Abstract
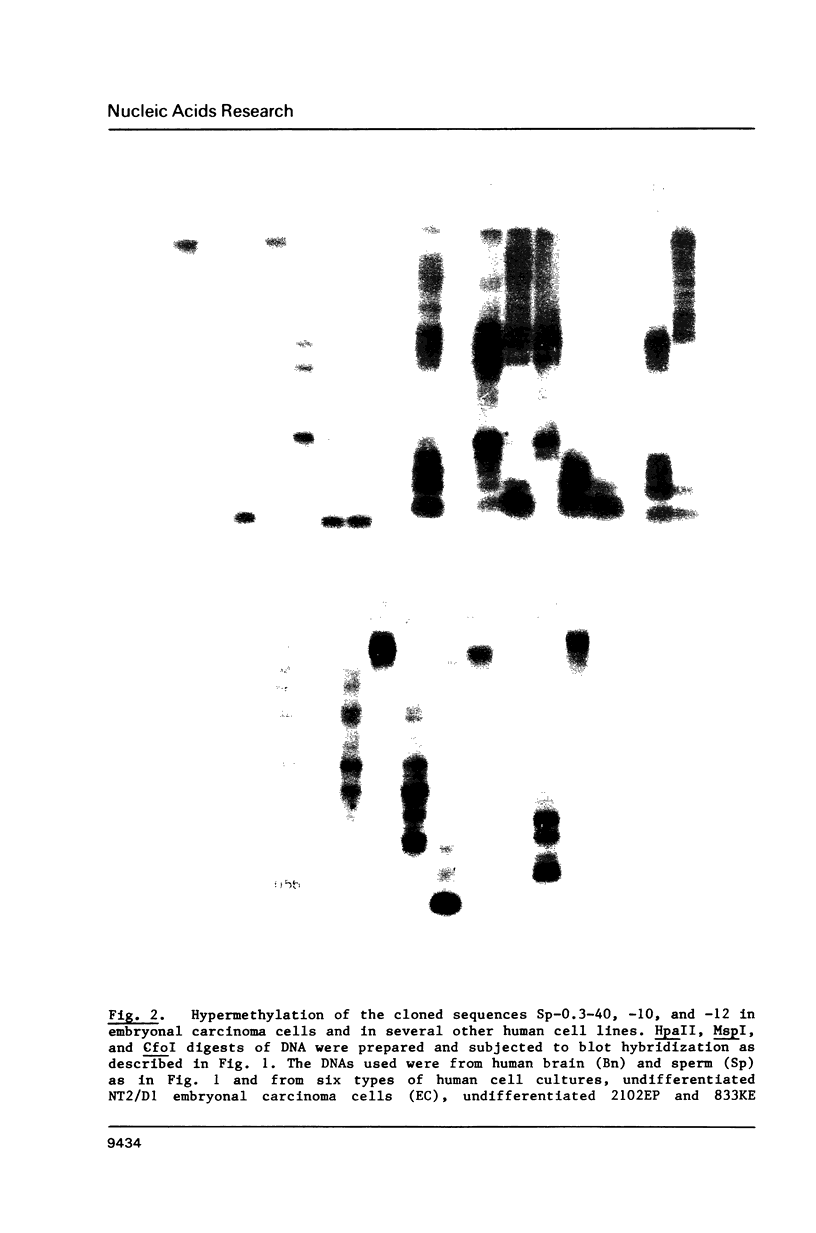
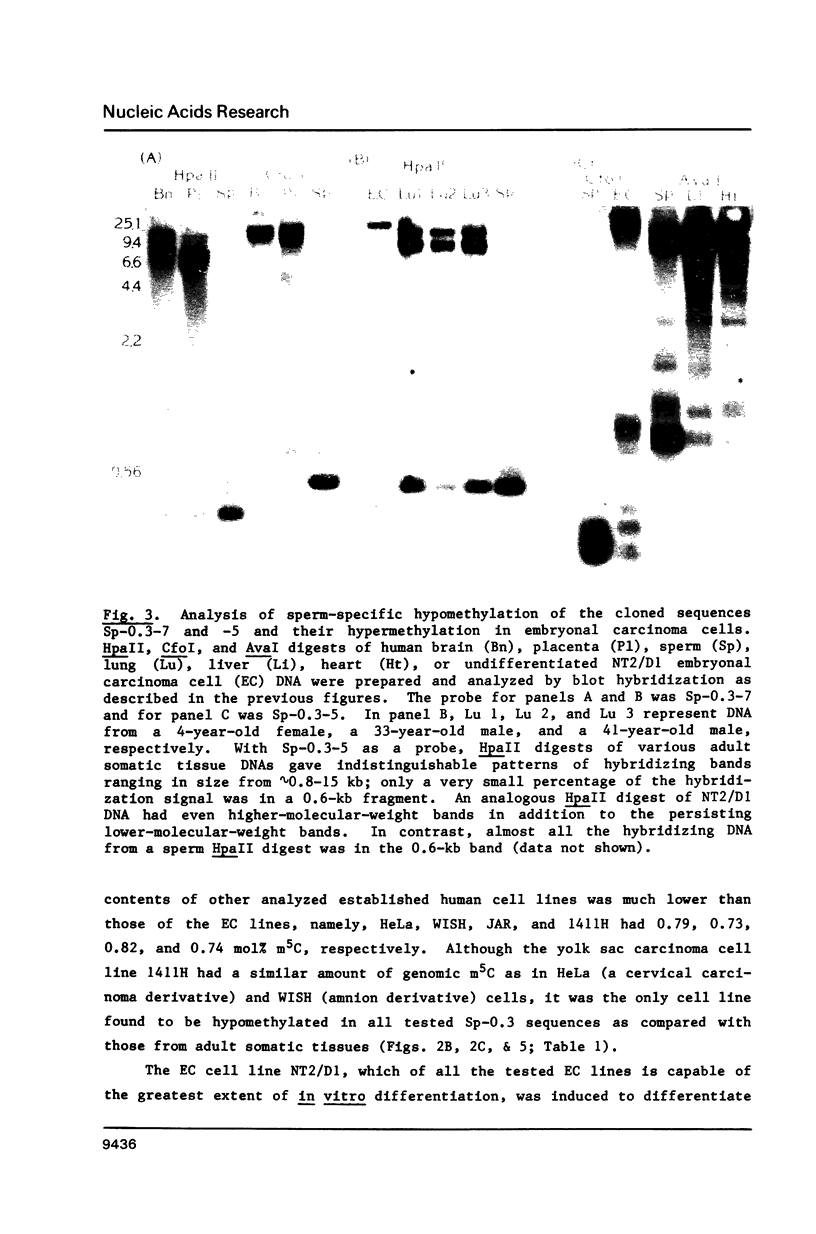
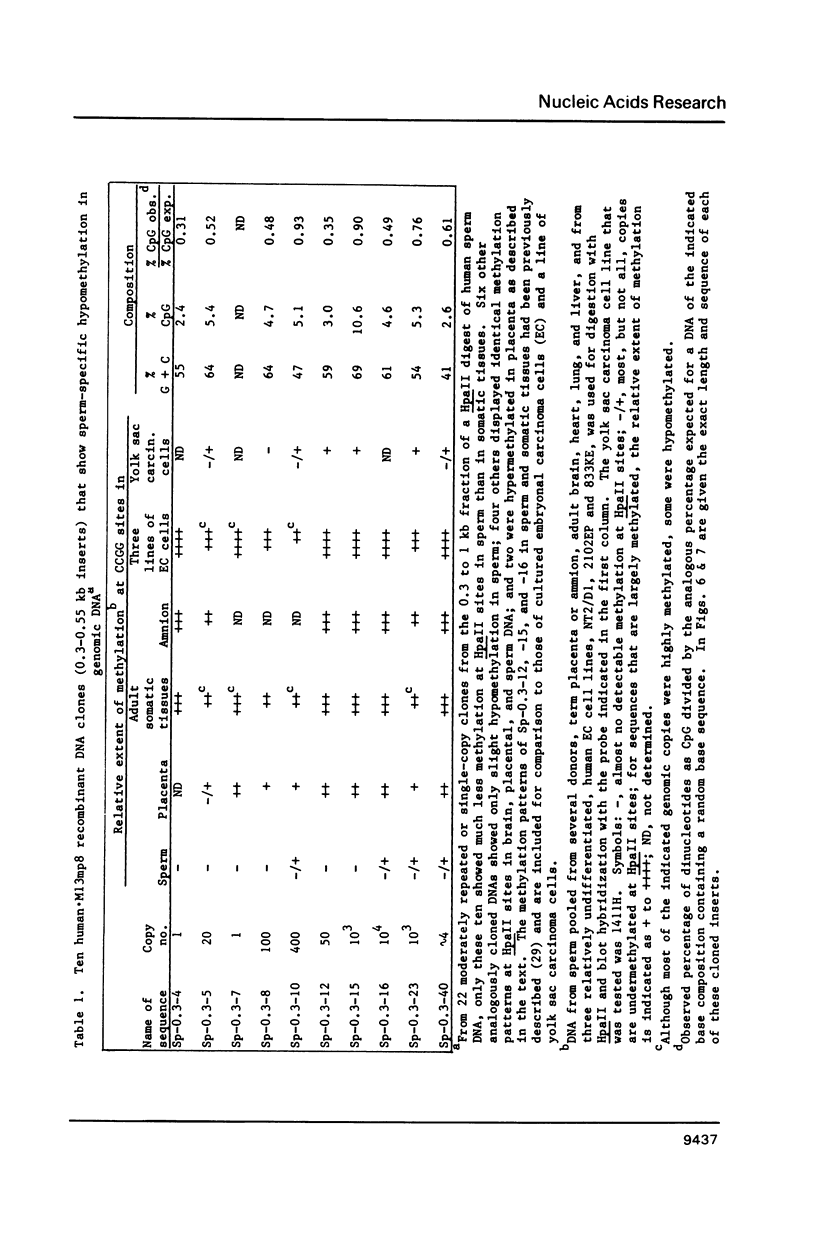
Certain human DNA sequences are much less methylated at CpG sites in sperm than in various adult somatic tissues. The DNA of term placenta displays intermediate levels of methylation at these sequences (Sp-0.3 sequences). We report here that pluripotent embryonal carcinoma (EC) cells derived from testicular germ cell tumors are hypermethylated at the three previously cloned Sp-0.3 sequences and seven newly isolated sequences that exhibit sperm-specific hypomethylation. In contrast to their hypermethylation in EC cells, the Sp-0.3 sequences are hypomethylated in a line of yolk sac carcinoma cells, which like placenta, represent an extraembryonic lineage. These DNA sequences, therefore, appear to be subject to coordinate changes in their methylation during differentiation, probably early in embryogenesis, despite their diversity in copy number (1 to 10(4] and primary structure. Two of these Sp-0.3 sequences are highly homologous to DNA sequences in human chromosomal regions that might be recombination hotspots, namely, a cryptic satellite DNA sequence at a fragile site and the downstream region of the beta-globin gene cluster.
Full text
PDF


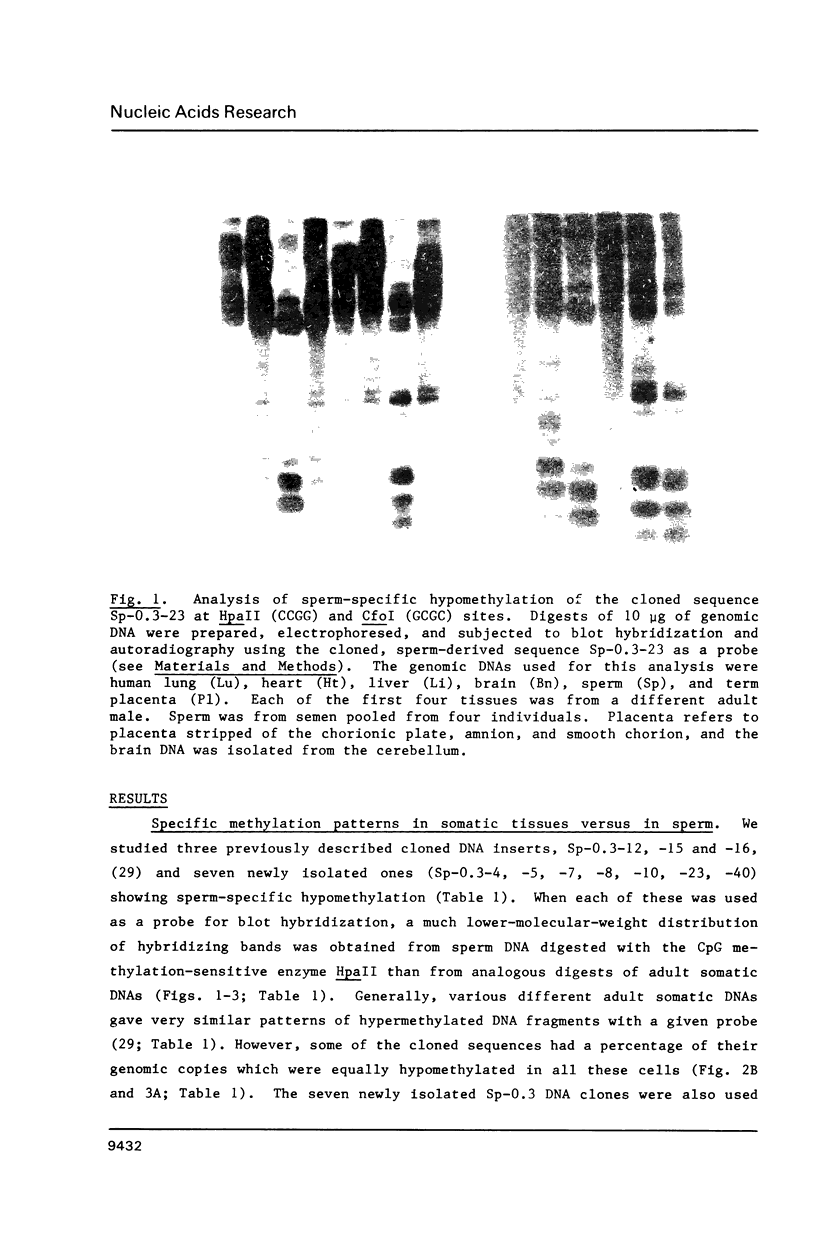














Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews P. W., Bronson D. L., Benham F., Strickland S., Knowles B. B. A comparative study of eight cell lines derived from human testicular teratocarcinoma. Int J Cancer. 1980 Sep 15;26(3):269–280. doi: 10.1002/ijc.2910260304. [DOI] [PubMed] [Google Scholar]
- Andrews P. W., Damjanov I., Simon D., Banting G. S., Carlin C., Dracopoli N. C., Føgh J. Pluripotent embryonal carcinoma clones derived from the human teratocarcinoma cell line Tera-2. Differentiation in vivo and in vitro. Lab Invest. 1984 Feb;50(2):147–162. [PubMed] [Google Scholar]
- Andrews P. W., Fenderson B., Hakomori S. Human embryonal carcinoma cells and their differentiation in culture. Int J Androl. 1987 Feb;10(1):95–104. doi: 10.1111/j.1365-2605.1987.tb00170.x. [DOI] [PubMed] [Google Scholar]
- Andrews P. W. Retinoic acid induces neuronal differentiation of a cloned human embryonal carcinoma cell line in vitro. Dev Biol. 1984 Jun;103(2):285–293. doi: 10.1016/0012-1606(84)90316-6. [DOI] [PubMed] [Google Scholar]
- Aujame L., Craig J., McBurney M. W. DNA methylation in differentiating mouse teratocarcinoma and erythroleukemia cells. Can J Biochem Cell Biol. 1984 Jul;62(7):584–591. doi: 10.1139/o84-078. [DOI] [PubMed] [Google Scholar]
- Bestor T. H., Hellewell S. B., Ingram V. M. Differentiation of two mouse cell lines is associated with hypomethylation of their genomes. Mol Cell Biol. 1984 Sep;4(9):1800–1806. doi: 10.1128/mcb.4.9.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busslinger M., deBoer E., Wright S., Grosveld F. G., Flavell R. A. The sequence GGCmCGG is resistant to MspI cleavage. Nucleic Acids Res. 1983 Jun 11;11(11):3559–3569. doi: 10.1093/nar/11.11.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke H. J., Schmidtke J., Gosden J. R. Characterisation of a human Y chromosome repeated sequence and related sequences in higher primates. Chromosoma. 1982;87(5):491–502. doi: 10.1007/BF00333470. [DOI] [PubMed] [Google Scholar]
- Cooper D. N., Errington L. H., Clayton R. M. Variation in the DNA methylation pattern of expressed and nonexpressed genes in chicken. DNA. 1983;2(2):131–140. doi: 10.1089/dna.1983.2.131. [DOI] [PubMed] [Google Scholar]
- Cooper D. N. Eukaryotic DNA methylation. Hum Genet. 1983;64(4):315–333. doi: 10.1007/BF00292363. [DOI] [PubMed] [Google Scholar]
- Deininger P. L., Jolly D. J., Rubin C. M., Friedmann T., Schmid C. W. Base sequence studies of 300 nucleotide renatured repeated human DNA clones. J Mol Biol. 1981 Sep 5;151(1):17–33. doi: 10.1016/0022-2836(81)90219-9. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Gama-Sosa M. A., Huang L. H., Midgett R. M., Kuo K. C., McCune R. A., Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982 Apr 24;10(8):2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M., Wang R. Y. 5-Methylcytosine in eukaryotic DNA. Science. 1981 Jun 19;212(4501):1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- Epstein N. D., Karlsson S., O'Brien S., Modi W., Moulton A., Nienhuis A. W. A new moderately repetitive DNA sequence family of novel organization. Nucleic Acids Res. 1987 Mar 11;15(5):2327–2341. doi: 10.1093/nar/15.5.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenderson B. A., Andrews P. W., Nudelman E., Clausen H., Hakomori S. Glycolipid core structure switching from globo- to lacto- and ganglio-series during retinoic acid-induced differentiation of TERA-2-derived human embryonal carcinoma cells. Dev Biol. 1987 Jul;122(1):21–34. doi: 10.1016/0012-1606(87)90328-9. [DOI] [PubMed] [Google Scholar]
- Flemington E., Bradshaw H. D., Jr, Traina-Dorge V., Slagel V., Deininger P. L. Sequence, structure and promoter characterization of the human thymidine kinase gene. Gene. 1987;52(2-3):267–277. doi: 10.1016/0378-1119(87)90053-9. [DOI] [PubMed] [Google Scholar]
- Gama-Sosa M. A., Midgett R. M., Slagel V. A., Githens S., Kuo K. C., Gehrke C. W., Ehrlich M. Tissue-specific differences in DNA methylation in various mammals. Biochim Biophys Acta. 1983 Jun 24;740(2):212–219. doi: 10.1016/0167-4781(83)90079-9. [DOI] [PubMed] [Google Scholar]
- Gama-Sosa M. A., Wang R. Y., Kuo K. C., Gehrke C. W., Ehrlich M. The 5-methylcytosine content of highly repeated sequences in human DNA. Nucleic Acids Res. 1983 May 25;11(10):3087–3095. doi: 10.1093/nar/11.10.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M., Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987 Jul 20;196(2):261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Gehrke C. W., McCune R. A., Gama-Sosa M. A., Ehrlich M., Kuo K. C. Quantitative reversed-phase high-performance liquid chromatography of major and modified nucleosides in DNA. J Chromatogr. 1984 Sep 28;301(1):199–219. doi: 10.1016/s0021-9673(01)89189-5. [DOI] [PubMed] [Google Scholar]
- HAYFLICK L. The establishment of a line (WISH) of human amnion cells in continuous cultivation. Exp Cell Res. 1961 Feb;23:14–20. doi: 10.1016/0014-4827(61)90059-3. [DOI] [PubMed] [Google Scholar]
- Hare J. T., Taylor J. H. One role for DNA methylation in vertebrate cells is strand discrimination in mismatch repair. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7350–7354. doi: 10.1073/pnas.82.21.7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henthorn P. S., Mager D. L., Huisman T. H., Smithies O. A gene deletion ending within a complex array of repeated sequences 3' to the human beta-globin gene cluster. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5194–5198. doi: 10.1073/pnas.83.14.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T. A. Induction of chromosome decondensation, sister-chromatid exchanges and endoreduplications by 5-azacytidine, an inhibitor of DNA methylation. Mutat Res. 1983 Jul;121(1):47–52. doi: 10.1016/0165-7992(83)90085-4. [DOI] [PubMed] [Google Scholar]
- Jablonka E., Goitein R., Marcus M., Cedar H. DNA hypomethylation causes an increase in DNase-I sensitivity and an advance in the time of replication of the entire inactive X chromosome. Chromosoma. 1985;93(2):152–156. doi: 10.1007/BF00293162. [DOI] [PubMed] [Google Scholar]
- Jähner D., Stuhlmann H., Stewart C. L., Harbers K., Löhler J., Simon I., Jaenisch R. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature. 1982 Aug 12;298(5875):623–628. doi: 10.1038/298623a0. [DOI] [PubMed] [Google Scholar]
- Keith D. H., Singer-Sam J., Riggs A. D. Active X chromosome DNA is unmethylated at eight CCGG sites clustered in a guanine-plus-cytosine-rich island at the 5' end of the gene for phosphoglycerate kinase. Mol Cell Biol. 1986 Nov;6(11):4122–4125. doi: 10.1128/mcb.6.11.4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet I., Lieman-Hurwitz J., Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986 Feb 28;44(4):535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- Klysik J., Stirdivant S. M., Singleton C. K., Zacharias W., Wells R. D. Effects of 5 cytosine methylation on the B-Z transition in DNA restriction fragments and recombinant plasmids. J Mol Biol. 1983 Jul 25;168(1):51–71. doi: 10.1016/s0022-2836(83)80322-2. [DOI] [PubMed] [Google Scholar]
- Kratzer P. G., Chapman V. M. X chromosome reactivation in oocytes of Mus caroli. Proc Natl Acad Sci U S A. 1981 May;78(5):3093–3097. doi: 10.1073/pnas.78.5.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar A. B., Paterson B. M., Weintraub H. Transfection of a DNA locus that mediates the conversion of 10T1/2 fibroblasts to myoblasts. Cell. 1986 Dec 5;47(5):649–656. doi: 10.1016/0092-8674(86)90507-6. [DOI] [PubMed] [Google Scholar]
- Libert F., Vassart G., Christophe D. Methylation and expression of the human thyroglobulin gene. Biochem Biophys Res Commun. 1986 Feb 13;134(3):1109–1113. doi: 10.1016/0006-291x(86)90365-7. [DOI] [PubMed] [Google Scholar]
- Lock L. F., Melton D. W., Caskey C. T., Martin G. R. Methylation of the mouse hprt gene differs on the active and inactive X chromosomes. Mol Cell Biol. 1986 Mar;6(3):914–924. doi: 10.1128/mcb.6.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel J. L., Chambon P. DNA methylation: organ specific variations in the methylation pattern within and around ovalbumin and other chicken genes. Nucleic Acids Res. 1979 Dec 20;7(8):2081–2103. doi: 10.1093/nar/7.8.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manes C., Menzel P. Demethylation of CpG sites in DNA of early rabbit trophoblast. Nature. 1981 Oct 15;293(5833):589–590. doi: 10.1038/293589a0. [DOI] [PubMed] [Google Scholar]
- McGrath J., Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984 May;37(1):179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- Perez-Stable C., Ayres T. M., Shen C. K. Distinctive sequence organization and functional programming of an Alu repeat promoter. Proc Natl Acad Sci U S A. 1984 Sep;81(17):5291–5295. doi: 10.1073/pnas.81.17.5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipsen J. N., Gruber M., Ab G. Expression-linked demethylation of 5-methylcytosines in the chicken vitellogenin gene region. Biochim Biophys Acta. 1985 Dec 18;826(4):186–194. doi: 10.1016/0167-4781(85)90005-3. [DOI] [PubMed] [Google Scholar]
- Ponzetto-Zimmerman C., Wolgemuth D. J. Methylation of satellite sequences in mouse spermatogenic and somatic DNAs. Nucleic Acids Res. 1984 Mar 26;12(6):2807–2822. doi: 10.1093/nar/12.6.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W., Collick A., Norris M. L., Barton S. C., Surani M. A. Genomic imprinting determines methylation of parental alleles in transgenic mice. Nature. 1987 Jul 16;328(6127):248–251. doi: 10.1038/328248a0. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Jones P. A. 5-methylcytosine, gene regulation, and cancer. Adv Cancer Res. 1983;40:1–30. doi: 10.1016/s0065-230x(08)60678-8. [DOI] [PubMed] [Google Scholar]
- Sanford J., Forrester L., Chapman V., Chandley A., Hastie N. Methylation patterns of repetitive DNA sequences in germ cells of Mus musculus. Nucleic Acids Res. 1984 Mar 26;12(6):2823–2836. doi: 10.1093/nar/12.6.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapienza C., Peterson A. C., Rossant J., Balling R. Degree of methylation of transgenes is dependent on gamete of origin. Nature. 1987 Jul 16;328(6127):251–254. doi: 10.1038/328251a0. [DOI] [PubMed] [Google Scholar]
- Schnedl W., Erlanger B. F., Miller O. J. 5-methylcytosine in heterochromatic regions of chromosomes in Bovidae. Hum Genet. 1976 Jan 28;31(1):21–26. doi: 10.1007/BF00270395. [DOI] [PubMed] [Google Scholar]
- Shenoy S., Daigle K., Ehrlich K. C., Gehrke C. W., Ehrlich M. Hydrolysis by restriction endonucleases at their DNA recognition sequences substituted with mismatched base pairs. Nucleic Acids Res. 1986 Jun 11;14(11):4407–4420. [PMC free article] [PubMed] [Google Scholar]
- Stein R., Sciaky-Gallili N., Razin A., Cedar H. Pattern of methylation of two genes coding for housekeeping functions. Proc Natl Acad Sci U S A. 1983 May;80(9):2422–2426. doi: 10.1073/pnas.80.9.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm K. S., Taylor J. H. Distribution of 5-methylcytosine in the DNA of somatic and germline cells from bovine tissues. Nucleic Acids Res. 1981 Sep 25;9(18):4537–4546. doi: 10.1093/nar/9.18.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland G. R., Parslow M. I., Baker E. New classes of common fragile sites induced by 5-azacytidine and bromodeoxyuridine. Hum Genet. 1985;69(3):233–237. doi: 10.1007/BF00293031. [DOI] [PubMed] [Google Scholar]
- Vanyushin B. F., Mazin A. L., Vasilyev V. K., Belozersky A. N. The content of 5-methylcytosine in animal DNA: the species and tissue specificity. Biochim Biophys Acta. 1973 Mar 28;299(3):397–403. doi: 10.1016/0005-2787(73)90264-5. [DOI] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. DNA methylation at a CCGG sequence in the large intron of the rabbit beta-globin gene: tissue-specific variations. Nucleic Acids Res. 1978 Dec;5(12):4631–4634. doi: 10.1093/nar/5.12.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. C., Manuelidis L. Sequence definition and organization of a human repeated DNA. J Mol Biol. 1980 Sep 25;142(3):363–386. doi: 10.1016/0022-2836(80)90277-6. [DOI] [PubMed] [Google Scholar]
- Yang R., Fristensky B., Deutch A. H., Huang R. C., Tan Y. H., Narang S. A., Wu R. The nucleotide sequence of a new human repetitive DNA consists of eight tandem repeats of 66 base pairs. Gene. 1983 Nov;25(1):59–66. doi: 10.1016/0378-1119(83)90167-1. [DOI] [PubMed] [Google Scholar]
- Zhang X. Y., Wang R. Y., Ehrlich M. Human DNA sequences exhibiting gamete-specific hypomethylation. Nucleic Acids Res. 1985 Jul 11;13(13):4837–4851. doi: 10.1093/nar/13.13.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ploeg L. H., Flavell R. A. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell. 1980 Apr;19(4):947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]