Abstract
Alternative splicing enables higher eukaryotes to increase their repertoire of proteins derived from a restricted number of genes. However, the possibility that functional diversity may also be augmented by splicing between adjacent genes has been largely neglected. Here, we show that the human melanocortin 1 receptor (MC1R) gene, a critical component of the facultative skin pigmentation system, has a highly complex and inefficient poly(A) site which is instrumental in allowing intergenic splicing between this locus and its immediate downstream neighbour tubulin-β-III (TUBB3). These transcripts, which produce two distinct protein isoforms localizing to the plasma membrane and the endoplasmic reticulum, seem to be restricted to humans as no detectable chimeric mRNA could be found in MC1R expressing mouse melanocytes. Significantly, treatment with the MC1R agonist α-MSH or activation of the stress response kinase p38-MAPK, both key molecules associated with ultraviolet radiation dermal insult and subsequent skin tanning, result in a shift in expression from MC1R in favour of chimeric MC1R-TUBB3 isoforms in cultured melanocytes. We propose that these chimeric proteins serve to equip melanocytes with novel cellular phenotypes required as part of the pigmentation response.
INTRODUCTION
The melanocortin 1 receptor (MC1R) is one of five G-protein coupled receptors belonging to the melanocortin subfamily, associated with the regulation of a variety of fundamental biological processes ranging from pigmentation and energy homeostasis to sexual health (1). MC1R is expressed on the cell surface of epidermal melanocytes located in the stratum basale of human skin and is an integral part of the constitutive and facultative pigmentation system (2). The role of human MC1R in facultative or adaptive pigmentation following ultraviolet radiation (UVR) exposure has been the subject of intensive investigation during the last few decades with more than 60 variant MC1R receptors identified, many associated with phenotypic traits such as red hair, fair skin (3–5) and skin cancer susceptibility (6–8).
MC1R acts between UVR stressed keratinocytes located in the stratum spinosum and the production and subsequent transfer of protective melanin from melanocytes. Although the melanocyte stimulating hormone α-MSH has long been implicated in skin pigmentation (1), a direct link between α-MSH, MC1R and UVR induced tanning has only recently been demonstrated (9). The binding of α-MSH to MC1R receptors triggers multiple signal transduction cascades, primarily dependent on elevated intracellular cAMP (10), resulting in the activation of a large number of genes (11) which together regulate several aspects of melanocyte biology such as proliferation, melanogenesis (12) and cell-cycle control (11). The activation of melanocytes by α-MSH and other paracrine factors also elicit functionally significant morphological changes. Critically, α-MSH binding to MC1R stimulates melanocyte dendricity (13–15) and so increases the number of individual keratinocytes a given melanocyte can contact. This is crucial in facilitating maximal transfer of melanin containing melanosomes along dendrites to target keratinocytes where the melanin is subsequently employed in the formation of photo-protective nuclear associated caps.
The gene encoding MC1R is located on the 3′-telomeric end of chromosome 16 in a locus that has a high density of genes. Consequently, only 2.5 kb of intergenic DNA separate MC1R from the downstream, tandem positioned Tubulin beta III gene (TUBB3). As with many G-protein coupled receptor genes, MC1R is expressed primarily as an intronless gene. We earlier reported that MC1R has an unusual cleavage and polyadenylation signal (PAS) constituting an AAUAAA hexamer, a degenerate downstream sequence element (DSE) adjacent to the cleavage site and finally two essential G-rich sequence elements (GRS) located further downstream (16). This arrangement is in contrast with normal bi-partite PAS comprising the hexameric sequence A(A/U)UAAA and a GU or U rich DSE in close proximity to the cleavage site (17).
Cleavage and polyadenylation is a highly regulated co-transcriptional process that not only equips the mRNA with a poly(A) tail essential for nuclear cytoplasmic export, stability and translation, but also triggers transcription termination downstream of genes (18). Importantly, alternative cleavage and polyadenylation, which occurs in half of all protein-encoding genes (19), is a significant cellular process that controls the expression of alternative mRNA isoforms in response to developmental and growth cues (20,21).
Here we present evidence that MC1R pre-mRNA is subject to complex alternative processing that depends on its unusual PAS. This allows readthrough transcription into the downstream-positioned TUBB3 locus which via alternative splicing results in the expression of at least two novel chimeric mRNAs that contain both the MC1R seven transmembrane receptor domain and a TUBB3 C-terminal extension. Strikingly, these chimeric transcripts are human specific being absent in mouse melanocytes. We demonstrate that these chimeric transcripts translate into proteins which localize to the plasma membrane and to the endoplasmic reticulum. Importantly, the expression levels of both MC1R and MC1R-TUBB3 chimeric mRNA can be modulated by stimulating discrete signalling pathways associated with the dermal pigmentation response to solar radiation.
MATERIALS AND METHODS
Cell culture and transfection
Cell lines HEK293 and HBL (human melanoma) were cultured in DMEM/10%FCS/pen-strep/glutamine, whilst cell lines B16 (mouse melanoma) and M14 (human melanoma) were cultured in RPMI/5%FCS/pen-strep/glutamine and RPMI/10%FCS/pen-strep/glutamine respectively. Melan-a and Melan-c cells were cultured in RPMI/10%FCS/pen-strep/glutamine supplemented with 200 nM TPA, Melan-c cells were further supplemented with 100 µM β-mercaptoethanol. HERMES-1 cells (which are transformed normal human melanocytes) (22) were cultured in RPMI/10%FCS/pen-strep/glutamine supplemented with 200 nM TPA, 200 pM Cholera toxin, 10 nM endothelin-1 and 10 ng/ml human stem cell factor. All transfection were carried out as described previously (16).
Plasmid construction
All human MC1R/TUBB3 locus constructs were cloned from HeLa genomic DNA, using pfu DNA polymerase amplification, and inserted into either pUC18/CMV or commercial pcDNA3.1-His expression vectors as previously described (16). The sequences of primers are available upon request. All constructs were verified by complete sequencing.
pCMTR includes a CMV promoter AflIII-EcoRI fused directly to the 5′-UTR of MC1R followed by the entire genomic locus for 16 kb to the 43rd codon of TUBB3 exon 2 fused in frame Xba I to the entire RFP cDNA, followed by a synthetic PAS SPA [synthetic poly(A) site] (23). In pΔCMTR the CMV promoter is removed by an AflIII-EcoRI digest. Constructs pCMTRΔi and pΔCMTRΔi are derived from pCMTR and pΔCMTR where TUBB3 intron is removed fusing exons 1 and 2 in frame by PCR. Construct pCΔiG is derived from pCMTR where the 3′-end of the TUBB3 first intron is fused 5′ to an AflII site 1-kb downstream of the MC1R PAS and 3′ to exon2/RFP. pCSΔ6ki1 was created by an XhoI TUBB3 intronic digest. pCΔG1G2 is based on the H36 construct [Dalziel et al. (16)]. In pCSPAΔ6ki1 the endogenous MC1R PAS replaced by the artificial SPA, inserted via BlpI- AflII. Constructs pE_MC1R, pE_Iso1 and pE_Iso2 were made by Pfu amplification from the appropriate cDNA and inserted EcoRI/XbaI(AvrII) into the commercial construct pcDNA3.1-v5His A (Invitrogen).
RNA extraction, RNase protection assay, 3′-RACE
RNA extraction, RNase protection (RP) and 3′RACE were performed as previously described in Dalziel et al. (16).
cAMP assay
All experiments were performed in duplicate and repeated at least three times. HEK293 cells were seeded into 12-well plates (Nunc) and grown overnight, then transfected (overnight) with 0.25 µg of a relevant pcDNA3.1His-MC1R construct. Next day, cells were serum starved for 1 h in pre-warmed DMEM alone, before being stimulated with NDP-MSH (10 nM for 1 h). Total cellular cAMP was then measured with a commercial kit (GE Healthcare) as per instructions.
Western blot, confocal microscopy
HEK293 cells transfected with pcDNA3.1-His MC1R variants were washed (PBS) and solubilized in 200 µl of solubilization buffer (50 mM Tris–HCl, pH 8.0, 1% Igepal CA-640, 1 mM EDTA, 0.1 mM PMSF and 10 mM iodoacetamide). The samples were subsequently vortexed for 1 h at 4°C, and centrifuged at 20 800g for 30 min. Supernatants were used for subsequent western blotting. Samples were incubated at room temperature for 30 min in sample buffer and subsequently loaded onto a 10% Tris–Glycine polyacrylamide gel and semi-dry transferred to nitrocellulose in Towbin buffer. Detection of MC1R variants was performed using the N19 antibody (SantaCruz) and ECL.
HEK293 cells, grown on coverslips and transfected with pcDNA3.1-His-MC1R variants, were washed in PBS, fixed with 4% paraformaldehyde, blocked with 2% BSA. If needed, cells were permeabilized with 0.5% Igepal-640. Cells were immunolabelled with the N19 anti-MC1R antibody (1:100) and the anti-goat Alexa488 secondary antibody (1:200). Photographs were taken in a Zeiss LSM 510 Meta system.
RESULTS
MC1R 3′-end processing requires a complex positional arrangement of four cis-elements
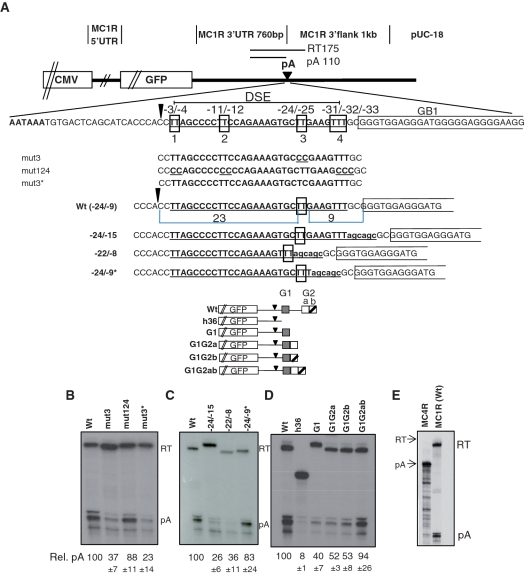
Earlier we used an MC1R minigene to study cleavage and polyadenylation in the context of an intronless gene (16). This analysis revealed that the MC1R PAS requires four sequence elements consisting of an AAUAAA hexamer, a DSE and two GRS one located adjacent to the DSE (GB1) and one positioned ∼400 nt downstream of the cleavage site (GB2). To further unravel the complexity of the MC1R PAS, we decided to analyse the positional requirements of nucleotides in the DSE and the GRS. We previously showed that a di-uridine at position −24/−25 downstream of the cleavage site is a critical component of the MC1R DSE (16). However, it was unclear whether other uridines, positioned at −3/−4, −11/−12 nt downstream of the cleavage site are also required for its function (Figure 1A). We thus created an additional construct ‘mut124′, which has uridines at positions −3/−4, −11/−12 and −31/−32/−33 substituted with cytidines (Figure 1A). This plasmid was transiently transfected into HEK293 cells and total RNA analysed by RNAse protection (RP) using an antisense riboprobe complementary to sequences overlapping the MC1R poly(A) site. Changing the uridines at these three positions had only a marginal effect on the cleavage at the MC1R poly(A) site (Figure 1B: lane mut124). In contrast, when both or a single uridine at positions −24/−25 are substituted, cleavage and polyadenylation efficiency is significantly reduced (Figure 1B: lanes mut3 and mut3*) confirming that the MC1R DSE surprisingly, only requires a di-uridine. We next analysed whether the UU needs to be positioned at a precise distance between both the hexamer and GB1 to be functional. To address this, we mutated the sequences surrounding the −24/−25 uridines altering the distance to the cleavage site and the downstream GB1. Increasing the distance between the UU and GB1 with the insertion of a 6-nt long spacer, severely reduced cleavage efficiency (Figure 1C: lane −24/−15). Cleavage levels were not affected if the distance between the UU and GB1 in the spacing construct is reverted to 9 nt by deleting the 6 nt adjacent to the UU (Figure 1C: lane −24/−9*). Remarkably, if a di-uridine is positioned 22 nt downstream of the hexamer and 8 nt upstream of GB1, cleavage was reduced by almost 3-fold (Figure 1C: lane −22/−8). These results indicate that the MC1R poly(A) site has an unusual DSE in which a di-uridine must be at a precise distance from both the hexamer and GB1.
Figure 1.
The MC1R poly(A) site has a complex sequence arrangement and is inefficient. (A) Outline of the MC1R parental minigene plasmid. The origin of sequences are indicated above the graph. CMV represents the CMV promoter, GFP represents the green fluorescence protein open reading frame and pA indicates the poly(A) site. The wild-type sequence of the MC1R PAS including the AATAAA hexamer, DSE and GB1 are indicated below the graph. The sequence alterations of mutant plasmids are shown below the wild-type sequence. The schematics of plasmids that have sequences between GB1 (G1, grey box) and GB2 (G2) fused are displayed below. GB2 has two distinct GRS a (white box) and b (white box with diagonal line). The position and length of the RP probe spanning the poly(A) site and the protected fragment lengths (RT, pA) are shown. (B–D) RP analysis of total RNA isolated from HEK293 cells transiently transfected with wild-type and mutant plasmids. Cleaved (pA) and non-cleaved readthrough (RT) protected fragments are indicated on the side of the gels. Quantitation of three independent transfections for each plasmid displayed as pA/RT ratio (rel. pA) with standard deviation indicated below each lane; the wt ratio is set to 100%. (E) RP comparing the levels of poly(A) readthrough (RT) and 3′-end processed transcripts (pA) between plasmids containing the MC4R 3′-UTR and poly(A) sequences (MC4R) and the MC1R wild-type minigene MC1R(wt) is shown.
As mentioned above, our earlier work also showed that the MC1R PAS requires two GRS elements which are separated by ∼400 nt. We speculated that GB2 may be brought into close proximity to the actual site of processing either by folding of the RNA sequence between GB1 and GB2 or by a complex interaction of multiple proteins that connect GB2 with the other elements. To test whether GB2 can function when positioned adjacent to the other cis-elements, we constructed three additional reporter constructs (Figure 1A, bottom graphs). In these plasmids the region between the two G-boxes was deleted and as a consequence GB1 is fused with either parts or the entire sequence of GB2. RP analysis of these constructs showed that GB2 can function when it is fused downstream of GB1 and that the entire sequence element of GB2 is required for optimal cleavage efficiency (Figure 1D).
From these results we conclude that the MC1R pre-mRNA contains an unusual PAS with rigid positional and structural arrangements of four individual sequence components, the hexamer, a di-uridine, GB1 and GB2. In addition, these data show that cleavage and polyadenylation at the MC1R 3′-end processing site is inefficient and results in significant amounts of uncleaved readthrough transcripts (RT; Figure 1B, C and D). This was particularly evident when poly(A) cleavage efficiency in the MC1R plasmid was compared to a similar reporter construct that contains the melanocortin 4 receptor (MC4R) 3′-UTR and 3′-flanking sequences including the PAS (24). In contrast to MC1R transfected cells, no readthrough can be detected by RP of total RNA transfected with the plasmid containing the MC4R PAS (Figure 1E).
MC1R transcripts that are not cleaved at the poly(A) site can create chimeric MC1R TUBB3 transcripts
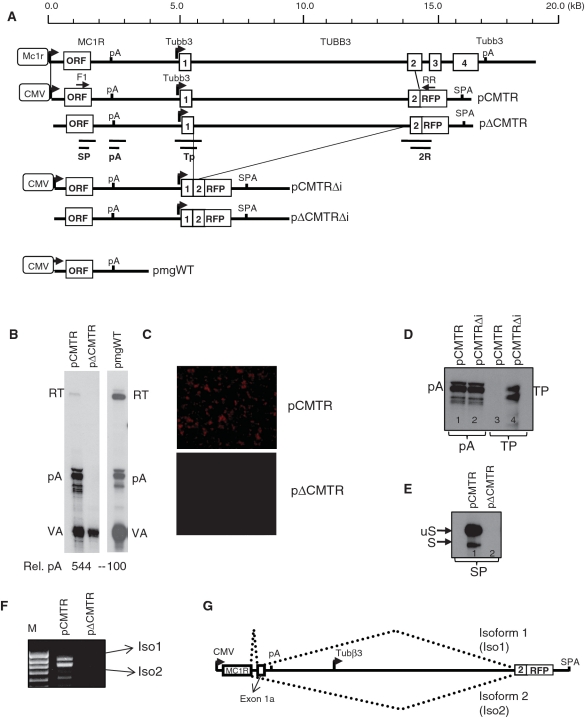
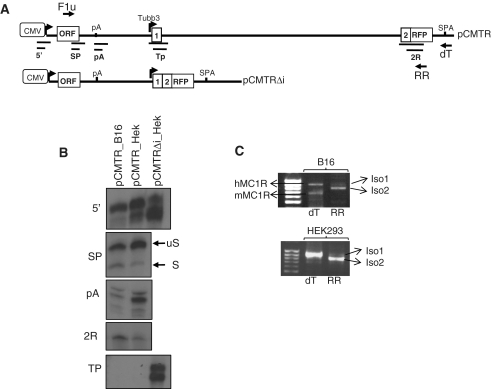
The above described analysis revealed the complex architecture of the 3′-end processing and showed that cleavage and polyadenylation at the MC1R poly(A) site is extremely inefficient. These observation indicated that either the complex MC1R PAS is suboptimal or that additional sequences, located downstream of GB2 (thus absent in the minigenes), afford efficient poly(A) cleavage in a genomic context. Consequently we created a new reporter gene (pCMTR) that contains the entire MC1R locus with 14 kb of sequence 3′ of the MC1R poly(A) site including exon1, intron1 and exon2 of the downstream positioned neuronally restricted TUBB3 gene (Figure 2A). The TUBB3 exon2 in this construct is fused in frame to the open reading frame (ORF) of the red fluorescent protein (RFP) located upstream of a synthetic poly(A) site (SPA) (Figure 2A). A second construct pΔCMTR, identical to pCMTR lacking the CMV promoter that drives transcription of the MC1R gene in the former construct was built. These plasmids not only enabled us to address whether additional downstream sequences are required for optimal cleavage but also to investigate how readthrough transcription at the MC1R poly(A) site affects the TUBB3 transcription locus. The latter is important since inefficient 3′-end processing and subsequent failure to terminate the transcribing RNA polymerase II (polII) can affect the expression of closely positioned downstream genes by transcriptional interference (25).
Figure 2.
Detection of chimeric transcripts representing mRNAs inter-spliced between MC1R and TUBB3. (A) Genomic map of the human MC1R-TUBB3 locus: MC1R promoter (MC1R open box with arrow), MC1R ORF (954 nt), the poly(A) site (pA), the TUBB3 promoter (Tubb3 arrow), exons 1, 2, 3, 4 (numbered open boxes) and the TUBB3 poly(A) site (Tubb3 pA) are depicted. Vertical lines indicate the 15-kb fragment cloned into the expression vectors pCMTR and pΔCMTR. Open box with ‘CMV’ represents CMV promoter driving transcription in the reporter plasmids. In frame fusion of exon 2 and the RFP ORF and insertion of the synthetic poly(A) site (SPA) are shown. The solid thick black line represents 5′-UTR, 3′-UTR, intergenic and intron sequences respectively. Deletions of intronic sequences resulting in pCMTRΔi and pΔCMTRΔi are indicated by thin lines. Position of PCR primers (F1 and RR) and the protected RP fragments for probes (SP), (pA), (Tp) and (2R) are depicted below the pΔCMTR construct. For comparison, the structure of the previously employed minigene pmgWT is indicated. (B) RP using the pA RP probe analysing levels of readthrough transcription (RT) isolated from HEK293 cells transfected with pCMTR and pΔCMTR are shown in the left panel. A representative RP showing readthrough observed with pmgWT is shown in the right panel. A single quantitation as a ratio pA/RT of RNA are shown. VA indicates RP against co-transfectional control, the viral polIII VA-transcript. (C) Fluorescent microscopy analysis of HEK293 cells transfected with pCMTR and pΔCMTR. (D) Deletion of the TUBB3 intron activates the TUBB3 promoter. Note that the two bands visible most likely represent more than one transcription initiation site as the same bands are visible in a CMV minus construct (data not shown). (E) RP analysis of total RNA isolated from HEK293 cells transiently transfected with pCMTR and pΔCMTR using the SP protection probe. (F) RT–PCR of total RNA isolated from HEK293 cells transiently transfected with pCMTR and pΔCMTR using oligo dT for RT and F1 and RR primers for PCR. (G) Graph of the splicing pattern resulting in the two chimeric transcripts Iso1 and Iso2.
RP analysis of RNA isolated from transfected HEK293 cells shows that significantly less uncleaved pre-mRNA is detected with the pCMTR construct than was previously observed with the minigenes (Figure 2B). This suggests that either additional sequences positioned downstream of GB2 may be essential in directing efficient cleavage and polyadenylation at the MC1R poly(A) site or that the readthrough transcripts from the pCMTR construct are more efficiently turned over compared to the uncleaved RNAs from the minigenes. However, as readthrough transcripts could still be detected in pCMTR transfected cells, we proceeded to analyse whether this affects the downstream positioned TUBB3 gene. We therefore compared RFP expression in cells transfected either with pCMTR or pΔCMTR. Surprisingly, cells transfected with the pCMTR constructs clearly expressed RFP but no fluorescence could be detected in cells transfected with the plasmid lacking the CMV promoter (Figure 2C). This indicates that the neuronally restricted TUBB3 promoter is inactive on the pΔCMTR plasmid when transfected into HEK293 cells. To verify whether potential readthrough at the MC1R poly(A) site somehow renders the neuronally restricted TUBB3 promoter active in the pCMTR context, we used an RP probe spanning the annotated TUBB3 promoter and flanking sequences (Figure 2A: TP). RP analysis of total RNA isolated from cells transfected with pCMTR indicated that the TUBB3 promoter is inactive (Figure 2D, lane 3). In contrast, a protected band representing transcription from the TUBB3 promoter is clearly visible when a construct (pCMTRΔi) lacking the 9.1-kb long first intron is transfected (Figure 2D, lane 4). This not only validated our finding that the TUBB3 promoter in the pCMTR is inactive but also showed that the TUBB3 intron1 is likely to play a role in the inactivation of its promoter on the plasmid and we are currently further investigating this phenomenon. RP analysis using a probe against the MC1R poly(A) site confirms that the RNA isolated from the pCMTR transfected cells is intact and that the failure to detect a band corresponding to transcription from the TUBB3 promoter is not due to degradation of the RNA in this sample or transfection efficiency (Figure 2D: lanes 1 and 2).
As no transcription from the TUBB3 promoter could be detected by RP in pCMTR, we speculated that the expression of RFP could be the result of a complex splicing reaction creating a transcript that originates from the CMV promoter and contains the TUBB3/RFP ORF. This is plausible since the inefficient MC1R PAS may result in the formation of a large fusion primary transcript containing the entire MC1R and TUBB3/RFP gene. Splicing of this pre-mRNA could then result in an mRNA capable of expressing RFP. To test this possibility we used an RP approach to identify potential 5′-splice site (5′-SS) located in the MC1R gene. We first used an RP probe spanning the entire 5′-UTR and analysed RNA isolated from pCMTR transfected cells but failed to detected any protected bands indicative of the presence of a 5′-SS (data not shown). In contrast, when we employed an RP probe complementary to sequences overlapping the MC1R ORF and 3′-UTR (Figure 2A: SP), we detected, in addition to the expected unspliced MC1R mRNA (uS), a shorter band (S) which could represent a spliced mRNA (Figure 2E). Interestingly, a minor spliced isoform of MC1R has been isolated from human testis (26). This isoform is created by a splicing event that uses a 5′-SS located at the end of the MC1R ORF and a 3′-splice site (3′-SS) in the MC1R 3′-UTR and then terminates at the MC1R PAS. This mRNA encodes an MC1R receptor with a short additional C-terminal extension.
The RP results allowed us to design specific RT–PCR primers to test whether RFP expression in pCMTR transfected cells is indeed the result of a splicing event. We therefore PCR amplified oligo dT primed cDNA of RNA isolated from pCMTR transfected HEK293 cells with a forward primer located in the MC1R ORF and a reverse primer complementary to sequences in the RFP ORF (Figure 2A: F1, RR). This PCR reactions resulted in two specific products, isoform 1 and 2 (Figure 2F). Subsequent re-amplification and sequencing of the two products confirmed two alternatively spliced chimeric MC1R-RFP mRNAs using the above described 5′-SS. Interestingly, isoform 1 is the result of two splicing events fusing the MC1R ORF to an exon, 1a, located in the MC1R 3′-UTR which is then joined in frame with exon2/RFP ORF (Figure 2G). Notably, the 3′-SS of exon 1a is identical to the above described isoform reported by (26). This transcript encodes an in frame MC1R-RFP protein explaining why pCMTR transfected cells are fluorescent. The second isoform is the result of a single splicing event fusing the 5′-SS to exon2/RFP creating a chimeric transcript where the RFP ORF is out of frame.
The above described analysis suggested that the MC1R poly(A) site may have evolved to be inefficient to allow a significant number of polymerases to read through the 3′-end processing site enabling the formation of alternatively spliced MC1R-TUBB3 chimeric transcripts. Thus, the reduction in readthrough RNAs in pCMTR transfected cells may not be due to the presence of additional auxiliary sequence elements that enhance the MC1R poly(A) site but instead, levels of readthrough RNAs are reduced because these transcripts are further processed and subjected to splicing.
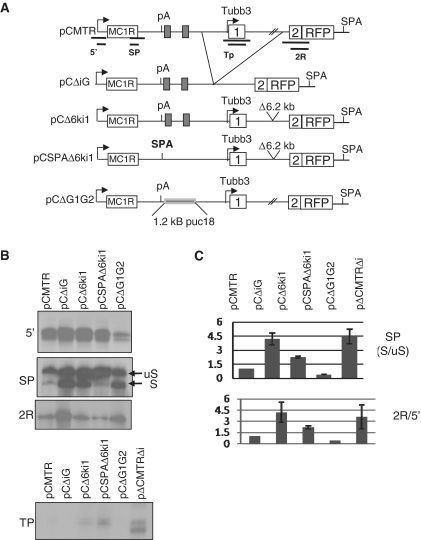
Modulating poly(A) site strength affects chimeric formation
The inefficient MC1R PAS may be a critical feature allowing alternative processing of transcripts in the MC1R TUBB3 locus. If this were true, artificial inactivation or strengthening of the MC1R poly(A) site should have opposing effects on the levels of chimeric transcripts that are produced. We therefore created four additional plasmids from the parental pCMTR construct (Figure 3A). First we deleted a fragment including the intergenic region (iG) and TUBB3 promoter, exon1 and most of intron1 resulting in the plasmid pCΔiG. This clone effectively brings the 3′-SS of the TUBB3 exon2 and the MC1R 5′-SS into close proximity. This novel sequence arrangement may favour splicing over cleavage and polyadenylation at the MC1R poly(A) site and tip the balance from MC1R transcripts towards MC1R-TUBB3 chimeric mRNAs. To simplify cloning of the replacement of the MC1R PAS with the strong synthetic SPA processing signal (23) a construct (pCΔ6ki1) was prepared where a 6.2-kb deletion of sequences in intron1 brings the TUBB3 3′-SS closer to the 5′-SS in the MC1R ORF. The MC1R AATAAA and the DSE in this plasmid was then replaced by SPA giving rise to the pCSPAΔ6ki1 construct. Finally, a fourth plasmid was built in which 1.2 kb of 3′-flanking sequences including GB1 to GB2 were replaced by 1.2 kb of sequence from pUC18. The MC1R 3′-end processing signal in this construct is similar to the earlier described H36 clone which dramatically reduces the cleavage efficiency of the MC1R PAS (16). The four plasmids were transiently transfected into HEK293 cells and quantitative effects on MC1R and MC1R-TUBB3 chimeric expression was assessed by RP using antisense probes complementary to sequences of the MC1R 5′-UTR (measuring total output), MC1R 5′-SS (measuring spliced versus unspliced mRNAs), TUBB3 promoter (measuring promoter activation) and RFP (measuring chimera production) (Figure 3A). The total transcriptional output from all four constructs was measured (Figure 3B: 5′ panel). Splicing at the MC1R 5′-SS dramatically increased in plasmids where the distance between the 5′-SS and the 3′-SS was reduced (Figure 3B: pCΔiG, pCΔ6ki1, panel SP). A similar increase in 5′-SS usage is also observed when the MC1R poly(A) site is inactivated by the replacement of the GB1 and GB2 by ‘neutral’ pUC18 sequences (Figure 3B: pCΔG1G2). In these constructs, the levels of RFP message was also significantly higher compared to transcripts analysed from pCMTR transfected cells (Figure 3B and C: 2R panel). The increase in RFP message in the pCΔiG and pCΔ6ki1 was not due to activation of the TUBB3 promoter which is evidenced by the lack of any protected bands when the TUBB3 promoter specific RP probe was employed (Figure 3B and C:TP panel). In contrast, when the inefficient MC1R poly(A) site was replaced by the strong synthetic SPA 3′-end processing signal, both splicing at the MC1R 5′-SS and the levels of RFP message were dramatically reduced compared to pCMTR and importantly its parental pCΔ6ki construct. This result suggests that the presence of an inefficient poly(A) site in the MC1R gene is essential in allowing the production of MC1R-TUBB3 chimeric transcripts. These results further imply that modulating the strength of the MC1R poly(A) site or the splice sites by potential trans-factors may be a process by which expression ratios between MC1R and the chimeric transcripts could be regulated.
Figure 3.
The complex arrangement of the MC1R PAS is critical for enabling intersplicing. (A) Schematic of the parental pCMTR plasmid and derived mutant constructs. The positions of RP probes used in the RP analysis are indicated below the pCMTR graph. The regions deleted in the various mutant clones are indicated by the branching lines connecting pCMTR and pCΔiG. The location of the 6.2-kb deleted in pCΔ6ki1 and pCSPAΔ6ki1 are shown. G-boxes are represented by filled boxes. The position of the substitution of the sequences encompassing G-box 1 (GB1) and 2 by 1.2 kb of ‘neutral’ pUC18 sequence is indicated in the pCΔG1G2 plasmid. (B) RPs of total RNA isolated from cells transfected with plasmids as indicated in each lane. The probes used in each RP are depicted on the side of the gels, note SP results in two fragments a spliced transcript (S) and a non-spliced transcript (uS). (C) Quantitation of three independent experiments for each probe. Quantitation of splicing efficiency is presented as the ratio between S/uS, the values of TP and 2R were normalized to the total amount of transcripts originating from the CMV promoter measured with the 5′-RP probe (2R/5′).
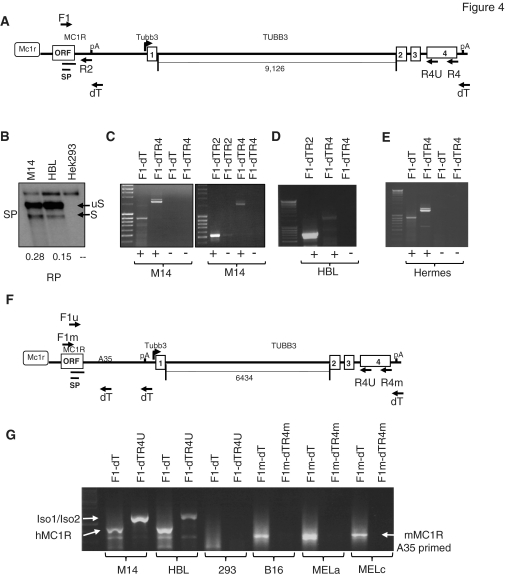
MC1R-TUBB3 chimeric transcripts are far more abundant in human than in mouse melanocytes
To establish the significance of the results described above, we investigated whether similar endogenous transcripts, as obtained by transfecting pCMTR into MC1R negative HEK293 cells, are expressed in mammalian melanocytes. We therefore isolated total RNA from two well-established human melanoma derived cell lines: M14 and HBL and performed an RP analysis with the above described antisense probe complementary to the region encompassing the MC1R 5′-SS. In this experiment, endogenous spliced and unspliced MC1R transcripts could be detected by RP in both melanoma derived M14 and HBL RNA but were absent in untransfected HEK293 control RNA (Figure 4B). This confirms that the 5′-SS identified in the MC1R ORF, is also used in a significant amount of endogenous MC1R transcripts. To verify if splicing of endogenous MC1R transcripts produces MC1R-TUBB3 chimera as detected in HEK293 cells transfected with pCMTR, we subjected total RNA isolated from both cell lines to RT–PCR analysis. RNA was reverse transcribed using oligo dT and cDNAs were subsequently amplified by PCR employing gene specific primers capable of amplifying MC1R and potential fusion transcripts as indicated in Figure 4A. In both melanoma derived cell lines M14 and HBL cells as well as in the immortalized melanocyte cell line HERMES-1 (22), MC1R and two chimeric transcript isoforms can be detected by RT–PCR (Figure 4C, D and E). The bands representing the two isoforms were gel isolated, re-amplified and sequenced. Sequencing confirmed that isoform 1 contains the ORF of the MC1R coding region fused to exon 1a located in the MC1R 3′-UTR which is joined to exon 2, 3 and 4 of the TUBB3 gene. This mRNA contains an uninterrupted ORF encoding a 797 amino acid long seven transmembrane MC1R receptor protein with a C-terminal TUBB3 extension (Supplementary Figure S1). The second isoform is a chimeric transcript that is identical to isoform 1 but skips the exon 1a in the MC1R 3′-UTR (Supplementary Figure S2). The consequence of this exon skipping is an mRNA with a shorter 432 amino acid long ORF that encodes for an MC1R receptor protein with a C-terminal extension that is out of frame with the human TUBB3 ORF. However, the 115 amino acid C-terminal extension of the MC1R receptor in isoform 2, in a ‘Blast’ search, showed some similarities with the chicken TUBB3 protein. From these results we concluded that human melanocytes express at least three MC1R isoform mRNAs, encoding the MC1R receptor and two additional receptors that contain C-terminal extensions of TUBB3.
Figure 4.
Detection of chimeric MC1R-TUBB3 transcripts in human and mouse melanocytes. (A) Diagram of the genomic human MC1R-TUBB3 context. The location of primers used in RT–PCR reactions and the RP probe SP are depicted. The length of the TUBB3 intron 1 is indicated below the main graph. (B) RP using the SP probe showing that 5′-splice site in the MC1R ORF is used in endogenous M14, HBL melanocytes. Numbers shown below the image are S/uS ratios derived from quantitation of each band in the gel. (C–D) RT–PCR of total RNA isolated from M14 and HBL melanoma derived melanocytes. Forward and reverse primers used in each PCR reaction are indicated for each lane on top of gels. The reverse primer names reflect the order in which they were used for RT and subsequent PCR. Note that 2R represents a gene specific reverse PCR primer complementary to sequences in the MC1R 3′-UTR resulting in a more discrete MC1R band compared to the adapter reverse primer ‘dT’ which is complementary to sequences at the 3′-end of the oligo dT primer used in the RT step. This also allows a more direct comparison between MC1R and chimeric transcript levels. (E) RT–PCR of total RNA isolated from the transformed melanocytes HERMES-1. +/− for all gels indicates presence or absence of reverse transcriptase in each reaction. (F) Diagram of the genomic mouse MC1R-TUBB3 locus. Location of primers used in the RT–PCR reactions are indicated and the distance of TUBB3 intron 1 is specified below the graph. A35 represents an annotated 35-nt long A stretch in the MC1R 3′-UTR which causes aberrant priming with the dT primer in the reverse transcription step. (G) RT–PCR analysis comparing MC1R and MC1R-TUBB3 chimeric expression in human (M14, HBL) melanocytes and mouse (B16, MELa, MELc) melanocytes. Iso1/Iso2 indicates the MC1R-TUBB3 chimera bands, MC1R indicates the RT–PCR amplified MC1R mRNA.
To verify whether chimeric transcripts were also expressed in melanocytes from furred animals we analysed total RNA isolated from B16, pigmented Melan-a melanocytes and the albino Melan-c melanocyte cell line. The MC1R and TUBB3 coding sequences between mouse and human are highly conserved and as a consequence, a 5′-SS with sequences identical to the human equivalent is also present in the mouse MC1R ORF. Surprisingly though, despite being able to easily amplify MC1R from all mouse derived cell lines (Figure 4G: F1mdT: B16, Melan-a, Melan-c) we were unable to detect any transcripts representing MC1R-TUBB3 fusion proteins. Notably, we unsuccessfully used multiple sets of reverse PCR primers complementary to sequences in the mouse TUBB3 exon 4. This included two PCR primers (Figure 4F: R4U and F1U) that are complementary to regions in MC1R and TUBB3 that are identical in sequence between mouse and humans. Although this primer pair was able to amplify human chimeric transcripts no such mRNAs were detected with RNA isolated from mouse melanocytes (data not shown). Furthermore, at best, only trace amounts of a band potentially representing chimeric transcripts could be detected from mouse total RNA compared to human even when we used a gene specific reverse primer R4U (Supplementary Figure S3). We therefore conclude that unlike human melanocytes, mouse derived melanocytes do not express significant amounts of detectable MC1R-TUBB3 chimeric mRNAs possibly due to the different poly(A) arrangement in the mouse MC1R gene (Supplementary Figure S4).
Mouse melanocytes can create MC1R-TUBB3 chimeric transcripts when a plasmid containing the human sequence is transfected
As shown above, mouse melanocytes appear to be unable to produce MC1R-TUBB3 significant amounts of chimeric transcripts. This was surprising given that the mouse TUBB3 intron 1 is significantly shorter than its human counterpart positioning the 5′-SS in the mouse ORF closer to the TUBB3 exon 2 3′-SS. According to the results presented above, this should favour splicing between MC1R and TUBB3 exons (Figure 3B). Two reasons could account for this observation: (i) mouse melanocytes either lack a potential trans-factor, or (ii) the sequences present in the mouse MC1R-TUBB3 locus do not support the alternative splicing event. To clarify this, we transfected the pCMTR plasmid into B16 mouse melanocytes and isolated RNA was subsequently analysed by RP using multiple RP probes (Figure 5A). Mouse melanocytes recognize the human MC1R poly(A) site less efficiently and show a higher degree of spliced versus unspliced transcripts relative to HEK293 cells. The increased level of splicing from the MC1R 5′-SS is mirrored by a similar increase in TUBB3 exon2-RFP containing RNA indicative of efficient intersplicing between MC1R and TUBB3 (Figure 5B). Importantly, this increase is not due to transcription from the TUBB3 promoter as is evidenced by RP (Figure 5B: TP panel) and the formation of chimeric transcripts was subsequently confirmed by RT–PCR (Figure 5C). Thus these results indicate that the mouse melanocytes are able to produce chimeric transcripts in the correct sequence environment. The lack of endogenous MC1R-TUBB3 chimeric transcripts in mouse melanocytes is therefore likely to be due to sequence constraints in the mouse rather than due to the lack of a specific trans-factor(s).
Figure 5.
Mouse melanocytes can generate MC1R-TUBB3 chimera from a plasmid bearing the human sequences. (A) Diagram of pCMTR and pCMTRΔi indicating the location of the RP probes used in the RP analysis presented. (B) RP of total RNA isolated from B16 mouse melanocytes transfected with pCMTR and pCMTRΔi using anti-sense riboprobes measuring total output (5′), MC1R 5′-SS usage (SP), MC1R poly(A) usage (pA), TUBB3 exon2/RFP containing transcripts (2R) and TUBB3 promoter activity (TP). (C) RT–PCR detecting MC1R-RFP/TUBB3 chimera in B16 mouse cells transfected with pCMTR, primers used are as described in Figure 2. Note, due to use of the universal forward primer (F1u) in the RT–PCR reaction, in B16 both endogenous mouse (mMC1R) and transfected human MC1R (hMC1R) are detected.
The relative levels of MC1R and MC1R-TUBB3 chimeric transcripts can be modulated by α-MSH and MKK6
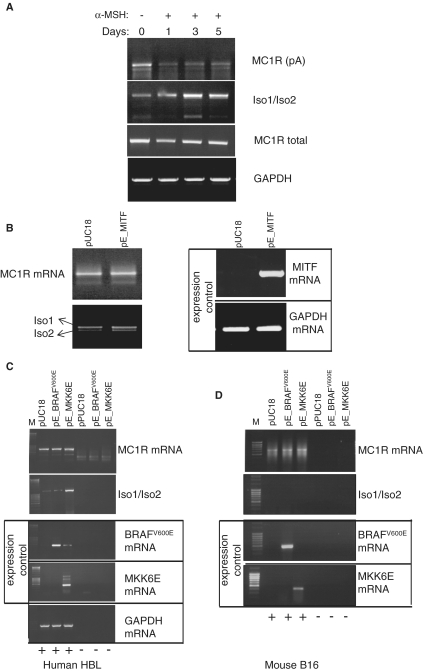
Our results presented above clearly show that human melanocytes express endogenous MC1R-TUBB3 chimeric transcripts, but it was unclear whether the expression of these alternatively processed isoforms can be related to facultative pigmentation in humans. When skin is exposed to solar radiation, the major signalling molecule α-MSH is secreted by keratinocytes. α-MSH binds MC1R and elevates intracellular cAMP levels in melanocytes activating expression of the microphthalmia-associated transcription factor (MITF) that controls the expression of multiple genes involved in melanogenesis (9). As α-MSH has been proposed to affect MC1R expression (12), we next tested whether exposure of human melanocytes to α-MSH has any impact on the expression patterns of MC1R and MC1R-TUBB3 chimeric transcripts. In this analysis we exposed human HBL melanoma derived melanocytes for 0, 1, 3 or 5 days to 10 nM α-MSH and subsequently monitored the expression pattern of both MC1R and the chimeric transcripts by RT–PCR. Exposure of the melanocytes to α-MSH for 3–5 days notably changed the levels of both MC1R and chimeric transcripts. Whereas the levels of MC1R transcripts dropped after exposure to α-MSH, the chimeric transcript levels dramatically increased over the same period (Figure 6A). A similar result was obtained when the melanocyte derived HERMES cell line was exposed to prolonged α-MSH treatment (Supplementary Figure S5). Interestingly, when a PCR was performed using the same HBL derived cDNA and a forward primer in the 5′-UTR and a reverse primer in MC1R ORF (detecting the total output MC1R and MC1R-TUBB3 transcripts) no significant change in transcript levels was observed between the different time points (Figure 6A: MC1R 5′-ORF). This implies that the overall output is not affected. Taken together, these results indicate that exposure of human melanocytes to α-MSH shifts the expression from predominantly MC1R, in the resting stage, to MC1R-TUBB3 chimera in the activated stage.
Figure 6.
Exposure of human melanocytes to α-MSH inversely affects expression levels of MC1R and MC1R-TUBB3 chimeric transcripts. (A) α-MSH treatment (10 nM) of HBL human melanoma cells results in a gradual increase in MC1R-TUBB3 chimera and reduced MC1R mRNA levels. RT–PCRs targeting all the above described transcripts originating from the MC1R locus were performed using total RNA isolated from HBL cells exposed to 10 nM α-MSH for 0, 1, 3 and 5 days, respectively (lanes 1–4) as indicated above the gel. The MC1R mRNA specific PCR is labelled: MC1R (pA) using PCR primers F1-dT, MC1R-TUBB3 chimera transcripts are denoted by (Iso1/Iso2) using PCR primers F1-R4, total transcripts from the MC1R locus (MC1R + MC1R-TUBB3) are labelled as (MC1R total) using PCR primers located in the 5′-UTR and the 5′-end of the MC1R ORF and GAPDH represents an additional control. (B) RT–PCR analysis of total RNA isolated from HBL cells transfected with a control pUC18 plasmid and a construct over expressing MITF (pE_MITF). Left panel showing results using primers amplifying MC1R mRNA and MC1R-TUBB3 chimera (Iso1/Iso2). Right panel, control RT–PCRs confirming over expression of MITF mRNA and GAPDH mRNA levels. (C) RT–PCR analysis of transfected HBL cells over expressing a constitutively active BRAF kinase (pE_BRAFV600E) or over expressing the constitutively active p38 specific MKK6 kinase (pE_MKK6E). RT–PCR results for MC1R specific primers (MC1R) (first panel) and chimera specific primers (Iso1/Iso2) (second panel) are shown. Control RT–PCRs confirming over expression of BRAF or MKK6 respectively are indicated by ‘expression control’ panel. A general control measuring unrelated GAPDH mRNA levels is presented (bottom panel). (+/−) indicates the presence or absence of reverse transcriptase in the RT reactions. (D) Identical analysis as shown in (C), but using mouse B16 melanocytes indicating that over expression of MKK6E does not result in the production of chimeric transcripts in mouse melanocytes.
Exposure of melanocytes to α-MSH not only regulates MITF expression but also activates the RAS-BRAF-ERK (27)-signalling cascade and stimulates the p38 MAPK-signalling pathway (28). In order to dissect which of the pathways stimulated by α-MSH may account for the observed ratio change of MC1R and MC1R-TUBB3 chimeras, we over expressed MITF (Figure 6B), constitutively active BRAFV600E (Figure 6C) and the constitutively active MKK6 mutant MKK6E (Figure 6C). Although all the mRNAs of the three factors could be over expressed in HBL melanocytes (Figure 6B and C: expression controls) only over expression of MKK6E had a significant impact on MC1R isoform expression. Over expression of MKK6E resulted in a dramatic increase in the chimeric isoforms but left MC1R levels unaffected (Figure 6C: Iso1/Iso2, lane: pE_MKK6E).
Since transfection of MKK6E dramatically increased MC1R-TUBB3 chimera formation in human melanocytes, we next tested whether we could force mouse melanocytes to create chimeric transcripts by over expressing MKK6E. As can be seen in Figure 6D, over expression of BRAFV600E or MKK6E failed to trigger chimera production in mouse melanocytes, further confirming that the mouse MC1R-TUBB3 locus somehow does not allow inter-splicing.
We thus conclude that the expression of chimeric MC1R-TUBB3 transcripts in human melanocytes is regulated in response to α-MSH and MC1R chimera formation is increased upon the activation of the p38-signalling pathway.
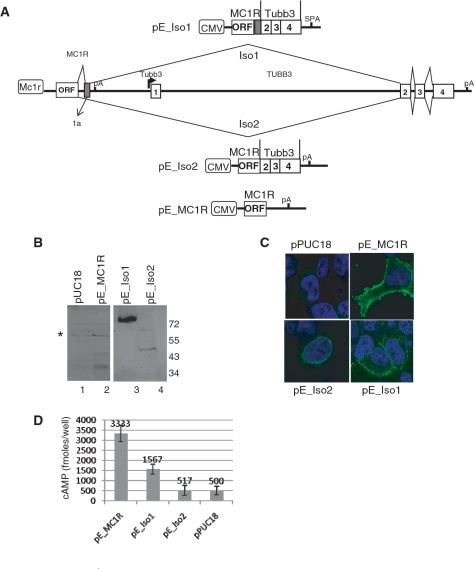
The two chimeric transcripts produce viable proteins and over expressed isoform 1 can be detected in the cell membrane
We have demonstrated that alternatively spliced mRNA isoforms containing MC1R and TUBB3 sequences are present in human melanocytes. However, we needed to clarify whether these transcripts produce detectable chimeric proteins and if so, to identify their subcellular localization. To that end, we cloned three expression plasmids encoding MC1R, the MC1R-TUBB3 fusion protein isoforms 1 (Iso1) and 2 (Iso2) (Figure 7A). Western blot analysis confirmed that MC1R and both chimeric proteins are produced in transiently transfected HEK293 cells (Figure 7B). Interestingly, the chimeric protein isoform 1 was consistently detected at much higher levels than either MC1R or isoform 2. Importantly, by using a MC1R specific antibody and confocal microscopy, we show that both MC1R and MC1R-TUBB3 isoform 1 are localized in the cell membrane of transfected HEK293 cells (Figure 7C). This is in stark contrast to the chimeric protein isoform 2, which appears to be retained in the ER or nuclear membrane.
Figure 7.
MC1R-TUBB3 chimeric proteins are viable and isoform 1 but not 2 correctly localizes to the cell membrane. (A) Genomic organization of the MC1R-TUBB3 locus is shown and the splicing pattern for both MC1R-TUBB3 chimeric mRNAs Iso1 and Iso2 are indicated. The new exon 1a present in Iso1 is indicated in the 3′-UTR of the MC1R gene. Above and below the genomic graph constructs used to over express MC1R (pE_MC1R), the chimeric isoform 1 (pE_Iso1) or chimeric isoform 2 (pE_Iso2) are detailed. (B) Western blot using the N19 antibody against MC1R confirming over expression of the proteins in HEK293 cells. Asterisk represents unspecific bands detected by N19. (C) Immunofluorescence analysis by confocal microscopy of HEK293 cells transfected with the expression plasmids and the pUC18 control vector using the N19 anti MC1R antibody (green staining). Nuclei are stained blue with DAPI. (D) cAMP assay using transiently transfected HEK293 cells exposed to 10 nM α-MSH; n = 3.
We next tested whether the TUBB3 C-terminal extension changes the receptor subunit’s ability to respond to α-MSH and elevate intracellular cAMP levels. We therefore expressed MC1R, MC1R-TUBB3 isoform 1 and 2 in HEK293 cells and assayed cAMP levels after exposure of the transfected cells to α-MSH. Whilst MC1R-TUBB3 chimeric isoform 1 has a notably reduced activity, isoform 2 failed to elevate cAMP levels in HEK293 cells exposed to α-MSH (Figure 7D).
From these experiments we conclude that in our system the chimeric MC1R-TUBB3 protein isoforms have a significantly reduced capacity to elevate intracellular cAMP levels in response to α-MSH and may thus contribute to the desensitization of the melanocytes for α-MSH.
DISCUSSION
By subjecting pre-mRNAs to alternative splicing and alternative 3′-end processing eukaryotic cells are able to generate a variety of mRNA isoforms and also expand their proteome which dramatically increases their capacity to respond to cellular and environmental cues.
In recent years alternative splicing and alternative polyadenylation (29) have been studied extensively. Alternative 3′-end pre-mRNA processing to date has focussed almost entirely on the generation of alternative transcripts from one particular gene. The field paid little or no attention to reports which suggest that alternative cleavage and polyadenylation coupled with alternative splicing is not limited to one gene but may frequently occur between genes positioned in tandem (30,31). Although such intergenic alternative poly(A) events are relatively well documented by bioinformatic studies only a handful of examples have been characterized at the molecular level (32–36).
The results presented in this paper provide dramatic evidence that alternative splicing combined with alternative polyadenylation can increase the available proteome even further by combining exons from different genes. Thus, ‘inter-splicing’ between closely in-tandem arranged genes may play a much more important role in eukaryotic gene expression than was previously thought.
Alternative pre-mRNA processing within the MC1R locus demonstrates that an inefficient and/or kinetically slow 3′-end processing site (Figures 1–3) is a prerequisite for inter-splicing between genes. Therefore, the regulation of suboptimal PAS between closely spaced genes may play a critical role in the regulation of poly(A) cleavage, subsequent transcription termination of pol II and ultimately control chimera formation.
Interestingly, we show that expression levels of the MC1R and the MC1R-TUBB3 chimeric transcripts can be modulated by artificially activating signalling pathways that are stimulated in melanocytes when skin is exposed to solar radiation. Prolonged exposure to α-MSH reduces MC1R mRNA levels but increases chimeric transcript levels (Figure 6). α-MSH exposure results in elevation of cAMP levels in melanocytes triggering a multifaceted response that includes the known activation of the transcription factor MITF, stimulation of the RAS/BRAF/ERK kinase pathway and induces the p38 stress response pathway. The sum of this signalling cascade is the activation of a large number of genes that are required for melanogenesis. Surprisingly, over expression of MITF and the constitutively active BRAFV600E kinase had little effect on either the MC1R or chimeric transcript levels. In contrast, expression of MKK6E, a kinase specifically associated with the p38 pathway, significantly changes the levels of chimeric transcripts but had no impact on MC1R mRNA levels. This suggests that the increase in chimeric mRNAs observed when cells were exposed to α-MSH can at least in part be explained by the activation of the p38 pathway. Modulation of chimeric transcript levels may be achieved at multiple steps including mRNA stability, regulation of MC1R poly(A) site efficiency and activation of alternative splicing. Intriguingly, activation of the p38 pathway has previously been shown to affect transcript stability, albeit mostly of mRNAs harbouring specific destabilizing A/U rich elements in their 3′-UTRs that are not obvious in the chimeric transcript (37,38). However, the reduction in MC1R transcripts observed when melanocytes are exposed to α-MSH must be the consequence of the activation of an additional unknown factor or pathway that triggers destabilization of the MC1R mRNA or increases intersplicing possibly by modulation the strength of the MC1R poly(A) site.
As mentioned above, expression levels of both MC1R and the chimeric transcripts are modulated by kinase pathways that are normally activated in human skin in response to the exposure to solar radiation. This finding makes it very likely that the chimeric receptors play an important role in the facultative pigmentation system in humans. The fact that both MC1R-TUBB3 chimeric receptors have a significantly reduced capacity to elevate intracellular cAMP levels upon exposure to α-MSH are important observations for their possible function (Figure 7). The reduction of MC1R transcripts and the concomitant increase of the chimeric mRNAs after a prolonged exposure to high levels of α-MSH may effectively desensitizes the melanocytes to α-MSH by forcing a switch in receptor isoform expression. MC1R is rapidly desensitized in the classical way by G protein-coupled receptor kinases and possibly subsequent internalization (39). However, a switch in receptor isoform expression would ensure that the MC1R receptor pool would not be replenished during the prolonged exposure of activated melanocytes to α-MSH and desensitize melanocytes once they are fully activated. This may prevent overstimulation of melanocytes during prolonged exposure of the skin to solar radiation. The importance of a complex α-MSH desensitization mechanism in human melanocytes may be a prerequisite of UVR induced skin pigmentation and thus explain why chimeric protein receptors are absent in mouse melanocytes.
However, the function of the chimeric MC1R-TUBB3 proteins may not be limited to a role in the desensitization process. In particular, given that MC1R has been shown to dimerize (40), heterodimerization between MC1R and either of the chimera or the chimeras themselves may result in alternative signalling capacities and events.
Finally the chimeric proteins themselves represent an intriguing link between a signalling receptor and a component of the cytoskeleton. This may be of particular importance as morphological changes, including the formation of dendrites during melanogenesis, is an essential process to afford UVR protection of human skin by tanning. Interestingly, TUBB3 is known to affect α/β tubulin dynamics and has been implicated in neuronal dendrite formation (41). We therefore propose that the MC1R-TUBB3 chimeric receptor may play a role in the rearrangement of the cytoskeleton that is required to initiate, propagate and guide the dendrites of activated melanocytes along an α-MSH gradient towards the keratinocytes. A critical role of the chimera during dendrite formation may also explain why the MC1R TUBB3 locus in mouse is unable to direct intersplicing between MC1R and TUBB3 (Figure 4) as tanning is not a necessary adaptation in furred skin.
SUPPLEMENTARY DATA
Supplementary data are available at NAR online.
FUNDING
Wellcome Trust (081083/Z/06/Z); E.P.A. Cephalosporin Fund (C069); Skaggs Scholarship TSRI (to M.K.); Pembroke College, BTP fellowship (to M.D.). Funding for open access charge: Wellcome Trust (081083/Z/06/Z).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank all the members of the Furger, Murphy and Proudfoot lab for valuable discussions. The author thank Nick Proudfoot for the critical reading of the article. The authors thank Nuno Nunes for technical assistance.
REFERENCES
- 1.Getting SJ. Targeting melanocortin receptors as potential novel therapeutics. Pharmacol. Therapeut. 2006;111:1–15. doi: 10.1016/j.pharmthera.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 2.Lin JY, Fisher DE. Melanocyte biology and skin pigmentation. Nature. 2007;445:843. doi: 10.1038/nature05660. [DOI] [PubMed] [Google Scholar]
- 3.García-Borrón JC, Sánchez-Laorden BL, Jiménez-Cervantes C. Melanocortin-1 receptor structure and functional regulation. Pigment Cell Res. 2005;18:393–410. doi: 10.1111/j.1600-0749.2005.00278.x. [DOI] [PubMed] [Google Scholar]
- 4.Lalueza-Fox C, Rompler H, Caramelli D, Staubert C, Catalano G, Hughes D, Rohland N, Pilli E, Longo L, Condemi S, et al. A melanocortin 1 receptor allele suggests varying pigmentation among Neanderthals. Science. 2007;318:1453–1455. doi: 10.1126/science.1147417. [DOI] [PubMed] [Google Scholar]
- 5.Rees JL. The genetics of sun sensitivity in humans. Am. J. Hum. Genet. 2004;75:739–751. doi: 10.1086/425285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Jakobsdottir M, Steinberg S, Gudjonsson SA, Palsson A, Thorleifsson G, et al. Two newly identified genetic determinants of pigmentation in Europeans. Nat. Genet. 2008;40:835–837. doi: 10.1038/ng.160. [DOI] [PubMed] [Google Scholar]
- 7.Valverde P, Healy E, Jackson I, Rees JL, Thody AJ. Variants of the melanocyte-stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat. Genet. 1995;11:328–330. doi: 10.1038/ng1195-328. [DOI] [PubMed] [Google Scholar]
- 8.Bishop DT, Demenais F, Iles MM, Harland M, Taylor JC, Corda E, Randerson-Moor J, Aitken JF, Avril M-F, Azizi E, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat. Genet. 2009;41:920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Orazio JA, Nobuhisa T, Cui R, Arya M, Spry M, Wakamatsu K, Igras V, Kunisada T, Granter SR, Nishimura EK, et al. Topical drug rescue strategy and skin protection based on the role of Mc1r in UV-induced tanning. Nature. 2006;443:340–344. doi: 10.1038/nature05098. [DOI] [PubMed] [Google Scholar]
- 10.Mountjoy KG, Robbins LS, Mortrud MT, Cone RD. The cloning of a family of genes that encode the melanocortin receptors. Science. 1992;257:1248–1251. doi: 10.1126/science.1325670. [DOI] [PubMed] [Google Scholar]
- 11.April CS, Barsh GS. Distinct pigmentary and melanocortin 1 receptor-dependent components of cutaneous defense against ultraviolet radiation. PLoS Genet. 2007;3:e9. doi: 10.1371/journal.pgen.0030009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annu. Rev. Genet. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- 13.Hunt G, Donatien PD, Lunec J, Todd C, Kyne S, Thody AJ. Cultured human melanocytes respond to MSH peptides and ACTH. Pigment Cell Res. 1994;7:217–221. doi: 10.1111/j.1600-0749.1994.tb00052.x. [DOI] [PubMed] [Google Scholar]
- 14.Hunt G, Todd C, Cresswell JE, Thody AJ. Alpha-melanocyte stimulating hormone and its analogue Nle4DPhe7 alpha-MSH affect morphology, tyrosinase activity and melanogenesis in cultured human melanocytes. J. Cell. Sci. 1994;107(Pt 1):205–211. doi: 10.1242/jcs.107.1.205. [DOI] [PubMed] [Google Scholar]
- 15.Wakamatsu K, Graham A, Cook D, Thody AJ. Characterisation of ACTH peptides in human skin and their activation of the melanocortin-1 receptor. Pigment Cell Res. 1997;10:288–297. doi: 10.1111/j.1600-0749.1997.tb00688.x. [DOI] [PubMed] [Google Scholar]
- 16.Dalziel M, Nunes NM, Furger A. Two G-rich regulatory elements located adjacent to and 440 nucleotides downstream of the core poly(A) site of the intronless melanocortin receptor 1 gene are critical for efficient 3′ end processing. Mol. Cell. Biol. 2007;27:1568–1580. doi: 10.1128/MCB.01821-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2010;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Proudfoot N. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayr C, Bartel DP. Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sviderskaya EV, Gray-Schopfer VC, Hill SP, Smit NP, Evans-Whipp TJ, Bond J, Hill L, Bataille V, Peters G, Kipling D, et al. p16/cyclin-dependent kinase inhibitor 2A deficiency in human melanocyte senescence, apoptosis, and immortalization: possible implications for melanoma progression. J. Natl Cancer Inst. 2003;95:723–732. doi: 10.1093/jnci/95.10.723. [DOI] [PubMed] [Google Scholar]
- 23.Levitt N, Briggs D, Gil A, Proudfoot NJ. Definition of an efficient synthetic poly(A) site. Genes Dev. 1989;3:1019–1025. doi: 10.1101/gad.3.7.1019. [DOI] [PubMed] [Google Scholar]
- 24.Nunes NM, Li W, Tian B, Furger A. A functional human Poly(A) site requires only a potent DSE and an A-rich upstream sequence. EMBO J. 2010;29:1523–1536. doi: 10.1038/emboj.2010.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greger IH, Demarchi F, Giacca M, Proudfoot NJ. Transcriptional interference perturbs the binding of Sp1 to the HIV-1 promoter. Nucleic Acids Res. 1998;26:1294–1301. doi: 10.1093/nar/26.5.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan CP, McKee KK, Weinberg DH, MacNeil T, Palyha OC, Feighner SD, Hreniuk DL, Van Der Ploeg LH, MacNeil DJ, Howard AD. Molecular analysis of a new splice variant of the human melanocortin-1 receptor. FEBS Lett. 1999;451:137–141. doi: 10.1016/s0014-5793(99)00525-6. [DOI] [PubMed] [Google Scholar]
- 27.Busca R, Abbe P, Mantoux F, Aberdam E, Peyssonnaux C, Eychene A, Ortonne J-P, Ballotti R. Ras mediates the cAMP-dependent activation of extracellular signal-regulated kinases (ERKs) in melanocytes. EMBO J. 2000;19:2900–2910. doi: 10.1093/emboj/19.12.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smalley K, Eisen T. The involvement of p38 mitogen-activated protein kinase in the alpha-melanocyte stimulating hormone (alpha-MSH)-induced melanogenic and anti-proliferative effects in B16 murine melanoma cells. FEBS Lett. 2000;476:198–202. doi: 10.1016/s0014-5793(00)01726-9. [DOI] [PubMed] [Google Scholar]
- 29.Lutz CS. Alternative polyadenylation: a twist on mRNA 3′ end formation. ACS Chem. Biol. 2008;3:609–617. doi: 10.1021/cb800138w. [DOI] [PubMed] [Google Scholar]
- 30.Akiva P, Toporik A, Edelheit S, Peretz Y, Diber A, Shemesh R, Novik A, Sorek R. Transcription-mediated gene fusion in the human genome. Genome Res. 2006;16:30–36. doi: 10.1101/gr.4137606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parra G, Reymond A, Dabbouseh N, Dermitzakis ET, Castelo R, Thomson TM, Antonarakis SE, Guigó R. Tandem chimerism as a means to increase protein complexity in the human genome. Genome Res. 2006;16:37–44. doi: 10.1101/gr.4145906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poulin F, Brueschke A, Sonenberg N. Gene fusion and overlapping reading frames in the mammalian genes for 4E-BP3 and MASK. J. Biol. Chem. 2003;278:52290–52297. doi: 10.1074/jbc.M310761200. [DOI] [PubMed] [Google Scholar]
- 33.Magrangeas F, Pitiot G, Dubois S, Bragado-Nilsson E, Cherel M, Jobert S, Lebeau B, Boisteau O, Lethe B, Mallet J, et al. Cotranscription and intergenic splicing of human galactose-1-phosphate uridylyltransferase and interleukin-11 receptor alpha-chain genes generate a fusion mRNA in normal cells. Implication for the production of multidomain proteins during evolution. J. Biol. Chem. 1998;273:16005–16010. doi: 10.1074/jbc.273.26.16005. [DOI] [PubMed] [Google Scholar]
- 34.Pradet-Balade B, Medema JP, Lopez-Fraga M, Lozano JC, Kolfschoten GM, Picard A, Martinez AC, Garcia-Sanz JA, Hahne M. An endogenous hybrid mRNA encodes TWE-PRIL, a functional cell surface TWEAK-APRIL fusion protein. EMBO J. 2002;21:5711–5720. doi: 10.1093/emboj/cdf565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Communi D, Suarez-Huerta N, Dussossoy D, Savi P, Boeynaems J-M. Cotranscription and Intergenic Splicing of HumanP2Y 11 and SSF1 Genes. J. Biol. Chem. 2001;276:16561–16566. doi: 10.1074/jbc.M009609200. [DOI] [PubMed] [Google Scholar]
- 36.Maeda K, Horikoshi T, Nakashima E, Miyamoto Y, Mabuchi A, Ikegawa S. MATN and LAPTM are parts of larger transcription units produced by intergenic splicing: intergenic splicing may be a common phenomenon. DNA Res. 2005;12:365–372. doi: 10.1093/dnares/dsi017. [DOI] [PubMed] [Google Scholar]
- 37.Lasa M, Mahtani KR, Finch A, Brewer G, Saklatvala J, Clark AR. Regulation of cyclooxygenase 2 mRNA stability by the mitogen-activated protein kinase p38 signaling cascade. Mol. Cell. Biol. 2000;20:4265–4274. doi: 10.1128/mcb.20.12.4265-4274.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winzen R, Kracht M, Ritter B, Wilhelm A, Chen CY, Shyu AB, Muller M, Gaestel M, Resch K, Holtmann H. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 1999;18:4969–4980. doi: 10.1093/emboj/18.18.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez-Mas J, Guillo LA, Zanna P, Jimenez-Cervantes C, Garcia-Borron JC. Role of G protein-coupled receptor kinases in the homologous desensitization of the human and mouse Melanocortin 1 receptors. Mol. Endocrinol. 2005;19:1035–1048. doi: 10.1210/me.2004-0227. [DOI] [PubMed] [Google Scholar]
- 40.Sanchez-Laorden BL, Sanchez-Mas J, Martinez-Alonso E, Martinez-Menarguez JA, Garcia-Borron JC, Jimenez-Cervantes C. Dimerization of the human melanocortin 1 receptor: functional consequences and dominant-negative effects. J. Invest. Dermatol. 2006;126:172–181. doi: 10.1038/sj.jid.5700036. [DOI] [PubMed] [Google Scholar]
- 41.Katsetos CD, Herman MM, Mork SJ. Class III beta-tubulin in human development and cancer. Cell Motil Cytoskeleton. 2003;55:77–96. doi: 10.1002/cm.10116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.