Abstract
Two G-quadruplex forming sequences, 5′-TGGGAG and the 17-mer sequence T30177, which exhibit anti-HIV-1 activity on cell lines, were modified using either locked nucleic acids (LNA) or via insertions of (R)-1-O-(pyren-1-ylmethyl)glycerol (intercalating nucleic acid, INA) or (R)-1-O-[4-(1-pyrenylethynyl)phenylmethyl]glycerol (twisted intercalating nucleic acid, TINA). Incorporation of LNA or INA/TINA monomers provide as much as 8-fold improvement of anti-HIV-1 activity. We demonstrate for the first time a detailed analysis of the effect the incorporation of INA/TINA monomers in quadruplex forming oligonucleotides (QFOs) and the effect of LNA monomers in the context of biologically active QFOs. In addition, recent literature reports and our own studies on the gel retardation of the phosphodiester analogue of T30177 led to the conclusion that this sequence forms a parallel, dimeric G-quadruplex. Introduction of the 5′-phosphate inhibits dimerisation of this G-quadruplex as a result of negative charge–charge repulsion. Contrary to that, we found that attachment of the 5′-O-DMT-group produced a more active 17-mer sequence that showed signs of aggregation—forming multimeric G-quadruplex species in solution. Many of the antiviral QFOs in the present study formed more thermally stable G-quadruplexes and also high-order G-quadruplex structures which might be responsible for the increased antiviral activity observed.
INTRODUCTION
G-quadruplex structures built of stacked guanine tetrads (Figure 1A) are exciting compounds for therapeutic strategies (1–3), since their existence in vivo may play an important role in controlling gene expression via recognition of these structures by proteins (4–7). For this reason, structural studies of G-quadruplex motifs are one of the ongoing tasks in nucleic acid chemical biology. Chemical modification of G-quadruplexes may lead to stabilization, rearrangement or total demolition of the guanosine assemblies. There have been a number of reports in which chemical modification of DNA monomers resulted in structural alterations and thus different biological activity of G-quadruplexes (8–11).
Figure 1.
(A) The G-quartet, represented by four guanine residues. Hydrogen bonds are in dashed lines. (B) Structure of LNA with torsion angles labelled, INA and TINA monomers. (C) Parallel tetramolecular G-quadruplex (5′- TGGGAG). The formation of G-quartets is indicated by dashed lines. (D) Anti-parallel G-quadruplex structure of T30177 having anti-parallel folding topology proposed by Jing N.J. and Hogan M.E. (20).
As a target we chose two G-rich sequences, 5′-TGGGAG and T30177, which are known inhibitors of human immunodeficiency virus type 1 (HIV-1) replication in culture (12–14). The 6-mer sequence, also known as Hotoda’s sequence, forms a tetramolecular parallel G-quadruplex (Figure 1C). It has been shown that attachment of aromatic substituents at the 5′-end resulted in enhancement of anti-HIV-1 activity (15–17). Alterations in the structure [shortness of the sequence, substitution of dG to dT (15–17) or to 8-aza-3-deaza-2′-deoxyguanosine (18)] which induce destabilization of the G-quadruplex structure have resulted in a drop in activity. The 17-mer sequence, T30177 (Zintevir, Figure 1D) has been extensively studied due to observed high inhibition of HIV-1 integrase (19–21). It has been proven that interaction of both 5′-TGGGAG and T30177 with the V3 loop of the envelope viral glycoprotein gp120 and, subsequent interference of a virus-to-cell transmission was a mechanism of action in a cell culture (12–14). We propose that both sequences can be used for biological evaluation of modified nucleic acid monomers in the G-quadruplexes.
Locked nucleic acids (LNA) is a class of oligonucleotide analogues containing one or more conformationally locked nucleotide monomers with a 2′-O,4′-C-methylene linkage (Figure 1B). The LNA sugar is locked in the N-type conformation, which is also the preferred conformation for ribonucleotides and can thus be considered as an RNA mimic (22–24). Moreover, LNA guanosines are incompatible with a syn conformation for the χ angle due to steric clashes between N3 and the protruding H3′. During anti-parallel quadruplex formation the guanosines alternate between anti and syn conformation, thus substitution of positions corresponding to syn by G-LNA should prevent its assembly into the anti-parallel conformation. Indeed, structural studies of some quadruplexes showed that substitution of nucleotides by LNA residues led to formation of G-quadruplexes with different folding topology (25,26), and different biological activity of thrombin binding aptamer, TBA (27–29) that was not necessarily correlated with thermal stability of G-quadruplexes (29). When LNA monomers were built into tetrameric parallel quadruplexes, which only have G-LNA in the anti conformation, the quadruplexes were stabilized (30,31). It has been shown that tetramolecular LNA-G-quadruplexes formed from relatively short TGGGT sequence have kinetic properties favouring quadruplex formation in comparison with DNA-G-quadruplexes [slower dissociation and faster association (32)].
Bulge insertions of (R)-1-O-(pyren-1-ylmethyl)glycerol (monomer P, Figure 1B) into oligodeoxynucleotides (ONs) (intercalating nucleic acids, INA) induced increased affinity for complementary ssDNA, but reduced affinity for an identical sequence of ssRNA (33). Twisted intercalating nucleic acids (TINA) are composed of bulged insertions of (R)-1-O-[4-(1-pyrenylethynyl)phenylmethyl]glycerol (monomer X, Figure 1B) into the middle of ONs. TINA has shown extraordinary high thermal stability for Hoogsteen-type, parallel triplexes and excellent discrimination between double-stranded DNA and single-stranded nucleic acids (34). Recently, we have shown that TINA insertion into G-rich sequences can be used to stabilize G-quadruplexes [TINA monomer on the top or the bottom of the G-quadruplex structures (35)]. Conversely, when TINA monomer was placed between adjacent guanines, G-quadruplex structures were destabilized (36,37). Therefore, we assume that both LNA as a nucleotide substituent and TINA/INA monomers as intercalating/aromatic residues represent valuable molecules for modulation of G-quadruplex folding topology. However, limited data is available about the impact of these monomers on the topology and biological activity of G-quadruplexes, which is vital for further exploration of chemically modified G-quadruplexes as potential pharmaceuticals.
Here, for the first time, we provide a detailed systematic study of incorporation of three different modifications LNA, INA and TINA (Figure 1B) in both terminal and internal positions of G-quadruplexes 5′-TGGGAG (15–17) and T30177 (38,39) and the effect on anti-HIV-1 activity. We found quadruplex forming oligonucleotides (QFOs) that were ca. 6–10 times more active than the wild-type quadruplex structures. To assess the topology and thermal stability of the formed G-quadruplexes we used circular dichroism (CD) spectroscopy and we used native polyacrylamide gel electrophoresis to evaluate the molecularity of complexes formed.
MATERIALS AND METHODS
Oligonucleotide synthesis
Incorporation of LNA and INA monomers into ONs was performed on a 0.2 μmol scale and was followed by purification of the ONs without the final deprotection of the 4,4′-dimethoxytrityl group (DMT-on ONs) by HPLC on Waters Xterra™ MS C18 column as described earlier (23,33). TINA containing ONs were obtained via postsynthetic Sonogashira solid-phase coupling reaction (34). Double treatment with Sonogashira reaction-mixture [1-ethynylpyrene (22.5 mM), Pd(PPh3)4 (7.5 mM), CuI (7.5 mM), dry dimethylformamide (DMF)/Et3N (3.5/1.5, 500 µl), 3 h] was applied for CPG-bound ONs containing insertions of (R)-1-O-(4-iodophenylmethyl)glycerol followed by purification procedure as described earlier (34). ONs after HPLC-purification were obtained as ammonium salts. The 5′-O-DMT group of ON4, ON7–ON17 was cleaved by 80% aq. AcOH (100 µL) followed by dilution with 50 mM NH4HCO3 (1 ml) and repurification on RP-HPLC. ONs 16–18 were resynthesised for the gel electrophoresis and CD analysis by Pentabase Aps (Søndersø, Denmark). Incorporation of the 5′-phosphate group was performed by solid-phase DNA automated synthesis using chemical phosphorylation reagent purchased from Glen research. For convenience in the text of the manuscript and on the pictures of native gels ONs possessing 5′-O-DMT group were labelled as  and DMT-off oligonucleotides as
and DMT-off oligonucleotides as 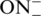 . MALDI-TOF analysis was performed on a Voyager Elite Biospectrometry Research Station from PerSeptive Biosystems. All DMT-off oligonucleotides gave satisfactory composition (see Supplementary Data, Table S1)
. MALDI-TOF analysis was performed on a Voyager Elite Biospectrometry Research Station from PerSeptive Biosystems. All DMT-off oligonucleotides gave satisfactory composition (see Supplementary Data, Table S1)
with purity over 90% that was verified by denaturing 20% PAGE (7M urea, samples were incubated for 10 min at 95°C prior to loading) and/or analytical ion-exchange chromatography using LaChrom system from Merck Hitachi on GenPak-Fax column (Waters) and basic buffer (gradient of 2M aq. NaCl in 0.01M aq. NaOH, pH 12), which was used to disrupt secondary structures of ONs. Oligonucleotide concentrations were determined by absorbance at 260 nm and the calculated single-strand extinction coefficients were based on a nearest neighbour model (the extinction coefficient for P and X monomers is 22 400 at 260 nm).
Viruses and cells
The inhibitory activity against HIV-1 infection was evaluated using MT-4 cells (40) as target cells and the HIV-1 strain HTLV-IIIB (41) as infectious virus. The virus was propagated in H9 (40) cells at 37°C, 5% CO2 using Roswell Park Memorial Institute medium (RPMI) 1640 with 10% heat-inactivated fetal calf serum (FCS) and antibiotics (growth medium). Culture supernatant was filtered (0.45 nm), aliquoted, and stored at −80°C until use. HIV-1 strain was obtained from the NIH AIDS Research and Reference Program.
Inhibition of HIV-1 replication
G-quadruplexes were examined for possible antiviral activity against HIV-1 using MT-4 cells as target cells. Before addition of the test dilutions of compounds cultures were preincubated for 30 min. Subsequently, MT-4 cells were incubated with virus (0.005 multiplicity of infection, MOI) and growth medium containing the test dilutions of ONs for six days in parallel with virus-infected and uninfected control cultures without compounds added. Expression of HIV in the cultures was indirectly quantified using the (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (42). Compounds that mediated less than 50% reduction of HIV expression at 1.0 µM were considered to be without biological activity. ONs were tested in parallel for cytotoxic effect in uninfected MT-4 culture containing the test dilutions of oligonucleotides as described above.
CD analysis
CD spectra were measured on a Chirascan spectropolarimeter from Applied Photophysics in 1.0 cm (6-mer sequences) or 0.1 cm (17-mer sequences) path length cuvettes. The spectra were recorded as an average of three scans with 0.25 s per point, at 1.0 nm bandwidth, at 20°C and normalized by subtraction of the background scan with the buffer. The molar ellipticity was calculated from the equation [θ] = 100θ/cl, where θ is a relative intensity, c is the concentration of an oligonucleotide and l is the path length of the cell in centimeters using software provided by Applied Photophysics. The CD measurements were performed at concentration of 6.7 µM (6-mer sequences) or 10 µM (17-mer sequences) in the 10 mM Na-phosphate buffer containing 135 mM NaCl and 5 mM KCl, pH 7.0. Samples were heated at 95°C for 5 min then slowly cooled down and left overnight at room temperature. CD melting experiments were performed on the same samples recording a spectrum in the range of 220–350 nm for unmodified and LNA containing ONs or in the range of 220–400 nm for INA/TINA ONs using a stepped ramping of 5.0°C from 20°C to 90°C and equilibration time of 300 s. The spectra were normalized by subtraction of the background scan of the buffer. In several cases the melting and annealing profiles were not superimposable, which means that dissociation/association processes were not in thermodynamic equilibrium. Partial deprotection of the 5′-O-DMT group can also occur at elevated temperatures and may result in irreproducibility of melting experiments (43). We indicated melting temperatures obtained in our experiments as T1/2 to distinguish thermodynamic melting temperatures, which are usually indicated as Tm, from non-thermodynamic values.
Gel electrophoresis
Non-denaturing gel electrophoresis experiments were performed on a 20 cm × 20 cm (0.5 mm thickness) 20% polyacrylamide (19:1 acrylamide:bisacrylamide ratio) gel in the RPMI 1640 buffer, which was also used for the running buffer (5.3 mM KCl, 100 mM NaCl, 25 mM HEPES, 6.7 mM NaH2PO4, 0.4 mM MgSO4, 1mM CaNO3, 23.8 mM NaHCO3, 10 mM glucose, pH 7.5 at 37°C). Gels were run for 4–4.5 h at 16W (for two gels) at 37°C. ONs were heated for 5 min at 90°C, then slowly cooled and left overnight at 37°C, 6-mer sequences at a concentration of 200 µM and 17-mer sequences at a concentration 100 µM (10 µl solutions) in RPMI 1640 buffer. Prior to loading on the gel to each sample had 2 µl 40% glycerol/dye mix added which was also kept at 37°C. After running gels were stained with Stains-all® and destained in H2O. ONs were also assessed in the native gels composed of 1 × TBE buffer (pH 8.4) supplemented with 150 mM KCl (6-mer sequences) or 10 mM KCl (17-mer sequences), which were run at temperature indicated for each gel (see Supplementary Data).
RESULTS
We synthesized ONs containing LNA and INA/TINA monomers on a DNA synthesizer as 5′-O-(4,4′-dimethoxytrityl)-on (5′-O-DMT-on) compounds. As reference compounds, the previously investigated unmodified G-rich sequences, 5′-DMT-on and off ON1 and ON7 were used (Table 1). ONs were checked in parallel for cytotoxic effect in uninfected MT-4 culture and 50% cytotoxic concentration for ON1 and ON7 was found to be over 10 µM, which is in agreement with previous studies (12–17). All the ONs from our work showed similar or lower cytotoxicity than ON1 and ON7 when evaluating the viability of the cells at 1.0 µM ON concentration (data not shown).
Table 1.
Oligonucleotides synthesized and their anti-HIV inhibition efficiency at 1.0 µMa
| Name | Sequence | DMT on (%) | DMT off (%) |
|---|---|---|---|
| ON1 | 5′- TGG GAG | 10 ± 3 | –b |
| ON2 | 5′- TGGLGLA GL | 65 ± 5 | – |
| ON3 | 5′- TGGLGGLG | 84 ± 8 | – |
| ON4 | 5′- PTGG GAG | – | 0 |
| ON5 | 5′- TPGGLGGLG | 2 | – |
| ON6 | 5′- TXGG GAG | 75 ± 6 | – |
| ON7 | 5′- GTG GTG GGT GGG TGG GT | 28 ± 2 | 10 |
| ON7p | 5′- p-GTG GTG GGT GGG TGG GT | – | – |
| ON8 | 5′- GTG GLTG GGLT GGGL TGG GLT | 84 ± 9 | 25 |
| ON9 | 5′- GTGL GTG GLGT GGLG TGGL GT | 16 | 19 |
| ON10 | 5′- GTG GTPG GGT GGG TGG GT | 77 ± 7 | 25 |
| ON11 | 5′- GTG GTPG GGT GGG TPGG GT | 76 ± 9 | 5 |
| ON12 | 5′- GTG GTG GGT GGG TPGG GT | 61 ± 7 | 18 |
| ON13 | 5′- GTG GTG GPGT GGG TGG GT | 72 ± 10 | 3 |
| ON14 | 5′- GTG GTG GGT GGPG TGG GT | 75 ± 2% | 15 |
| ON15 | 5′- GTG GTG GGTP GGG TGG GT | 100 ± 8c | 4 |
| ON16 | 5′- PGTG GTG GGTP GGG TGG GT | – | 90 ± 6 |
| ON17 | 5′- PGTG GTG GGT GGG TGG GTP | – | 89 ± 12 |
| ON18 | 5′- GTG GTPG GPGT GGG TPGG GT | 0 | – |
| ON19 | 5′- GTG GLTPG GPGLT GGGL TPGG GLT | 0 | – |
| ON20 | 5′- GTG GLTPG GGLT GGGL TPGG GLT | 0 | – |
| ON21 | 5′- GLTG GLTPG GGLT GGGL TPGG GLT | 0 | – |
| ON22 | 5′- GLTGG TPG GPGT GGG TPGG GTL | 16 | – |
| ON23 | 5′- GLTGG TPG GGT GGG TPGG GTL | 63 ± 4 | – |
| ON24 | 5′- GLTGG TG GGT GGG TGG GTL | 0 | – |
| ON25 | 5′- GTG GTXG GGT GGG TXGG GT | 87 ± 8 | - |
| ON26 | 5′- GTG GTXG GXGT GGG TXGG GT | 78 ± 8 | - |
aP represents INA monomer, X represents TINA monomer, GL and TL represents LNA corresponding monomers, p represents phosphate group.
bThe test was not performed.
cNo effect was observed at 0.1 µM.
Hotoda’s sequence
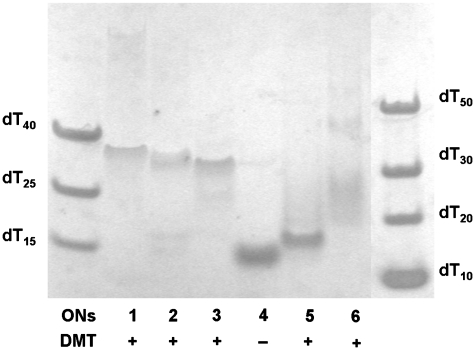
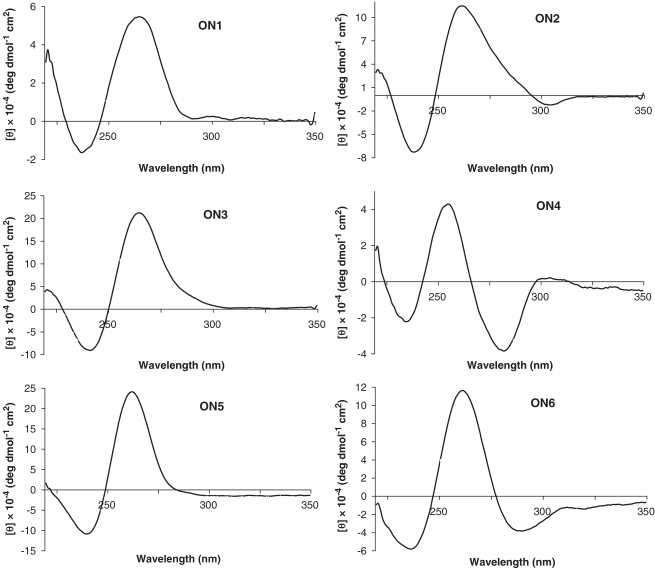
The ON forming a parallel tetrameric G-quadruplex, 5′-TGGGAG (Figure 1C), was modified by LNA guanosines (G-LNAs). It has been proposed that the most stable parallel quadruplex is obtained when substituting every guanosine to the LNA analogue except the 5′-terminal one (31). The glycosidic angle of the guanosine at the 5′-termini is very close to the syn domain for a closely related sequence (31). Based on this we synthesized ON2 substituting each dG with G-LNA except for the 5′-terminal one (Table 1) and enhancement of the anti-HIV-1 inhibition efficiency of 10% for ON1 to 65% for ON2 was observed at 1.0 µM. Substitution of dA and the next-neighbouring dG in the structure of quadruplex ON1 by G-LNAs resulted in a higher activity for ON3 (84%) than for ON2 (65%). In Figure 2, it is apparent that complexes formed by ON2 and ON3 are composed of four strands and migrate comparable to ON1 in between the 30- and 40-mer components of the ladder. The DMT-group is responsible for the slightly slower migration of these complexes as it might be expected for the G-quadruplex composed of four 6-mer QFOs (6 × 4 = 24-mer). G-quadruplex ON3 had negligible changes in CD spectra even after prolonged incubation at 90°C for 30 min which suggest very high thermal stability (>90°C, Supplementary Figures S1 and S2). The native complex ON1 had a T1/2 value of 55.0°C while a T1/2 value for ON2 was 75°C. CD profiles of ON1–ON3 (Figure 3) are characteristic for a parallel G-quadruplex structure (maxima at 265 nm and minima at 235–240 nm) (15,44). Another interesting finding is that the unmodified sequence ON1 has one more slowly migrating band on the gel above dT50. This band is seen more clearly on the native 20% PAGE which was run in 1 × TBE buffer at 4°C (see Supplementary Figure SI). This is an indication of a G-quadruplex formation that is presumably composed of eight strands when at relatively high oligonucleotide concentration (150–200 µM, see discussion for 17-mer sequences below).
Figure 2.
Nondenaturing 20% polyacrylamide gel, RPMI 1640 buffer (also used for the running buffer) run at 37°C. 5′-O-DMT-on compounds are marked as +, DMT-off as −. DNA ladder is composed of oligothymidylates of different length as indicated on the picture.
Figure 3.
CD spectra of compounds ON1–ON6, all possess 5′-O-DMT group except ON4.
Earlier reports on the 5′-modified 5′-TGGGAG assemblies (15–17) prompted our studies on G-quadruplexes possessing insertions of pyrene bearing moieties, INA (P) and TINA (X). Insertion of the INA moiety into Hotoda’s sequence resulted in inactive compounds. From native PAGE (Figure 2) we concluded that the totally inactive species (ON4 and ON5) failed to produce tetrameric G-quadruplex structures. Instead, both these complexes migrated similar to dT15, which is an indication of formation of duplexes. It is important to mention that the CD spectrum of ON5 might lead to the erroneous conclusion that parallel G-quadruplex was formed, because of its similarity with CD spectra of ON1–ON3 (Figure 3). Contrary to this, the CD profile of ON4 was drastically different from a standard parallel G-quadruplex profile showing a positive band at 255 nm and negative bands at 235 and 282 nm. Therefore, we can conclude that INA monomer cannot mimic the 5′-O-DMT group in the Hotoda’s sequence despite the large pyrene moiety present in the structure of INA. It is interesting to note the conversion from a very active LNA based G-quadruplex ON3 to totally inactive compound ON5 upon insertion of INA monomer on the top of the G-quartets’ stack. TINA-containing ON6 exists in an equilibrium involving several species, which led to the smear between dT20–dT50 of the ladder on native PAGE (Figure 2) and caused the undefined T1/2 value from its CD melting (Supplementary Figure S2). The CD spectrum of ON6 indicated the presence of parallel G-quadruplexes in the mixture.
The dissociation and reassociation of the tetrameric G-quadruplexes were followed by CD spectroscopy, recording a change in molar ellipticity at 265 nm with respect to change in temperature (Supplementary Figure S2). All the complexes show hysteresis between melting and annealing profiles; however, LNA and TINA containing active G-quadruplexes are formed considerably faster than ON1 at the temperature ramp used in these experiments (see details in Experimental section).
T30177 sequence and its alterations using LNA, INA/TINA
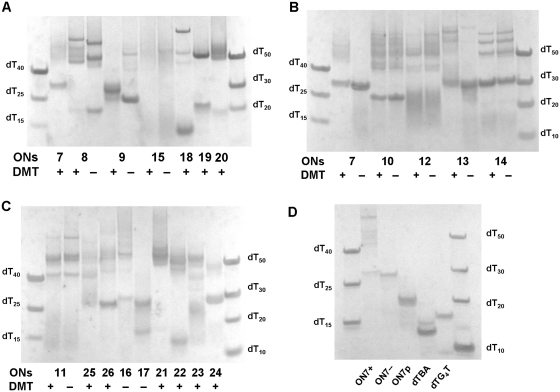
We used the non-thioated analogue of T30177 (d(G*TGGTGGGTGGGTGGG*T, * represents phosphothioate internucleosidic linkage) and we observed that introduction of the 5′-O-DMT group in the unmodified sequence ON7 led to increased inhibition efficiency despite the fact that T1/2 values were very similar for DMT-on and DMT-off sequences, 58°C and 60°C, respectively (Supplementary Figures S4 and S5). For the sequences containing a lipophilic moiety at the 5′-end a smeared band was observed, migrating above dT40 in the native PAGE as observed for DMT-on ON7 (ON7+, Figure 5B), while DMT-off ON7 (ON7−, Figure 5B) had a retardation similar to the 30-mer component of the ladder. This indicates that under these conditions ON7- does indeed exist as a dimer, which is in agreement with the results of electrospray ionization mass-spectrometry (ESI-MS) (45,46), and attachment of the DMT group led to the formation of multimeric species, presumably a mixture of tri- and tetrameric structures. Analysing earlier reports on T30177 and its phosphodiester analogue in which native PAGEs were performed (38,47) we suspected that enzymatic 5′-phosphorylation led to the formation of a G-quadruplex that migrates at a similar rate to dTBA on the native gel, which in turn led to the conclusion that sequence T30177 is a unimolecular G-quadruplex. To test this theory we synthesized the phosphorylated oligomer ON7p by chemical addition of a phosphate group at the 5′-end. As shown in Figure 5D the mobility of ON7− and ON7p was significantly different in the native gel. The band of ON7p migrated similar to dT20, thus confirming that 5′-phosphorylation prevents dimerisation of the phosphodiester analogue of T30177, perhaps due to the negative charge–charge repulsion. The CD spectra of ON7p and ON7+ confirm the formation of a parallel G-quadruplex for both QFOs (Supplementary Figure S3). In our case dTBA had significantly lower mobility on the gel (between dT10 and dT15) in comparison with ON7 or ON7p (Figure 5D and Supplementary Figure S6). It should be noted that dTBA sequence was not phosphorylated at the 5′-position and our sequences did not possess internucleosidic phosphothioate linkages. This might explain the difference in the mobility of ON7p and dTBA in our gels in comparison with earlier reports (38,47).
Figure 5.
(A–C) Nondenaturing 20% polyacrylamide gel, RPMI 1640 buffer (also used for the running buffer) run at 37°C. 5′-O-DMT-on compounds are marked as +, DMT-off as −. DNA ladder is composed of oligothymidylates of different length as indicated on each picture. (D) Non-denaturing 20% polyacrylamide gel, 1 × TBE supplemented with 10 mM KCl (also used for the running buffer) run for 4.5 h at 10W at 37°C.
5′-O-DMT-on sequence ON8 possessing four G-LNAs instead of dGs showed almost three times higher anti-HIV-1 activity (84%) in comparison with DMT-on ON7 (28%). To our astonishment this sequence had marginal changes in the intensity of CD signals during temperature increase from 20°C to 90°C and an overall parallel stranded topology (+ve ellipticity at 263 nm and −ve ellipticity at 242 nm) was remained even at elevated temperatures (Supplementary Figures S4 and S5). Another remarkable observation is that ON8+ had a significantly slower movement in the gel and migrated as a mixture of several bands above dT40 (Figure 5A). Formation of a band with retardation similar to dT20 was observed only after cleavage of the DMT-group (ON8−), which was also accompanied with the loss of antiviral activity (25% versus 84% for ON8+). Interestingly, another LNA-containing sequence ON9, with different position for the LNA modifications, both as DMT-on and off, did not exhibit high inhibition efficiency and migrated on the gel between the 25- and 30-mer components of the ladder showing a T1/2 value of 72°C.
INA monomer (P) was inserted in different positions in the 17-mer sequence (ON10–ON18). Generally, with the exception of sequence ON18 possessing three INA moieties, more active compounds with inhibition efficiency of 61–100% were obtained by insertion of INA in comparison with ON7+. According to the gel mobility assay (Figure 5) multistranded species with retardation slower than the 40-mer component of the ladder were observed in addition to the band(s) migrating at the level of dT20–dT30 (ON10–ON14, ON16). It is interesting to note that removal of the DMT group had a significant effect only on the mobility of ON13 (dimeric complex was observed). This might mean that the presence of the lipophilic moiety at the 5′-end is important not only because it leads to the formation of multistranded G-quadruplexes but also because it might result in crucial interactions with a viral glycoprotein. For the most active compound ON15 we observed a poorly visible smear of bands above dT40 with some species present around dT15. When more concentrated samples were run on the native gel we observed a considerable amount of oligonucleotides that did not penetrate into the gel (data not shown). This suggests that DMT-on ON15 exists in equilibria of multiple high-order structures and some of them show very high antiviral activity. In contrast to other INA containing ONs in this series the intensity of a CD spectrum of ON15 was considerably lower, however characteristic features of parallel stranded G-quadruplexes similar to ON7 were present for all these sequences (ON10−ON17, Figure 4 and Supplementary Figure S3). Thermal denaturing profile of ON15 (Supplementary Figure S4) has no distinctive melting point which again confirms a presence of multiple G-quadruplexes. For ON18 a positive peak is observed at ca. 295 nm which is indicative of an existence of anti-parallel G-quadruplexes in the mixture (Figure 4). We also note that for ON13–ON14 for which INA was inserted exclusively in the middle of the G-stretch no signs of an anti-parallel quadruplex formation were observed. For several compounds (ONs 11, 13,15,19,21,23) a weak Cotton signal derived from the pyrene moiety was detected in the region of 340–380 nm (Supplementary Figure S3). The T1/2 values of INA containing sequences were not as high as for LNA-modified 17-mers, i.e 54°C (ON10), 53°C (ON14), 68°C (ON17, Supplementary Figure S4). Sequence ON17 possessing INA moieties at the 3′- and 5′-ends without 5′-O-DMT group did not show signs of the formation of multistranded species on the native gel, the major band migrated between the 25- and 30-mers of the ladder and even though 89% inhibition efficiency was observed. This is quite an interesting result, suggesting that anti-HIV-1 activity can be accomplished by different quadruplex structures and hence, does not depend heavily on a certain conformation.
Figure 4.
CD spectra of compounds ON7–ON10, ON18 and ON26, all possess 5′-O-DMT group.
In continuation of our study we tried to combine the design of active and inactive LNA and INA containing sequences based on T30177 (ON19–ON23). Unfortunately, despite the formation of G-quadruplexes, including the appearance of multistranded species on the native gel, and high thermal stability (as for ON20, Supplementary Figure S4) only one sequence showed a substantial antiviral activity (ON23). Apparently, its activity is due to internal insertion of INA moieties as in the sequence ON11 and not substitution of terminal nucleotides by LNA nucleotides, as in ON24. Compounds ON19–ON21 in which LNA and INA were mixed in internal positions of the 17-mer sequence and ON5 and showed no activity. This indicates that the flexibility is lost for allowing the formation of antiviral quadruplexes.
Introduction of the third P in the middle of the 17-mer (ON18) prevented formation of the G-quadruplex observed for ON11 (the major band on the gel migrated below the 15-mer component of the ladder) and loss of antiviral activity was observed. It is interesting to note a difference for ON25, ON26 in which INA was substituted by TINA moiety with the initial compounds ON11 and ON18. Activity of ON11 was maintained in ON25 though less distinct bands were observed for ON25 on the native gel (Figure 5). In contrast to ON18 three TINA insertions led to an active compound (ON26, 78%) that migrated differently on the gel, i.e. a discrete band corresponding to the dimeric complex was observed in addition to some slower moving bands. CD spectra for ON18 and ON26, though of low intensity, were not significantly different (Figure 4); both of them showed several peaks with positive ellipticity at 260 and 295 nm suggesting a mixture of parallel and anti-parallel G-stretches in quadruplexes. These results emphasise the difference in activity and topology that might be obtained upon insertion of structurally very similar intercalating units in G-quadruplexes. Further discussion of the impact of LNA and INA/TINA insertions on the T30177 G-quadruplex structure needs the availability of well defined G-quadruplex structures.
DISCUSSION
One of the most challenging and yet fascinating features of G-quadruplexes is the very low predictability of folding topology and molecularity (48) which is affected upon perturbation of the system by chemical modifications. We decided several years ago to investigate the impact of LNA and INA modifications on anti-HIV-1 activity of two sequences, the 6-mer Hotoda’s sequence and the phosphodiester analogue of T30177. Our data show that anti-HIV activity of the original Hotoda’s sequence possessing 5′-O-DMT group can be further improved by incorporation of LNA or TINA moieties. Significantly higher thermal stability, faster association and higher inhibition efficiency were observed for G-quadruplexes possessing G-LNAs (ON2 and ON3) in comparison with the reference ON1. Improved anti-HIV-1 activity of the TINA containing sequence ON6 could be related to the formation of multiple G-quadruplex species, which could be quickly formed and also be stable in physiological conditions.
As a stepping stone for the design of LNA substitutions in the T30177 17-mer sequence we used an earlier proposed NMR structure, which was described as an anti-parallel unimolecular G-quadruplex similar to the dTBA motif (Figure 1D). Because G-LNA prefers the anti conformation around the glycosidic bond, we placed G-LNAs in those positions corresponding to an anti configuration in the supposed anti-parallel G-quadruplex structure, hence compatible with the native structure so that the more stable G-quadruplex should have been formed (sequence ON8). We have also placed G-LNAs in the opposite (‘wrong’) positions where LNA puckering would be incompatible with the anti conformation (ON9). In this case, we aimed to destroy formation of G-quadruplexes. Moreover, INA monomers and, later, TINA monomers were incorporated in different positions in G-stretches so as to be placed at the top, bottom or between G-tetrads of the supposed anti-parallel G-quadruplex. It soon became obvious that the structure of T30177, its analogues T30695, T30923 and our own sequences do not match with the structure proposed in early reports (38,49,50). According to CD spectra (Figure 4 and Supplementary Figure S3), three insertions of INA or TINA (ON18, ON22 and ON26) were required in order to induce formation of a substantial amount of an anti-parallel G-quadruplex.
Several research groups (9,51,52) has also pointed out the discrepancy between CD profiles recorded for T30177, T30695 (38), their phosphodiester analogues (9,51) and an anti-parallel topology suggested earlier by nuclear magnetic resonance, NMR (20,38,49,53) (Figure 1D). For anti-parallel G-quadruplexes one can expect to see a positive ellipticity maximum at 295 nm and a negative minimum at 265 nm. Instead CD spectra showed a positive peak at 263 nm and a negative minimum at 240 nm which is characteristic for parallel stranded G-quadruplexes (54–56). This empirical interpretation of CD spectra associated with the relative orientation of strands is an oversimplification (57); however it can be applied for G-quadruplexes composed of unmodified guanosines. These sequences were also found to inhibit HIV integrase (19,20,39,53). Another inhibitor of HIV-1 integrase, the 16-mer G-rich sequence d(GGGGTGGGAGGAGGGT), designated 93del, was studied by NMR and was found to form an inter-locked dimer with parallel orientation of G-stretches (52) and an inner tetrad having a mixture of 3 anti and one syn guanine bases. The CD spectrum of 93del had a positive peak at 260 nm and small positive peak at ca. 295 nm, which is probably due to the mixed anti–syn tetrad. Recently, the dimeric structure for T30923 was confirmed by NMR (58), and ESI-MS provided evidence of dimeric G-quadruplexes formation for T30923 and the phosphodiester analogue of T30177 in ammonium acetate solutions (45,46). Based on retardation of ON7− on the native gel shown in the present work and literature reports discussed above we argue that the unmodified DMT-off sequence ON7, which is a reference compound in our study, forms a parallel dimeric G-quadruplex. This also means that dGs prefer anti conformation of the glycosidic angles in the native structure. Therefore, substitution of dG by G-LNA should lead to similar folding and a more stable quadruplex structure and consequently to an anti-HIV-1 active compound. We also used intercalating moieties in the structure of ON7, given the ability of INA/TINA to stabilize G-quadruplexes via stacking interactions with G4-tetrads (35,59).
Clearly the molecularity of G-quadruplexes based on the sequence of ON7− is highly susceptible to changes as a result of chemical modifications. Our experiment shows that 5′-phosphate capping can prevent dimerisation not only tetramolecular quadruplexes as it has been demonstrated for d(G5CTA) (60) but also unimolecular quadruplexes if their sequences possess guanosine(s) at the 5′-end. It also means that the commonly used enzymatic introduction of the P32 as a 5′-phosphate for visualization of G-quadruplexes on a native gel can lead to the incorrect conclusion about molecularity of these complexes, which happened for T30177 at the time of their first structural analysis in 1990′s. Other methods, such as staining of gels with stains-all, gel filtration chromatography (61) or ESI-MS should be used for stoichiometry determination of G-quadruplexes in solution. It seems that it is a general feature of many G-quadruplexes to form dimeric or multimeric structures (60,62–68). Capping of sequences possessing guanosine(s) at 3′- or 5′-ends by a phosphate (60) or thymidine (64,66) prevents dimerisation or oligomerization of G-quadruplexes. From that point of view the phosphodiester analogue of T30177 (ON7−) is an important example for researchers dealing with biologically active G-quadruplexes, showing how commonly used techniques can lead to incorrect interpretation of the structure.
Anti-HIV-1 activity accompanied with the formation of multimeric complexes is one of the most striking observations for the following sequences: ON8+, INA/TINA containing sequences ON10–ON16, ON25, ON26 and even for reference sequences ON1 and ON7 possessing 5′-O-DMT group. We cannot completely eliminate the likelihood of small amounts of impurities being responsible for formation of slow-moving bands on the native gel. However, the existence of complexes composed of presumably three- or four 17-mer strands for several active sequences, including INA containing compounds, might indicate that these high-order structures can account for increased antiviral activity, especially for the sequence ON8+. Multi-stranded species are usually formed at relatively high concentrations. Once they are formed, dilution of the samples does not considerably change the ratio of these species in solution. Since all the sequences were obtained as ammonium salts, which triggers the formation of G-quadruplexes, and were tested in HIV assay by several dilutions of stock solutions we assume that multi-stranded species visible on the native gel were present during evaluation of antiviral activity (see Supplementary Figure S6).
We assume that the preliminary target for the formed G-quadruplexes was the V3 loop of the envelope viral glycoprotein gp120 as has been shown earlier (12–14). This might mean that multimeric species, especially those ones formed by ON8+, fit better within the glycoprotein structure as hence new drugs should be designed with these properties leading to better inhibition efficiency. Aromatic moieties were found to be essential for activity, which was earlier observed for Hotoda’s sequence (15). The effect of lipophilic moieties can be ascribed to increased thermal stability, faster association of G-quadruplexes, facilitation of aggregation (formation of multimeric species) or non-specific interactions with glycoproteins in the active site.
Large aromatic molecules might play a significant role in a G-quadruplex aggregation. In this work, we have seen that the DMT-group triggers formation of more complex quadruplexes: ON7+ versus ON7−, ON8+ versus ON8−, ON13+ versus ON13− (Figure 5). Aggregate formation is a well established fact for duplexes possessing organic chromophores like porphyrins (69,70) and perylenes (71,72) at 5′ termini. Consequently, aggregation driven by the DMT-group can be expected for G-quadruplexes. It is important to mention that pyrene-containing G-rich sequences did not show higher cytotoxicity than native and LNA modified quadruplexes. The use of intercalating moieties not only INA and TINA should be encouraged in the screening of already known G-quadruplexes or G-rich aptamers in order, either to find out those species responsible for the activity, or to discover more active and selective G-quadruplexes. As we have demonstrated in this article, one can envisage an opportunity in synthesizing a library of QFOs by encompassing LNA or INA/TINA monomers in biologically active ONs with G-rich sequences where the secondary structure is still unknown. We believe that this approach will be applied more often in future and we will see more G-quadruplex structures revised and revisited. Altogether these findings expand our knowledge about anti-HIV-1 G-quadruplexes; their modifications with LNA, INA/TINA; and can be useful in the molecular engineering of novel modified G-rich oligonucleotides with improved biological activity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Financial support from the Marsden grant administrated by the Royal Society of New Zealand (grant MAU0704) and Massey University is gratefully acknowledged. Nucleic Acid Center is funded by The Danish National Research Foundation for studies on nucleic acid chemical biology. Funding for open access charge: IFS (Massey University).
Conflict of interest statement. None declared.
REFERENCES
- 1.Mergny JL, Helene C. G-quadruplex DNA: A target for drug design. Nat. Med. 1998;4:1366–1367. doi: 10.1038/3949. [DOI] [PubMed] [Google Scholar]
- 2.Neidle S, Read MA. G-quadruplexes as therapeutic targets. Biopolymers. 2000;56:195–208. doi: 10.1002/1097-0282(2000)56:3<195::AID-BIP10009>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 3.Gatto B, Palumbo M, Sissi C. Nucleic acid aptamers based on the G-quadruplex structure: therapeutic and diagnostic potential. Curr. Med. Chem. 2009;16:1248–1265. doi: 10.2174/092986709787846640. [DOI] [PubMed] [Google Scholar]
- 4.Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 5.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl Acad. Sci. USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang C, Zhang XH, Ratliff R, Moyzis R, Rich A. Crystal-structure of 4-stranded Oxytricha telomeric DNA. Nature. 1992;356:126–131. doi: 10.1038/356126a0. [DOI] [PubMed] [Google Scholar]
- 8.Sacca B, Lacroix L, Mergny JL. The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides. Nucleic Acids Res. 2005;33:1182–1192. doi: 10.1093/nar/gki257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dapic V, Abdomerovic V, Marrington R, Peberdy J, Rodger A, Trent JO, Bates PJ. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 2003;31:2097–2107. doi: 10.1093/nar/gkg316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dapic V, Bates PJ, Trent JO, Rodger A, Thomas SD, Miller DM. Antiproliferative activity of G-quartet-forming oligonucleotides with backbone and sugar modifications. Biochemistry. 2002;41:3676–3685. doi: 10.1021/bi0119520. [DOI] [PubMed] [Google Scholar]
- 11.D’Onofrio J, Petraccone L, Martino L, Di Fabio G, Iadonisi A, Balzarini J, Giancola C, Montesarchio D. Synthesis, biophysical characterization, and anti-HIV activity of glyco-conjugated G-quadruplex-forming oligonucleotides. Bioconjugate Chem. 2008;19:607–616. doi: 10.1021/bc7003395. [DOI] [PubMed] [Google Scholar]
- 12.Este JA, Cabrera C, Schols D, Cherepanov P, Gutierrez A, Witvrouw M, Pannecouque C, Debyser Z, Rando RF, Clotet B, et al. Human immunodeficiency virus glycoprotein gp120 as the primary target for the antiviral action of AR177 (Zintevir) Mol. Pharmacol. 1998;53:340–345. doi: 10.1124/mol.53.2.340. [DOI] [PubMed] [Google Scholar]
- 13.Urata H, Kumashiro T, Kawahata T, Otake T, Akagi M. Anti-HIV-1 activity and mode of action of mirror image oligodeoxynucleotide analogue of Zintevir. Biochem. Biophys. Res. Commun. 2004;313:55–61. doi: 10.1016/j.bbrc.2003.11.094. [DOI] [PubMed] [Google Scholar]
- 14.Cherepanov P, Este JA, Rando RF, Ojwang JO, Reekmans G, Steinfeld R, David G, De Clercq E, Debyser Z. Mode of interaction of G-quartets with the integrase of human immunodeficiency virus type 1. Mol. Pharmacol. 1997;52:771–780. doi: 10.1124/mol.52.5.771. [DOI] [PubMed] [Google Scholar]
- 15.Hotoda H, Koizumi M, Koga R, Kaneko M, Momota K, Ohmine T, Furukawa H, Agatsuma T, Nishigaki T, Sone J, et al. Biologically active oligodeoxyribonucleotides. 5. (1) 5′-End-substituted d(TGGGAG) possesses anti-human immunodeficiency virus type 1 activity by forming a G-quadruplex structure. J. Med. Chem. 1998;41:3655–3663. doi: 10.1021/jm970658w. [DOI] [PubMed] [Google Scholar]
- 16.Koizumi M, Koga R, Hotoda H, Momota K, Ohmine T, Furukawa H, Agatsuma T, Nishigaki T, Abe K, Kosaka T, et al. Biologically active oligodeoxyribonucleotides––IX. Synthesis and anti-HIV-1 activity of hexadeoxyribonucleotides, TGGGAG, bearing 3′- and 5′-end-modification. Bioorg. Med. Chem. 1997;5:2235–2243. doi: 10.1016/s0968-0896(97)00161-2. [DOI] [PubMed] [Google Scholar]
- 17.Hotoda H, Koizumi M, Koga R, Momota K, Ohmine T, Furukawa H, Nishigaki T, Kinoshita T, Kaneko M, Kimura S, et al. Biologically active oligodeoxyribonucleotides. 4. Anti-HIV-1 activity of TGGGAG having hydrophobic substituent at its 5′-end via phosphodiester linkage. Nucleosides Nucleotides. 1996;15:531–538. [Google Scholar]
- 18.Jaksa S, Kralj B, Pannecouque C, Balzarini J, De Clercq E, Kobe J. How a modification (8-aza-3-deaza-2′-deoxyguanosine) influences the quadruplex structure of Hotoda’s 6-mer TGGGAG with 5′- and 3′-end modifications. Nucleosides Nucleotides Nucleic Acids. 2004;23:77–88. doi: 10.1081/ncn-120027819. [DOI] [PubMed] [Google Scholar]
- 19.Mazumder A, Neamati N, Ojwang JO, Sunder S, Rando RF, Pommier Y. Inhibition of the human immunodeficiency virus type I integrase by guanosine quartet structures. Biochemistry. 1996;35:13762–13771. doi: 10.1021/bi960541u. [DOI] [PubMed] [Google Scholar]
- 20.Jing NJ, Hogan ME. Structure-activity of tetrad-forming oligonucleotides as a potent anti-HIV therapeutic drug. J. Biol. Chem. 1998;273:34992–34999. doi: 10.1074/jbc.273.52.34992. [DOI] [PubMed] [Google Scholar]
- 21.Pinskaya MD, Brodin P, Romanova EA, Volkov EM, Mouscadet JF, Gottikh MB. Inhibition of the human immunodeficiency virus type 1 DNA integration by modified oligonucleotides. Mol. Biol. 2000;34:888–894. [PubMed] [Google Scholar]
- 22.Obika S, Nanbu D, Hari Y, Andoh J, Morio K, Doi T, Imanishi T. Stability and structural features of the duplexes containing nucleoside analogues with a fixed N-type conformation, 2′-O,4′-C-methyleneribonucleosides. Tetrahedron Lett. 1998;39:5401–5404. [Google Scholar]
- 23.Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J. LNA (Locked Nucleic Acids): synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 1998;54:3607–3630. [Google Scholar]
- 24.Singh SK, Nielsen P, Koshkin AA, Wengel J. LNA (locked nucleic acids): synthesis and high-affinity nucleic acid recognition. Chem. Commun. 1998:455–456. [Google Scholar]
- 25.Dominick PK, Jarstfer MB. A conformationally constrained nucleotide analogue controls the folding topology of a DNA G-quadruplex. J. Am. Chem. Soc. 2004;126:5050–5051. doi: 10.1021/ja039192z. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen JT, Arar K, Petersen M. Solution structure of a locked nucleic acid modified quadruplex: introducing the V4 folding topology. Angew. Chem., Int. Ed. 2009;48:3099–3103. doi: 10.1002/anie.200806244. [DOI] [PubMed] [Google Scholar]
- 27.Randazzo A, Esposito V, Ohlenschlager O, Ramachandran R, Virgilio A, Mayol L. Structural studies on LNA quadruplexes. Nucleosides Nucleotides Nucleic Acids. 2005;24:795–800. doi: 10.1081/ncn-200060279. [DOI] [PubMed] [Google Scholar]
- 28.Virno A, Randazzo A, Giancola C, Bucci M, Cirinoc G, Mayol L. A novel thrombin binding aptamer containing a G-LNA residue. Bioorg. Med. Chem. 2007;15:5710–5718. doi: 10.1016/j.bmc.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 29.Bonifacio L, Church FC, Jarstfer MB. Effect of locked-nucleic acid on a biologically active G-quadruplex. A structure-activity relationship of the thrombin aptamer. Int. J. Mol. Sci. 2008;9:422–433. doi: 10.3390/ijms9030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Randazzo A, Esposito V, Ohlenschlager O, Ramachandran R, Mayol L. NMR solution structure of a parallel LNA quadruplex. Nucleic Acids Res. 2004;32:3083–3092. doi: 10.1093/nar/gkh629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nielsen JT, Arar K, Petersen M. NMR solution structures of LNA (locked nucleic acid) modified quadruplexes. Nucleic Acids Res. 2006;34:2006–2014. doi: 10.1093/nar/gkl144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petraccone L, Erra E, Randazzo A, Giancola C. Energetic aspects of locked nucleic acids quadruplex association and dissociation. Biopolymers. 2006;83:584–594. doi: 10.1002/bip.20591. [DOI] [PubMed] [Google Scholar]
- 33.Christensen UB, Pedersen EB. Intercalating nucleic acids containing insertions of 1-O-(1-pyrenylmethyl)glycerol: stabilisation of dsDNA and discrimination of DNA over RNA. Nucleic Acids Res. 2002;30:4918–4925. doi: 10.1093/nar/gkf624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Filichev VV, Pedersen EB. Stable and selective formation of Hoogsteen-type triplexes and duplexes using twisted intercalating nucleic acids (TINA) prepared via postsynthetic Sonogashira solid-phase coupling reactions. J. Am. Chem. Soc. 2005;127:14849–14858. doi: 10.1021/ja053645d. [DOI] [PubMed] [Google Scholar]
- 35.Cogoi S, Paramasivam M, Filichev V, Geci I, Pedersen EB, Xodo LE. Identification of a new G-quadruplex motif in the KRAS promoter and design of pyrene-modified G4-decoys with antiproliferative activity in pancreatic cancer cells. J. Med. Chem. 2009;52:564–568. doi: 10.1021/jm800874t. [DOI] [PubMed] [Google Scholar]
- 36.Paramasivam M, Cogoi S, Filichev VV, Bomholt N, Pedersen EB, Xodo LE. Purine twisted-intercalating nucleic acids: a new class of anti-gene molecules resistant to potassium-induced aggregation. Nucleic Acids Res. 2008;36:3494–3507. doi: 10.1093/nar/gkn242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boutorine AS, Doluca O, Filichev VV. Optimization of the sequence of twisted intercalating nucleic acids (TINA) forming triple helix with the polypurine tract of the proviral HIV DNA. Nucleic Acids Symp. Ser. 2009;53:139–140. doi: 10.1093/nass/nrp070. [DOI] [PubMed] [Google Scholar]
- 38.Jing NJ, Rando RF, Pommier Y, Hogan ME. Ion selective folding of loop domains in a potent anti-HIV oligonucleotide. Biochemistry. 1997;36:12498–12505. doi: 10.1021/bi962798y. [DOI] [PubMed] [Google Scholar]
- 39.Ojwang JO, Buckheit RW, Pommier Y, Mazumder A, Devreese K, Este JA, Reymen D, Pallansch LA, Lackmansmith C, Wallace TL, et al. T30177, an oligonucleotide stabilized by an intramolecular guanosine octet, is a potent inhibitor of laboratory strains and clinical isolates of human-immunodeficiency-virus type-1. Antimicrob. Agents Chemother. 1995;39:2426–2435. doi: 10.1128/aac.39.11.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harada S, Koyanagi Y, Yamamoto N. Infection of Htlv-Iii/Lav in Htlv-I-carrying cells Mt-2 and Mt-4 and application in a plaque-assay. Science. 1985;229:563–566. doi: 10.1126/science.2992081. [DOI] [PubMed] [Google Scholar]
- 41.Popovic M, Sarngadharan MG, Read E, Gallo RC. Detection, isolation, and continuous production of cytopathic retroviruses (Htlv-Iii) from patients with AIDS and Pre-AIDS. Science. 1984;224:497–500. doi: 10.1126/science.6200935. [DOI] [PubMed] [Google Scholar]
- 42.Richman D, Shih CK, Lowy I, Rose J, Prodanovich P, Goff S, Griffin J. Human-immunodeficiency-virus type-1 mutants resistant to nonnucleoside inhibitors of reverse-transcriptase arise in tissue-culture. Proc. Natl Acad. Sci. USA. 1991;88:11241–11245. doi: 10.1073/pnas.88.24.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D'Onofrio J, Petraccone L, Erra E, Martino L, Di Fabio G, De Napoli L, Giancola C, Montesarchio D. 5′-Modified G-quadruplex forming oligonucleotides endowed with anti-HIV activity: Synthesis and biophysical properties. Bioconjugate Chem. 2007;18:1194–1204. doi: 10.1021/bc070062f. [DOI] [PubMed] [Google Scholar]
- 44.Petraccone L, Erra E, Esposito V, Randazzo A, Mayol L, Nasti L, Barone G, Giancola C. Stability and structure of telomeric DNA sequences forming quadruplexes containing four G-tetrads with different topological arrangements. Biochemistry. 2004;43:4877–4884. doi: 10.1021/bi0300985. [DOI] [PubMed] [Google Scholar]
- 45.Li HH, Yuan G. Collision-induced dissociation of dimeric G-quadruplexes of HIV-1 integrase inhibitors and their complexes by tandem-in-time mass spectrometry. Eur. J. Mass. Spec. 2009;15:731–737. doi: 10.1255/ejms.1033. [DOI] [PubMed] [Google Scholar]
- 46.Li HH, Yuan G, Du DM. Investigation of formation, recognition, stabilization, and conversion of dimeric G-quadruplexes of HIV-1 integrase inhibitors by electrospray ionization mass spectrometry. J. Am. Soc. Mass Spec. 2008;19:550–559. doi: 10.1016/j.jasms.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 47.Rando RF, Ojwang J, Elbaggari A, Reyes GR, Tinder R, McGrath MS, Hogan ME. Suppression of human-immunodeficiency-virus type-1 activity in-vitro by oligonucleotides which form intramolecular tetrads. J. Biol. Chem. 1995;270:1754–1760. doi: 10.1074/jbc.270.4.1754. [DOI] [PubMed] [Google Scholar]
- 48.Dailey MM, Miller MC, Bates PJ, Lane AN, Trent JO. Resolution and characterization of the structural polymorphism of a single quadruplex-forming sequence. Nucleic Acids Res. 2010;38:4877–4888. doi: 10.1093/nar/gkq166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jing NJ, Gao XL, Rando RF, Hogan ME. Potassium-induced loop conformational transition of a potent anti-HIV oligonucleotide. J. Biomol. Struct. Dyn. 1997;15:573–585. doi: 10.1080/07391102.1997.10508967. [DOI] [PubMed] [Google Scholar]
- 50.Jing N, De Clercq E, Rando RF, Pallansch L, Lackman-Smith C, Lee S, Hogan ME. Stability-activity relationships of a family of G-tetrad forming oligonucleotides as potent HIV inhibitors - A basis for anti-HIV drug design. J. Biol. Chem. 2000;275:3421–3430. doi: 10.1074/jbc.275.5.3421. [DOI] [PubMed] [Google Scholar]
- 51.Porumb H, Monnot M, Fermandjian S. Circular dichroism signatures of features simultaneously present in structured guanine-rich oligonucleotides: A combined spectroscopic and electrophoretic approach. Electrophoresis. 2002;23:1013–1020. doi: 10.1002/1522-2683(200204)23:7/8<1013::AID-ELPS1013>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 52.Phan AT, Kuryavyi V, Ma JB, Faure A, Andreola ML, Patel DJ. An interlocked dimeric parallel-stranded DNA quadruplex: A potent inhibitor of HIV-1 integrase. Proc. Natl Acad. Sci. USA. 2005;102:634–639. doi: 10.1073/pnas.0406278102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jing NJ, Marchand C, Liu J, Mitra R, Hogan ME, Pommier Y. Mechanism of inhibition of HIV-1 integrase by G-tetrad-forming oligonucleotides in vitro. J. Biol. Chem. 2000;275:21460–21467. doi: 10.1074/jbc.M001436200. [DOI] [PubMed] [Google Scholar]
- 54.Balagurumoorthy P, Brahmachari SK. Structure and stability of human telomeric sequence. J. Biol. Chem. 1994;269:21858–21869. [PubMed] [Google Scholar]
- 55.Balagurumoorthy P, Brahmachari SK, Mohanty D, Bansal M, Sasisekharan V. Hairpin and parallel quartet structures for telomeric sequences. Nucleic Acids Res. 1992;20:4061–4067. doi: 10.1093/nar/20.15.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hardin CC, Henderson E, Watson T, Prosser JK. Monovalent cation induced structural transitions in telomeric DNAs - G-DNA folding intermediates. Biochemistry. 1991;30:4460–4472. doi: 10.1021/bi00232a013. [DOI] [PubMed] [Google Scholar]
- 57.Masiero S, Trotta R, Pieraccini S, De Tito S, Perone R, Randazzo A, Spada GP. A non-empirical chromophoric interpretation of CD spectra of DNA G-quadruplex structures. Org. Biomol. Chem. 2010;8:2683–2692. doi: 10.1039/c003428b. [DOI] [PubMed] [Google Scholar]
- 58.Li HH, Yuan G. Investigation of formation of dimeric G-quadruplex of HIV-1 integrase inhibitor by nuclear magnetic resonance. Chinese Chem. Lett. 2008;19:1108–1110. [Google Scholar]
- 59.Cogoi S, Paramasivan M, Xodo LE, Filichev VV, Pedersen EB. The effect of INA [(R)-1-O-(1-pyrenylmethyl)glycerol] insertions on the structure and biological activity of a G-quadruplex from a critical KRAS G-rich sequence. Nucleosides Nucleotides Nucleic Acids. 2007;26:1641–1643. doi: 10.1080/15257770701549087. [DOI] [PubMed] [Google Scholar]
- 60.Uddin MK, Kato Y, Takagi Y, Mikuma T, Taira K. Phosphorylation at 5′ end of guanosine stretches inhibits dimerization of G-quadruplexes and formation of a G-quadruplex interferes with the enzymatic activities of DNA enzymes. Nucleic Acids Res. 2004;32:4618–4629. doi: 10.1093/nar/gkh766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Phan AT, Gueron M, Leroy JL. Investigation of unusual DNA motifs. Methods Enzymol. 2001;338:341–371. doi: 10.1016/s0076-6879(02)38228-4. [DOI] [PubMed] [Google Scholar]
- 62.Protozanova E, Macgregor RB. Frayed wires: a thermally stable form of DNA with two distinct structural domains. Biochemistry. 1996;35:16638–16645. doi: 10.1021/bi960412d. [DOI] [PubMed] [Google Scholar]
- 63.Sen D, Gilbert W. Novel DNA superstructures formed by telomere-like oligomers. Biochemistry. 1992;31:65–70. doi: 10.1021/bi00116a011. [DOI] [PubMed] [Google Scholar]
- 64.Wang Y, Patel DJ. Guanine residues in d(T2AG3) and d(T2G4) form parallel-stranded potassium cation stabilized G-quadruplexes with antiglycosidic torsion angles in solution. Biochemistry. 1992;31:8112–8119. doi: 10.1021/bi00150a002. [DOI] [PubMed] [Google Scholar]
- 65.Krishnan-Ghosh Y, Liu DS, Balasubramanian S. Formation of an interlocked quadruplex dimer by d(GGGT) J. Am. Chem. Soc. 2004;126:11009–11016. doi: 10.1021/ja049259y. [DOI] [PubMed] [Google Scholar]
- 66.Lu M, Guo Q, Kallenbach NR. Structure and stability of sodium and potassium complexes of dT4G4 and dT4G4T. Biochemistry. 1992;31:2455–2459. doi: 10.1021/bi00124a003. [DOI] [PubMed] [Google Scholar]
- 67.Mergny JL, De Cian A, Ghelab A, Sacca B, Lacroix L. Kinetics of tetramolecular quadruplexes. Nucleic Acids Res. 2005;33:81–94. doi: 10.1093/nar/gki148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sket P, Plavec J. Tetramolecular DNA quadruplexes in solution: insights into structural diversity and cation movement. J. Am. Chem. Soc. 2010;132:12724–12732. doi: 10.1021/ja104889t. [DOI] [PubMed] [Google Scholar]
- 69.Mammmana A, Pescitelli G, Asakawa T, Jockusch S, Petrovic AG, Monaco RR, Purrello R, Turro NJ, Nakanishi K, Ellestad GA, et al. Role of environmental factors on the structure and spectroscopic response of 5′-DNA-porphyrin conjugates caused by changes in the porphyrin–porphyrin interactions. Chem. Eur. J. 2009;15:11853–11866. doi: 10.1002/chem.200902029. [DOI] [PubMed] [Google Scholar]
- 70.Onoda A, Igarashi M, Naganawa S, Sasaki K, Ariyasu S, Yamamura T. Circular dichroism of neutral zinc porphyrin-oligonucleotide conjugates modified with flexible linker. Bull. Chem. Soc. Jpn. 2009;82:1280–1286. [Google Scholar]
- 71.Zheng Y, Long H, Schatz GC, Lewis FD. Duplex and hairpin dimer structures for perylene diimide–oligonucleotide conjugates. Chem. Commun. 2005:4795–4797. doi: 10.1039/b509754a. [DOI] [PubMed] [Google Scholar]
- 72.Baumstark D, Wagenknecht HA. Perylene bisimide dimers as fluorescent ‘Glue’ for DNA and for base-mismatch detection. Angew. Chem., Int. Ed. 2008;47:2612–2614. doi: 10.1002/anie.200705237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.