formation of a HIV-1 RNA/eIF3/40S ternary complex. Interestingly enough, the RNPs observed in RRL with HIV-1 gag IRES, but not with the globin RNA, sediment at the exact same position as the putative ternary complex, suggesting that the eIF3–40S–IRES complex is accumulated in RRL. Finally, it is of note that while several initiation complexes can form on HIV-1 RNA in RRL, the mobility of the binary complex obtained with purified components is in favor of the presence of a single site of 40S binding in HIV-1 gag IRES, in agreement with the 1:1 stoichiometry observed with HIV-2 gag IRES by binding assays (see above).
Figure 6.
Mobility of ribosomal complexes during centrifugation through 10–30% linear sucrose density gradients. (A) Fractionation by sucrose density gradient of initiation complexes formed after incubation of capped and polyadenylated globin mRNA (open circles) or HIV-1 (closed circles) RNA with RRL pre-treated with GMP-PNP. (B) Fractionation by sucrose density gradient centrifugation of capped and globin mRNA in the absence (open circles) or presence (closed circles) of excess purified eIF3 and 40S subunits. (C) Fractionation by sucrose density gradient centrifugation of HIV-1 RNA (open circles) in the presence of excess 40S subunits (triangles), eIF3 (squares) or eIF3 and 40S subunits (rounds). Arrows indicate whether the RNA is free or associated with 40S subunits and/or eIF3. The percent of RNA bound represents the mean of at least two independent experiments.
DISCUSSION
Unusual initiation complex composition on the HIV-2 gag IRES
We have previously characterized the lentiviral gag IRES as an atypical IRES (13,15), and as such it is predicted to exhibit a specific molecular mechanism. Determining the initiation factor requirements of an IRES appears to be a reliable way to define to which IRES class it belongs to (19). We analyzed the presence of most of the core initiation factors on purified 48S complexes paused on the HIV-2 gag IRES, and found that all were present, except for eIF4E and eIF1. This is reminiscent of the factors required for picornavirus type I and II IRESes (37) which recruit initiation complexes through direct binding of the central domain of eIF4G (38,39). The absence of eIF4E correlates with the absence of inhibition observed with an eIF4G/eIF4E interaction inhibitor, and with previous experiments in which we showed that the C-terminal fragment of the eIF4G central domain, lacking the eIF4E binding domain, was sufficient to promote internal entry of ribosomes onto the HIV-2 gag open reading frame (13,14). Although it has been shown in many cases that eIF4E is not functionally required during IRES mediated translation, here, we show for the first time that eIF4E is physically absent from complexes recruited onto IRESes in RRL. Although one could expect eIF4E to be carried along with eIF4G, our results suggest that either the eIF4F complex is not preformed in a cellular environment, or else, that the entry of eIF4G into the initiation complex destabilizes its interaction with eIF4E. While these results could reflect a specific recruitment of eIF4G by the IRES, we were unable to detect any direct specific interaction between the two components, highlighting a functional difference between the HIV gag IRES and the type I and type II IRESes.
In the cellular 43S ribosomal complex, eIF1 binds a region of the 40S ribosome close to the P site (40), and remains bound to the 40S particle until recognition of the initiation codon and the subsequent hydrolysis of the eIF2 bound GTP (41–43). Consequently, it is present in GMP-PNP blocked initiation complexes. The absence of eIF1 in the initiation complexes recruited on the HIV gag IRES may be particularly relevant in the context of an RNA on which translation starts at a 5′-terminal AUG. Indeed, it has been shown that the lack of eIF1 abrogates the scanning ability of the ribosome, thus enabling initiation complexes to assemble onto 5′-terminal AUG codons (44). The absence of eIF1 suggests that the HIV gag IRES and eIF1 may bind to the same region of the 40S subunit, so that IRES binding competes out eIF1. Here we hypothesize that the eviction of eIF1 would clamp the ribosome into a closed conformation so that it initiates at the 5′terminal codon. This would be in contrast to the HCV IRES, the binding of which induces a conformational rearrangement in the 40S subunit similar to what is observed upon eIF1 binding (45).
Formation of a ternary complex on the IRES core as a first step towards initiation
In this study, we systematically investigated the possibility that some components of the initiation complex might directly contact the IRES. We found that binary and ternary complexes efficiently form with the 40S ribosomal subunit and eIF3. This is also true for a genomic-like construct, because the presence of the 5′-UTR upstream from the IRES affects neither the eIF3 nor the 40S binding (N. Chamond, unpublished results). The observed cross-linking pattern that identified the eIF3 b/c, d and e/f subunits as directly contacting the HIV-2 gag IRES, is very similar to what has been observed with HCV and CSFV IRESes (32). In the latter case the cross linked proteins were identified as eIF3 a, b/c, d and f subunits (32), and mass spectroscopy analysis of the eIF3/IRES complex suggested that eIF3 f, g and h were responsible for the direct binding (33). Also similar to type III IRESes, the lentivirus gag IRES tightly binds to the small subunit of the ribosome. However, no obvious structural similarity between type III and lentivirus IRESes exists that would allow us to extrapolate some common mechanistic function. Both eIF3 and the 40S subunit interact with the conserved core of the IRES lying between P2 and P5. It is of note that the core does not contain any of the initiation codons (except AUG2 in the case of HIV-2), and that the core is far downstream of AUG1. Interestingly, both binding sites overlap, but are not identical, as P5 is necessary for ribosome binding but is dispensable in the case of eIF3.
Formation of the HIV IRES/40S/eIF3 ternary complex is likely to be an early event leading to initiation. Thus, mutations disrupting the binding of the 40S subunit, and/or eIF3 are predicted to affect translation. We have previously reported that mutations affecting the P2–P5 core structure impair Gag expression in vitro (15). Here we show that P2 and P4 deletions affect the recruitment of eIF3 and the 40S subunit, this correlates with the strongly reduced translation efficiency of P2 point mutants or P4 deleted constructs (15). However, one of our observations suggests that the formation of the ternary complex may not be the rate limiting step of the initiation process on this IRES. This could explain why mutants affected for the ternary complex formation retain significant translation efficiency as compared to the wild type. Indeed, in RRL we observed the accumulation of a complex that sediments at the same position as the eIF3/40S/IRES complex. This could reflect that the recruitment of eIF3 and the 40S subunit is followed by a rate limiting step that governs the translation kinetics. The slow step could be the transfer of the complexes onto the initiation codons, as much less 48S than ternary complexes are formed in RRL. The respective efficiency of the 48S and the ternary complex formation suggests that some of the fast migrating species observed on sucrose gradient profile contain a 48S complex and a ternary complex on a same IRES molecule rather than two 48S complexes as proposed previously (15).
Taken together, our results lead to a unique model in which the 40S ribosome and eIF3 are recruited at a unique site on the IRES, and from there are dispatched to each of the three initiation codons. Thus, if several initiation complexes are recruited by one RNA molecule (15), this can only be done sequentially and not concomitantly. The mechanism(s) involved in the transfer of this complex necessitate(s) the intervention of other initiation factors. In this respect, the requirement for eIF4A could suggest a scanning event from the ‘landing’ site to the AUG(s) downstream. However, this cannot be invoked in the case of translation initiation from the 5′-terminal initiation triplet. In this case, the helicase could instead trigger an RNA structural rearrangement leading to the repositioning of the initiation complex onto the AUG. Alternatively, the 3D structure could bring the initiation codon close the 40S recruitment site. Nonetheless, in any of those hypotheses, after transfer of the initial complex onto one of the initiation codons, the IRES is able to recruit a second 40S subunit as several complexes can assemble onto a single mRNA. Whilst parts of the mechanism remain to be unraveled, this study elucidates the first step of an original mechanism to initiate translation initiation. The structural and functional conservation of the gag open reading frame among the fast-evolving primate lentiviruses strongly suggests an important role of this IRES in the virus life cycle; rendering the RNA structure of the coding region an attractive therapeutic target.
Although local structures such as pseudoknots involved in frameshifting (46) have been identified, this is the first time that such a large structure within an open reading frame is found to control its own translation. This is rather puzzling on a mechanistic point of view since it is located downstream to the initiation site, and puts an important evolutionary constraint on the coding region. Exploring the RNA secondary structure of the whole HIV genome, Watts et al. found that the level of folding of an RNA region is inversely correlated with the compaction of the protein segment it encodes for (47). Along with our work this clearly shows that mRNA is not to be considered as a string of beads bearing the codon sequences, but that its structure also encodes a wealth of information that influences the translation process itself.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique Action Thématique et Incitative sur Programme (ATIP); Agence Nationale pour la Recherche sur le S.I.D.A (ANRS–2008-358); Agence Nationale pour la Recherche (ANR–BLAN06-1_146425); SIDACTION (AI20-1-01577). Funding for open access charge: ANRS–2008-358.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank J. Pelletier and L. Lindqvist for generous gift of hippuristanol, S. Butcher and L. Roberts for critical reading of the manuscript and N. Ulryck for technical help.
REFERENCES
- 1.Balvay L, Lastra ML, Sargueil B, Darlix JL, Ohlmann T. Translational control of retroviruses. Nat. Rev. Microbiol. 2007;5:128–140. doi: 10.1038/nrmicro1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butsch M, Boris-Lawrie K. Destiny of unspliced retroviral RNA: ribosome and/or virion? J. Virol. 2002;76:3089–3094. doi: 10.1128/JVI.76.7.3089-3094.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 1996;54:1–34. doi: 10.1016/s0079-6603(08)60359-1. [DOI] [PubMed] [Google Scholar]
- 4.Dirac AM, Huthoff H, Kjems J, Berkhout B. Regulated HIV-2 RNA dimerization by means of alternative RNA conformations. Nucleic Acids Res. 2002;30:2647–2655. doi: 10.1093/nar/gkf381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.James L, Sargueil B. RNA secondary structure of the feline immunodeficiency virus 5'UTR and Gag coding region. Nucleic Acids Res. 2008;36:4653–4666. doi: 10.1093/nar/gkn447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geballe AP, Gray MK. Variable inhibition of cell-free translation by HIV-1 transcript leader sequences. Nucleic Acids Res. 1992;20:4291–4297. doi: 10.1093/nar/20.16.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miele G, Mouland A, Harrison GP, Cohen E, Lever AM. The human immunodeficiency virus type 1 5' packaging signal structure affects translation but does not function as an internal ribosome entry site structure. J. Virol. 1996;70:944–951. doi: 10.1128/jvi.70.2.944-951.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parkin NT, Cohen EA, Darveau A, Rosen C, Haseltine W, Sonenberg N. Mutational analysis of the 5' non-coding region of human immunodeficiency virus type 1: effects of secondary structure on translation. EMBO J. 1988;7:2831–2837. doi: 10.1002/j.1460-2075.1988.tb03139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braddock M, Thorburn AM, Chambers A, Elliott GD, Anderson GJ, Kingsman AJ, Kingsman SM. A nuclear translational block imposed by the HIV-1 U3 region is relieved by the Tat-TAR interaction. Cell. 1990;62:1123–1133. doi: 10.1016/0092-8674(90)90389-v. [DOI] [PubMed] [Google Scholar]
- 10.Brasey A, Lopez-Lastra M, Ohlmann T, Beerens N, Berkhout B, Darlix JL, Sonenberg N. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J. Virol. 2003;77:3939–3949. doi: 10.1128/JVI.77.7.3939-3949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buck CB, Shen X, Egan MA, Pierson TC, Walker CM, Siliciano RF. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J. Virol. 2001;75:181–191. doi: 10.1128/JVI.75.1.181-191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohlmann T, Lopez-Lastra M, Darlix JL. An internal ribosome entry segment promotes translation of the simian immunodeficiency virus genomic RNA. J. Biol. Chem. 2000;275:11899–11906. doi: 10.1074/jbc.275.16.11899. [DOI] [PubMed] [Google Scholar]
- 13.Herbreteau CH, Weill L, Decimo D, Prevot D, Darlix JL, Sargueil B, Ohlmann T. HIV-2 genomic RNA contains a novel type of IRES located downstream of its initiation codon. Nat. Struct. Mol. Biol. 2005;12:1001–1007. doi: 10.1038/nsmb1011. [DOI] [PubMed] [Google Scholar]
- 14.Ricci EP, Herbreteau CH, Decimo D, Schaupp A, Datta SA, Rein A, Darlix JL, Ohlmann T. In vitro expression of the HIV-2 genomic RNA is controlled by three distinct internal ribosome entry segments that are regulated by the HIV protease and the Gag polyprotein. RNA. 2008;14:1443–1455. doi: 10.1261/rna.813608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weill L, James L, Ulryck N, Chamond N, Herbreteau CH, Ohlmann T, Sargueil B. A new type of IRES within gag coding region recruits three initiation complexes on HIV-2 genomic RNA. Nucleic Acids Res. 2010;38:1367–1381. doi: 10.1093/nar/gkp1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ricci EP, Soto Rifo R, Herbreteau CH, Decimo D, Ohlmann T. Lentiviral RNAs can use different mechanisms for translation initiation. Biochem. Soc. Trans. 2008;36:690–693. doi: 10.1042/BST0360690. [DOI] [PubMed] [Google Scholar]
- 17.Chamond N, Locker N, Sargueil B. The different pathways of HIV genomic RNA translation. Biochem. Soc. Trans. 2010;38 doi: 10.1042/BST0381548. [DOI] [PubMed] [Google Scholar]
- 18.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2009;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kieft JS. Viral IRES RNA structures and ribosome interactions. Trends Biochem. Sci. 2008;33:274–283. doi: 10.1016/j.tibs.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Locker N, Easton LE, Lukavsky PJ. Affinity purification of eukaryotic 48S initiation complexes. RNA. 2006;12:683–690. doi: 10.1261/rna.2227906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sargueil B, Hampel KJ, Lambert D, Burke JM. In vitro selection of second site revertants analysis of the hairpin ribozyme active site. J. Biol. Chem. 2003;278:52783–52791. doi: 10.1074/jbc.M306703200. [DOI] [PubMed] [Google Scholar]
- 22.Sargueil B, McKenna J, Burke JM. Analysis of the functional role of a G.A sheared base pair by in vitro genetics. J. Biol. Chem. 2000;275:32157–32166. doi: 10.1074/jbc.M005591200. [DOI] [PubMed] [Google Scholar]
- 23.Pisarev AV, Unbehaun A, Hellen CU, Pestova TV. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol. 2007;430:147–177. doi: 10.1016/S0076-6879(07)30007-4. [DOI] [PubMed] [Google Scholar]
- 24.Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryder SP, Recht MI, Williamson JR. Quantitative analysis of protein-RNA interactions by gel mobility shift. Methods Mol. Biol. 2008;488:99–115. doi: 10.1007/978-1-60327-475-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Locker N, Easton LE, Lukavsky PJ. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 2007;26:795–805. doi: 10.1038/sj.emboj.7601549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Locker N, Lukavsky PJ. A practical approach to isolate 48S complexes: affinity purification and analyses. Methods Enzymol. 2007;429:83–104. doi: 10.1016/S0076-6879(07)29005-6. [DOI] [PubMed] [Google Scholar]
- 28.Bachler M, Schroeder R, von Ahsen U. StreptoTag: a novel method for the isolation of RNA-binding proteins. RNA. 1999;5:1509–1516. doi: 10.1017/s1355838299991574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, Gross JD, Degterev A, Yuan J, Chorev M, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128:257–267. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 30.Lindqvist L, Oberer M, Reibarkh M, Cencic R, Bordeleau ME, Vogt E, Marintchev A, Tanaka J, Fagotto F, Altmann M, et al. Selective pharmacological targeting of a DEAD box RNA helicase. PLoS One. 2008;3:e1583. doi: 10.1371/journal.pone.0001583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buratti E, Tisminetzky S, Zotti M, Baralle FE. Functional analysis of the interaction between HCV 5'UTR and putative subunits of eukaryotic translation initiation factor eIF3. Nucleic Acids Res. 1998;26:3179–3187. doi: 10.1093/nar/26.13.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU. Specific interaction of eukaryotic translation initiation factor 3 with the 5' nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J. Virol. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou M, Sandercock AM, Fraser CS, Ridlova G, Stephens E, Schenauer MR, Yokoi-Fong T, Barsky D, Leary JA, Hershey JW, et al. Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc. Natl Acad. Sci. USA. 2008;105:18139–18144. doi: 10.1073/pnas.0801313105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Damoc E, Fraser CS, Zhou M, Videler H, Mayeur GL, Hershey JW, Doudna JA, Robinson CV, Leary JA. Structural characterization of the human eukaryotic initiation factor 3 protein complex by mass spectrometry. Mol. Cell Proteomics. 2007;6:1135–1146. doi: 10.1074/mcp.M600399-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Kozak M. Influence of mRNA secondary structure on binding and migration of 40S ribosomal subunits. Cell. 1980;19:79–90. doi: 10.1016/0092-8674(80)90390-6. [DOI] [PubMed] [Google Scholar]
- 36.Kolupaeva VG, Unbehaun A, Lomakin IB, Hellen CU, Pestova TV. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA. 2005;11:470–486. doi: 10.1261/rna.7215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pestova TV, Hellen CU, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CU. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc. Natl Acad. Sci. USA. 2009;106:9197–9202. doi: 10.1073/pnas.0900153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolupaeva VG, Pestova TV, Hellen CU, Shatsky IN. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J. Biol. Chem. 1998;273:18599–18604. doi: 10.1074/jbc.273.29.18599. [DOI] [PubMed] [Google Scholar]
- 40.Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev. 2003;17:2786–2797. doi: 10.1101/gad.1141803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheung YN, Maag D, Mitchell SF, Fekete CA, Algire MA, Takacs JE, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch AG. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev. 2007;21:1217–1230. doi: 10.1101/gad.1528307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maag D, Fekete CA, Gryczynski Z, Lorsch JR. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon Mol. Cell. 2005;17:265–275. doi: 10.1016/j.molcel.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 44.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fraser CS, Hershey JW, Doudna JA. The pathway of hepatitis C virus mRNA recruitment to the human ribosome. Nat. Struct. Mol. Biol. 2009;16:397–404. doi: 10.1038/nsmb.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giedroc DP, Cornish PV. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res. 2009;139:193–208. doi: 10.1016/j.virusres.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baugh JM, Pilipenko EV. 20S proteasome differentially alters translation of different mRNAs via the cleavage of eIF4F and eIF3. Mol. Cell. 2004;16:575–586. doi: 10.1016/j.molcel.2004.10.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.