Abstract
Non-coding (nc) RNAs are increasingly recognized to play important regulatory roles in eukaryotic gene expression. The highly abundant and essential 7SK ncRNA has been shown to negatively regulate RNA Polymerase II transcription by inactivating the positive transcription elongation factor b (P-TEFb) in cellular and Tat-dependent HIV transcription. Here, we identify a more general, P-TEFb-independent role of 7SK RNA in directly affecting the function of the architectural transcription factor and chromatin regulator HMGA1. An important regulatory role of 7SK RNA in HMGA1-dependent cell differentiation and proliferation regulation is uncovered with the identification of over 1500 7SK-responsive HMGA1 target genes. Elevated HMGA1 expression is observed in nearly every type of cancer making the use of a 7SK substructure in the inhibition of HMGA1 activity, as pioneered here, potentially useful in therapy. The 7SK-HMGA1 interaction not only adds an essential facet to the comprehension of transcriptional plasticity at the coupling of initiation and elongation, but also might provide a molecular link between HIV reprogramming of cellular gene expression-associated oncogenesis.
INTRODUCTION
Small non-coding (nc) RNAs are increasingly recognized to play crucial roles in both, pre- and post-transcriptional regulation of gene expression (1). They may act to directly affect components of the chromatin remodeling (2), the transcription (3) and translation (4) apparatuses. Also, ncRNAs have been reported to affect protein stability, localization and post-translational modification (5,6).
Among the best studied ncRNAs involved in pre-transcriptional regulation is the 7SK RNA. 7SK is a RNA Polymerase III transcript of 331 nt in mammals, belonging to the highly abundant small nuclear (sn) RNAs (7). It is stabilized by a γ-monomethylephosphate-GTP cap on its 5′-end, which is synthesized by the methylephosphate capping enzyme (MePCE) (8,9). The La-related protein LARP7 has been shown to associate with the 3′-poly(U) region of 7SK RNA, thereby contributing to the stability of the RNA as well (10). Recent studies revealed that MePCE and LARP7 act cooperatively to maintain the integrity of 7SK snRNP (11).
7SK RNA is evolutionary highly conserved among vertebrates (12) and is also found in higher invertebrates, such as Drosophila melanogaster (13). Since its discovery in 1976 (14), it took a quarter century until two groups discovered a key function of 7SK RNA in the regulation of gene transcription elongation (15,16): 7SK RNA and HEXIM1 or HEXIM2 bind to the positive transcription elongation factor b (P-TEFb) and mask its cyclin-dependent kinase-9 activity, required for cyclin T1 or T2-mediated phosphorylation of the C-terminal domain of RNA Polymerase II. Thus, P-TEFb's role in RNA Polymerase II transcription elongation is abolished. This mechanism is thought to establish negative regulatory control of the RNA Polymerase II elongation activity as a function of 7SK RNA nuclear concentrations for P-TEFb-dependent gene expression. Several P-TEFb target genes have been identified to date (KLK3, IL8, CYP1A1, MCL1, CKM, SLC2A4, NR4A1) (17–21), and P-TEFb is known to play a role in transcription elongation from the HIV long terminal repeat (22,23). With little surprise it was recently possible to also place 7SK RNA at the center of Tat-dependent transcription complexes (24). The mechanism of 7SK RNA binding to P-TEFb and CDK9 inhibition is well characterized and understood (25). Loops 1, 3 and 4 of the predicted secondary structure of 7SK RNA (see Figure 1A) are thereby directly involved in P-TEFb binding, and serve as a molecular scaffold for the assembly of the inactive P-TEFb complex. When P-TEFb is released from 7SK RNA to activate RNA Polymerase II transcription elongation, these hairpin regions are covered by different heterogenous nuclear ribonucleoproteins (hnRNPs) of distinct compositions, what is thought to provide a pool of inactive 7SK RNA (26–28). Loop 2 of the 7SK RNA is neither required nor involved in P-TEFb, MePCE or LARP7 interactions. A crucial question remains with respect to the regulation of this 7SK-dependent inactivation process. 7SK RNA, as a RNA Polymerase III transcribed gene, is thought to be expressed at robust and constant levels through most phases of the cell cycle and in most embryonic and adult tissues (29). While 7SK RNA may serve as a buffer to fine-tune P-TEFb to constant activity, it is conceivable that 7SK RNA transcription itself or the stability, localization and availability of the RNA are more tightly controlled by yet unidentified mechanisms. In this respect, the finding that P-TEFb inhibition is not sufficient to explain the physiological relevance of 7SK RNA is of great importance. A knockdown of 7SK RNA in HeLa cells identified P-TEFb-independent regulatory phenomena (30). When 7SK RNA concentrations are diminished to less than 5%, P-TEFb target genes are expressed at a higher rate, as expected due to the higher processivity of the RNA Polymerase II apparatus. But also gene-expression not regulated by P-TEFb or even expression from minimal, plasmid-coded promoters is both positively and negatively altered with changing intracellular concentrations of 7SK RNA. These observations clearly point to a yet unidentified second function in transcription regulation exerted by the 7SK snRNA.
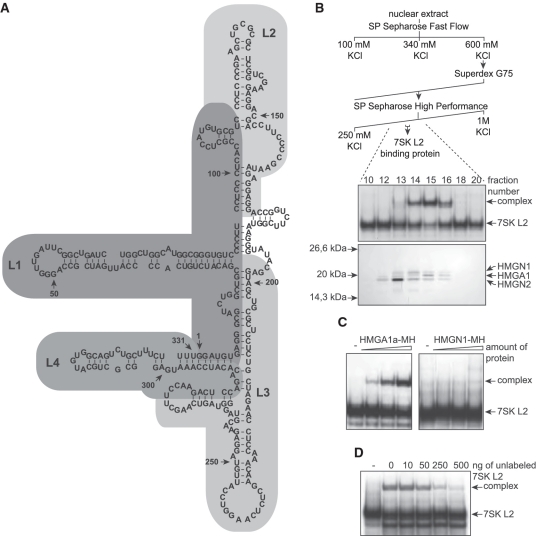
Figure 1.
Purification and identification of a 7SK L2 binding protein from HeLa nuclear extract. (A) According to its predicted secondary structure (76), wild-type 7SK RNA can be subdivided into four stem-loop structures: loop1 (L1), loop2 (L2), loop3 (L3) and loop4 (L4). (B) HeLa nuclear extract was fractionated by cation exchange chromatography (SP-Sepharose Fast Flow), gel filtration (Superdex G75 16/60) and cation exchange chromatography (SP-Sepharose High Performance), obtaining fractions with a specific 7SK L2 binding activity (upper scheme). The 7SK L2 binding activity of the fractions obtained after the final SP-Sepharose step was tested by EMSAs (middle panel). The protein content of each of these fractions was analyzed by silver staining after SDS–PAGE and most likely 7SK binding candidate proteins were identified by tandem mass spectrometry as indicated (lower panel). (C) Increasing amounts of purified HMGA1a-myc/His (MH) (left panel) and HMGN1-MH (right panel) were tested for 7SK L2 binding activity compared with 7SK L2 RNA alone (−). Two nanogram of 32P-labeled 7SK L2 RNA were incubated with ∼2, 4 and 6 ng of purified HMGA1a-MH (HMGN1-MH) and the resulting complexes were analyzed in EMSAs. (D) Binding of 2 ng of 32P-labeled 7SK L2 RNA to ∼4 ng of purified HMGA1a-MH was competed with increasing amounts of unlabeled 7SK L2 RNA. The resulting complexes were analyzed in EMSAs compared with 7SK L2 RNA alone (−).
We therefore set out to identify additional 7SK interacting proteins in an effort to unveil such P-TEFb-independent mechanism of gene regulation. We thereby used the loop 2 substructure of 7SK RNA (Figure 1A) as a bait, as this is the only substructure not involved in P-TEFb, MePCE or LARP7 binding. We report that the chromatin factor and transcription regulatory hub HMGA1 is bound by 7SK RNA, which in turn affects its DNA binding and regulatory activities for over 1500 target genes.
MATERIALS AND METHODS
Cell culture and preparation of extracts
Culturing conditions and transfections
HEK293 and HeLa cells were cultured as described previously (31). Transfections were either performed by the calciumphosphate coprecipitation method (32) or by FugeneHD (Roche) according to the manufacturer's instructions.
Extract preparations
The preparation of nuclear extract and total cellular extract was performed as described previously (33). To obtain extracts for co-immunoprecipitation experiments, cells were reconstituted in isotonic IP-lysis buffer (50 mM Tris/HCl, pH 7.4, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 1% TRITON™ X-100, protease inhibitor) and homogenized in a dounce-homogenizer before cellular debris was removed by high speed centrifugation (16.000g). The extract was subsequently dialyzed against IP-buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)/KOH, pH 7.9, 100 mM KCl, 0.2 mM EDTA, 20% glycerol).
Chromatin isolation
Fractionation of cytoplasmic, soluble nuclear and chromatin-bound proteins was carried out as descried previously (34). Cells were resuspended in two packed cell volumes of lysis buffer (10 mM HEPES/KOH, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 340 mM sucrose, 1 mM dithiothreitol (DTT), 0.1% TRITON™ X-100, 10% glycerol, protease inhibitor) and incubated for 15 min on ice. Nuclei were pelleted by centrifugation (1.300g) and the supernatant containing cytoplasmic proteins was cleared by centrifugation (16.000g). After washing once with lysis buffer lacking TRITON™ X-100, the soluble nuclear proteins were extracted by resuspending the nuclei in an equal volume of extraction buffer (3 mM EDTA, 0.2 mM ethyleneglycoltetraacetic acid (EGTA), 1 mM DTT, protease inhibitor) and incubating for 30 min on ice. Chromatin was pelleted by centrifugation (1.700g) and the soluble nuclear fraction was removed before the chromatin fraction was washed once with extraction buffer. Chromatin-bound proteins were eluted by incubation with SDS–PAGE loading buffer for 10 min at 95°C. The protein concentration of each fraction was detected as described previously (35).
Fluorescence microscopy
Mitochondria were stained with Mito-FluorTMRed 594 (Invitrogen) as recommended by the manufacturer. Fluorescence microscopy was performed using a TCS SP2 confocal microscope (Leica) and the obtained data were analyzed using Confocal Software TCS SP2 (Leica).
Molecular biology and RNA biochemistry
Western blot analyses and antibodies
SDS–PAGE and silver staining was performed as described previously (36). Antibodies used in western blot analyses were M2 anti-FLAG antibody (Sigma), anti-HMGA1 antibody (Thermo Fisher Scientific), anti-RAR-α antibody (Thermo Fisher Scientific), anti-LARP7 antibody (ProteinTech) or anti-MePCE antibody (ProteinTech) with the corresponding secondary horseradish peroxidase-coupled antibody, either anti-rabbit antibody (Cell Signaling) or anti-mouse antibody (Sigma) as recommended by the manufacturer. Western blot signals were quantified using ImageJ (Version 1.42q) (37).
Immunoprecipitations
Immunoprecipitations of FLAG-fusion proteins were performed with M2 anti-FLAG agarose (Sigma) as recommended by the manufacturer. Briefly, anti-FLAG agarose was incubated with extracts containing FLAG fusion proteins in IP buffer (20 mM HEPES/KOH, pH 7.9, 100 mM KCl, 0.2 mM EDTA, 20% glycerol) for 2 h at 4°C. The agarose was pelleted by centrifugation and the supernatant was removed. After washing three times with the 20-fold volume of IP washing buffer (20 mM HEPES/KOH, pH 7.9, 100 mM KCl, 0.2 mM EDTA, 0.025% Tween 20), the HMGA1a-FLAG bound compounds were eluted in two steps with the 5-fold volume of IP buffer containing increasing concentrations of KCl (500 mM and 1 M), before the FLAG fusion protein was eluted with the 5-fold volume of elution buffer (100 mM glycine/HCl, pH 3.5). To detect RNA in the elution steps by northern blot analyses, these fractions were extracted with phenole/chloroforme and the RNA was subsequently precipitatied with ethanol. In case the FLAG fusion protein was purified for downstream EMSAs, the elution fraction at pH 3.5 was subsequently dialyzed against IP buffer before use in EMSAs.
Purification of myc-His fusion proteins
HMGA1a-MH was purified using Ni-NTA agarose (Qiagen) as recommended by the manufacturer. The extract containing myc-His fusion proteins was adjusted to 400 mM KCl and 5 mM imidazole, before it was incubated with Ni-NTA agarose for 2 h at 4°C. The Ni-NTA agarose was pelleted by centrifugation and the supernatant was removed. After washing twice with the 20-fold volume of washing buffer (20 mM HEPES/KOH, pH 7.9, 400 mM KCl, 1 mM MgCl2, 20 mM imidazole, 0.2 mM EDTA), the bound proteins were eluted by four elution steps with the 5-fold volume of elution buffer (20 mM HEPES/KOH, pH 7.9, 400 mM KCl, 1 mM MgCl2, 250 mM imidazole, 0.2 mM EDTA). The elution fractions were subsequently dialyzed against IP buffer before use in EMSAs.
Electrophoretic mobility shift assays
For electrophoretic mobility shift assays (EMSAs), 32P-labeled or Fam6-labeled RNA or DNA was denatured for 5 min at 75°C and renatured by cooling slowly down to room temperature. The nucleic acid was incubated with immuno-purified protein in EMSA-buffer (10 mM HEPES/KOH, pH 7.9, 100 mM KCl, 5 mM MgCl2, 0.25 mM DTT, 0.1 mM EDTA, 10% glycerol and 2 μg yeast tRNA per lane) for 10 min at 37°C and subsequently for 10 min on ice. The resulting complexes were separated by 10% native PAGE and visualized by autoradiography (32P) or at UV light (Fam6). For competition experiments in EMSAs, a double-stranded DNA oligonucleotide with the sense sequence 5′-GGA AGG AAA CCA AGG GGC GGA CCC AAA AAA CTG GAG-3′ was used. The double-stranded, Fam6-labeled IL2-Rα promoter fragment used for competition experiments in EMSAs had the sense sequence 5′-ACC GCA AAC TAT ATT GTC ATC AAA AAA AAA AAA AAA AAA ACA CTT CCT ATA TTT GAG ATG-3′.
Affinity purification with oligo dT-cellulose
For affinity purification with poly-adenylated 7SK L2 RNA, oligo dT-cellulose (Boehringer) was prepared as recommended by the manufacturer. Briefly, the freshly renatured poly-adenylated RNA was incubated for 20 min at 4°C with oligo dT-cellulose equilibrated against oligo-dT wash-buffer (10 mM HEPES/KOH, pH 7.9, 100 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 0.25 mM DTT). Cellular extract containing HMGA1a-FLAG fusion protein was applied to the column containing the loaded oligo-dT cellulose and the flowthrough was collected. After washing with the 25-fold volume of oligo-dT wash-buffer, the RNA-bound components were eluted by addition of one volume of oligo-dT wash buffer containing 1μg/μl RNaseA. The protein contents of each fraction were analyzed in western blot experiments.
Total RNA preparation
Total RNA from transfected or non-transfected cells was either prepared as described previously (38) or using the RNeasy Midi Kit (Qiagen) as recommended by the manufacturer.
In vitro transcription
In vitro transcription analyses were performed either using nuclear extract from HeLa cells (33) or using T7 RNA Polymerase (NEB) as recommended by the manufacturer. To label RNA with 32P, the reaction mixture contained a 10-fold excess of 32P-labeled α-uridine-5′-triphosphate.
Northern blot analyses
Northern blot analyses were performed as described previously (31) using specific 32P-labeled antisense RNA probes complementary to the first 89 nt of wild-type 7SK or U6 RNA. To detect EBER2 7SK L2, EBER2 7SK L2 m137, EBER2 7SK L2 m122/m137 RNA, EBER2-Lb RNA as well as wild-type EBER2 RNA a RNA probe complementary to the first 77 nt of wild-type EBER2 RNA was utilized.
Chromatographic extract fractionation
Approximately 100 mg of nuclear extract from HeLa cells was fractionated by cation exchange chromatography (SP-Sepharose Fast Flow, Amersham), gel filtration (Superdex G75 16/60) and cation exchange chromatography (SP-Sepharose High Performance, Amersham) as described by the manufacturer. After loading with the extract, the SP-Sepharose Fast Flow was washed extensively with wash-buffer (20 mM HEPES/KOH, pH 7.9, 100 mM KCl, 0.2 mM EDTA) and the flowthrough was collected. The bound proteins were eluted in two steps with wash buffer containing increasing concentrations of KCl (340 mM and 600 mM). The fraction containing the 7SK L2 binding protein was loaded on a gel filtration column (Superdex G75 16/60) to separate the components by size. The elution was performed with wash buffer and the fractions containing the 7SK L2 binding protein were loaded on a column containing SP-Sepharose High Performance. After washing with wash buffer, the bound proteins were eluted by a linear KCl-gradient (from 250 mM to 1 M) in wash-buffer (see Figure 1B). The fractions were subsequently dialyzed against IP buffer (20 mM HEPES/KOH, pH 7.9, 100 mM KCl, 0.2 mM EDTA, 20% glycerol).
Mass spectrometry
Visible silver-stained protein bands on SDS–PAGE were cut out and proteins were in gel digested according to Shevchenko et al. (39). Eluted peptides were analyzed by liquid chromatography (LC) coupled tandem mass spectrometry (MS/MS) on a Q-Tof Ulitma (Waters) under standard conditions. Proteins were identified by database search of the peptide fragment spectra using MASCOT as search engine (39).
RNA structure prediction
The secondary structure of RNA was calculated by RNAstructure (Version 4.4) (40).
Quantitative PCR analyses
qRT-PCR analyses were performed using SuperScript™ 2 Reverse Transcriptase (Invitrogen), Lightcycler (Roche) and Fast Start DNA MasterPLUS SYBR Green I reaction mix (Roche) as recommended by the manufacturer. Primers used to quantify the housekeeping gene ubiquitin were 5′-GTT GAG ACT TCG TGG TGG TG-3′ (sense) and 5′-TCT CGA CGA AGG CGA CTA AT-3′ (antisense). Primers used to quantify endogenous HMGA1 were 5′-TCC CAG CCA TCA CTC TTC-3′ (sense) and 5′-CTC CTT CTG ACT CCC TAC C-3′ (antisense).
Transcriptome analyses
For microarray analyses, RNA amplification, labeling, hybridization and detection were performed following the protocols supplied by Applied Biosystems using the corresponding kits (Applied Biosystems, ProdNo: 4339628 and 4336875). The data obtained were analyzed as described previously (31,41–44). Transcriptome data, annotated to MIAME I+II standards, were deposited in the public database MACE using Accession Nos.: 2970966830 (HMGA1), and 2979879726 (7SK) (http://mace.ihes.fr).
Probability calculation
To calculate the random expectation to obtain a given number of probes returning statistically significant fold changes in response to HMGA1 and EBER2 7SK L2, a hypergeometric distribution was used:
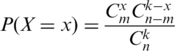 |
where m is the number of possible successes, n is the total number of elements, k is the sample size and x is the number of successes in the sample.
Constructs used in this study
EBER2 7SK L2 fusion transcripts
The EBER2 gene from −171 nt to +226 nt containing the complete viral promoter sequence was cloned into the EcoRI and HindIII restriction sites of PUc18 vector (Fermentas). This clone was used to construct all EBER2 7SK L2 RNA chimeras. To obtain the EBER2 7SK L2 construct, the sequence from +81 nt to +140 nt of wild-type EBER2 RNA was exchanged by the sequence from +113 nt to +154 nt of wild-type 7SK RNA. To obtain the EBER2 7SK L2 m137 construct, nucleotides +104 to +106 (CGG) of the EBER2 7SK L2 construct were mutated to GCC. In the construct EBER2 7SK L2 m122/m137, in addition the nucleotides +89 to +91 (CCG) were mutated to GGC. To obtain the EBER2-Lb construct, nucleotides +80 to +145 of wild-type EBER2 RNA were deleted.
HMGA1a-FLAG, HMGA1a 3xΔA/T-FLAG, HMGA1aΔA/T1-FLAG, HMGA1aΔA/T2-FLAG and HMGA1aΔA/T3-FLAG
To obtain a vector for the expression of FLAG fusion proteins, the hybridized oligonucleotides 5′-CTA GAG GGC GAC TAC AAA GAC GAT GAC GAC AAA GGA TGA A-3′ (sense) and 3′-TC CCG CTG ATG TTT CTG CTA CTG CTG TTT CCT ACT TGG CC-5′ (antisense) were inserted into the XbaI and AgeI restriction sites of the vector pcDNA4/TO/myc-his B (Invitrogen). The coding sequence of wild-type HMGA1a from +1 nt to +321 nt was inserted into the PstI and XbaI restriction sites of the vector mentioned above. To obtain the HMGA1a 3xΔA/T-FLAG construct, nucleotides +82, +178 and +256 (A,C,A) were mutated to G in order to change the codons at these positions from arginine to glycine. To obtain the single mutants HMGA1aΔA/T1-FLAG, HMGA1aΔA/T2-FLAG and HMGA1aΔA/T3-FLAG, the single nucleotides +82, +178 or +256 were mutated to G, respectively.
HMGA1a-myc/His (MH)
The coding sequence of wild-type HMGA1a from +1 nt to +321 nt was inserted into the PstI and XbaI restriction sites of the vector pcDNA4/TO/myc-his B (Invitrogen).
ShRNA knockdown of HMGA1
To knock down the expression of endogenous HMGA1, we used the expression of short hairpin RNA containing a functional siRNA sequence targeting HMGA1 mRNA (45). The full sequence of the HMGA1 targeting shRNA was 5′-CAA CUC CAG GAA GGA AAC CAA GCG AUU GGU UUC CUU CCU GGA GUU G-3′. For control, a non-targeting (scramble) shRNA was used with the sequence 5′-AAC AGU CGC GUU UGC GAC UGG GCG ACC AGU CGC AAA CGC GAC UGU U-3′. For downstream transcriptome analyses, these shRNAs were expressed using a vector coding for the corresponding shRNA under control of the 7SK promoter. To verify the functionality of the shRNAs, we used a vector coding for both—the corresponding shRNA under control of a H1 promoter as well as the HMGA1a-GFP fusion protein under control of a CMV promoter. Effects of the corresponding shRNA on the expression of HMGA1a-GFP fusion protein were followed by fluorescence microscopy.
Localization studies of HMGA1a
To study the localization of HMGA1a-GFP fusion proteins during a simultaneous expression of EBER2 7SK L2 RNA chimeras, a vector was used that codes for both the corresponding EBER2 7SK L2 RNA chimera as well as the HMGA1a-GFP fusion protein under control of a CMV promoter. To obtain such an expression vector, the coding sequence of HMGA1a from +1 nt to +321 nt was inserted into the XhoI and KpnI restriction sites of the peGFP N1 vector (clontech). Subsequently, the coding sequence as well as the complete promoter region of the corresponding RNA chimera from −171 nt to +206 nt was inserted into the EcoR109I restriction site of the obtained HMGA1a-GFP expression vector. The expression of the EBER2 7SK L2 RNA chimeras was verified by northern blot analyses and the expression of HMGA1a-GFP fusion protein was verified by fluorescence microscopy. Localization studies were performed by confocal microscopy.
RESULTS
Fractionation of cellular proteins probed for 7SK interaction
In order to identify additional, specific interaction partners of the human 7SK RNA, we concentrated on the sole predicted stem-loop structure (loop 2, comprising 110–172 nt of the wild-type 7SK RNA, Figure 1A) that is neither involved in the regulation of P-TEFb activity (46,47) nor in the interaction with MePCE or LARP7 (9,10). HeLa nuclear extracts were fractionated by cation exchange chromatography (SP-Sepharose Fast Flow) into three fractions eluting at 100 mM, 340 mM and 600 mM KCl. Each fraction was probed for 7SK L2 RNA-binding activity in EMSAs, revealing that the 600 mM KCl fraction contained a protein binding to 7SK L2. Following, the compounds of this fraction were separated by size via gel filtration (Superdex G75). The resulting fractions containing the 7SK L2 binding protein were loaded on a second cation exchange chromatography (SP-Sepharose High Performance) and proteins were eluted applying a linear KCl gradient from 250 mM KCl to 1 M KCl (Figure 1B, upper scheme). Each fraction was analyzed for the presence of 7SK L2 RNA-binding proteins in EMSAs (Figure 1B, middel panel). Almost homogeneous protein fractions with 7SK RNA L2-binding activity were obtained (Fractions 14–16 from the second cation exchange chromatography; Figure 1B, middel panel). Gradient polyacrylamide gel electrophoresis of these fractions revealed three potential candidates for the 7SK L2-binding protein (Figure 1B, lower panel). LC–MSMS analysis of the visible proteins revealed that they belong to the high mobility group proteins HMGN1, HMGA1 and HMGN2 (see Table 1). Alternative splicing of the HMGA1 gene transcript leads to three different splice variants HMGA1a (11.7 kDa), HMGA1b (10.6 kDa) and HMGA1c (19.7 kDa) (48,49). HMGA1b and c differ from the major variant by an internal deletion of 11 amino acids or a frame-shift before the third DNA-binding motif, respectively. In the LC–MSMS analysis of the protein band matching, the elution profile of the 7SK L2 binding protein of only the splice variants a and b were detected (Table 1). To include also the 11 amino acids lacking in HMGA1b compared to HMGA1a as potential interaction site, for all following experiments recombinant HMGA1a protein or HMGA1a cDNA was used. Recombinant variants of the two most likely candidates, HMGA1 and HMGN1, were screened for 7SK L2 binding in EMSAs. While HMGN1 did not show any complex formation with 7SK L2 RNA, we were able to identify HMGA1 as the 7SK L2-binding protein (Figure 1C). Furthermore, increasing amounts of unlabeled 7SK L2 RNA compete with the labeled 7SK L2 RNA for binding to HMGA1 in EMSAs, confirming the specificity of the complex (Figure 1D).
Table 1.
HMG proteins identified by their corresponding peptides in LC–MSMS
| Protein | MWr | MWr calc. | Peptide | Position |
|---|---|---|---|---|
| HMGN1 | 810.54 | 810.50 | LSAKPPAK | 24–31 |
| 946.50 | 946.51 | KVSSAEGAAK | 4–15 | |
| 1116.60 | 1116.54 | QAEVANQETK | 62–71 | |
| 1301.72 | 1301.65 | VSSAEGAAKEEPK | 6–18 | |
| 1462.95 | 1462.89 | LSAKPPAKVEAKPK | 24–37 | |
| 1676.79 | 1676.74 | TEESPASDEAGEKEAK | 83–98 | |
| HMGA1a/b | 816.35 | 816.43 | SSQPLASK | 8–15 |
| 982.44 | 982.50 | EPSEVPTPK | 47–55 | |
| 1201.53 | 1201.63 | SSQPLASKQEK | 8–18 | |
| 1391.69 | 1391.75 | EPSEVPTPKRPR | 47–58 | |
| 1731.77 | 1731.86 | SSQPLASKQEKDGTEK | 8–23 | |
| HMGA1a | 1592.77 | 1592.38 | KQPPVSPGTALVGSQK | 31–46 |
| 2557.17 | 2557.38 | KQPPVSPGTALVGSQKEPSEVPTPK | 31–55 | |
| HMGA1b | 1560.77 | 1560.85 | KQPPKEPSEVPTPK | 31–44 |
| HMGN2 | 1216.53 | 1216.64 | KAEGDAKGDKAK | 5–16 |
| 1225.60 | 1225.69 | AKVKDEPQRR | 15–24 | |
| 1388.49 | 1388.65 | TDQAQKAEGAGDAK | 77–90 | |
| 1583.81 | 1583.94 | LSAKPAPPKPEPKPK | 28–42 |
After SDS–PAGE and silver staining (see Figure 1B, lower panel), protein bands were excised from the gel, digested enzymatically and the resulting peptides were analyzed by LC–MSMS. The experimentally determined relative molecular weight (MWr), the calculated relative molecular weight (MWr calc.), the amino acid sequence of the corresponding peptide as well as the position of this peptide within the corresponding protein are indicated. The corresponding proteins were identified by database search of the peptide fragment spectra using MASCOT as search engine.
HMGA1 specifically binds 7SK RNA in vitro
We disrupted the secondary structure of the terminal stem-loop region of the wild-type 7SK RNA by mutation of three nucleotides (137–139 nt; Figure 2A; 7SK L2 m137). EMSAs of the mutated RNA revealed a strong reduction of HMGA1a binding which was not recovered by compensatory mutations of the complementary strand (122–124 nt; Figure 2A; 7SK L2 m122/m137), though such a rescue should partially restore the original secondary structure. HMGA1a binding thus occurs via the terminal stem-loop structure of L2 and the interaction is furthermore specific to either the tertiary structure or the sequence of the 7SK RNA. As increasing amounts of unlabeled 7SK L2 RNA compete with labeled 7SK L2 RNA for HMGA1a binding (Figure 1D), these results taken together confirm the specificity of that RNA–protein interaction.
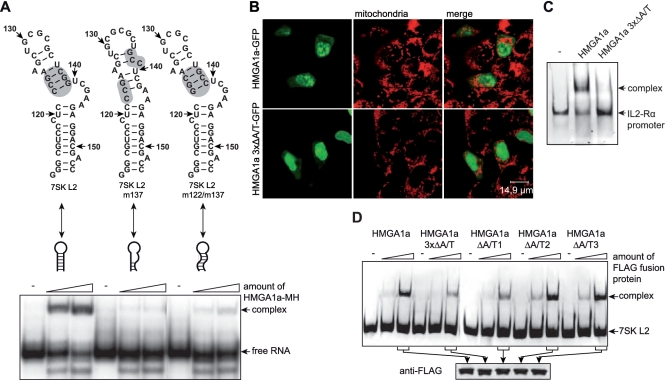
Figure 2.
The 7SK-HMGA1 interaction is specific for the secondary structure of 7SK L2 and the A/T-hook motif 1 of HMGA1a. (A) The secondary structure of 7SK L2, 7SK L2 m137 and 7SK L2 m122/m137 RNA was calculated using RNAstructure 4.4 software (40). The nucleotides are numbered corresponding to their position in wild-type 7SK RNA. The region mutated in 7SK L2 m137 as well as 7SK L2 m122/m137 RNA is marked in grey (upper scheme). Two nanogram of each of these 32P-labeled RNAs were tested for HMGA1a-Myc/His (MH) binding activity in EMSAs using increasing amounts of purified HMGA1a-MH (∼4 and 8 ng), in comparison with the corresponding RNA alone (−) (lower panel). (B) The intracellular localization of wild-type HMGA1a-GFP and the mutant HMGA1a 3xΔA/T-GFP containing three A/T-hook motifs with disrupted DNA binding activity was analyzed by confocal microscopy. Mitochondria were stained to visualize the cell body. (C) The IL2-Rα promoter consisting of the basepairs −136 to −6 of the human IL2-Rα gene was used to compare the DNA-binding activity of HMGA1a-FLAG with HMGA1a 3xΔA/T-FLAG. EMSAs were performed with 100 ng of Fam6-labeled DNA and ∼100 ng of each protein in comparison to the DNA fragment alone (−). (D) Hundred nanogram of Fam6-labeled 7SK L2 RNA were incubated with increasing amounts (∼50 and 100 ng) of immuno-purified HMGA1a-FLAG, the A/T-hook mutant HMGA1a 3xΔA/T-FLAG or single A/T-hook motif mutants (HMGA1a ΔA/T1-FLAG, HMGA1a ΔA/T2-FLAG, HMGA1a ΔA/T3-FLAG). The resulting complexes were analyzed in EMSAs in comparison to the RNA alone (-) (upper panel). The amount of protein used in these analyses was verified by western blot analyses (lower panel).
7SK RNA interacts with HMGA1 through the 1st A/T hook
HMGA1a is a DNA binding protein containing three A/T hook motifs (50). The GFP fusion protein of a HMGA1 mutant (HMGA1a 3xΔA/T) carrying single R→G transitions in all three A/T hooks, which have been previously shown to abrogate DNA binding (51), shows a strongly decreased chromatin-associated nuclear distribution compared with the wild-type GFP fusion protein (Figure 2B).
We verified this loss of DNA-binding activity of HMGA1a 3xΔA/T compared with wild-type HMGA1a in EMSAs with a promoter fragment (bp −136 to −76) of the interleukin 2 receptor α gene (IL2-Rα), which has been previously shown to contain defined HMGA1-binding sites (52,53).
EMSAs with 7SK L2 RNA revealed that HMGA1a 3xΔA/T shows not only a decreased DNA-binding activity but also a significantly reduced ability to interact with 7SK L2 RNA (Figure 2C). Furthermore, EMSAs of 7SK L2 RNA with HMGA1 versions containing mutations of each single A/T-hook motif (HMGA1a ΔA/T1, HMGA1a ΔA/T2 and HMGA1a ΔA/T3) identify the first A/T-hook motif of HMGA1a as the site of interaction with 7SK RNA (Figure 2C, upper panel), since the same reduction in binding affinity is observed with the single HMGA1a ΔA/T1 mutant as with the triple mutant. Moreover, mutation of either one of the remaining A/T hooks does not reduce 7SK-binding in EMSAs when compared to the wild-type HMGA1 protein (Figure 2C, upper panel). Load controls thereby confirm the use of equal amounts of HMGA1 and mutant proteins in the reactions (Figure 2C, lower panel).
HMGA1 co-purifies specifically with 7SK RNA
To verify a specific interaction of full length 7SK RNA with HMGA1a in vitro, we immuno-precipitated purified HMGA1a-FLAG in the presence of in vitro transcribed 7SK, U6 and EBER2 RNA. After extensive washing, the protein-bound RNA was eluted with increasing concentrations of KCl (500 mM for E1 and 1 M for E2), before the FLAG fusion protein was eluted at pH 3.5 (E3). The RNA contents of each immuno-precipitation step were analyzed by denaturing PAGE (Figure 3A), revealing that only 7SK RNA was detectable in the elution fractions containing increasing concentrations of KCl. Neither U6 nor EBER2 snRNAs were obtained. Thus, not only the L2 substructure but also the entire 7SK RNA can bind specifically to HMGA1, which in turn does not interact with the other control RNAs tested.
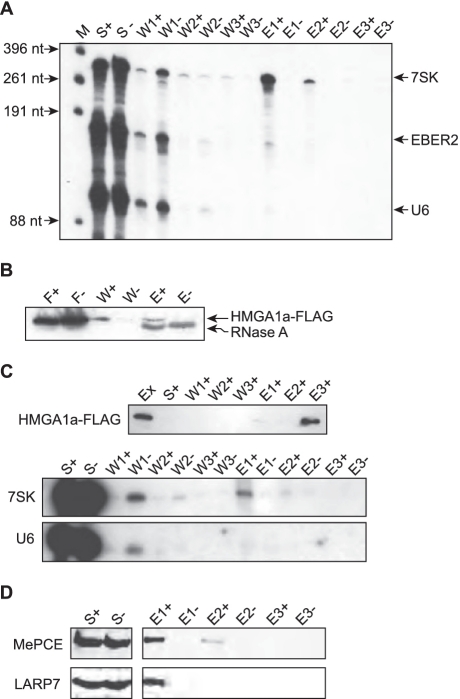
Figure 3.
Full length 7SK RNA binds to HMGA1a-FLAG and endogenous 7SK RNA is associated with HMGA1a-FLAG in living cells. (A) Full length T7-transcribed, 32P-labeled 7SK RNA was co-immunoprecipitated with purified HMGA1a-FLAG in vitro by anti-FLAG agarose. The supernatant (S) was removed and after washing three times (W), the bound RNA was eluted (E) using increasing concentrations of KCl (500 mM for E1 and 1 M for E2), before HMGA1a-FLAG was eluted at pH 3.5 (E3). Every step was analyzed for RNA by denaturing gel electrophoresis. T7-transcribed, 32P-labeled U6 RNA and viral EBER2 RNA as well as an immunoprecipitation lacking HMGA1a-FLAG (−) were used for control. (B) Oligo dT-cellulose was loaded with poly-adenylated 7SK L2 RNA. Cellular extract containing HMGA1a-FLAG (+) was added and after extensive washing (W), the RNA-bound proteins were eluted by RNaseA digest (E). Unloaded oligo dT-cellulose treated with cellular extract containing HMGA1a-FLAG was used in a parallel procedure for control (−). Flowthrough (F), washing steps (W) and elution steps (E) were tested for HMGA1a-FLAG by western blot analyses. (C) Endogenous 7SK RNA was co-immunoprecipitated with HMGA1a-FLAG from HEK293 cellular extract by anti-FLAG agarose. After washing three times (W), the protein-bound RNA was eluted (E) using increasing concentrations of KCl (500 mM for E1 and 1M for E2), before the HMGA1a-FLAG was eluted at pH 3.5 (E3). Extract (Ex), supernatant (S), washing steps and elution steps were tested for HMGA1a-FLAG by western blot analyses. Co-immunoprecipitated 7SK RNA was visualized by northern blot analyses (middle panel). U6 RNA (lower panel) as well as an immunoprecipitation of HEK293 extract lacking HMGA1a-FLAG (−) was used for control. (D) MePCE and LARP7 co-immunoprecipitate with 7SK RNA and HMGA1a-FLAG. The fractions of the immunoprecipitation shown in (C) were analyzed for MePCE and LARP7 by western blot analyses.
Vice versa, oligo-dT cellulose was first loaded with polyadenylated 7SK L2 RNA as bait and then incubated with cellular extracts containing the recombinant HMGA1a-FLAG fusion protein, efficiently enriched HMGA1 protein from a HEK293 cellular extract. This is evident from the western blot analysis of the eluted fraction, obtained after extensive washing and RNase A digestion (Figure 3B).
7SK RNA interacts with HMGA1 in vivo
To prove the interaction in a cellular context, HMGA1a-FLAG was immunoprecipitated from transiently transfected HEK293 cells (Figure 3C, upper panel), and co-purified RNAs were analyzed in northern blots utilizing RNA probes specific for 7SK or U6 RNA (Figure 3C, lower panels). It is evident that significant amounts of cellular 7SK RNA were released but not spliceosomal U6 RNA, which served as an internal control. Hence, the interaction between 7SK RNA and HMGA1 in vitro remains detectable even in the presence of high amounts of both competing cellular proteins and competing cellular RNAs. Since the majority of cellular 7SK RNA is known to be associated with LARP7 by its 3′-poly(U) tail (10) and MePCE by its 5′-end (8,9), we tested each fraction of this co-immunoprecipitation for the presence of these proteins. Indeed, we could confirm the association of LARP7 as well as MePCE with the HMGA1-7SK RNA complex. Both proteins elute at 500 mM KCl from the HMGA1a-FLAG fusion protein—in the same fraction as 7SK RNA (Figure 3D).
HMGA1a was described as the ‘hub’ of nuclear function for being involved in the majority of decisive nuclear events through its chromatin and gene expression regulation activities (48,49,54). The finding that HMGA1 specifically interacts with 7SK RNA via its A/T-hook 1 DNA binding motif points to a regulatory function of this novel snRNA–protein complex.
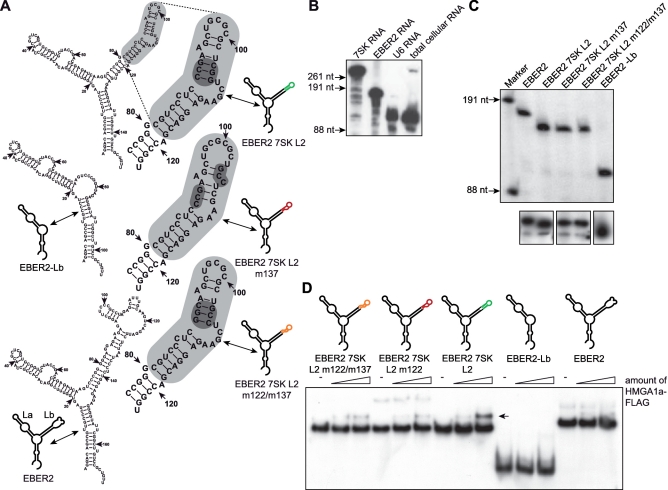
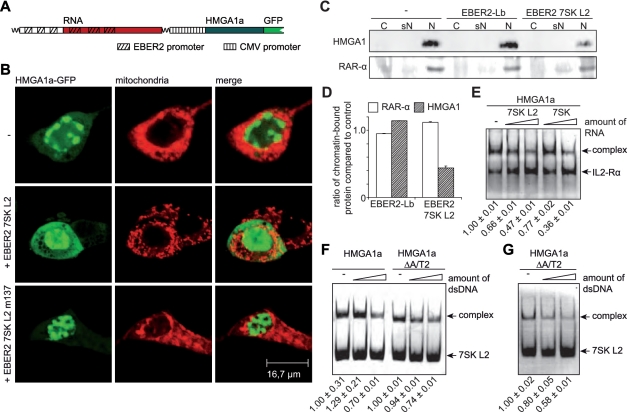
In order to unveil that biological function, we controlled the amount of free endogenous HMGA1a by over-expression of the 7SK L2 substructure. For that, the promoter of the viral EBER2 snRNA gene was used, which is one of the strongest RNA Polymerase III promoters known, resulting in more than 107 copies of this 172 nt long nuclear transcript per infected cell (55,56) (Figure 4A; EBER2 RNA). RNA chimeras of EBER2 RNA (1−80 nt and 141–172 nt) and the 7SK L2 substructure (113–154 nt; EBER2 7SK L2) as well as the two mutant 7SK RNA L2 substructures (EBER2 7SK L2 m137 and EBER2 7SK L2 m122/m137) were generated (Figure 4A). Those constructs exclude any effects on P-TEFb activity as the chimeras lack the 7SK substructures L1, L3 and L4 required for P-TEFb binding (46,47) (Figure 1A). EBER2 7SK L2 chimeras express to high levels, as checked by both in vitro transcription analysis and northern blot analysis (Figure 4C). The specificity of the antisense RNA probes used in these northern blot analyses as well as in the experiments shown in Figure 3C was verified in northern blot analysis using in vitro transcripts as well as total cellular RNA (Figure 4B). Furthermore, EMSAs verified that HMGA1a binding characteristics of the 7SK L2 substructures are entirely retained in the context of the EBER2 chimeras (Figure 4D). Thus neither the EBER2 backbone (31) influences the affinity of the L2 RNAs for HMGA1a nor do parts or the entire EBER2 RNA bind to HMGA1a.
Figure 4.
EBER2 7SK L2 fusion RNAs express at high levels and conserve HMGA1 binding activity. (A) The secondary structures of EBER2-Lb RNA, EBER2 7SK L2 RNA, EBER2 7SK L2 m137 RNA, EBER2 7SK L2 m122/m137 RNA as well as EBER2 wild-type RNA was calculated using RNAstructure 4.4 software (40). The 7SK L2 substructure is highlighted in grey and the region mutated in 7SK L2 m137 as well as 7SK L2 m122/m137 RNA is marked in dark grey. (B) The RNA probes used to detect 7SK, U6 and EBER2 RNA in northern blot analyses were tested for specificity. Northern blot analyses were performed with T7-transcribed 7SK, U6 and EBER2 RNA as well as with total cellular RNA using equal amounts of the 32P-labeled probes targeting 7SK, U6 and EBER2 RNA. (C) The transcription rate of EBER2 7SK L2, EBER2 7SK L2 m137, EBER2 7SK L2 m122/m137 RNA as well as of EBER2-Lb RNA was compared with wild-type EBER2 RNA in in vitro transcription analyses using HeLa nuclear extract (upper panel). The in vivo expression was checked in northern blot analyses using a 32P-labeled probe specific for the 5-region of EBER2 RNA (lower panel). (D) Two nanogram of 32P-labeled EBER2 7SK L2, EBER2 7SK L2 m137, EBER2 7SK L2 m122/m137, EBER2-Lb and wild-type EBER2 RNA were incubated with increasing amounts of immuno-purified HMGA1a-FLAG fusion protein (∼4 and 8 ng). The resulting complexes were analyzed in EMSAs compared with the corresponding RNAs alone (−). The HMGA1a-binding affinity of the corresponding RNAs is visualized as color code (green = high affinity, orange = medium affinity, red = low affinity). The arrow points at the resulting complex of EBER2 7SK L2 RNA with HMGA1a-FLAG.
7SK RNA competes with DNA for binding to HMGA1
High HMGA1 expression levels have been reported for both, embryonic and transformed neoplastic cells (57–61). The protein is readily detectable in the human adenovirus five transformed embryonic cell line HEK293, using immunocytochemistry as well as RT–PCR analysis. Recombinant HMGA1a-GFP, co-expressed from the same plasmid as the EBER2 7SK L2 m137 RNA mutant in HEK293 cells (Figure 5A), shows a nuclear, chromatin-associated localization (Figure 5B, lower panels), comparable to HMGA1a-GFP alone (Figure 5B, upper panels). Simultaneous expression of EBER2 7SK L2 RNA and HMGA1a-GFP results in a less defined nuclear localization, concomitant with a significantly increased accumulation of the protein within the cytoplasm (Figure 5B, middle panels). To verify the decrease of chromatin-bound endogenous HMGA1 upon 7SK L2 over-expression, a chromatin preparation was performed from HeLa cells over-expressing EBER2 7SK L2 RNA or EBER2-Lb RNA. The amounts of chromatin-bound HMGA1 were compared with non-transfected cells in western blot analyses (Figure 5C, upper panel). The quantification of the western blot signals revealed a reduction of the amount of chromatin-bound endogenous HMGA1 of about 50% upon EBER2 7SK L2 RNA expression (Figure 5D). The amount of chromatin-bound RAR-α, which served as an internal control in these experiments did not change significantly among the different conditions (Figure 5D). These data corroborate the hypothesis of a competitive mechanism of HMGA1a binding either to DNA or 7SK RNA. Indeed, using EMSAs, competitive binding of 7SK RNA and DNA to HMGA1 can be demonstrated, as the presence of increasing amounts of non-labeled dsDNA reduces the amount of complex formed by HMGA1a and 7SK L2 RNA (Figure 5F and G). Interestingly, the competition can be specifically traced to the 7SK interacting A/T hook 1, as it also readily occurs in the HMGA1a ΔA/T2, leaving only A/T-hook 1 available for DNA and RNA interaction, since the A/T-hooks 1 and 2 have been shown previously to be the main mediators of DNA binding (Figure 5F and G) (51). Hence, the competition also does not rely on unspecific steric hindrance. EMSAs with the promoter region (bp −136 to −76) of the interleukin 2 receptor alpha (IL2-Rα) gene, which has been shown previously to contain several HMGA1 binding sites (52,53), and HMGA1a-FLAG in the presence of increasing amounts of non-labeled 7SK L2 RNA confirm the competition of 7SK L2 RNA with DNA for HMGA1 binding (Figure 5E). The competition could also be shown by the addition of increasing amounts of full length 7SK RNA (Figure 5E).
Figure 5.
7SK RNA competes with DNA for HMGA1 binding. (A) Schematic drawing of the constructs used for simultaneous expression of EBER2 7SK L2 RNA chimeras and HMGA1a-GFP fusion protein. The corresponding promoter elements are highlighted. (B) The intracellular localization of HMGA1a-GFP alone (−) as well as in co-expression with EBER2 7SK L2 RNA or EBER2 7SK L2 m137 RNA was analyzed by confocal microscopy. Mitochondria were stained to visualize the cell body. (C) 7SK L2 over-expression leads to a reduction of chromatin-bound HMGA1. HeLa cells over-expressing EBER2-Lb RNA and EBER2 7SK L2 RNA as well as non-transfected control cells were fractionated into cytoplasmic (C), soluble nuclear (sN) and chromatin-bound nuclear fraction (N). The presence of HMGA1 in each fraction was tested in western blot analyses. RAR-α was used as a control. (D) The western blot signals of the chromatin-bound fraction (N) in C were quantified using ImageJ software (37) and the ratios of the corresponding signals after expression of EBER2 7SK L2 RNA or EBER2-Lb RNA and the signal of non-transfected cells were calculated. (E) The IL2-Rα promoter fragment described in Figure 2C was used to study competition between DNA and 7SK L2 RNA or full length 7SK RNA for HMGA1a-binding. Hundred nanogram of the Fam6-labeled promoter fragment were incubated with ∼100 ng of HMGA1a-FLAG in the absence (−) and presence of increasing amounts (500 ng and 1 μg) of 7SK L2 and full length 7SK RNA. EMSAs were performed to analyze the resulting complexes. The fractions of shifted DNA were quantified from three independent experiments, normalized to the shifted fraction in absence of competitor (lane 1) and indicated at the bottom of the panel. (F) Hundred nanogram of Fam6-labeled 7SK L2 RNA were incubated with 100 ng of HMGA1a-FLAG as well as the mutant HMGA1aΔA/T2-FLAG in presence of increasing amounts (500 ng and 1 μg) of dsDNA. The resulting complexes were analyzed in EMSAs. The fractions of shifted RNA were quantified from three independent experiments, normalized, and indicated as in (E). (G) Hundred nanogram of Fam6-labeled 7SK L2 RNA were incubated with ∼100 ng of HMGA1aΔA/T2-FLAG in the presence of increasing amounts (500 ng and 1 μg) of dsDNA containing a specific HMGA1 binding motif (77). The resulting complexes were analyzed in EMSAs. The fractions of shifted RNA were quantified from three independent experiments, normalized and indicated as in (E).
HMGA1 target genes are affected in their expression by 7SK RNA
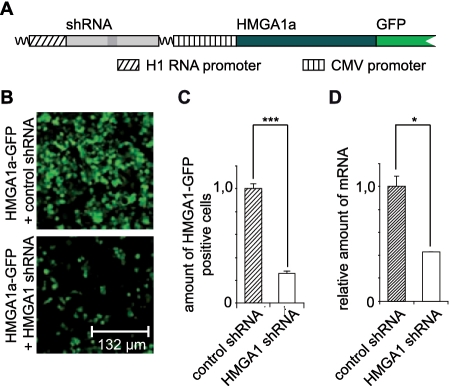
We next determined HMGA1 regulated genes in the same cell line by comparing transcriptome profiles obtained after short hairpin (sh) RNA-mediated knockdown of HMGA1 and control shRNA transfected cells. shRNA was designed according to a published, functional siRNA, targeting HMGA1 (45) and controlled by the strong cellular 7SK promoter. Expression of that construct resulted in a significant knockdown of the endogenous HMGA1 mRNA (Figure 6D). The knockdown was also verified on the protein level using, however, a vector that allows the simultaneous expresion of both the shRNA as well as HMGA1 fused to GFP (Figure 6A). The amount of HMGA1a-GFP positive cells after co-expression of HMGA1 targeting shRNA was compared with that after co-expression of control shRNA, revealing a highly significant knockdown of the HMGA1a-GFP fusion protein (Figure 6B and C).
Figure 6.
Knockdown of endogenous and exogenous HMGA1. (A) Schematic drawing of the constructs used to verify HMGA1 expression knockdown on protein level. The expression of the corresponding shRNA (HMGA1 targeting or control) was controlled by the H1 RNA promoter. The target gene (HMGA1) was expressed as GFP-fusion protein under control of the CMV promoter from the same vector as the shRNA. (B and C) HMGA1 expression in HEK293 cells was knocked down by shRNA in comparison to non-targeting control shRNA. HMGA1a-GFP was co-expressed with either control or HMGA1 shRNA using plasmids coding for both, shRNA and GFP fusion protein (see A). HMGA1a-GFP positive cells were counted via fluorescence microscopy. ***P < 0.01 in a Student's t-test. The error bars show the SD. (D) The expression of endogenous HMGA1 in HEK293 cells was knocked down by shRNA in comparison to non-targeting control shRNA using a vector that allows the expression of either shRNA under control of the 7SK RNA promoter. The knockdown of endogenous HMGA1 mRNA was verified by quantitative PCR analyses. *P < 0.05 in a Student's t-test. The error bars show the SD.
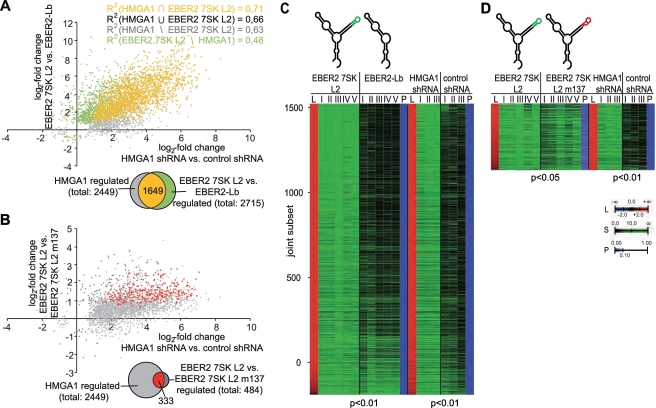
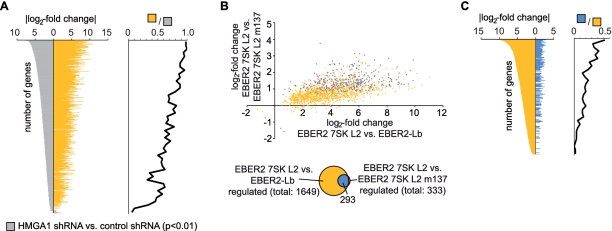
Microarray analysis of RNA from HEK293 cells with knockdown of endogeneous HMGA1 using three independent biological replicates revealed a total of 2449 genes regulated by HMGA1 in a statistically significant way (P < 0.01). Most of these genes (2436 or 99%) were found to be up-regulated upon depletion of HMGA1, which is in line with the previous observations (48). Similarly, the relative effect of EBER2 7SK L2 RNA chimera expression on HEK293 gene transcription as compared with the EBER2-L2 backbone alone was determined using five independent biological replicates. A total of 2715 genes were identified to be regulated in a statistically significant manner (P < 0.01), when compared with the control. In all 1649, or 61%, is also statistically significantly (P < 0.01) regulated by HMGA1 (Figure 7A). This common subset corresponds to 67% of the 2449 HMGA1 target genes. Such an extraordinary concordance (random expectation P < 4.9 × 10−324, hypergeometric distribution) between HMGA1 targets and 7SK L2-responsive genes can only be explained by a strong and a highly relevant physiological interaction between both molecules. Importantly, albeit most genes are repressed in their expression by HMGA1, we also identified several genes that are positively regulated by HMGA1 and 7SK RNA (Figure 7A). An even stronger indication that 7SK and HMGA1 transcriptional effects are closely related stems from the observation that the Pearson correlation coefficient for the statistically significant co-regulated subset (R2=0.71) is comparable to the joint set of genes regulated by either HMGA1 or 7SK L2 (R2=0.66), and even to the subsets specific to either molecule (R2=0.63, R2=0.48) in these completely independent experiments (Figure 7A and C). Albeit not provable due to the limits of statistical testing, the statistical correlations indicate that the HMGA1 and the 7SK RNA-induced transcriptome profiles are not only tightly linked but identical. The high correlation between gene expression downstream of HMGA1 and 7SK L2 also confirms the quality of the experimental techniques used that were previously thoroughly tested to minimize unspecific effects from the EBER2 backbone used for over-expression of the 7SK L2 substructure (31). To further investigate the joint gene regulatory activity of HMGA1 and 7SK RNA, we also compared HEK293 transcriptome profiles using the EBER2 7SK L2 wild-type versus the EBER2 7SK L2 m137 mutant, which shows significantly lowered HMGA1 binding activity (Figure 2A). Again 333 out of 484, or 69%, of the 7SK L2 regulated genes (P < 0.05) were found to be HMGA1 targets (Figure 7B and D). Thus even when comparing the 7SK L2 substructure to the 7SK L2 m137 mutant, a large and perfectly concordant set of HMGA1 targets can be identified. Due to the residual binding of the L2-mutant to HMGA1 (Figure 2A), the relative effect is diminished and harder to appreciate statistically. The common subset of genes differentially regulated after HMGA1 knockdown as well as after expression of EBER2 7SK L2 RNA in comparison to EBER2-Lb RNA (Figure 7A; yellow) is especially enriched in genes, which are strongly affected by the HMGA1 knockdown (Figure 8A). In all, 293 (88%) out of the 333 genes of the common subset differentially expressed after expression of both, EBER2 7SK L2 RNA in comparison to EBER2 7SK L2 m137 RNA and the HMGA1 knockdown (Figure 7B; red), also belong to the common subset of genes differentially expressed after expression of both, EBER2 7SK L2 RNA in comparison to EBER2-Lb RNA and HMGA1 knockdown (Figure 8B). Even though not affected in a significant manner in every case, the expression of the genes belonging to both subsets clearly appears to be linked (Figure 8B). The overlap of both subsets (Figure 8B; blue) is enriched in genes, which are strongly affected among the common subset differentially regulated after HMGA1 knockdown as well as after expression of EBER2 7SK L2 RNA in comparison to EBER2-Lb RNA (Figure 8C).
Figure 7.
The 7SK–HMGA1a interaction impacts the HMGA1a-controlled gene expression. (A and B) The log2-fold change of gene expression after over-expression of EBER2 7SK L2 RNA (y-axis) is compared with that after a shRNA-mediated knockdown of HMGA1 (x-axis). Either the over-expression of EBER2-Lb RNA (A) or the over-expression of EBER2 7SK L2 m137 RNA (B) were chosen for control. Genes significantly regulated by both conditions are marked in yellow (red). Those, which are solely regulated significantly by the shRNA-mediated knockdown of HMGA1 are marked in grey and those solely regulated by the 7SK L2 RNA are marked in green (dark grey). (C and D) Heat maps of the joint subset of statistically significant (at least P < 0.05) changes in gene expression between HEK293 cells over-expressing EBER2 7SK L2 RNA compared with EBER2-Lb (C) EBER2 7SK L2 m137 RNA (D) and HEK293 cells with a shRNA-mediated HMGA1-knockdown compared with HEK293 cells expressing scramble shRNA. L indicates the log2-fold change in expression, S indicates the signal, P indicates the P-value and the roman numbers indicate independent biological replicates.
Figure 8.
Comparison of the common subsets of genes regulated differentially between EBER2 7SK L2 and the two different controls (EBER2-Lb RNA and EBER2 7SK L2 m137 RNA) as well as HMGA1 knockdown and control. (A) The common subset of genes expressed differentially after HMGA1 knockdown compared to the control were ordered by their log2-fold change (2449 genes in Figure 7A; grey). Those genes also expressed differentially after EBER2 7SK L2 RNA expression in comparison to the expression of EBER2-Lb RNA are plotted in yellow (1649 genes in Figure 7A). A density histogram for the enrichment of the yellow versus grey gene sets (bin-size 50 genes)is plotted against the log2-fold change in gene expression. (B) The common subset of differential gene expression after EBER2 7SK L2 RNA expression compared to EBER2-Lb RNA expression and HMGA1 knockdown (yellow) is compared to the common subset of differential gene expression after EBER2 7SK L2 RNA expression compared to EBER2 7SK L2 m137 RNA expression and HMGA1 knockdown (red). Genes significantly regulated in both subsets are marked in blue. For these subsets the log2-fold change of gene expression in each condition is plotted on the x- and y-axis respectively. (C) As (A), but for the yellow and blue (see (B)) subsets.
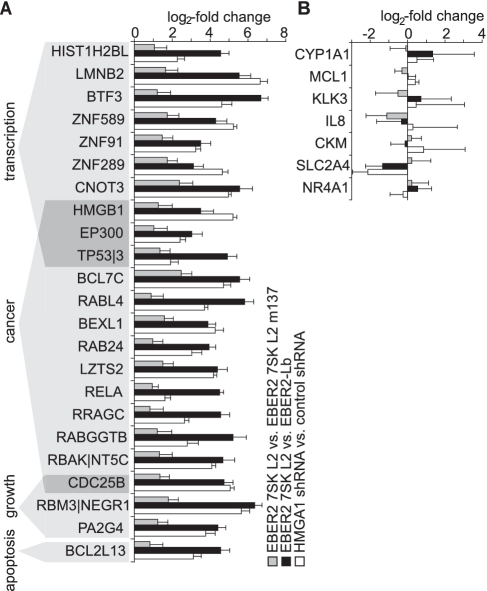
Among the genes regulated differentially by both, the 7SK L2 substructure and the HMGA1-knockdown were various genes with a linkage to major cellular functions that are known to be regulated by HMGA1 (60–64). The expression of several transcription factors (e.g. BTF3, CNOT3, EP300), cancer-related proteins (e.g. TPT1, BCL7C, RELA), apoptosis-related proteins (e.g. BCL2L13) and members of the histone family (e.g. HIST1H2BL) was found to be significantly increased by the over-expression of 7SK L2 domain as well as by the HMGA1-knockdown (Figure 9A). Previous studies had already identified for instance MAP2K2, IGFBP2, SOX4, GNAZ, STK6, CCND3, LAMA1, ACSL3, ARL3, COL6A1 (65), COX2 (64), IFNAR (63) and HLA-A (62), as HMGA1 target genes which we show here are also to be 7SK regulated genes. Due to the limitation of over-expression of solely the L2 substructure of 7SK RNA, P-TEFb regulated genes were not supposed to be targeted by our approach, which is confirmed when verifying the expression of known P-TEFb target genes (17–21) (Figure 9B).
Figure 9.
Genes with HMGA1 connected major cellular functions are regulated by the 7SK-HMGA1 interaction. (A) Different genes, up-regulated by both, over-expression of EBER2 7SK L2 RNA either compared with over-expression of EBER2 7SK L2 m137 RNA (grey bars) or compared with over-expression of EBER2-Lb RNA (black bars) and shRNA-mediated knockdown of HMGA1 (white bars), sorted by the functions of their gene products. The error bars show the SD. (B) The expression of P-TEFb-regulated genes (17–21) is not affected at the chosen conditions.
DISCUSSION
HMGA1 is crucially involved in the vast majority of essential nuclear processes (48,49,54) as well as in neoplastic transformation and tumor progression (57–61). A specific, functional interaction of 7SK RNA with the first A/T-hook of HMGA1a via its highly conserved loop2 affects HMGA1-DNA binding and consequently promotes positive or negative regulatory activity. 7SK RNA hence provides direct regulatory control of HMGA1 activity. This is, to our knowledge, the first report of a direct affector for HMGA1 function not acting at the transcriptional or translational level. As HMGA1 has both positive and negative effects on transcription regulation by other transcription factors such as the estrogen receptor (ER, positive) or the retinoblastoma protein (RB) and p53 which are negatively regulated, the amounts of free 7SK RNA directly affect a large number of transcription programs and cell cycle check-points by regulating HMGA1 function. The transcriptional programs of HMGA1 and 7SK RNA that we identify here and show to be virtually identical will provide rich information on the multitude of gene programs at the control of HMGA1 in transformed cells once analyzed in detail.
The disruption of the HMGA1-7SK snRNA complex by siRNA-mediated depletion of 7SK RNA in HeLa cells has led to the observation of significant effects on P-TEFb-independent genes without explanation of the underlying mechanism (30). The fact that 7SK RNA provides regulatory control of HMGA1 function is likely the explanation for the unexpected effects on gene expression independent of P-TEFb (30), as HMGA1 is known to interact with AT-rich sequences present in the minimal promoter-driven reporters used.
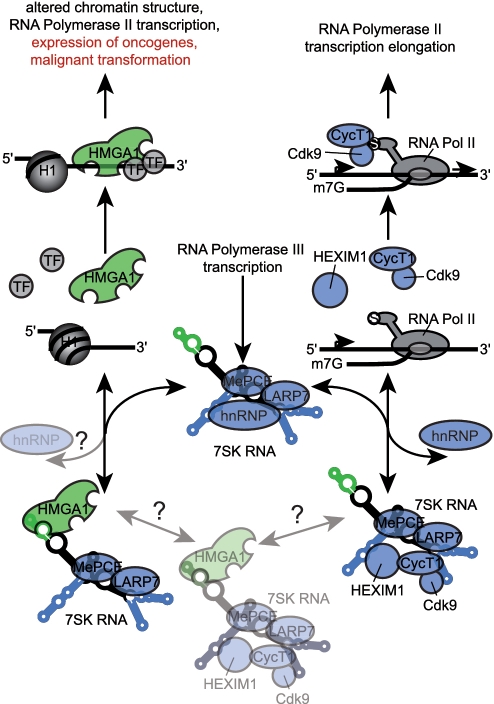
As 7SK RNA has a positive regulatory effect on the large majority of HMGA1 controlled genes, and a negative regulatory effect on elongation of P-TEFb-dependent gene expression, 7SK RNA establishes plasticity between transcription initiation and transcription elongation. The relative amounts of free 7SK RNA in the nucleus, and the opposite effects 7SK RNA has on both transcription regulators indicates yet another intricate mechanism of fine-tuning within the transcription apparatus. A potential 7SK snRNP containing both P-TEFb and HMGA1 would raise important questions concerning possible interlocking functions of HMGA1 and P-TEFb in gene expression regulation and would directly link the modification of chromatin structure (by HMGA1) to gene transcription elongation (by T-TEFb). Furthermore, also possible effects of the hnRNPs, which have been shown to replace P-TEFb from 7SK RNA, on the HMGA1-7SK interaction have to be elucidated. In any case, the connection between P-TEFb-dependent and HMGA1-dependent signaling via 7SK RNA which we uncovered here represents a new molecular switch (Figure 10). The role that P-TEFb and 7SK RNA play in Tat-dependent HIV transcription (24,66–69) thereby might also provide a molecular explanation for the HIV-associated B-cell lymphoma and Kaposi sarcoma incident rates (70–73). Increased titration of nuclear 7SK RNA into Tat-dependent transcription regulatory complexes will, according to our observations, directly increase HMGA1 activity. As increased HMGA1 activity is associated with virtually every known type of cancer, the switch function of 7SK RNA might indeed explain why 7SK RNA is an essential gene.
Figure 10.
Schematic drawing of the regulatory switch function of 7SK RNA. P-TEFb consisting of Cyclin T1 (CycT1) and the cyclin-dependent kinase 9 (Cdk9) activates the RNA Polymerase II (RNA Pol II) transcription elongation reaction by phosphorylation of a serine (S) in the C-terminal domain of RNA Pol II. The interaction of the RNA Polymerase III transcribed 7SK snRNA with P-TEFb and HEXIM1 via its loop 1, loop 3 and loop 4 substructures (blue) leads to a decreased kinase activity of Cdk9 and thus to an inactivation of P-TEFb. Recently, the 7SK RNA P-TEFb interaction has been shown to be central to Tat-dependent HIV transcription. 7SK RNA released from the 7SK/P-TEFb snRNP is trapped by different hnRNPs, which is thought to provide a pool of inactive 7SK RNA. The methylephosphate capping enzyme (MePCE) and the La-related protein 7 (LARP7) have been shown to be stably associated with 7SK RNA. The non-histone chromosomal protein HMGA1, assigned as a ‘hub’ of nuclear function, alters chromatin structure being an antagonist of histone 1 (H1). In addition, it also alters the affinity of several transcription factors (TF) for DNA. That way HMGA1 impacts the expression of many RNA Polymerase II transcribed genes connected to essential cellular processes as well as several oncogenes, leading in case of increased amounts of HMGA1 to malignant transformation. We identified the loop 2 substructure of 7SK RNA (green) to specifically interact with HMGA1 and we identified more than 1500 HMGA1-dependent genes to be controlled by this interaction. Our results point to a switch function of 7SK RNA between RNA Polymerase II transcription elongation and alteration of chromatin structure. Thereby, the 7SK RNA might be central to the plasticity between transcription initiation and elongation, an equilibrium likely to be affected by HMGA1 over-expression in cancer and HIV-driven reprogramming of cellular transcription through the increased titration of 7SK RNA into HMGA1 or P-TEFb complexes, respectively. Interestingly, this competitive link between major transcription regulation complexes established by 7SK RNA might also be a molecular explanation for the development of HIV infection-associated lymphomas and Kaposi sarcomas.
Interestingly, also is the fact that the 7SK-HMGA1 complex represents a novel link between RNA Polymerase II and III gene expression programs. While examples of RNA Polymerase III transcripts affecting RNA Polymerase II-driven gene expression and RNA processing, such as the U6 RNA and its role in splicing (74), exist, the observation of a RNA Polymerase III transcribed gene directly regulating RNA Polymerase II transcription initiation on such a large scale, raises important questions as to the functional inter-dependence of both transcription machines. Furthermore, renewed interest should be accorded to the question of regulatory phenomena in RNA Polymerase III transcription (75). As non-translated RNAs are rarely quantified on widely used microarray technology, and also for technical reasons are only seldom captured in SAGE analysis, we know too little about variations in gene expression patterns of RNA Polymerase III transcripts especially in oncogenesis and viral infection.
Considering the direct connection between the derepression of HMGA1 and cancerogenesis (57–61), over-expression of the 7SK L2 substructure might establish a powerful tool to control the activity of endogenous HMGA1 in physiopathology. From our works it comes that regulating 7SK RNA expression will directly, positively or negatively affect HMGA1 activity (Figure 10). The plasticity between the control of transcription elongation by P-TEFb and of chromatin and transcription regulation by HMGA1 established by 7SK snRNA might be highly relevant for HIV-induced reprogramming of cellular and viral gene expression, and furthermore raises important questions regarding the control of RNA Polymerase III gene transcription (75). The intertwined regulation via 7SK RNA of the control of transcription elongation (by P-TEFb) and of the control of gene activity (by HMGA1) assigns a major role to that snRNA as a molecular switch for nuclear key functions (Figure 10).
FUNDING
Centre National de la Recherche Scientifique (CNRS); the Region Nord; the Genopole Evry (all to A.B.); Deutsche Forschungsgemeinschaft (BE 531/19-3, to B.-J.B.). Funding for open access charge: Genopole Every (to A.B.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Axel Witt for providing the northern blot probes specific for 7SK, U6 and EBER2 RNA. We thank Christina Klein-Schmidt for help with confocal microscopy. We are grateful to Brendan Bell for his helpful comments on the manuscript. Hélène Léger and Nicolas Tchitchek are thanked for instructions on the use of the microarray and relevant statistical analysis technologies.
REFERENCES
- 1.Morey C, Avner P. Employment opportunities for non-coding RNAs. FEBS Lett. 2004;567:27–34. doi: 10.1016/j.febslet.2004.03.117. [DOI] [PubMed] [Google Scholar]
- 2.Malecova B, Morris KV. Transcriptional gene silencing through epigenetic changes mediated by non-coding RNAs. Curr. Opin. Mol. Ther. 2010;12:214–222. [PMC free article] [PubMed] [Google Scholar]
- 3.Barrandon C, Spiluttini B, Bensaude O. Non-coding RNAs regulating the transcriptional machinery. Biol. Cell. 2008;100:83–95. doi: 10.1042/BC20070090. [DOI] [PubMed] [Google Scholar]
- 4.Cannell IG, Kong YW, Bushell M. How do microRNAs regulate gene expression? Biochem. Soc. Trans. 2008;36:1224–1231. doi: 10.1042/BST0361224. [DOI] [PubMed] [Google Scholar]
- 5.Grudnik P, Bange G, Sinning I. Protein targeting by the signal recognition particle. Biol. Chem. 2009;390:775–782. doi: 10.1515/BC.2009.102. [DOI] [PubMed] [Google Scholar]
- 6.Ikegami K, Ohgane J, Tanaka S, Yagi S, Shiota K. Interplay between DNA methylation, histone modification and chromatin remodeling in stem cells and during development. Int. J. Dev. Biol. 2009;53:203–214. doi: 10.1387/ijdb.082741ki. [DOI] [PubMed] [Google Scholar]
- 7.Gurney T, Jr, Eliceiri GL. Intracellular distribution of low molecular weight RNA species in HeLa cells. J. Cell. Biol. 1980;87:398–403. doi: 10.1083/jcb.87.2.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta S, Busch RK, Singh R, Reddy R. Characterization of U6 small nuclear RNA cap-specific antibodies. Identification of gamma-monomethyl-GTP cap structure in 7SK and several other human small RNAs. J. Biol. Chem. 1990;265:19137–19142. [PubMed] [Google Scholar]
- 9.Shumyatsky GP, Tillib SV, Kramerov DA. B2 RNA and 7SK RNA, RNA Polymerase III transcripts, have a cap-like structure at their 5′ end. Nucleic Acids Res. 1990;18:6347–6351. doi: 10.1093/nar/18.21.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He N, Jahchan NS, Hong E, Li Q, Bayfield MA, Maraia RJ, Luo K, Zhou Q. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol. Cell. 2008;29:588–599. doi: 10.1016/j.molcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xue Y, Yang Z, Chen R, Zhou Q. A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP. Nucleic Acids Res. 38:360–369. doi: 10.1093/nar/gkp977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gursoy HC, Koper D, Benecke BJ. The vertebrate 7S K RNA separates hagfish (Myxine glutinosa) and lamprey (Lampetra fluviatilis) J. Mol. Evol. 2000;50:456–464. doi: 10.1007/s002390010048. [DOI] [PubMed] [Google Scholar]
- 13.Gruber AR, Koper-Emde D, Marz M, Tafer H, Bernhart S, Obernosterer G, Mosig A, Hofacker IL, Stadler PF, Benecke BJ. Invertebrate 7SK snRNAs. J. Mol. Evol. 2008;66:107–115. doi: 10.1007/s00239-007-9052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zieve G, Penman S. Small RNA species of the HeLa cell: metabolism and subcellular localization. Cell. 1976;8:19–31. doi: 10.1016/0092-8674(76)90181-1. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 16.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 17.Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA Polymerase II. Mol. Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- 18.Lam LT, Pickeral OK, Peng AC, Rosenwald A, Hurt EM, Giltnane JM, Averett LM, Zhao H, Davis RE, Sathyamoorthy M, et al. Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anti-cancer drug flavopiridol. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-10-research0041. RESEARCH0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee DK, Duan HO, Chang C. Androgen receptor interacts with the positive elongation factor P-TEFb and enhances the efficiency of transcriptional elongation. J. Biol. Chem. 2001;276:9978–9984. doi: 10.1074/jbc.M002285200. [DOI] [PubMed] [Google Scholar]
- 20.Nojima M, Huang Y, Tyagi M, Kao HY, Fujinaga K. The positive transcription elongation factor b is an essential cofactor for the activation of transcription by myocyte enhancer factor 2. J. Mol. Biol. 2008;382:275–287. doi: 10.1016/j.jmb.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian Y, Ke S, Chen M, Sheng T. Interactions between the aryl hydrocarbon receptor and P-TEFb. Sequential recruitment of transcription factors and differential phosphorylation of C-terminal domain of RNA Polymerase II at cyp1a1 promoter. J. Biol. Chem. 2003;278:44041–44048. doi: 10.1074/jbc.M306443200. [DOI] [PubMed] [Google Scholar]
- 22.Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 23.Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews MB, Price DH. Trans.cription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol. Cell. 38:439–451. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diribarne G, Bensaude O. 7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor. RNA Biol. 2009;6:122–128. doi: 10.4161/rna.6.2.8115. [DOI] [PubMed] [Google Scholar]
- 26.Barrandon C, Bonnet F, Nguyen VT, Labas V, Bensaude O. The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes. Mol. Cell. Biol. 2007;27:6996–7006. doi: 10.1128/MCB.00975-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Herreweghe E, Egloff S, Goiffon I, Jady BE, Froment C, Monsarrat B, Kiss T. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J. 2007;26:3570–3580. doi: 10.1038/sj.emboj.7601783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hogg JR, Collins K. RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA. 2007;13:868–880. doi: 10.1261/rna.565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paule MR, White RJ. Survey and summary: transcription by RNA Polymerases I and III. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haaland RE, Herrmann CH, Rice AP. siRNA depletion of 7SK snRNA induces apoptosis but does not affect expression of the HIV-1 LTR or P-TEFb-dependent cellular genes. J. Cell. Physiol. 2005;205:463–470. doi: 10.1002/jcp.20528. [DOI] [PubMed] [Google Scholar]
- 31.Eilebrecht S, Pellay FX, Odenwalder P, Brysbaert G, Benecke BJ, Benecke A. EBER2 RNA-induced transcriptome changes identify cellular processes likely targeted during Epstein Barr Virus infection. BMC Res. Notes. 2008;1:100. doi: 10.1186/1756-0500-1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graham FL, van der Eb AJ. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 33.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA Polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wysocka J, Reilly PT, Herr W. Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol. Cell. Biol. 2001;21:3820–3829. doi: 10.1128/MCB.21.11.3820-3829.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 36.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 37.Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- 38.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 39.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 40.Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]
- 41.Noth S, Benecke A. Avoiding inconsistencies over time and tracking difficulties in Applied Biosystems AB1700/Panther probe-to-gene annotations. BMC Bioinformatics. 2005;6:307. doi: 10.1186/1471-2105-6-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noth S, Brysbaert G, Benecke A. Normalization using weighted negative second order exponential error functions (NeONORM) provides robustness against asymmetries in comparative transcriptome profiles and avoids false calls. Genomics Proteomics Bioinformatics. 2006;4:90–109. doi: 10.1016/S1672-0229(06)60021-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noth S, Brysbaert G, Pellay FX, Benecke A. High-sensitivity transcriptome data structure and implications for analysis and biologic interpretation. Genomics Proteomics Bioinformatics. 2006;4:212–229. doi: 10.1016/S1672-0229(07)60002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilhelm E, Kornete M, Targat B, Vigneault-Edwards J, Frontini M, Tora L, Benecke A, Bell B. TAF6delta orchestrates an apoptotic transcriptome profile and interacts functionally with p53. BMC Mol. Biol. 11:10. doi: 10.1186/1471-2199-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liau SS, Ashley SW, Whang EE. Lentivirus-mediated RNA interference of HMGA1 promotes chemosensitivity to gemcitabine in pancreatic adenocarcinoma. J. Gastrointest. Surg. 2006;10:1254–1262. doi: 10.1016/j.gassur.2006.06.011. discussion 1263. [DOI] [PubMed] [Google Scholar]
- 46.Egloff S, Van Herreweghe E, Kiss T. Regulation of Polymerase II transcription by 7SK snRNA: two distinct RNA elements direct P-TEFb and HEXIM1 binding. Mol. Cell. Biol. 2006;26:630–642. doi: 10.1128/MCB.26.2.630-642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Herreweghe E, Egloff S, Goiffon I, Jady BE, Froment C, Monsarrat B, Kiss T. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J. 2007;26:3570–3580. doi: 10.1038/sj.emboj.7601783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reeves R, Beckerbauer L. HMGI/Y proteins: flexible regulators of transcription and chromatin structure. Biochim. Biophys. Acta. 2001;1519:13–29. doi: 10.1016/s0167-4781(01)00215-9. [DOI] [PubMed] [Google Scholar]
- 49.Reeves R. Molecular biology of HMGA proteins: hubs of nuclear function. Gene. 2001;277:63–81. doi: 10.1016/s0378-1119(01)00689-8. [DOI] [PubMed] [Google Scholar]
- 50.Reeves R, Nissen MS. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J. Biol. Chem. 1990;265:8573–8582. [PubMed] [Google Scholar]
- 51.Harrer M, Luhrs H, Bustin M, Scheer U, Hock R. Dynamic interaction of HMGA1a proteins with chromatin. J. Cell Sci. 2004;117:3459–3471. doi: 10.1242/jcs.01160. [DOI] [PubMed] [Google Scholar]
- 52.Reeves R, Elton TS, Nissen MS, Lehn D, Johnson KR. Posttranscriptional gene regulation and specific binding of the nonhistone protein HMG-I by the 3′ untranslated region of bovine interleukin 2 cDNA. Proc. Natl Acad. Sci. USA. 1987;84:6531–6535. doi: 10.1073/pnas.84.18.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reeves R, Leonard WJ, Nissen MS. Binding of HMG-I(Y) imparts architectural specificity to a positioned nucleosome on the promoter of the human interleukin-2 receptor alpha gene. Mol. Cell. Biol. 2000;20:4666–4679. doi: 10.1128/mcb.20.13.4666-4679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gerlitz G, Hock R, Ueda T, Bustin M. The dynamics of HMG protein-chromatin interactions in living cells. Biochem. Cell. Biol. 2009;87:127–137. doi: 10.1139/O08-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lerner MR, Andrews NC, Miller G, Steitz JA. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc. Natl Acad. Sci. USA. 1981;78:805–809. doi: 10.1073/pnas.78.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dumpelmann E, Mittendorf H, Benecke BJ. Efficient transcription of the EBER2 gene depends on the structural integrity of the RNA. RNA. 2003;9:432–442. doi: 10.1261/rna.2176603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chiappetta G, Avantaggiato V, Visconti R, Fedele M, Battista S, Trapasso F, Merciai BM, Fidanza V, Giancotti V, Santoro M, et al. High level expression of the HMGI (Y) gene during embryonic development. Oncogene. 1996;13:2439–2446. [PubMed] [Google Scholar]
- 58.Chiappetta G, Bandiera A, Berlingieri MT, Visconti R, Manfioletti G, Battista S, Martinez-Tello FJ, Santoro M, Giancotti V, Fusco A. The expression of the high mobility group HMGI (Y) proteins correlates with the malignant phenotype of human thyroid neoplasias. Oncogene. 1995;10:1307–1314. [PubMed] [Google Scholar]
- 59.Cleynen I, Van de Ven WJ. The HMGA proteins: a myriad of functions (Review) Int. J. Oncol. 2008;32:289–305. [PubMed] [Google Scholar]
- 60.Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat. Rev. Cancer. 2007;7:899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- 61.Hess JL. Chromosomal translocations in benign tumors: the HMGI proteins. Am J. Clin. Pathol. 1998;109:251–261. doi: 10.1093/ajcp/109.3.251. [DOI] [PubMed] [Google Scholar]
- 62.Abdulkadir SA, Krishna S, Thanos D, Maniatis T, Strominger JL, Ono SJ. Functional roles of the transcription factor Oct-2A and the high mobility group protein I/Y in HLA-DRA gene expression. J. Exp. Med. 1995;182:487–500. doi: 10.1084/jem.182.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bonnefoy E, Bandu MT, Doly J. Specific binding of high-mobility-group I (HMGI) protein and histone H1 to the upstream AT-rich region of the murine beta interferon promoter: HMGI protein acts as a potential antirepressor of the promoter. Mol. Cell. Biol. 1999;19:2803–2816. doi: 10.1128/mcb.19.4.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ji YS, Xu Q, Schmedtje JF., Jr Hypoxia induces high-mobility-group protein I(Y) and transcription of the cyclooxygenase-2 gene in human vascular endothelium. Circ Res. 1998;83:295–304. doi: 10.1161/01.res.83.3.295. [DOI] [PubMed] [Google Scholar]
- 65.Martinez Hoyos J, Fedele M, Battista S, Pentimalli F, Kruhoffer M, Arra C, Orntoft TF, Croce CM, Fusco A. Identification of the genes up- and down-regulated by the high mobility group A1 (HMGA1) proteins: tissue specificity of the HMGA1-dependent gene regulation. Cancer Res. 2004;64:5728–5735. doi: 10.1158/0008-5472.CAN-04-1410. [DOI] [PubMed] [Google Scholar]
- 66.Barboric M, Yik JH, Czudnochowski N, Yang Z, Chen R, Contreras X, Geyer M, Matija, Peterlin B, Zhou Q. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res. 2007;35:2003–2012. doi: 10.1093/nar/gkm063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sedore SC, Byers SA, Biglione S, Price JP, Maury WJ, Price DH. Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucleic Acids Res. 2007;35:4347–4358. doi: 10.1093/nar/gkm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Y, Liu XY, De Clercq E. Role of the HIV-1 positive elongation factor P-TEFb and inhibitors thereof. Mini. Rev. Med. Chem. 2009;9:379–385. doi: 10.2174/1389557510909030379. [DOI] [PubMed] [Google Scholar]
- 69.Yik JH, Chen R, Pezda AC, Samford CS, Zhou Q. A human immunodeficiency virus type 1 Tat-like arginine-rich RNA-binding domain is essential for HEXIM1 to inhibit RNA Polymerase II transcription through 7SK snRNA-mediated inactivation of P-TEFb. Mol. Cell. Biol. 2004;24:5094–5105. doi: 10.1128/MCB.24.12.5094-5105.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cadogan M, Dalgleish AG. HIV induced AIDS and related cancers: chronic immune activation and future therapeutic strategies. Adv Cancer Res. 2008;101:349–395. doi: 10.1016/S0065-230X(08)00409-0. [DOI] [PubMed] [Google Scholar]
- 71.Carbone A, Gloghini A. AIDS-related lymphomas: from pathogenesis to pathology. Br. J. Haematol. 2005;130:662–670. doi: 10.1111/j.1365-2141.2005.05613.x. [DOI] [PubMed] [Google Scholar]
- 72.Greene W, Kuhne K, Ye F, Chen J, Zhou F, Lei X, Gao SJ. Molecular biology of KSHV in relation to AIDS-associated oncogenesis. Cancer Treat Res. 2007;133:69–127. doi: 10.1007/978-0-387-46816-7_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martellotta F, Berretta M, Vaccher E, Schioppa O, Zanet E, Tirelli U. AIDS-related Kaposi's sarcoma: state of the art and therapeutic strategies. Curr. HIV Res. 2009;7:634–638. doi: 10.2174/157016209789973619. [DOI] [PubMed] [Google Scholar]
- 74.Valadkhan S. snRNAs as the catalysts of pre-mRNA splicing. Curr. Opin. Chem. Biol. 2005;9:603–608. doi: 10.1016/j.cbpa.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 75.Kenneth NS, White RJ. Regulation by c-Myc of ncRNA expression. Curr. Opin. Genet. Dev. 2009;19:38–43. doi: 10.1016/j.gde.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 76.Luo Y, Kurz J, MacAfee N, Krause MO. C-myc deregulation during transformation induction: involvement of 7SK RNA. J. Cell. Biochem. 1997;4:313–327. doi: 10.1002/(sici)1097-4644(199702)64:2<313::aid-jcb13>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 77.Manabe T, Katayama T, Tohyama M. HMGA1a recognition candidate DNA sequences in humans. PLoS ONE. 2009;4:e8004. doi: 10.1371/journal.pone.0008004. [DOI] [PMC free article] [PubMed] [Google Scholar]