Abstract
DNA lesions cause stalling of DNA replication forks, which can be lethal for the cell. Homologous recombination (HR) plays an important role in DNA lesion bypass. It is thought that Rad51, a key protein of HR, contributes to the DNA lesion bypass through its DNA strand invasion activity. Here, using model stalled replication forks we found that RAD51 and RAD54 by acting together can promote DNA lesion bypass in vitro through the ‘template-strand switch’ mechanism. This mechanism involves replication fork regression into a Holliday junction (‘chicken foot structure’), DNA synthesis using the nascent lagging DNA strand as a template and fork restoration. Our results demonstrate that RAD54 can catalyze both regression and restoration of model replication forks through its branch migration activity, but shows strong bias toward fork restoration. We find that RAD51 modulates this reaction; by inhibiting fork restoration and stimulating fork regression it promotes accumulation of the chicken foot structure, which we show is essential for DNA lesion bypass by DNA polymerase in vitro. These results indicate that RAD51 in cooperation with RAD54 may have a new role in DNA lesion bypass that is distinct from DNA strand invasion.
INTRODUCTION
DNA replication progression can be stalled by a variety of DNA lesions or by anomalous DNA secondary structures (1–4). Disintegration of DNA replication forks may lead to cell death or genome rearrangements that are responsible for cancer and other genetic diseases (5). The effects of DNA lesions on replication fork progression vary significantly depending on which DNA strand these lesions are formed on. Lesions on the lagging DNA strand template can be skipped over and new Okazaki fragments re-initiated downstream of the lesion allowing the lesion to be repaired after completion of DNA replication (6,7). In contrast, lesions on the leading DNA strand template are more likely to block the replication fork progression. In response to this threat, DNA damage tolerance pathways have evolved to enable DNA polymerase to bypass lesions on the leading DNA strand template. Genetic studies in Saccharomyces cerevisiae identified two major pathways responsible for DNA lesion bypass: the system of post replication repair (PRR) and homologous recombination (HR) encoded by the Rad6/Rad18 and the Rad52 epistasis groups, respectively (6,8–12). In addition, as recent data indicate, lesion bypass can occur by re-initiation of the leading strand synthesis downstream of a lesion that blocks replication fork progression (13).
HR plays an important role in both the repair of DNA double-strand breaks (DSB) and in DNA lesion bypass (14–18). The mechanisms of DSB repair have been intensively investigated during the recent decade. It was found that DSBs are first exonucleolytically processed to generate single-stranded DNA (ssDNA) tails that are targeted by HR enzymes including Rad51 and several auxiliary proteins (19,20). Rad51 possesses DNA strand exchange activity that is thought to be critically important for DSB repair. Rad51 promotes a search for homologous sequences and then DNA strand exchange to form Joint Molecules (D-loops) that serve as a template for DSB repair (17,21–23). Rad54, a member of the Snf2 family of DNA-dependent ATPases (24–27), stimulates Rad51 DNA strand exchange activity (28–31). Rad54 also performs several additional functions; it translocates on dsDNA in an ATPase-dependent manner (32), as well as promotes chromatin remodeling (33), Rad51 displacement from dsDNA (34), and branch migration of Holliday junctions (35–38).
Stalling of the replication fork at a lesion on the leading strand template results in uncoupling of leading and lagging strand synthesis; DNA synthesis continues only on the undamaged lagging template strand generating ssDNA gaps that may activate HR (39). Similar to DSBs, stalled replication forks can be repaired through a mechanism that relies on Rad51 DNA strand exchange activity [reviewed in ref. (40)]. Rad51 promotes invasion of the ssDNA gap into homologous sister chromatids, forming a joint molecule that is used as a template for DNA repair synthesis followed by restoration of replication forks through endonucleolytic resolution of the recombination intermediates (41,42).
Alternatively, it was suggested that DNA lesion bypass can occur through the template strand switch mechanism [reviewed in ref. (40)]. This mechanism includes conversion of the fork to a Holliday junction (known as the ‘chicken foot’ structure) by branch migration (6,43). In the chicken foot structure, an elongated lagging strand provides a template for extension of the leading strand beyond the point of the replication block on the parental template. After extension of the leading strand by DNA polymerase, the chicken foot structure can be reset by reverse branch migration into the fork, in which the lesion is bypassed. In Escherichia coli, HR proteins RecG, RuvABC and RecA catalyze fork regression, restoration and Holliday junction resolution (6,44–46). In humans, RecQ helicaes, WRN, BLM, and RecQ5β, and a Fanconi anemia protein, FANCM, promote conversion of a replication fork to the ‘chicken foot’ structure in vitro (47–50). Saccharomyces cerevisiae Rad5, a member of the Rad6/Rad18 pathway, and its human ortholog HLTF is also capable of promoting regression of a model replication fork (51,52).
We previously showed that Rad54 protein promotes branch migration of Holliday junctions (35,36,38). Here we investigated the ability of human RAD54 to promote regression of model replication forks in vitro. Our results show that RAD54 protein promotes both regression and restoration of model stalled replication forks. However, RAD54 showed a strong preference for fork restoration, as only a small amount of the chicken foot accumulated under equilibrium conditions. We then found that RAD51 specifically stimulates RAD54 in such a way that that it changes the equilibrium toward accumulation of the chicken foot structure. Furthermore, we demonstrate that RAD51-mediated accumulation of the chicken foot structure is essential for DNA lesion bypass by DNA polymerase in vitro. These data indicate that RAD51 may have a new role in DNA lesion bypass that is distinct from DNA strand invasion. Our current results suggest that RAD54 and RAD51 by acting together may promote error-free lesion bypass through the template switch mechanism.
MATERIALS AND METHODS
Proteins and DNA
Human RAD51, BLM and RAD54 proteins were purified as described (30,36). DNA polymerase I Klenow Fragment was purchased from New England Biolabs. All oligonucleotides used in this study (Supplementary Tables S1 and S2) were purchased from IDT, Inc. and further purified, labeled and stored as described previously (53). dsDNA substrates were prepared by annealing of equimolar amounts of oligonucleotides, as described in (53).
Regression of a model replication fork
A 32P-labeled tailed DNA #71/2* (here and below 32P-labeled strands are marked by the asterisk) and non-labeled tailed DNAs #117/1 were prepared by annealing, as described in ref. (53). To produce model replication forks a 32P-labeled tailed DNA #71/2* (32 nM, molecules) was mixed with non-labeled tailed DNA #117/1 (48 nM, molecules) and incubated in buffer A containing 25 mM Tris–acetate, pH 7.5, 2 mM ATP, 5 mM magnesium acetate, 2 mM DTT, BSA (100 µg/ml), 15 mM phosphocreatine and creatine phosphokinase (30 units/ml) for 15 min at 37°C. Then, RAD54 or BLM were added in indicated concentrations to the reaction mixtures to initiate regression of replication forks. Spontaneous backgrounds were determined under identical conditions, except that RAD54 or BLM were replaced with their storage buffers. At the indicated time points, aliquots were withdrawn, and the reaction was stopped by addition of 1.5% SDS and proteinase K (800 µg/ml). A 0.10 volume of loading buffer (70% glycerol, 0.1% bromophenol blue) was added to reaction mixtures, and the DNA products were analyzed by electrophoresis in 8% polyacrylamide gels in TBE buffer (89 mM Tris–borate, pH 8.3, and 1 mM EDTA) at 135 V for 1.5 h. Gels were dried on DEAE-81 paper (Whatman), and the DNA products were quantified using a Storm 840 PhosphorImager (GE Healthcare).
Regression of replication fork containing heterologous bases
To prepare model stalled replication forks containing heterogous bases, 32P-labeled DNA intermediate #117/1* (32 nM, molecules) was mixed with one of the non-labeled tailed DNAs intermediates (48 nM, molecules) and incubated in buffer A for 15 min at 37°C. Resulting forks (Figure 2A) contained 0, 1, 2, 4 or 8 heterologous bases (Supplementary Table S3). Fork regression reactions were started by addition of either RAD54 (100 nM) or BLM (50 nM) followed by incubation at 37°C for the indicated time periods. The DNA products were analyzed by electrophoresis in 8% polyacrylamide gels and visualized, as described in previous section.
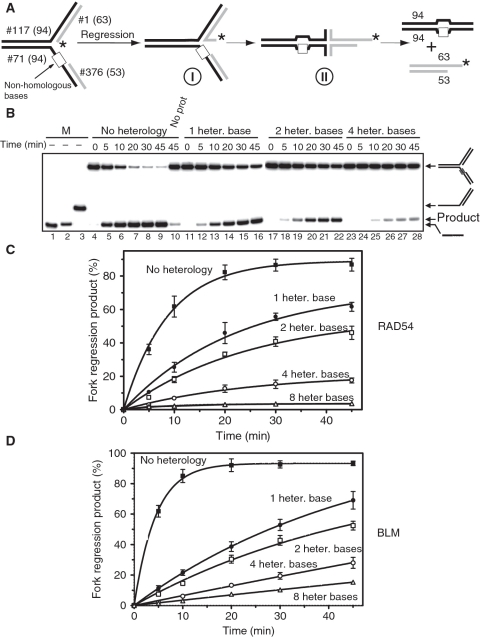
Figure 2.
RAD54 and BLM promote regression of replication forks containing heterologous bases. (A) The experimental scheme. Model replication forks contain regions of heterology of 1, 2, 4 or 8 bases (denoted by the white box) on the leading DNA strand template. The asterisk indicates the 32P-label. (B) Fork regression was initiated by addition of RAD54 (100 nM). The DNA products were analyzed by electrophoresis in an 8% polyacrylamide gel. Lanes 1–3 show DNA migration markers. Lane 10 shows the DNA products of spontaneous regression of a fully homologous replication fork in the absence of RAD54. Numbers of heterologous bases are indicated above the gel. (C) The kinetics of replication fork regression by RAD54 (100 nM) or (D) by BLM (50 nM) presented as a graph. The error bars indicate SEM.
Branch migration of partial X-junctions and X-junctions containing heterologous bases
To prepare model partial X-junctions and X-junctions containing heterologous bases, 32P-labeled DNA intermediates (32 nM, molecules) were mixed with one of the non-labeled tailed DNAs intermediates (48 nM, molecules) and incubated in buffer A for 15 min at 37°C. Resulting X-junctions (Supplementary Figure S2A and S2C) contained between 1 and 14 heterologous bases (Supplementary Table S4). Branch migration reactions were started by addition of RAD54 (100 nM) followed by incubation at 30°C for the indicated time periods. The DNA products were analyzed by electrophoresis in 8% polyacrylamide gels and visualized, as described in previous section.
Regression of replication forks in the presence of RAD51
Replication forks were prepared by annealing of 32P-labeled tailed DNA intermediate #117/1* (32 nM, molecules) with non-labeled tailed DNA #379/376 (48 nM, molecules) in buffer A for 15 min at 37°C. Then RAD51 in the indicated concentrations was added followed by a 10 min incubation. Fork regression was initiated by addition of RAD54 (100 nM) or BLM (50 nM) and carried out for 15 min at 37°C. Products were analyzed by electrophoresis in 8% polyacrylamide gels in TBE buffer and quantified using a Storm 840 PhosphorImager (GE Healthcare), as described above.
The effect of RAD54, BLM and RAD51 on the equilibrium between the replication forks and the X-structure
To prepare the model replication fork structure that can regress to form the chicken foot structure 32P-labeled forked DNA intermediate #290/342* (32 nM, molecules) was mixed with non-labeled tailed DNA #340/341 (48 nM, molecules) in buffer A for 15 min at 37°C. To prepare the chicken foot structure that can be restored to the model replication fork 32P-labeled tailed DNA #341/342* (32 nM, molecules) was mixed with non-labeled tailed DNA #340/290 (48 nM, molecules) in buffer A for 15 min at 37°C. The reactions were initiated by addition of RAD54 (100 nM) or BLM (50 nM) and carried out for indicated time periods followed at 37°C. When indicated, RAD51 (200 nM, or indicated otherwise) was added for 10 min before addition of RAD54 or BLM. The DNA products were analyzed by electrophoresis in 8% polyacrylamide gels and quantified using a Storm 840 PhosphorImager (GE Healthcare).
In vitro DNA lesion bypass through the template switch mechanism
The replication fork repair was carried out in two ways: (i) starting from the replication fork and (ii) starting from the chicken foot structure. In both structures the leading template strand (#290) contained an isocytosin (iso-C) residue that mimics a replication blocking lesion.
The replication fork structure was prepared by mixing 32P-labeled tailed DNA #290/417* (32 nM, molecules) with non-labeled tailed DNA #340/341 (48 nM, molecules) in buffer A containing dATP, dGTP, dCTP and dTTP (33 µM each) for 15 min at 37°C. The chicken foot structure was prepared by mixing 32P-labeled forked DNA #341/342* (32 nM, molecules) with non-labeled forked DNA #340/290 (48 nM, molecules) under the same conditions. The proteins were added to replication forks or chicken foot structures in the following order and for the indicated periods of time: DNA polymerase I Klenow Fragment (10 ng/ml) for 10 min, RAD51 (200 nM) or RAD51 storage buffer for 10 min, and finally RAD54 (100 nM) or BLM (50 nM) for 30 min. The reactions were carried out at 37°C and were terminated by ethanol precipitation. The DNA products were dissolved in formamide containing 0.1% bromphenol blue and 0.1% xylene cyanol, heated for 3 min at 95°C, and analyzed by electrophoresis in 15% denaturing polyacrylamide gels containing 7 M urea.
Results
RAD54 promotes fork regression
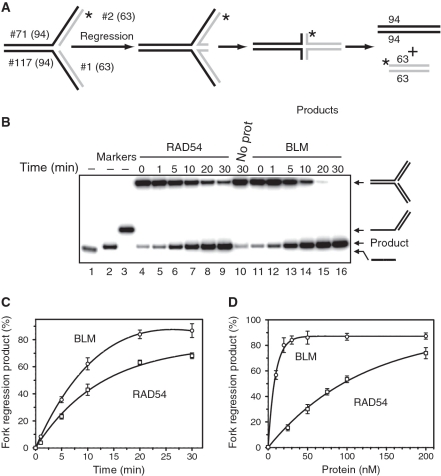
Since human RAD54 binds specifically to various branched DNA structures and promotes the branch migration of Holliday junctions (35,38), we wished to examine whether it can catalyze regression of a model replication fork. A model fork was constructed by annealing of two synthetic tailed dsDNA substrates, #71/2* and #117/1. Regression of the resulting fork would lead to formation of the Holliday junction (chicken foot structure); further branch migration of the chicken foot structure would produce two linear dsDNA products detectable by gel electrophoresis (Figure 1A). Indeed, we found that RAD54 promotes efficient regression of a model replication fork in a time dependent manner (Figure 1B, Lanes 3–8; and Figure 1C, open squares). We compared RAD54 fork regression activity with that of the BLM helicase as it was already known to promote fork regression in vitro (47). We found that these two proteins promote fork regression with similar rates; however an approximately 10-fold lower concentration of BLM was required to achieve the same extent of fork regression (Figure 1C and D). Different stoichiomeric requirements for RAD54 and BLM could be attributed, at least partially, to different oligomeric states of the active BLM and RAD54 complexes. We previously showed that RAD54 forms large, likely dodecameric, complexes during branch migration of Holliday junctions on oligonucleotide substrates (38). While the structure of the BLM active complex during branch migration is currently unknown, electron microscopy studies demonstrated formation of smaller BLM complexes, hexamers and tetramers (54).
Figure 1.
Regression of model replication forks by RAD54 and BLM. (A) The experimental scheme. The asterisk marks the 32P-label. The numbers indicate the numbers and length (nt) of the individual strands in DNA substrates and products. (B) Analysis of fork regression by RAD54 and BLM by electrophoresis in an 8% polyacrylamide gel. Reactions were initiated by adding RAD54 (100 nM) (Lanes 4–9), BLM (10 nM) (Lanes 11–16), or storage buffer (Lane 10) to the DNA fork substrate (#71/2* + #117/1; see Supplementary Table S1). 32P-ssDNA (#2*) (Lane 1), 32P-dsDNA (#1/2*) and 32P-tailed DNA (#71/2*) (Lanes 1–3, respectively) were used as migration markers. (C) The kinetics of fork regression promoted by RAD54 or BLM presented as a graph. (D) The effect of RAD54 or BLM concentrations on the efficiency of fork regression. The reactions were initiated by addition of RAD54 or BLM (in the indicated concentrations) and were carried out for 15 min at 37°C. The spontaneous background was determined in identical reactions, except that RAD54 and BLM were replaced by their respective storage buffers, and was subtracted from the data. The experiments were repeated at least three times. The error bars indicate standard error of the mean (SEM).
RAD54 can promote regression of forks containing heterology
The replication machinery stalls at DNA lesions that impair base pairing. These lesions may also inhibit replication fork regression because of their inability to pair with bases on the adjacent DNA arm. We tested whether RAD54 can promote fork regression on the substrates that contain such lesions. To mimic the effect of DNA lesions on fork regression we constructed a fork in which heterologous bases were placed in the leading strand template in front of the primer strand (Figure 2A).
We investigated the effect of the heterology length on the fork regression by RAD54 using DNA substrates containing heterologous bases (from one to eight) (Figure 2A). The efficiency of fork regression was reduced with the increase in the heterology length; with heterology regions of 1, 2, 4 and 8 bases reducing the initial rate of fork regression by 3-, 4-, 11- and 26-fold over substrates that lacked heterology, respectively (Figure 2B–D). Since bypass of the heterology region requires unwinding of one or several DNA base pairs, it was conceivable that BLM, which unlike RAD54, does possess canonical helicase activity, would show greater efficiency in regression of forks containing heterology than RAD54. Surprisingly, for BLM the decrease in the rate of replication fork regression by heterology was even more significant than for RAD54; with the 1, 2, 4 and 8 base heterology causing 7-, 9-, 25- and 47-fold decrease in rate, respectively. We also found that mismatches placed at the proximity of the junctions have a more significant inhibitory effect on the fork regression by both RAD54 and BLM than mismatches placed a distance from the junction (Supplementary Figure S1).
We also examined the length of heterology RAD54 can bypass using Holliday junction (X-junction) and partial Holliday junction (PX-junction) substrates (Supplementary Figure S2). Similar to the reaction on the fork substrates, a 4-strand reaction on X-junctions becomes undetectable when the region of heterology reached 8 bases (Supplementary Figure S2B). As expected, RAD54 can bypass a longer heterology, up to 14 bases, in the three-strand reaction on PX-junctions which is energetically easier to accomplish than the four-strand reaction because the four-strand reaction requires the simultaneously disruption of two DNA duplexes whereas only one duplex is disrupted in the three-strand reaction (Supplementary Figure S2D).
Thus, RAD54 or BLM can promote regression of replication forks through short regions of heterology, albeit at a reduced rate. Even though RAD54 lacks helicase activity, it can still tolerate a limited number of unpaired bases to bypass the vast majority of DNA lesions it would encounter during fork regression.
RAD51 stimulates fork regression by RAD54, but not by BLM
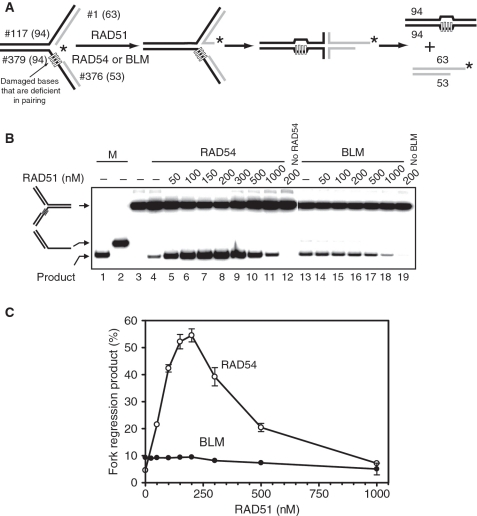
RAD51, a major protein of HR, interacts physically and functionally with both RAD54 and BLM; moreover RAD51 is known to stimulate various activities of RAD54 including branch migration (25,37,55). Here, we asked whether RAD51 can stimulate fork regression promoted by RAD54 and BLM. To test the effect of RAD51 we used the model replication fork containing four heterologous bases on the leading DNA strand template, which mimic a DNA lesion (Figure 3A). We found that RAD51 strongly stimulates RAD54-dependent fork regression (Figure 3B and C). The maximal stimulation, ∼10-fold, was achieved at 200 nM RAD51. This corresponds to a stochiometric ratio of two RAD51 monomers per one RAD54 (Figure 3B, Lanes 3–10; Figure 3C). In contrast, under tested conditions RAD51 had no significant effect on BLM-dependent fork regression (Figure 3B, Lanes 12–17; Figure 3C). Similarly, we tested if RAD51 stimulated the branch migration of PX-junctions by BLM and found that RAD51 had no stimulatory effect (data not shown). Thus, our current data demonstrate specific cooperation between RAD54 and RAD51 in regression of replication forks.
Figure 3.
RAD51 stimulates fork regression promoted by RAD54, but not by BLM. (A) The experimental scheme. The leading DNA strand template contains four heterologous bases. The asterisk indicates the 32P-label. (B) To promote fork regression RAD54 (100 nM) or BLM (50 nM) were incubated with the replication fork (#117/1* + #379/376) in the presence of RAD51 in indicated concentrations. Lanes 1 and 2 show DNA migration markers. In controls (Lanes 3, 4, 12, 13, 19), storage buffers were added instead of RAD51, RAD54, or BLM. The DNA products of the reaction were analyzed by electrophoresis in an 8% polyacrylamide gel. (C) The data from (B) are presented as graphs. The error bars indicate SEM.
RAD54 catalyzes both regression and restoration of replication forks
After completion of DNA repair synthesis, the chicken foot structure must be converted back to a replication fork. This can be achieved by branch migration in the direction opposite to that of fork regression (Figure 4A). We wished to examine whether RAD54 can catalyze restoration of replication forks. We designed a set of four oligonucleotides that can be annealed by two alternative ways to produce either the replication fork or the chicken foot structure (Figure 4A and C). The terminal regions of the arms were mutually heterologous preventing their complete separation; therefore the replication fork and the chicken foot structure would exist in equilibrium. This DNA substrate allowed us to compare the efficiency of fork restoration with that of fork regression. To mimic a DNA lesion that stalls the replication fork, we incorporated a single isocytosine (iso-C) residue (56) in the oligonucleotide that represents the template of the leading DNA strand (Figure 4A).
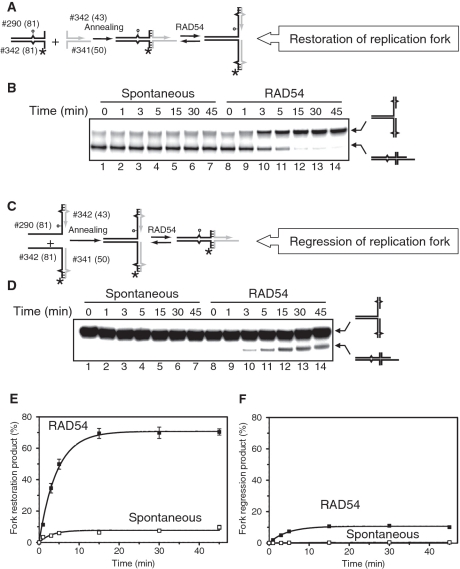
Figure 4.
RAD54 promotes restoration of replication forks more efficiently than fork regression. (A) The scheme of the chicken foot structure preparation and its regression to the replication fork. The circle indicates the position of iso-C that mimics a lesion on the leading DNA strand template. Unpaired single DNA bases are shown by carets. Hatched regions denote heterologous DNA terminal regions that prevent complete strand separation during fork regression. (B) RAD54 (100 nM) (Lanes 8–14) were incubated with the chicken foot structure (#341/342* + #340/290) (32 nM, molecules) for the indicated periods of time. In Lanes 1–7, RAD54 was replaced with storage buffer. The DNA products were analyzed by electrophoresis in an 8% polyacrylamide gel. (C) The scheme of the replication fork preparation and its regression to the chicken foot structure. (D) Spontaneous and Rad54-driven regression of replication forks. RAD54 (100 nM) (Lanes 8–14) were mixed with the replication fork structure (#290/342* + #340/341) (32 nM, molecules) and incubated for indicated periods of time. In Lane 1–7 RAD54 was replaced with storage buffer. Products of the reaction were analyzed by electrophoresis in an 8% polyacrylamide gel. (E and F) The data from (B) and (D) are presented as graphs. Background amounts of the DNA products (B and D, Lanes 1 and 8) were subtracted from the data before plotting. The error bars indicate SEM.
We found that RAD54 can use the chicken foot structure to promote restoration of the replication fork with a high efficiency; the reaction was almost completed in 5 min (Figure 4B and E). In contrast, we observed a significantly lower extent of the chicken foot structure formation when the replication fork was used as a substrate for RAD54; only 10% of the fork was converted after 15 min, with no further increase within a 45-min course of reaction (Figure 4C and F). In the absence of RAD54, the rates of both spontaneous fork regression and restoration were significantly slower; however, like in the RAD54-driven reaction the fork restoration occurred with a higher rate than the fork regression (Figure 4B, Lanes 1–7 and Figure 4C, Lanes 1–7).
Thus, both in the presence or absence of RAD54, the equilibrium between the chicken foot structure and the replication fork is shifted toward the fork indicating greater stability of the later structure. Therefore, formation of the chicken foot structure may represent the rate-limiting step of the template switch mechanism of DNA lesion bypass.
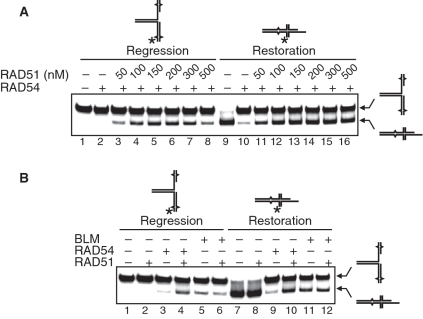
RAD51 promotes accumulation of the chicken foot structure
We demonstrated above that RAD51 stimulates RAD54-dependent fork regression (Figure 3). Here, we wished to examine the effect of RAD51 on the equilibrium between the chicken fork structure and the replication fork in the RAD54-catalyzed reaction. Using a model replication fork as a substrate we found that RAD51 stimulated formation of the chicken fork structure by RAD54 (Figure 5A, Lanes 2–7). Surprisingly, using the chicken foot structure as a substrate we found that RAD51 inhibited conversion of this structure to the fork by RAD54 (Figure 5A, Lanes 10–15). Thus, RAD51 causes an accumulation of the chicken foot structure. We also tested the effect of RAD51 on the BLM-driven reactions. Interestingly, BLM, similar to RAD54, promotes restoration of the replication fork with a greater efficiency than its regression (Figure 5B, Lanes 5 and 11). However, we found no stimulation of either fork regression or fork restoration reactions under tested conditions (Figure 5B, compare Lanes 5 and 6; 11 and 12). These data show specificity in RAD51 stimulation of RAD54; namely, RAD51 promotes accumulation of the chicken foot structure.
Figure 5.
RAD51 stimulates RAD54-driven fork regression and inhibits fork restoration. (A) The effect of RAD51 concentration on a RAD54-dependent fork regression and restoration. RAD54 (100 nM) was mixed with the replication fork (#290/342* + #340/341) (32 nM, molecules) (Lanes 2–7) or the chicken foot structure (#341/342* + #340/290) (32 nM, molecules) (Lanes 10–15) in the presence of indicated concentrations of RAD51, or in the absence of RAD51 (Lanes 1 and 9). In the control reaction, the chicken fork structure was incubated in the absence of both RAD54 and RAD51 (Lane 8). (B) The effect of RAD54 (100 nM), RAD51 (200 nM) and BLM (50 nM) on the regression and restoration of the replication fork. Replication fork (#290/342* + #340/341) (32 nM, molecules) (Lanes 1–6) or chicken foot structure (#341/342* + #340/290) (32 nM, molecules) (Lanes 7–12) were mixed with indicated proteins. DNA products were analyzed by electrophoresis in an 8% polyacrylamide gel.
DNA lesion bypass through the template strand switch mechanism in vitro
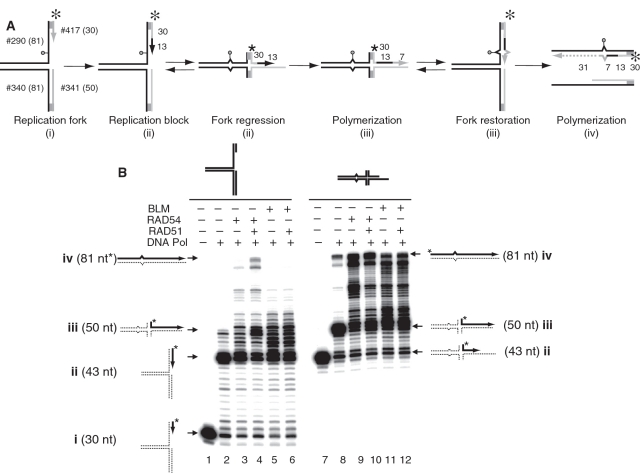
To examine the role of RAD54, BLM and RAD51 in DNA lesion bypass we wished to model this process in vitro. As a representative generic DNA polymerase, we employed DNA polymerase I Klenow Fragment (3′–5′ exo−). As a substrate we used a model replication fork containing a modified base, iso-C, incorporated in the leading DNA strand template (Figure 6A). Iso-C blocks DNA synthesis, since DNA polymerase cannot incorporate any base residue against iso-C, except isoguanine (iso-G) (57). We found that DNA polymerase alone could extend the leading DNA strand 13 nt up to the iso-C position (product ii, 43 nt) (Figure 6B, compare Lanes 1 and 2), but failed to bypass it (Figure 6B, Lane 2). Addition of RAD54 (Figure 6B, Lane 3) or BLM (Figure 6B, Lane 5) stimulated iso-C bypass, likely by promoting fork regression, and resulted in formation of the product iii (50 nt). However, in the presence of RAD54 or BLM only small amounts of a full-length product iv (81 nt) was formed. Addition of RAD51 to the DNA polymerase reaction containing RAD54 increased accumulation of the fully extended product of DNA polymerization (Figure 6B, Lane 4). In contrast, but as expected from previous results (Figure 5B) RAD51 had no stimulatory effect on the reaction containing BLM (Figure 6B, Lane 6).
Figure 6.
In vitro reconstitution of the DNA lesion bypass through the template switch mechanism. (A) The scheme of the lesion bypass via a mechanism that involves fork regression, DNA synthesis using the resulted chicken foot structure as a template, and fork restoration. The circle indicates the position of iso-C that mimics a DNA lesion at the stalled replication fork. The asterisk indicates the 32P-label. Shaded regions denote heterologous DNA terminal branches that prevent complete strand separation during fork regression. The numbers indicate the numbers and length (nt) of the DNA fragments in the substrates and the products of the reaction. (B) DNA polymerase reactions were carried out using DNA polymerase I Klenow Fragment (10 ng/ml) and the replication fork (#290/417* + #340/341) (32 nM, molecules) (left) or the chicken foot structure (#341/342* + #340/290) (32 nM, molecules) (right) as templates. RAD54 (100 nM), BLM (50 nM) and RAD51 (200 nM) were added to the reactions as indicated in ‘Materials and Methods’ section. The products of DNA synthesis were analyzed by electrophoresis in a 15% denaturing polyacrylamide gel.
If RAD51stimulates lesion bypass by promoting accumulation of the chicken foot structure, then no stimulatory effect of RAD51 should be expected when the chicken foot structure is used as a template for DNA polymerase (Figure 6B, Lanes 7–12). Using this substrate, we found that RAD54 or BLM efficiently stimulates formation of the full-length product iv (81 nt) of DNA synthesis, likely by promoting restoration of the replication fork from the chicken foot structure after its partial extension by DNA polymerase (Figure 6B, Lanes 9 and 11). In contrast, we found that with the chicken foot structure RAD51 had no stimulatory effect on the formation of the full-length product iv (81 nt) in the presence of either BLM or RAD54 (Figure 6B, compare Lanes 9 and 10; and Lanes 11 and 12). Taken together, these data indicate that RAD51 stimulates the RAD54-dependent bypass of DNA lesions by promoting regression of replication forks into the chicken foot structure.
Discussion
Our results demonstrate the feasibility of DNA lesion bypass by DNA polymerase through the template switch mechanism promoted by the RAD54 and RAD51 proteins. RAD54, a DNA branch migration protein, is capable of both promoting regression and restoration of a model replication fork. Importantly, RAD51, by shifting the equilibrium in this process toward the fork regression, increases the efficiency of DNA lesion bypass by DNA polymerase.
Multiple DNA repair systems have evolved to remove DNA lesions prior to genome replication (58). Nevertheless, at every cycle of DNA replication the DNA polymerase encounters unrepaired lesions that may block replication fork progression (1). Genetic data indicate that replication forks can bypass DNA lesions in the process known as DNA damage tolerance. In eukaryotes, DNA damage tolerance is carried out by HR and Rad6/Rad18 pathway (59).
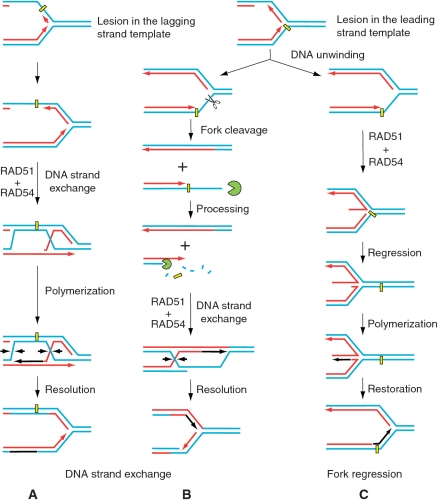
Several models have been proposed to explain how HR can promote DNA lesion bypass (40). In these models, the daughter-strand gaps resulted from replication fork blocks or ssDNA tails produced by the cleavage of stalled replication forks by structure-specific nucleases, e.g. Mus81 (41,60,61), can be repaired through joint molecule formation with the undamaged sister chromatid, in a process that depends on DNA strand exchange activity of Rad51 (Figure 7A and B). However, recent studies underscore the importance of the branch migration activity of HR proteins in DNA repair (36,40). While DNA strand exchange activity is pivotal to the function of HR, our current data demonstrate that the concerted action of RAD51 and RAD54 provides an alternative means for DNA lesion bypass by DNA polymerase via the template strand switch mechanism (Figure 7C).
Figure 7.
Possible role of RAD54 and RAD51 proteins in DNA lesion bypass by replication forks. (A and B) Stalled replication forks can be rescued by HR via a mechanism that relies on DNA strand exchange activity of RAD51. RAD51 and auxiliary proteins including RAD54 promote pairing of either the daughter-strand gaps (A) or ssDNA tails produced as a result of fork cleavage (B) with undamaged chromatid. (C) Alternatively, stalled DNA replication can be restored through a mechanism that involves the DNA template strand switch. The important step of this mechanism involving fork regression and formation of the chicken foot structure may be carried out by the RAD51 and RAD54 proteins.
In addition to Rad54 (35), several eukaryotic proteins have been reported recently to promote branch migration of Holliday junctions or Holliday junction-like structures including model replication forks in vitro. In humans, these proteins include members of the RecQ helicase family, WRN, BLM, and RecQ5β (47–49), and a Fanconi anemia helicase FANCM (62). S. cerevisiae Rad5, a member of the Rad6/Rad18 pathway, and its human ortholog HLTF is also capable of promoting regression of a model replication fork (51,52). Recent data indicate multiple and non-redundant functions of these proteins, e.g. RAD54 and BLM, in mammalian cells (63).
Further experiments are needed to better understand the specific functions of these branch migration proteins in DNA damage tolerance. However, our current results demonstrate that branch migration activity alone is necessary, but not sufficient to generate chicken fork structures in amounts required for an efficient DNA lesion bypass through the template-strand switch mechanism, as both RAD54 and BLM show a strong bias toward fork restoration. Similarly, in protein-independent reaction the equilibrium is also shifted toward the replication fork. In our experiments, RAD51 specifically stimulates the regression of replication forks by RAD54 and inhibits reverse regression of the resulting chicken foot structure to the replication fork. Such an arrangement is required to achieve DNA lesion bypass by DNA polymerase. The mechanism of this selectivity of RAD51 remains to be investigated. However, the specific interaction between RAD51 and RAD54 plays a central role in this selectivity, as RAD51 does not stimulate a BLM-dependent fork regression. The ability of RAD51 to bind ssDNA–dsDNA junctions (64) may stimulate RAD54 loading onto the fork junction promoting its conversion into the chicken-fork structure (37). In contrast, such stimulation of loading may not occur with the chicken foot structure, as the ssDNA region in this structure is not adjacent to the junction. Consequently, RAD51 stimulates only RAD54-dependent fork regression, but not fork restoration. Thus, while many proteins that possess branch migration activity could likely promote both replication fork regression and restoration, a specific bias toward fork regression of branch migration enzymes or their auxiliary protein partners seems to be especially important for stimulation of lesion bypass by DNA polymerase. In addition to protein factors, the propensity of the chicken foot structure formation could also be increased by positive supercoiling in DNA at the replication fork (65), however the supercoil status of stalled replication forks is currently unknown.
Recently, it was demonstrated that in mammalian cells RAD51 has distinct early and late roles in the repair of hydroxyurea-stalled replication forks (66). It was suggested that while at the late stage RAD51 mediates the repair of collapsed forks through the canonical ssDNA invasion mechanism, at the early step it promotes DNA lesion bypass through formation of the chicken fork structure. Our current biochemical study provides a strong support for this early role of RAD51; it indeed can promote formation of the chicken foot structure in vitro by acting together with RAD54 branch migration protein. Also, formation Rad51-dependent Holliday junctions was previously observed in sgs1 yeast cells during replication on the MMS-damaged DNA, although the molecular mechanism is unknown (67).
Overall, previous and our current data suggest RAD51 and RAD54 proteins may be involved in stalled replication fork restart by three different mechanisms, which can be used depending on a particular type of DNA lesion or availability of other supplementary enzymes. First, RAD51 and RAD54 may participate in the repair of lagging (not shown) or leading strand template lesion (Figure 7A) by promoting DNA strand exchange between ssDNA gap in the daughter strand and homologous sister chromatid. Second, both proteins may also be required for the repair of a stalled fork that was first cleaved by structure-specific endonucleases following DNA double strand processing by exonucleases (Figure 7B). In this case, RAD51 and RAD54 promote D-loop formation, and later catalyse branch migration of Holliday junctions that are resolved by structure-specific nucleases to restore the replication fork. Finally, our current results suggest that RAD54, in coordination with RAD51, provides lesion bypass through the DNA template switch mechanism that involves fork regression and restoration (Figure 7C).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health Grant (CA100839 to A.V.M., AG033484-01 to M.J.R.); Leukemia and Lymphoma Society Scholar Award (1054-09 to A.V.M.). Funding for open access charge: National Institutes of Health Grant (CA100839 to A.V.M.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank A. Kuzminov, O. Mazina and F. Huang for the comments and discussion.
REFERENCES
- 1.Rothstein R, Michel B, Gangloff S. Replication fork pausing and recombination or ‘gimme a break’. Genes Dev. 2000;14:1–10. [PubMed] [Google Scholar]
- 2.Budzowska M, Kanaar R. Mechanisms of dealing with DNA damage-induced replication problems. Cell Biochem. Biophys. 2009;53:17–31. doi: 10.1007/s12013-008-9039-y. [DOI] [PubMed] [Google Scholar]
- 3.Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 4.Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 6.Michel B, Boubakri H, Baharoglu Z, LeMasson M, Lestini R. Recombination proteins and rescue of arrested replication forks. DNA Repair. 2007;6:967–980. doi: 10.1016/j.dnarep.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 7.Wu L, Hickson ID. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu. Rev. Genet. 2006;40:279–306. doi: 10.1146/annurev.genet.40.110405.090636. [DOI] [PubMed] [Google Scholar]
- 8.Hanawalt PC. Paradigms for the three rs: DNA replication, recombination, and repair. Mol. Cell. 2007;28:702–707. doi: 10.1016/j.molcel.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 9.Kuzminov A. DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc. Natl Acad. Sci. USA. 2001;98:8461–8468. doi: 10.1073/pnas.151260698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kogoma T. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuzminov A. Collapse and repair of replication forks in Escherichia coli. Mol. Microbiol. 1995;16:373–384. doi: 10.1111/j.1365-2958.1995.tb02403.x. [DOI] [PubMed] [Google Scholar]
- 12.Unk I, Hajdu I, Blastyak A, Haracska L. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair. 2010;9:257–267. doi: 10.1016/j.dnarep.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 13.Heller RC, Marians KJ. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 2006;439:557–562. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- 14.Neale MJ, Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wyman C, Ristic D, Kanaar R. Homologous recombination-mediated double-strand break repair. DNA Repair. 2004;3:827–833. doi: 10.1016/j.dnarep.2004.03.037. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Ann. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 18.Krogh BO, Symington LS. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 19.Mimitou EP, Symington LS. DNA end resection: many nucleases make light work. DNA Repair. 2009;8:983–995. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc. Natl Acad. Sci. USA. 2008;105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci. 1998;3:D570–D603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 22.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 23.Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl Acad. Sci. USA. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thoma NH, Czyzewski BK, Alexeev AA, Mazin AV, Kowalczykowski SC, Pavletich NP. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nat. Struct. Mol. Biol. 2005;12:350–356. doi: 10.1038/nsmb919. [DOI] [PubMed] [Google Scholar]
- 25.Mazin AV, Mazina OM, Bugreev DV, Rossi MJ. Rad54, the motor of homologous recombination. DNA Repair. 2010;9:286–302. doi: 10.1016/j.dnarep.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan TL, Kanaar R, Wyman C. Rad54, a Jack of all trades in homologous recombination. DNA Repair. 2003;2:787–794. doi: 10.1016/s1568-7864(03)00070-3. [DOI] [PubMed] [Google Scholar]
- 27.Heyer WD, Li X, Rolfsmeier M, Zhang XP. Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 2006;34:4115–4125. doi: 10.1093/nar/gkl481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 29.Mazin AV, Alexeev AA, Kowalczykowski SC. A novel function of Rad54 protein. Stabilization of the Rad51 nucleoprotein filament. J. Biol. Chem. 2003;278:14029–14036. doi: 10.1074/jbc.M212779200. [DOI] [PubMed] [Google Scholar]
- 30.Mazina OM, Mazin AV. Human Rad54 protein stimulates DNA strand exchange activity of hRad51 protein in the presence of Ca2+ J. Biol. Chem. 2004;279:52042–52051. doi: 10.1074/jbc.M410244200. [DOI] [PubMed] [Google Scholar]
- 31.Sigurdsson S, Van Komen S, Petukhova G, Sung P. Homologous DNA pairing by human recombination factors Rad51 and Rad54. J. Biol. Chem. 2002;277:42790–42794. doi: 10.1074/jbc.M208004200. [DOI] [PubMed] [Google Scholar]
- 32.Amitani I, Baskin RJ, Kowalczykowski SC. Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol. Cell. 2006;23:143–148. doi: 10.1016/j.molcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Alexeev A, Mazin A, Kowalczykowski SC. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat. Struct. Biol. 2003;10:182–186. doi: 10.1038/nsb901. [DOI] [PubMed] [Google Scholar]
- 34.Li X, Zhang XP, Solinger JA, Kiianitsa K, Yu X, Egelman EH, Heyer WD. Rad51 and Rad54 ATPase activities are both required to modulate Rad51-dsDNA filament dynamics. Nucleic Acids Res. 2007;35:4124–4140. doi: 10.1093/nar/gkm412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bugreev DV, Mazina OM, Mazin AV. Rad54 protein promotes branch migration of Holliday junctions. Nature. 2006;442:590–593. doi: 10.1038/nature04889. [DOI] [PubMed] [Google Scholar]
- 36.Bugreev DV, Hanaoka F, Mazin AV. Rad54 dissociates homologous recombination intermediates by branch migration. Nat. Struct. Mol. Biol. 2007;14:746–753. doi: 10.1038/nsmb1268. [DOI] [PubMed] [Google Scholar]
- 37.Rossi MJ, Mazin AV. Rad51 protein stimulates the branch migration activity of Rad54 protein. J. Biol. Chem. 2008;283:24698–24706. doi: 10.1074/jbc.M800839200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mazina OM, Rossi MJ, Thomaa NH, Mazin AV. Interactions of human rad54 protein with branched DNA molecules. J. Biol. Chem. 2007;282:21068–21080. doi: 10.1074/jbc.M701992200. [DOI] [PubMed] [Google Scholar]
- 39.Pages V, Fuchs RP. Uncoupling of leading- and lagging-strand DNA replication during lesion bypass in vivo. Science. 2003;300:1300–1303. doi: 10.1126/science.1083964. [DOI] [PubMed] [Google Scholar]
- 40.Mazloum N, Holloman WK. Brh2 promotes a template-switching reaction enabling recombinational bypass of lesions during DNA synthesis. Mol. Cell. 2009;36:620–630. doi: 10.1016/j.molcel.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 2007;14:1096–1104. doi: 10.1038/nsmb1313. [DOI] [PubMed] [Google Scholar]
- 42.Osman F, Whitby MC. Exploring the roles of Mus81-Eme1/Mms4 at perturbed replication forks. DNA Repair. 2007;6:1004–1017. doi: 10.1016/j.dnarep.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 43.Higgins NP, Kato K, Strauss B. A model for replication repair in mammalian cells. J. Mol. Biol. 1976;101:417–425. doi: 10.1016/0022-2836(76)90156-x. [DOI] [PubMed] [Google Scholar]
- 44.Robu ME, Inman RB, Cox MM. RecA protein promotes the regression of stalled replication forks in vitro. Proc. Natl Acad. Sci. USA. 2001;98:8211–8218. doi: 10.1073/pnas.131022698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seigneur M, Bidnenko V, Ehrlich SD, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 46.McGlynn P, Lloyd RG. Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell Biol. 2002;3:859–870. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]
- 47.Ralf C, Hickson ID, Wu L. The Bloom’s syndrome helicase can promote the regression of a model replication fork. J. Biol. Chem. 2006;281:22839–22846. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- 48.Machwe A, Xiao L, Groden J, Orren DK. The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry. 2006;45:13939–13946. doi: 10.1021/bi0615487. [DOI] [PubMed] [Google Scholar]
- 49.Kanagaraj R, Saydam N, Garcia PL, Zheng L, Janscak P. Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 2006;34:5217–5231. doi: 10.1093/nar/gkl677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gari K, Decaillet C, Delannoy M, Wu L, Constantinou A. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc. Natl Acad. Sci. USA. 2008;105:16107–16112. doi: 10.1073/pnas.0804777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blastyak A, Pinter L, Unk I, Prakash L, Prakash S, Haracska L. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol. Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blastyak A, Hajdu I, Unk I, Haracska L. Role of double-stranded DNA translocase activity of human HLTF in replication of damaged DNA. Mol. Cell. Biol. 2010;30:684–693. doi: 10.1128/MCB.00863-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bugreev DV, Mazina OM, Mazin AV. Analysis of branch migration activities of proteins using synthetic DNA substrates. Nature Protocols. 2006 September 1 (doi:2010.1038/nprot.2006.2217; epub ahead of print) [Google Scholar]
- 54.Karow JK, Newman RH, Freemont PS, Hickson ID. Oligomeric ring structure of the Bloom's syndrome helicase. Curr. Biol. 1999;9:597–600. doi: 10.1016/s0960-9822(99)80264-4. [DOI] [PubMed] [Google Scholar]
- 55.Bugreev DV, Mazina OM, Mazin AV. Bloom syndrome helicase stimulates RAD51 DNA strand exchange activity through a novel mechanism. J. Biol. Chem. 2009;284:26349–26359. doi: 10.1074/jbc.M109.029371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson SC, Sherrill CB, Marshall DJ, Moser MJ, Prudent JR. A third base pair for the polymerase chain reaction: inserting isoC and isoG. Nucleic Acids Res. 2004;32:1937–1941. doi: 10.1093/nar/gkh522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Switzer CY, Moroney SE, Benner SA. Enzymatic recognition of the base pair between isocytidine and isoguanosine. Biochemistry. 1993;32:10489–10496. doi: 10.1021/bi00090a027. [DOI] [PubMed] [Google Scholar]
- 58.Friedberg EC, Walker GC, Siede W, Wood RD, Ellenberger T. DNA repair and Mutagenesis. Washington, DC: ASM Press; 2006. [Google Scholar]
- 59.Gangavarapu V, Prakash S, Prakash L. Requirement of RAD52 group genes for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell Biol. 2007;27:7758–7764. doi: 10.1128/MCB.01331-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whitby MC, Osman F, Dixon J. Cleavage of model replication forks by fission yeast Mus81-Eme1 and budding yeast Mus81-Mms4. J. Biol. Chem. 2003;278:6928–6935. doi: 10.1074/jbc.M210006200. [DOI] [PubMed] [Google Scholar]
- 61.Mazina OM, Mazin AV. Human Rad54 protein stimulates human Mus81-Eme1 endonuclease. Proc. Natl Acad. Sci. USA. 2008;105:18249–18254. doi: 10.1073/pnas.0807016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol. Cell. 2008;29:141–148. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- 63.Chu WK, Hanada K, Kanaar R, Hickson ID. BLM has early and late functions in homologous recombination repair in mouse embryonic stem cells. Oncogene. 2010;29:4705–4714. doi: 10.1038/onc.2010.214. [DOI] [PubMed] [Google Scholar]
- 64.Mazin AV, Zaitseva E, Sung P, Kowalczykowski SC. Tailed duplex DNA is the preferred substrate for Rad51 protein-mediated homologous pairing. EMBO J. 2000;19:1148–1156. doi: 10.1093/emboj/19.5.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Postow L, Ullsperger C, Keller RW, Bustamante C, Vologodskii AV, Cozzarelli NR. Positive torsional strain causes the formation of a four-way junction at replication forks. J. Biol. Chem. 2001;276:2790–2796. doi: 10.1074/jbc.M006736200. [DOI] [PubMed] [Google Scholar]
- 66.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.