Abstract
The multistep kinetics through which DNA-binding proteins bind their targets are heavily studied, but relatively little attention has been paid to proteins leaving the double helix. Using single-DNA stretching and fluorescence detection, we find that sequence-neutral DNA-binding proteins Fis, HU and NHP6A readily exchange with themselves and with each other. In experiments focused on the Escherichia coli nucleoid-associated protein Fis, only a small fraction of protein bound to DNA spontaneously dissociates into protein-free solution. However, if Fis is present in solution, we find that a concentration-dependent exchange reaction occurs which turns over the bound protein, with a rate of kexch = 6 × 104 M−1s−1. The bacterial DNA-binding protein HU and the yeast HMGB protein NHP6A display the same phenomenon of protein in solution accelerating dissociation of previously bound labeled proteins as exchange occurs. Thus, solvated proteins can play a key role in facilitating removal and renewal of proteins bound to the double helix, an effect that likely plays a major role in promoting the turnover of proteins bound to DNA in vivo and, therefore, in controlling the dynamics of gene regulation.
INTRODUCTION
Many small proteins, such as transcription factors, bind the DNA double helix so as to control chromosome function and dynamics. Understanding the kinetic pathways through which such proteins find their binding sites is a subject of intense study, with 3D and 1D diffusion processes having been discussed theoretically (1–3) and investigated experimentally (4–6). However, release of proteins from DNA is also important for regulation of chromosome function. In the absence of active processes (e.g. chromatin remodeling factors), release of proteins from DNA is generally assumed to be governed by their off-rates (or for a complex, by a dissociation timescale for the complex). The influence of proteins in solution on the turnover of transcription factors has not been a subject of careful study. Here, we show that proteins in solution can play a crucial role in mediating exchange kinetics through rapid turnover of proteins which would otherwise remain bound to the double helix on a long timescale, and present evidence that this is a general phenomenon.
Our study focuses on the Escherichia coli nucleoid-associated protein Fis, which acts as both a specific regulatory protein for a diverse set of DNA reactions and a general chromosome-compacting factor (7–10). Cellular Fis levels are drastically up-regulated upon nutrient upshifts and remain very high in rapid growth conditions, making Fis along with HU the most abundant DNA binding proteins in rapidly growing E. coli cells (both at approximately 30 000 dimers per cell) (11,12). Fis binds DNA in a relatively sequence-neutral manner, with complexes forming on random DNA fragments in vitro beginning with a Kd of ∼1 nM and with formation of a fully coated DNA fragment at 20 ± 10 nM (13,14). At higher Fis concentrations, increasing numbers of Fis dimers associate with the Fis-DNA filaments to form high-order complexes. In the presence of a large excess of DNA, Fis binds selectively to AT-rich sites with low nanomolar binding constants (15). These specific binding sites, which exhibit only a modest relationship to each other at the primary sequence level (15–19), are most often found in intergenic regions (16,20) and can function to positively or negatively regulate transcription (21–24). During slow growth or as cells adapt to starvation conditions, cellular Fis levels are low and thus primarily high affinity Fis binding sites are expected to be bound. However, since Fis levels in rapidly growing cells can exceed 50 µM, much of the Fis under these conditions must be bound non-specifically throughout the chromosome. Fis acts to compact DNA in vitro by introducing bends ranging up to 90° per Fis dimer complex (25–28) and by formation and stabilization of loops between high-order complexes (14,29).
We also study two other sequence-independent DNA binding and bending proteins. HU also compacts DNA and participates in many DNA transactions (7,8,30). NHP6A, a member of the HMGB family of DNA-binding proteins, is an abundant constituent of Saccharomyces cerevisiae chromosomes that functions to organize chromatin and control transcription (31,32). Fis, HU and NHP6A are structurally unrelated and bind and bend DNA by very different mechanisms (28,33,34).
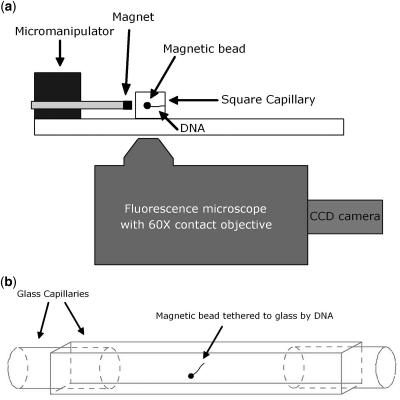
Previous single molecule mechanical measurements quantitatively demonstrated the bending and looping abilities of Fis, but did not elucidate the binding or release dynamics (14,29). Mechanical studies of Fis, HU, HMGB1 and NHP6A have also suggested that once these proteins bind DNA they remain bound for long times in protein-free solution (14,35,36). Such mechanical measurements do not allow for simple analyses of the stability of protein–DNA complexes in protein-free solution, whether or not bound protein exchanges with solution-phase protein, or the nature of that exchange if it indeed occurs. To address these questions, we developed a novel combined magnetic tweezers-fluorescence microscope allowing extension of single DNA molecules in the focal plane (37) of an inverted epifluorescence microscope (schematic, Figure 1A). In addition to being able to apply precisely calibrated forces to extend individual DNA molecules, this system allows direct measurement of DNA length as well as visualization of fluorescently labeled proteins bound to the DNA. Traditional magnetic tweezers are able to make mechanical measurements of DNA–protein interactions, but are limited in their ability to detect absolute numbers of proteins bound to DNA due to their relatively indirect method of estimating protein binding from extension changes (38).
Figure 1.
Schematic of instrument and sample cell. (a) DNA is tethered between the wall of a square glass capillary tube and a paramagnetic bead. Force is applied using a permanent magnet to extend the DNA in the focal plane of a fluorescence microscope (force is to the left in schematic). Images are acquired using an EMCCD camera (see ‘Materials and Methods’ section for details). (b) Schematic of sample cell assembled from glass capillaries. Force is out of the page normal to the facing surface of the cell.
MATERIALS AND METHODS
Protein preparation
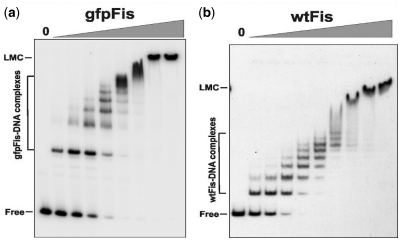
The GFP F64L S65T coding region from pRJ1510 (39) was inserted into the MluI site at codon 5 of fis using PCR to generate gfpFis. The resulting sequence beginning at the fis N-terminus is: ATG TTC GAA CAA•GCA AGT AAA gfp coding sequence TAC AAA•CGC GTA followed by the remainder of fis. In vitro gel shift assays performed with purified protein as described in ref. (14), except that only 100 µg/ml BSA was present in the binding buffer, showed that gfpFis had similar DNA binding properties to wtFis (Figure 6). Formation of the first bound complex by gfpFis on a 150 bp DNA fragment from the coding region the S. cerevisiae MET14 gene occurred with an approximate Kd of 3.1 ± 1.4 nM as compared with 1.7 ± 0.5 nM for wtFis, and formation of the fully coated DNA complex [seven Fis dimers, see ref. (14)] occurred with Kd’s of 27.5 ± 7.8 and 32.0 ± 3.6 nM, respectively. Moreover, gfpFis forms the high-order ‘low mobility complex’ (14) in a similar manner as wtFis. Purified gfpFis also cooperatively binds with the phage λ Xis protein with similar properties as wtFis (C.V. Papagiannis and R.C. Johnson, unpublished data).
Figure 6.
Non-specific DNA binding by gfpFis and wtFis evaluated by gel mobility shift experiments. gfpFis (a) and wtFis (b) were incubated with a 150 bp 32P-labeled fragment from the S. cerevisiae MET14 gene and subjected to electrophoresis in a native 5% polyacrylamide gel. 0 designates no added protein followed by 2-fold increasing amounts of protein beginning at 1.3 nM for gfpFis and 1.1 nM for wtFis. Complexes containing from 1 to 6–8 dimers of Fis are formed with increasing amounts of added protein in both cases. The calculated Kd for the first bound complex was 3.1 ± 1.4 and 1.7 ± 0.5 nM and for the fully-coated DNA complex [seven Fis dimers, see ref. (14)] was 27.5 ± 7.8 and 32.0 ± 3.6 nM for gfpFis (n = 3) and wtFis (n = 5), respectively. At ≥70 nM both proteins also form a high-order complex referred to as the low-mobility complex (LMC). The slower relative migrations of the gfpFis complexes are consistent with the MW difference of the dimeric proteins: 76.6 kDa for gfpFis and 22.8 kDa for wtFis.
For purification, gfpFis cloned into pET11a (pRJ2010) was expressed in RJ3387 [BL21 (DE3) fis::kan-767 endA8::tet] by overnight induction at 15°C in LB with 1 mM IPTG. Harvested cells resuspended in FB [50 mM HEPES (pH 7.5), 5 mM DTT, 1 mM EDTA and 10% glycerol] + 0.5 M NaCl were lysed by passage through a French Press, and DNA was removed from the clarified lysates after the NaCl was increased to 0.75 M by addition of 0.35% polyethyleneimine (Sigma-Aldrich, St Louis, MO, USA). After centrifugation at 30 000g, gfpFis was precipitated from the supernatant with 476 g/l ammonium sulfate, resuspended in FB and dialyzed against FB + 0.15 M NaCl. gfpFis was purified to near homogeneity by FPLC chromatography on a Bioscale S20 (Bio-Rad, Hercules, CA, USA) column followed by a Superdex 200 column (GE Healthcare, Piscataway, NJ, USA) run in FB + 1 M NaCl. Wild-type Fis was purified in a similar manner except that a Superdex 75 column was used. Fis proteins were stored in FB buffer containing 1 M NaCl and 50% glycerol at −20°C. Purification of NHP6A and NHP6Agfp was described in (36,39) and E. coli HU in (36).
Flow cell and DNA tethering
Experiments were performed in 1 mm square glass capillary tubes (VitroCells, VitroCom, Mountain Lakes, NJ, USA) with a total volume in the experimental region of 10–20 μl. The capillaries were cleaned using an 8% Helmanex solution (Helma Worldwide, Müllheim, Germany) for 10–15 min, rinsed extensively with ethanol (99%) and distilled water, then incubated with 50 μg/ml antidigoxygenin (Roche Diagnostics, Indianapolis, IN, USA) for 50 min. In order to reduce the likelihood of Fis binding to the glass, the capillaries were incubated for 20 min with 0.5–1 mg/ml casein (Sigma-Aldrich, St Louis, MO, USA). Linear 48.5 kbp λ-DNA (Promega, Madison, WI, USA) was labeled on one end (left) with biotin, and on the other (right) with digoxygenin as previously described (37). The labeled DNA was incubated with 2.8 μm streptavidin coated paramagnetic beads (Dynabeads M-280, Invitrogen, Grand Island, NY, USA) and subsequently added to the flow cell and allowed to incubate for 20 min. For the images in Figure 2, the flow cell was prepared as usual but the DNA was added and allowed to bind to the antidigoxygenin-labeled capillary for 20 min. The sample was then washed with buffer containing 0.5 mg/ml casein and beads were then added and allowed to incubate for 20 min. This process reduces the amount of excess DNA bound to the bead improving imaging.
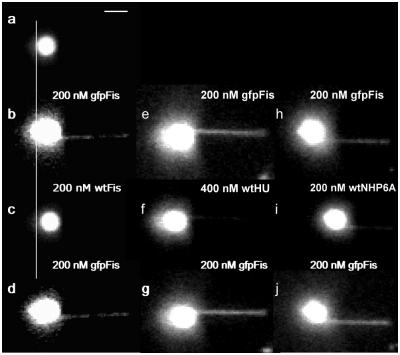
Figure 2.
Sequences of Fis exchange images. DNA is tethered to a glass surface on the right with the magnetic bead on the left and an applied force of 0.7 pN. (a) Image of bead tethered by a single λ-DNA molecule prior to addition of Fis. (b) Image of complex formed on DNA by addition of 200 nM gfpFis, obtained after washing with 1 ml of buffer. (c) Same tether as in (b) after introducing 200 nM wtFis. Exchange occurs on the time-scale required to flow the new solution into the cell (∼30 s). Vertical line highlights the length difference (a two-pixel shift to the right) indicating Fis is bound. (d) Reintroduction of gfpFis replaces wtFis. Image was obtained after buffer wash as above. (e–g) Separate experiment showing exchange between gfpFis and wtHU. (h–j) Similar experiment showing exchange between gfpFis and wtNHP6A. Note the incomplete exchange evident in panels (f and i), both along the tether and the DNA coils bound to the bead, which is likely due in part to differing affinities of the solution phase and bound proteins, as discussed in the text. Scale bar (top) is ∼3 μm.
Magnetic tweezers and fluorescence microscopy
A magnetic tweezers apparatus was designed to extend DNA in the focal plane of an inverted epifluorescence microscope (Figure 1). Four 0.5′′ cube NdFeB magnets were mounted on a micromanipulator (MP-285, Sutter Instrument Company, Novato, CA, USA) perpendicular to the optical axis of a 60× oil-immersion microscope objective (NA = 1.25, PlanApo, Olympus, Melville, NY). Fluorescence microscopy was performed using a Polychrome V (Till Photonics, Gräfelfing, Germany) as the excitation light source and a green fluorescent protein filter set (Semrock, Rochester, NY, USA). Images were acquired with an iXon EMCCD camera (Andor, South Windsor, CT, USA) cooled to −50°C. Data were collected with software created using LabView 6.1 (National Instruments, Austin, TX, USA).
Protein exchange experiments
All buffers were 20 mM HEPES, 100 mM k-glutamate and 0.5 mM EDTA, pH 7.6. For the protein buffers, 0.5 mg/ml casein was added to help prevent protein adsorption to the sides of the flow cell, 10 mM DTT was added to limit photo-oxidation of the fluorophores and 5% glycerol was added to reproduce the conditions of previous experiments as well as to aid in the prevention of Fis adsorbing to the glass. All experiments were performed at room temperature (22°C). Tethered DNA molecules were extended and the applied force measured using the technique previously reported (37). The force was typically measured for three different extensions to determine if the force-extension response matched the worm-like chain model to ensure that a single molecule was tethered.
Once a single tether was found, gfpFis was introduced at a concentration of 200 nM. This concentration was chosen as it is known not to loop DNA, at least at the forces used in these experiments (≥0.7 pN) (14). In all experiments, the gfpFis was allowed to incubate for 3 min to mimic the conditions of previous single-molecule Fis-DNA experiments (14,29), and then the sample was washed with 1 ml of buffer prior to imaging (0.5 s exposure for all images). For the visual Fis exchange experiments (Figure 2), after the gfpFis–DNA complex was imaged 200 nM wtFis was introduced, allowed to incubate for 3 min, washed with 1 ml buffer and then imaged. The final step was to reintroduce 200 nM gfpFis, imaging as above. Introduction and incubation of proteins was done at 1.5–3 pN in order to avoid destruction of the tether. Prior to imaging, the force was reduced to 0.7 pN.
For the exchange rate experiments, gfpFis was introduced at 200 nM, incubated and washed as above. Subsequently, wtFis was introduced at concentrations of 5, 10, 25 or 50 nM, the force reduced to 0.7 pN and the exchange monitored by imaging every 30 s (1 s exposure to excitation light due to delay in remote control of the light source).
All data were analyzed using Igor Pro 6.0.1. To monitor the disappearance of gfpFis fluorescence, the ‘Image Line Profile’ function in Igor was used to measure a region of the gfpFis-DNA complex near the tether point to avoid measuring fluorescence from the bead, providing an average fluorescence intensity measure for the region. A region to the right of the tether, analyzed in the same manner and using the same line profile, was used for background measurements and this data was subtracted from the ‘raw’ data obtained from the tether. To determine the rate of fluorescence disappearance and, therefore, the rate of protein exchange, the best fit to the data was obtained using an exponential with a baseline offset. Two types of controls were performed in order to determine the effect of bleaching and/or spontaneous protein dissociation. For these, gfpFis was added at 200 nM, allowed to incubate and washed, but no wtFis was introduced. One control was done with 30 s imaging intervals and the other with 2 min intervals allowing us to distinguish between protein loss and bleaching.
Experiments with other proteins were carried out in a similar fashion, substituting the concentrations of HU and NHP6A noted in the ‘Results’ section.
RESULTS
Measurements of exchange of Fis bound to DNA with DNA-binding proteins in solution
In order to visualize exchange of Fis bound to λ-DNA with Fis in solution, a single λ-DNA tether, in buffer containing 100 mM k-glutamate, was first identified (Figure 2a). Note that only the intrinsically fluorescent paramagnetic bead is visible in Figure 2a as the DNA is not fluorescently labeled. Next, a 200 nM solution of gfpFis was introduced into the sample cell. After a buffer wash, gfpFis is clearly seen to be coating the length of the DNA, as well as the few excess DNA coils bound to the bead (Figure 2b). Fis bound to the DNA coils is evident from the increase in, and asymmetry of the bead fluorescence (compare Figure 2b to a). We emphasize that the buffer wash removes all free gfpFis from the flow cell (based on the low level of background fluorescence we estimate a free gfpFis concentration <0.1 nM) and that the gfpFis visible in Figure 2b is stably bound to the DNA tether.
Introduction of 200 nM wtFis resulted in rapid, complete removal of gfpFis from both the tethered DNA and the bead-bound DNA (Figure 2c). Notably, this step allows for a direct measure of the length of the DNA, which is shorter than that measured prior to introducing any Fis into the sample (compare Figure 2c to a). Since 200 nM Fis mildly compacts DNA (14), this length decrease (not clearly visible in Figure 2b and d due to fluorescence from gfpFis bound to excess DNA coils bound to the bead as mentioned above) indicates that wtFis is bound in panel 2c and therefore, that it exchanged with the gfpFis. Reintroduction of 200 nM gfpFis resulted in exchange with wtFis as evidenced by the renewed fluorescence signal from both the DNA tether and the DNA on the bead (Figure 2d).
We have found this type of facilitated exchange of DNA-bound protein with solution-phase protein to be common among nonspecific DNA-binding proteins as shown in experiments employing the E. coli nucleoid-associated protein HU and the HMGB chromatin protein NHP6A from S. cerevisiae. In Figure 2, panels e–g show the exchange between gfpFis and wtHU. As with the Fis exchange experiment discussed above, a DNA tether was allowed to incubate with a 200 nM solution of gfpFis and the excess washed away (Figure 2e). A 400 nM solution of wtHU was then introduced and exchange was observed (Figure 2f). Reintroduction of 200 nM gfpFis resulted in its exchange with the bound HU (Figure 2g). The remaining fluorescence associated with the excess DNA coils bound to the bead present in Figure 2f indicates that not all the gfpFis has exchanged.
A similar experiment performed using gfpFis and wtNHP6A further generalizes the exchange phenomenon (Figure 2 h–j). Again, a complex was formed on λ-DNA from 200 nM gfpFis then allowed to exchange with 200 nM wtNHP6A. This exchange has clearly occurred as evidenced by the drastically shortened tether in Figure 2i, which is expected from NHP6A (36). The data in Figure 2j shows that gfpFis exchanges readily with wtNHP6A. Another experiment using NHP6Agfp and wtNHP6A showed that NHP6A will also exchange with itself (Figure 5a).
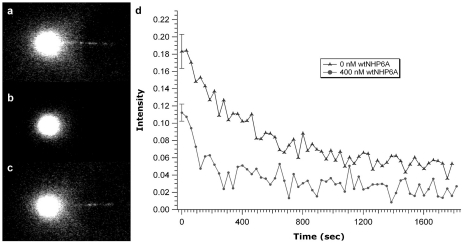
Figure 5.
NHP6A exchange data. (a) Complex formed from 200 nM NHP6Agfp. (b) Addition of 200 nM wtNHP6A resulted in complete exchange with the bound NHP6Agfp. (c) Re-addition of 200 nM NHP6Agfp resulted in exchange with bound wtNHP6A. (d) NHP6A exchange curves. Each curve is the average of several data sets. Because most of the exchange occurs so quickly (i.e. before the first image could be acquired), the data is presented in terms of absolute fluorescence showing a clear acceleration of off rate after addition of 400 nM wtNHP6A (bottom curve) compared to 0 nM wtNHP6A (top curve). The error in the initial fluorescence point is the standard error of the mean for several data sets.
Measurements of rates of exchange of DNA-bound protein with solution-phase protein
To more precisely examine the dynamics of Fis turnover, 200 nM gfpFis was allowed to bind to a λ-DNA. The solution was then exchanged with Fis-free buffer, and fluorescence images were subsequently acquired every 30 s for 30 min. Analysis of these images reveals an ∼20% reduction of fluorescence intensity along the DNA suggesting that 20% of the protein dissociates into protein-free solution over a ∼1800 s time course (Figure 3a, hour-glass markers). The data are well fit by an exponential decay, with a baseline offset, to a final level ∼80% of the initial fluorescence intensity.
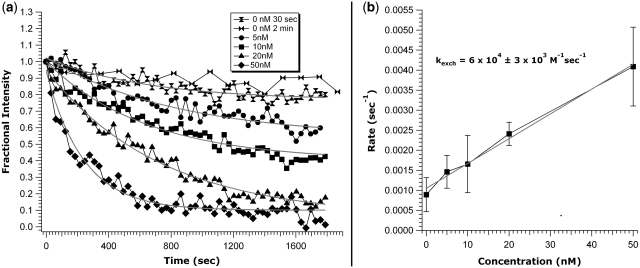
Figure 3.
Fis exchange curves and exchange rate. (a) DNA was incubated with 200 nM gfpFis, and a 1 ml buffer wash was performed prior to addition of wtFis. Images were acquired every 30 s, except for the series denoted by bowties. Note the distinctive trend to higher rates as the wtFis concentration is increased from 5 to 50 nM. Data are normalized to show the fraction of protein exchanged. Overlap of the control curves (top two curves, 30 s and 2 min acquisition intervals) indicates that bleaching contributes negligibly to the measured exchange rates. Solid lines are fits to the data. The 0 nM, 2 min fit is omitted for clarity. (b) Linear exchange rate trend as a function of wtFis concentration obtained from the curve fits in (a). Data error is standard error and error for the fit is ±1 SD.
To verify that the 20% fluorescence reduction was not due to photobleaching, the experiment was repeated on the same time scale, but with images acquired at 2 min intervals, reducing the total exposure to fluorescence excitation by 75%. The resulting time series coincides with the series acquired at 30 s intervals, indicating that photobleaching contributes negligibly, if at all, to the fluorescence decay (Figure 3a, bowties).
The data presented in Figure 2b–d indicates that DNA-bound Fis exchanges readily with Fis in solution. In order to analyze the nature of this exchange reaction, a series of similar experiments were carried out by introducing wtFis at 5, 10, 20 and 50 nM (Figure 3a, circles, squares, triangles and diamonds, respectively) to a sample containing a complex formed from 200 nM gfpFis. For all of the wtFis concentrations used, the fluorescence dropped below the 80% level seen for dissociation into protein-free solution within ∼300 s, and there is a clear trend to faster exchange rate with increasing concentration.
Exchange rates at each wtFis concentration were obtained by fitting the exchange curves to an exponential with a baseline offset. Plotting the rates, obtained from the average of fits to several data sets, as a function of concentration resulted in a linear relationship (Figure 3b), with error bars representing the standard error of the mean. A linear fit to the measured rates results in an exchange rate constant kexch = (6.0 ± 0.3) × 104 M−1 s−1 (error in fit is 1SD). The baseline offset was observed to progressively decrease with increased wtFis concentration, approaching zero for the highest wtFis concentrations studied (20 and 50 nM, see Figure 3a).
A similar analysis of the Fis/HU exchange reaction (Figure 2e and f) was performed to test whether the exponential time course and its linear dependence on solution concentration is a more general phenomenon. In this case, a series of wtHU concentrations was used instead of wtFis (Figure 4a). Again acceleration of the off-rate and gradual reduction of the final fluorescence level is seen with increasing wtHU concentration. The data were fit to an exponential with a baseline offset, obtaining a rate constant of kexch = (2.7 ± 0.5) × 103 M−1 s−1 (Figure 4b).
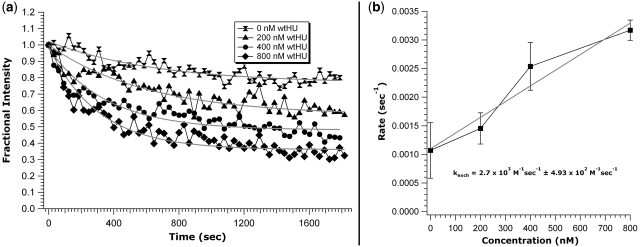
Figure 4.
Exchange curves for different concentrations of wtHU. (a) Experiments were performed exactly as those for Figure 3 using wtHU instead of wtFis. While less pronounced than the gfpFis/wtFis exchange, there is a clear trend toward higher rates with increasing wtHU concentration. (b) The gfpFis/wtHU exchange rate trend also exhibits a linear relationship with an exchange rate constant of (2.7 ± 0.5) × 103 M−1s−1. Data error is standard error and error for the fit is ±1 SD.
To further generalize the effect of concentration on protein off-rate, we performed a third set of experiments using NHP6Agfp and wtNHP6A. Image data in Figure 5a–c shows exchange between NHP6Agfp and wtNHP6A similar to the gfpFis/wtFis exchange presented in Figure 2a–d. This exchange also exhibits a concentration-dependent exchange rate evident from the data in Figure 5d, which was obtained in the same manner as for the Fis and Fis/HU exchange curves. However, in the case of NHP6A, the exchange occurs very rapidly. Indeed, most exchange occured prior to acquisition of the first image. In this case, we used absolute fluorescence and considered only the fluorescence of the initial data point which is significantly lower for the 400 nM wtNHP6A than for the 0 nM wtNHP6A (Figure 5d bottom and top curves, respectively). Unfortunately, the fluorescence of the NHP6Agfp is too dim to reliably extract a rate from the exchange data (note the relation of DNA-bound NHP6Agfp to the background in Figure 5a and c).
DISCUSSION
Dissociation of Fis bound to DNA is accelerated by exchange with DNA-binding proteins in solution
We demonstrate that Fis proteins bound to DNA readily exchange with other solution-phase proteins under physiological salt conditions (100 mM k-glutamate). The replacement of gfpFis by wtFis shown in Figure 2a–d is not a consequence of differing affinities because the reverse exchange (replacement of wtFis with gfpFis, Figure 2c and d), occurs on the same time scale. Moreover, gel-shift experiments employing small DNA fragments demonstrate that the two proteins non-specifically bind DNA with similar properties (Figure 6 and ‘Material and Methods’ section). In addition, mechanical measurements indicate that the overall binding, compaction and looping properties of gfpFis and wtFis are nearly indistinguishable. This is reasonable since the gfp has been fused at the Fis N terminus, distant from the C-terminal DNA-binding domain. The gfpFis/wtFis exchange reaction can, therefore, be considered to involve proteins with nearly equivalent DNA-binding properties.
We have shown that this type of facilitated exchange of DNA-bound protein with solution-phase protein is common among nonspecific DNA-binding proteins as shown in experiments employing the E. coli nucleoid-associated protein HU and the HMGB chromatin protein NHP6A from S. cerevisiae. In Figure 2, panels e–g show the exchange between gfpFis and wtHU, and panels h–j show exchange between gfpFis and wtNHP6A.
These experiments clearly demonstrate that proteins bound non-specifically to DNA readily exchange with other solution-phase DNA binding proteins, and that this process not only occurs between homotypic but also between heterotypic DNA-binding proteins. We note that the heterotypic exchange was suggested by experiments using proteins inducing different mechanical responses but also that those experiments were not definitive as to the completeness of the release of bound protein (40).
Dissociation rate of DNA-bound protein depends on concentration of solution-phase protein
Previous micromanipulation experiments suggested that once wtFis binds to DNA it remains bound in protein-free solution (14), and similar effects have been reported for other DNA-binding proteins (35,36). Here, we show that 80% of gfpFis bound to λ-DNA remains bound after 30 min, and that the 20% reduction in fluorescence is due to loss of protein into solution and not photobleaching (Figure 3a, hour-glass and bowtie markers). If photobleaching were contributing to the observed fluorescence decay, a slower rate would be expected when imaging at 2 min intervals (Figure 3a, bowties) as the excitation light is only on during image acquisition (∼1 s per image).
Gel-shift experiments on short DNA fragments (<500 bp) indicate that Fis concentrations of 20 ± 10 nM result in complete coating of random DNA fragments (14). These singly coated Fis-DNA filaments have a Fis dimer bound about every 21 bp. At 200 nM (the beginning concentration for the current experiments) gel-purified Fis complexes exhibit binding densities that are at least twice those of singly coated complexes, and increasing numbers of Fis dimers continue to bind to the high-order complexes in the presence of greater concentrations of Fis. The spontaneous dissociation of a subset of Fis from DNA into protein-free solution observed in the present work is consistent with loss of Fis dimers that are more loosely associated with the complexes through protein–protein interactions, leaving most of the DNA bound by Fis dimers and interspersed patches of high-order complexes. These transitions are likely to be occurring within cells during changes in growth conditions, where for example, Fis is present at concentrations up to 50 μM in early exponential growth but falls to much lower amounts in late exponential phase and during slow growth (11,12). The exponential time course of spontaneous Fis dissociation (Figure 3a) implies that release of this fraction is not a cooperative process, but occurs as individual proteins randomly escape into solution. For a cooperative process, the time course should show a lag time, i.e. exactly the opposite curvature of that exhibited in Figure 3a (hour-glass and bowtie markers).
More interesting is how DNA-bound protein behaves in the presence of solution-phase protein. Noting that there is no apparent protein loss since solution-phase protein exchanges with bound protein, the rate of removal of the bound Fis is greatly accelerated by proteins in solution as the exchange occurs. Further, the rate of exchange depends on the concentration of free protein in solution. For exchange of gfpFis with wtFis, using the series of wtFis concentrations 5, 10, 20 and 50 nM (Figure 3a, circles, squares, triangles and diamonds, respectively), the rate increased with increasing concentration resulting in an exchange rate constant of kexch = (6.0 ± 0.3) × 104 M−1 s−1 (Figure 3b), a rate well below the diffusion limit which is expected to be on the order of 109 M−1 s−1. Since the fluorescence dropped below the 80% level seen for dissociation into protein-free solution within ∼300 s for all of the wtFis concentrations used, and there is a clear trend to faster exchange rate with increasing concentration, it is clear that intrinsic dissociation kinetics (i.e., off-rates) do not fully govern this protein exchange process. Concentration-dependent exchange of a DNA-bound protein for the same species in solution was recently demonstrated for single-strand binding (SSB) protein complexes bound to short, homopolymeric single-stranded DNA (41), but that work did not address the effect of other protein species, with binding affinities different from the bound protein, on the rate of exchange. To our knowledge, the present study provides the first report of concentration-dependent exchange of proteins bound to double-helix DNA in a non-cooperative manner.
Proteins of different types display concentration-dependent exchange kinetics
A more striking result is that the exchange of DNA-bound protein with a different species of solution-phase proteins also has a concentration-dependent exchange rate. Exchange of DNA-bound gfpFis with the E. coli nucleoid protein HU exhibits an increased rate of exchange for wtHU concentrations of 200, 400 and 800 nM (Figure 4a, triangles, circles and diamonds, respectively). While there is a clear trend to greater exchange rate with increasing concentration of solution-phase wtHU as for the gfpFis/wtFis exchange, the rate extracted is an order of magnitude slower, kexch = (2.7 ± 0.5) × 103 M−1 s−1 (Figure 4b). The combination of higher concentrations of wtHU being needed to trigger exchange (an order of magnitude higher than the concentrations of wtFis needed) with an exchange rate an order of magnitude lower illustrates that the binding affinities of the bound and solution-phase proteins (Kd’s for Fis and HU are ∼1 and ≥10 nM, respectively) affect the exchange rate.
We have further generalized the effect of concentration on protein off-rate using NHP6Agfp and wtNHP6A (Figure 5). Figure 5a–c show data that demonstrates the exchange between NHP6Agfp and wtNHP6A both at 200 nM. The concentration dependence was demonstrated by using 0 and 400 nM wtNHP6A (Figure 5d). In this case, because of the dimness of the NHP6Agfp fusion protein and the fact that a large fraction of the exchange reaction had already occurred prior to acquisition of the first image, absolute fluorescence had to be used instead of the relative fluorescence used for Fis and HU. Using data averaged for several experiments, we therefore only considered the first data points which clearly differ in intensity with the error representing the standard error of the mean for the several data sets used. Also because of the rapidity of the exchange and the dimness of the NHP6Agfp, a rate constant could not be reliably extracted from the NHP6A exchange data.
Possible physical origin of exchange reaction: ‘micro-dissociation’ events
We have shown that a variety of DNA-binding proteins display remarkable kinetics: although they remain stably bound for many minutes to the double helix, they display strikingly fast ‘exchange’ reactions when in the presence of proteins in solution. The involvement of the solution-phase protein in this exchange process is made clear by the linear dependence of the exchange reaction rate on solution protein concentration.
It is important to note that the exchange process is not diffusion limited, since the measured rate constant is five orders of magnitude below the diffusion limit which is on the order of, roughly, 109 M−1 s−1. Instead, we hypothesize that the exchange rate is limited by a chemical bond-breaking step whereby transient dissociation of proteins from the DNA allows for replacement by the solution-phase species.
This can be illustrated by noting that the intrinsic, or ‘macroscopic’, off-rate of the fraction of Fis which is able to spontaneously leave the double helix is approximately koff,macro = 1 × 10−3 s−1 (Figure 3b), corresponding to a macroscopic dissociation lifetime of ∼1000 s [koff,macro is enhanced by the possibility of rebinding to the extended DNA molecule, but this effect grows slowly, ∝ ln(L) with a molecule of length L for the extended geometry studied here]. However, ‘micro-dissociation’ events, where a protein releases some or all of the chemical interactions holding it to its binding site, but remains localized near the double helix by long-range electrostatic interactions of range r ∼ 1 nm (the electrostatic screening length), likely occur at a much higher rate koff,micro. We hypothesize that the exchange reaction involves these fast micro-dissociation events.
For exchange to occur, one requires a solution-phase protein to be near enough to the transiently opened binding site for it to take the place of the original protein. A rough estimate for the probability of a solution-phase protein being present within a distance r of the open site is just cr3, for concentration c, and therefore one can write the total rate of exchange as:
| (1) |
This relation can be used to estimate the micro-dissociation rate koff,micro = kexch/r3. For our observed kexch and r = 1 nm, we estimate koff,micro = 9 × 104 s−1, almost 108 times faster than the koff,macro associated with complete dissociation of protein away from DNA in protein-free solution, but still far below the diffusion limit for fluctuation of a protein-sized molecule (roughly kBT/ηr3 > 109 s−1 for r = 1 nm, where η is the viscosity of water).
The micro-dissociation rate can be related to the energy barrier ΔE associated with breaking of protein–DNA contacts to allow protein exchange, via the formula
| (2) |
where η = 1 × 10−3 Pa s is the viscosity of water; kBT = 4 × 10−21 J is the unit of thermal energy at T = 300 K; and r = 1 nm is the range of micro-dissociation as well as the radius of the proteins of interest here. Our estimated koff,micro = 105 s−1 gives ΔE = 10 kBT (6 kcal/mol), consistent with the energy cost of substantial breakage of several protein–DNA contacts. While this estimated ΔE indicates that our thermally excited micro-dissociation picture is reasonable, it is important to bear in mind the possibility, however unlikely given that exchange occurs between Fis and other proteins such as HU and NHP6A, that specific chemical interactions between bound and unbound proteins promote the exchange reactions that we have observed. However, we emphasize that the micro-dissociation scenario does not, strictly speaking, require the formation of a complex containing both proteins involved in an exchange reaction (41), but instead requires only sufficient breaking of bound protein–DNA contacts to occur so that the solvated protein can ‘steal’ the transiently open binding position.
We are, therefore, led to the physical picture that non-specific DNA-binding proteins, even when apparently stably bound to the double helix, are continuously undergoing micro-dissociation fluctuations where chemical contacts are lost, but where the protein remains near its binding position, most likely as a result of long-ranged electrostatic forces (range of the Debye screening length is ∼1 nm for physiological conditions). If no proteins are nearby in solution, the most likely outcome is rebinding of the original protein at the original or a nearby DNA site. However, if a solution-phase protein is nearby, it can exchange with the original protein by an essentially diffusion-limited reaction. An implication of this is a strong dependence of this type of exchange reaction on electrostatic interactions whose strength and range both depend on local salt concentration. Further experimental and theoretical work needs to be carried out to understand this process.
Low concentrations of solution-phase protein only partially remove bound proteins
An important feature of the concentration-dependent turnover kinetics that we have observed is that the concentration-mediated exchange reactions cause exchange of only a subset of previously bound proteins, i.e. the decay of bound protein proceeds to a final constant ‘offset’ (final levels of decays shown in Figures 3a and 4a). Furthermore, this final level of bound protein is progressively reduced with increasing solution protein concentration. It is unlikely that this is a mass action effect because the (dark) wtFis protein in solution is in vast excess over the small number of DNA molecules present in the sample. As the exchange process proceeds there will be little or no depletion of bulk wtFis concentration. Instead, this effect is most likely due to heterogeneity of the degree to which the bound proteins are localized to their binding positions; i.e. the binding affinity of the bound protein to a preferred sequence. In our visualizations of gfpFis–DNA complexes inhomogeneity of binding is clearly visible (Figure 2). The dependence of exchange rate on DNA sequence provides a simple explanation for the partial exchange at low concentrations and is straightforward to describe within the framework of the micro-dissociation picture outlined above. Sequence-dependent binding affinities lead to sequence-dependent off-rates, and therefore to sequence-dependent micro-dissociation rates [through sequence variation of the ΔE in Equation (2)]. As a consequence of sequence-dependent micro-dissociation there will be widely varying rates of exchange at different sequence positions. We plan to study this systematically via observation of exchange kinetics at different positions along DNA of defined and varied sequence composition.
Implications for measurements of binding affinities
The equilibrium constant for binding of a ligand to a substrate is related to the ratio of the binding reaction rate constant k and the off rate koff by
| (3) |
This relation is often considered to apply to protein–DNA interactions, where the protein is considered to be a ligand, and the substrate a DNA molecule. A crucial result of this study is the observation that koff may depend on the concentration of the ligand, which in this case is solvated versions of the bound protein, most simply via the result shown in Figure 3b,
| (4) |
where the off-rate at zero concentration is koff,0. Furthermore, the off-rate may depend on concentrations of other species of molecules, as our experiments with exchange reactions between different proteins show (Figure 4b). The concentration-dependence of koff is of potential importance for interpreting experiments aimed at measurements of either the equilibrium constant or the kinetic rates.
Consider a measurement of equilibrium constant KD for a particular pair of DNA and protein molecules using a series of measurements of equilibrated binding fractions at different protein concentrations (e.g. using EMSA), and a second measurement done by measuring the on-rate at one concentration, and the off-rate at a different concentration (possibly zero, as might be done in a Biacore-type off-rate measurement). In the latter experiment, the off-kinetics are observed at lower concentration than in the former experiment, with the result that the two experiments will lead to different affinity measurements. In the cases studied in this article, the latter experiment would involve much lower off-rates than the former, and would result in a much higher affinity measurement. Concentration-dependent off-rates may explain inconsistencies in affinities obtained from different experiments, particularly inconsistencies between equilibrium binding and kinetics measurements (42,43).
While these studies were performed under physiological salt conditions (i.e. 100 mM k-glutamate), higher salt conditions are expected to accelerate the process further, while low salt conditions will likely slow the process. Under both higher and lower salt conditions we still expect solution-phase protein to accelerate release of protein bound to DNA.
It is likely that protein–DNA exchange reactions also involve competition between nucleic acid molecules for binding to a protein molecule, essentially the inverse of the reactions examined in this, article. Facilitated exchange by DNA has been documented in bulk solution experiments for Fis (25) as well as a number of other DNA-binding proteins (36,44–49). Methods similar to those described in this article could be also employed to study this reaction.
Implications for kinetics of gene regulation
This report clearly establishes that solution-phase protein hugely accelerates release of protein that is otherwise relatively stably bound to dsDNA. The type of exchange reaction analyzed here is likely to occur for a wide variety of DNA-binding proteins, and we imagine it to play a major role in renewal of binding of transcription factors to their targets in vivo where protein concentration is large and often strongly modulated. A prime example where this occurs is for the Fis protein that is the focus of this study. Fis is produced in large quantities in the E. coli cell following a shift from poor to rich growth medium (11,12); the shift in cellular Fis concentration from nearly undetectable to ∼50 μM levels is likely to trigger rapid replacement of a wide spectrum of proteins previously bound to the chromosome with Fis. This article shows that the rate at which this occurs will depend on the intracellular Fis concentration, and not just on the off-rates associated with spontaneous dissociation of the previously bound proteins.
Our results also suggest a possible explanation for an effect observed in a recent study of HMGB protein binding to duplex DNA (50). It was found that whereas excess HMGB protein over the DNA probe failed to form a discrete complex band in a gel mobility shift assay, increasing concentrations of unlabeled DNA relative to excess protein resulted in the formation of a novel complex consisting of more than one DNA fragment. A possible explanation is that the excess protein present without the competitor was promoting dissociation, but when the free HMGB concentration was effectively reduced by addition of competitor, a complex stable to electrophoresis could form.
Implications of the rapid exchange kinetics we have demonstrated for systems biological analysis of transcription networks are profound. As shown in Figures 3b and 4b, direct exchange reactions of proteins with DNA apparently stably bound by other proteins are likely to be dominant, that is, dissociation rates measured in vitro (which are measurements of koff,macro) are likely to be significantly lower than the actual rates at which protein-binding profiles can actually be modified. Given the variation of amount and rate of exchange of proteins bound to dsDNA for different proteins that we have observed, our demonstrated concentration-dependent exchange mechanism can permit rapid replacement of otherwise stably bound transcription factors by a new species of protein.
FUNDING
National Institutes of Health (grant number GM038509 to R.C.J.); National Science Foundation (grant numbers DMR-0715099, PHY-0852130 and DMR-0520513 to J.M.]; National Institutes of Health (grant U54CA143869-01 to J.M.); Chicago Biomedical Consortium with support from The Searle Funds at The Chicago Community Trust (to J.M.) Funding for open access charge: NSF-PHY-0852130.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Yana Bernatavichute, Jenna McCracken, Daniel Yoo and Roxanne Isaksson for construction of gfpFis and purification of proteins.
REFERENCES
- 1.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu T, Shklovskii BI. How a protein searches for its specific site on DNA: the role of intersegment transfer. Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys. 2007;76:051909. doi: 10.1103/PhysRevE.76.051909. [DOI] [PubMed] [Google Scholar]
- 3.von Hippel PH, Berg OG. Facilitated target location in biological systems. J. Biol. Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 4.Gowers DM, Wilson GG, Halford SE. Measurement of the contributions of 1D and 3D pathways to the translocation of a protein along DNA. Proc. Natl Acad. Sci. USA. 2005;102:15883–15888. doi: 10.1073/pnas.0505378102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorman J, Greene EC. Visualizing one-dimensional difusion of proteins along DNA. Nat. Struct. Mol. Biol. 2008;15:768–774. doi: 10.1038/nsmb.1441. [DOI] [PubMed] [Google Scholar]
- 7.Dame RT. The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol. Microbiol. 2005;56:858–870. doi: 10.1111/j.1365-2958.2005.04598.x. [DOI] [PubMed] [Google Scholar]
- 8.Johnson RC, Johnson LM, Schmidt JW, Gardner JF. Major nucleoid proteins in the structure and function of the Escherichia coli chromosome. In: Higgins NP, editor. The Bacterial Chromosome. Washington, DC: ASM Press; 2005. pp. 65–132. [Google Scholar]
- 9.Travers A, Muskhelishvili G. Bacterial chromatin. Curr. Opin. Genet. Dev. 2005;15:507–514. doi: 10.1016/j.gde.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Dorman CJ. Nucleoid-associated proteins and bacterial physiology. Adv. Appl. Microbiol. 2009;67:47–64. doi: 10.1016/S0065-2164(08)01002-2. [DOI] [PubMed] [Google Scholar]
- 11.Ball CA, Osuna R, Ferguson KC, Johnson RC. Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J. Bacteriol. 1992;174:8043–8056. doi: 10.1128/jb.174.24.8043-8056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Talukder AA, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bétermier M, Galas DJ, Chandler M. Interaction of Fis protein with DNA: bending and specificity of binding. Biochimie. 1994;76:958–967. doi: 10.1016/0300-9084(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 14.Skoko D, Yoo D, Bai H, Schnurr B, Yan J, McLeod SM, Marko JF, Johnson RC. Mechanism of chromosome compaction and looping by the Escherichia coli nucleoid protein Fis. J. Mol. Biol. 2006;364:777–798. doi: 10.1016/j.jmb.2006.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finkel SE, Johnson RC. The Fis protein: it's not just for DNA inversion anymore. Mol. Microbiol. 1992;6:3257–3265. doi: 10.1111/j.1365-2958.1992.tb02193.x. [DOI] [PubMed] [Google Scholar]
- 16.Cho BK, Knight EM, Barrett CL, Palsson BO. Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts. Genome Res. 2008;18:900–910. doi: 10.1101/gr.070276.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hengen PN, Bartram SL, Stewart LE, Schneider TD. Information analysis of Fis binding sites. Nucleic Acids Res. 1997;25:4994–5002. doi: 10.1093/nar/25.24.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao Y, Feldman-Cohen LS, Osuna R. Functional characterization of the Escherichia coli Fis-DNA binding sequence. J. Mol. Biol. 2008;376:771–785. doi: 10.1016/j.jmb.2007.11.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ussery D, Larsen TS, Wilkes KT, Friis C, Worning P, Krogh A, Brunak S. Genome organisation and chromatin structure in Escherichia coli. Biochimie. 2001;83:201–212. doi: 10.1016/s0300-9084(00)01225-6. [DOI] [PubMed] [Google Scholar]
- 20.Grainger DC, Hurd D, Goldberg MD, Busby SJ. Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res. 2006;34:4642–4652. doi: 10.1093/nar/gkl542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dorman CJ, Deighan P. Regulation of gene expression by histone-like proteins in bacteria. Curr. Opin. Genet. Dev. 2003;13:179–184. doi: 10.1016/s0959-437x(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 22.McLeod SM, Johnson RC. Control of transcription by nucleoid proteins. Curr. Opin. Microbiol. 2001;4:152–159. doi: 10.1016/s1369-5274(00)00181-8. [DOI] [PubMed] [Google Scholar]
- 23.Paul BJ, Ross W, Gaal T, Gourse RL. rRNA transcription in Escherichia coli. Annu. Rev. Genet. 2004;38:749–770. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- 24.Travers A, Muskhelishvili G. DNA microloops and microdomains: a general mechanism for transcription activation by torsional transmission. J. Mol. Biol. 1998;279:1027–1043. doi: 10.1006/jmbi.1998.1834. [DOI] [PubMed] [Google Scholar]
- 25.Pan CQ, Finkel SE, Cramton SE, Feng JA, Sigman DS, Johnson RC. Variable structures of Fis-DNA complexes determined by flanking DNA-protein contacts. J. Mol. Biol. 1996;264:675–695. doi: 10.1006/jmbi.1996.0669. [DOI] [PubMed] [Google Scholar]
- 26.Perkins-Balding D, Dias DP, Glasgow AC. Location, degree, and direction of DNA bending associated with the Hin recombinational enhancer sequence and Fis-enhancer complex. J. Bacteriol. 1997;179:4747–4753. doi: 10.1128/jb.179.15.4747-4753.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson JF, Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988;16:9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stella S, Cascio D, Johnson RC. The shape of the DNA minor groove directs binding by the DNA-bending protein Fis. Genes Dev. 2010;24:814–826. doi: 10.1101/gad.1900610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skoko D, Yan J, Johnson RC, Marko JF. Low-force DNA condensation and discontinuous high-force decondensation reveal a loop-stabilizing function of the protein Fis. Phys. Rev. Lett. 2005;95:208101. doi: 10.1103/PhysRevLett.95.208101. [DOI] [PubMed] [Google Scholar]
- 30.Oberto J, Nabti S, Jooste V, Mignot H, Rouviere-Yaniv J. The HU regulon is composed of genes responding to anaerobiosis, acid stress, high osmolarity and SOS induction. PLoS ONE. 2009;4:e4367. doi: 10.1371/journal.pone.0004367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stillman DJ. Nhp6: a small but powerful effector of chromatin structure in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2010;1799:175–180. doi: 10.1016/j.bbagrm.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dowell NL, Sperling AS, Mason MJ, Johnson RC. Chromatin-dependent binding of the S. cerevisiae HMGB protein Nhp6A affects nucleosome dynamics and transcription. Genes Dev. 2010;24:2031–2042. doi: 10.1101/gad.1948910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swinger KK, Lemberg KM, Zhang Y, Rice PA. Flexible DNA bending in HU-DNA cocrystal structures. EMBO J. 2003;22:3749–3760. doi: 10.1093/emboj/cdg351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masse JE, Wong B, Yen YM, Allain FH, Johnson RC, Feigon J. The S. cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding. J. Mol. Biol. 2002;323:263–284. doi: 10.1016/s0022-2836(02)00938-5. [DOI] [PubMed] [Google Scholar]
- 35.McCauley M, Hardwidge PR, Maher LJ, 3rd, Williams MC. Dual binding modes for an HMG domain from human HMGB2 on DNA. Biophys. J. 2005;89:353–364. doi: 10.1529/biophysj.104.052068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skoko D, Wong B, Johnson RC, Marko JF. Micromechanical analysis of the binding of DNA-bending proteins HMGB1, NHP6A, and HU reveals their ability to form highly stable DNA-protein complexes. Biochemistry. 2004;43:13867–13874. doi: 10.1021/bi048428o. [DOI] [PubMed] [Google Scholar]
- 37.Yan J, Skoko D, Marko JF. Near-field-magnetic-tweezer manipulation of single DNA molecules. Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys. 2004;70:011905. doi: 10.1103/PhysRevE.70.011905. [DOI] [PubMed] [Google Scholar]
- 38.Strick TR, Allemand JF, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 39.Yen YM, Roberts PM, Johnson RC. Nuclear localization of the Saccharomyces cerevisiae HMG protein NHP6A occurs by a Ran-independent nonclassical pathway. Traffic. 2001;2:449–464. doi: 10.1034/j.1600-0854.2001.20703.x. [DOI] [PubMed] [Google Scholar]
- 40.Skoko D. 2006. Single-molecule manipulation study of DNA-organizing proteins. PhD Thesis, University of Illinois-Chicago, Chicago. [Google Scholar]
- 41.Kunzelmann S, Morris C, Chavda AP, Eccleston JF, Webb MR. Mechanism of interaction between single-stranded DNA binding protein and DNA. Biochemistry. 2010;49:843–852. doi: 10.1021/bi901743k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugimura S, Crothers DM. Stepwise binding and bending of DNA by Escherichia coli integration host factor. Proc. Natl Acad. Sci. USA. 2006;103:18510–18514. doi: 10.1073/pnas.0608337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang SW, Nash HA. Comparison of protein binding to DNA in vivo and in vitro: defining an effective intracellular target. EMBO J. 1995;14:6292–6300. doi: 10.1002/j.1460-2075.1995.tb00319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doucleff M, Clore GM. Global jumping and domain-specific intersegment transfer between DNA cognate sites of the multidomain transcription factor Oct-1. Proc. Natl Acad. Sci. USA. 2008;105:13871–13876. doi: 10.1073/pnas.0805050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fried MG, Crothers DM. Kinetics and mechanism in the reaction of gene regulatory proteins with DNA. J. Mol. Biol. 1984;172:263–282. doi: 10.1016/s0022-2836(84)80026-1. [DOI] [PubMed] [Google Scholar]
- 46.Iwahara J, Zweckstetter M, Clore GM. NMR structural and kinetic characterization of a homeodomain diffusing and hopping on nonspecific DNA. Proc. Natl Acad. Sci. USA. 2006;103:15062–15067. doi: 10.1073/pnas.0605868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruusala T, Crothers DM. Sliding and intermolecular transfer of the lac repressor: kinetic perturbation of a reaction intermediate by a distant DNA sequence. Proc. Natl Acad. Sci. USA. 1992;89:4903–4907. doi: 10.1073/pnas.89.11.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lieberman BA, Nordeen SK. DNA intersegment transfer, how steroid receptors search for a target site. J. Biol. Chem. 1997;272:1061–1068. doi: 10.1074/jbc.272.2.1061. [DOI] [PubMed] [Google Scholar]
- 49.Kozlov A, Lohman TM. Kinetic mechanism of direct transfer of Escherichia coli SSB tetramers between single-stranded DNA molecules. Biochemistry. 2002;41:11611–11627. doi: 10.1021/bi020361m. [DOI] [PubMed] [Google Scholar]
- 50.Zimmerman J, Maher LJ., III Transient HMGB protein interactions with B-DNA duplexes and complexes. Biochem. Biophys. Res. Commun. 2008;371:79–84. doi: 10.1016/j.bbrc.2008.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]