Abstract
The 3′ untranslated regions (3′UTRs) of eukaryotic genes regulate mRNA stability, localization and translation. Here, we present evidence that large numbers of 3′UTRs in human, mouse and fly are also expressed separately from the associated protein-coding sequences to which they are normally linked, likely by post-transcriptional cleavage. Analysis of CAGE (capped analysis of gene expression), SAGE (serial analysis of gene expression) and cDNA libraries, as well as microarray expression profiles, demonstrate that the independent expression of 3′UTRs is a regulated and conserved genome-wide phenomenon. We characterize the expression of several 3′UTR-derived RNAs (uaRNAs) in detail in mouse embryos, showing by in situ hybridization that these transcripts are expressed in a cell- and subcellular-specific manner. Our results suggest that 3′UTR sequences can function not only in cis to regulate protein expression, but also intrinsically and independently in trans, likely as noncoding RNAs, a conclusion supported by a number of previous genetic studies. Our findings suggest novel functions for 3′UTRs, as well as caution in the use of 3′UTR sequence probes to analyze gene expression.
INTRODUCTION
The 3′ untranslated regions (3′UTRs) of messenger RNAs (mRNAs) affect the expression of eukaryotic genes by regulating mRNA translation, stability and subcellular localization (1). 3′UTRs are typically defined by cDNA cloning, which shows they are contiguous with the upstream protein-coding region in the mRNA. The length of 3′UTRs has undergone a massive expansion during metazoan evolution, with annotated 3′UTRs in human and mouse rivaling the average size of protein-coding sequences and in some cases exceeding 10 kb (2,3). Furthermore, 3′UTRs are highly conserved and contain some of the most conserved elements within the mammalian genome (4). Together, these observations suggest that 3′UTRs have assumed an increasingly important role in the evolution of the eukaryotic genome.
The control of mRNA expression by 3′UTRs is mediated by trans-acting factors, including RNA-binding proteins and microRNAs (miRNAs), which interact with cis-regulatory elements within the 3′UTR (1). The post-transcriptional regulation mediated by 3′UTRs is crucial for the correct spatial and temporal expression of the protein encoded by the mRNA. Indeed, the importance of regulation by 3′UTRs was recently highlighted by the finding that 3′UTRs are reduced in length in proliferating cells, which in some cases was shown to mediate an increased expression of the associated mRNA (5). Interestingly, the analysis of transcription start sites has suggested that transcription may also be initiated from within 3′UTR sequences, and therefore act as a source of independent transcripts (6) that may exhibit expression patterns different from their upstream protein-coding sequences.
Here, we show that the 5′ termini of many RNAs map within 3′UTRs of genes in human, mouse and fly, and verify the separate and developmentally regulated expression of 3′UTR-associated RNAs (which we have termed uaRNAs) by a range of in silico and molecular biology approaches, including in situ hybridization (ISH). Furthermore, we present evidence that a portion of these distinctively expressed 3′UTRs arises by post-transcriptional processing rather than new transcription initiation. Our results, supported by previous genetic studies of several individual genes, suggest that there is trans-acting embedded genetic information in 3′UTRs with potential biological function.
MATERIALS AND METHODS
Capped analysis gene expression/serial analysis of gene expression analysis
Analyses were performed using RefSeq (7) gene annotations and the hg18, mm8 and dm3 genome assemblies provided within the UCSC Genome Browser (8). Human and mouse capped analysis gene expression (CAGE) retrieved from RIKEN (http://fantom3.gsc.riken.jp/) and fruity fly Serial Analysis of Gene Expression (SAGE) tags retrieved from MachiBase (9) were mapped to the genome with ZOOM requiring exact and unique matches (10). Syntenic locations of mouse 3′UTR CAGE tags in the human genome were identified using the LiftOver utility (8). Mouse CAGE tags that mapped to the same site as human CAGE tags were defined as conserved.
Full-length cDNA analysis
Full-length human and mouse cDNA sequences were retrieved from RIKEN (http://fantom3.gsc.riken.jp/). Putative uaRNAs were identified by intersecting 5′ cDNA coordinates with RefSeq-annotated 3′UTRs. The CRITICA algorithm (11) was used to identify non-protein-coding from the RIKEN FANTOM3 full-length mouse cDNA library as described previously (12).
UaRNA transcription initiation analysis
Deep sequencing tags derived from H3K4me1, H3K4me2, H3K4me3 and H3K27ac and RNAPII immunoprecipitation for resting CD4+ cells (13) were obtained from the NCBI short read archive (accession ID SRA000234 and SRA000287) and mapped to the human genome (hg18) with ZOOM requiring exact and unique matches (10). To determine enrichment of chromatin marks with uaRNA or mRNA initiation sites, the relative mapping position of sequencing tags to the nucleotide associated with the highest CAGE tag frequency within the 3′UTR or promoter was plotted over a ±50-nt window. CAGE tags spanning exon–exon junctions (EEJs) were identified by mapping tags without a perfect match to the genome to EEJ sequences, which comprise 20 nt on either side of the splice site, located within RefSeq-annotated 3′UTRs.
CAGE expression analysis
To determine the dynamic expression of 3′UTR CAGE tags across eight mouse tissues (embryo, lung, liver, visual cortex, somatosensory cortex, cerebellum and hippocampus) (14) and six time points during the differentiation of the human THP1 myelomonocytic leukemia cell line (15), we summed the total normalized CAGE tag frequency for each 3′UTR. The 500 genes that contained the highest frequency of 3′UTR CAGE tags were clustered using the Cluster utility (16). For human genes, 3′UTR CAGE tag frequency was normalized to the median across the time series. CAGE tag frequencies were log transformed and visualized as a heat map. CAGE tag frequencies in 3′UTRs were compared to the CAGE tag frequency in the promoter for the gene subset. Promoter expression levels were defined as the sum of CAGE tags within the promoter region (±50-nt window around RefSeq-annotated transcription start site). The ratio of promoter and 3′UTR expression levels were calculated and visualized as a heat map alongside the expression clusters.
In situ hybridization
Section in situ hybridization (ISH) on paraffin-embedded, sectioned at 7 μm, whole-mouse embryos was performed as described previously (17). The genomic coordinates and length of the different ISH probes used are shown in the Supplementary Data.
RESULTS
Identification of 3′UTRs with independent expression
To survey 3′UTRs with evidence of independent expression, we used publicly available CAGE (capped analysis of gene expression) and cDNA libraries that were generated from a wide variety of embryonic and adult mouse tissues (18,19). CAGE uses the 5′ cap of RNA transcripts to identify the first 20–25 nt of polyadenylated RNAs. We found 175 916 (13% of the total) CAGE tags in mouse and 57 400 (5.2%) CAGE tags in human mapped to 3′UTRs (Supplementary Figure S1). The difference in the proportion of 3′UTR CAGE tags mapping to 3′UTRs in mouse and human may reflect differences in the tissue sources represented in the libraries. In total, we found 4960 mouse genes and 1518 human genes contained at least one high confidence CAGE mapping site (defined by at least three tags mapping to the same 5′ nt) within their 3′UTRs (Supplementary Table S1). These genes were not enriched for any gene ontology classes, suggesting this phenomenon is a common characteristic of mammalian genomes.
With the exception of gene promoters, the density of CAGE tags was higher in 3′UTRs than in other genomic regions, being enriched 136-fold relative to mouse intergenic regions and 1.6-fold relative to mouse coding regions. CAGE tags within 3′UTRs were generally organized into clusters, rather than being evenly distributed. These clusters ranged from a broad distribution over many nucleotides to a single peak, where a large number of tags mapped to a single nucleotide. Consistent with a previous report (20), we also observed enrichment for a GGG motif at the 5′ end of 3′UTR CAGE tags, indicating underlying sequence specificity (Supplementary Figure S1).
We next examined the conservation of 3′UTR CAGE sites between human and mouse. We identified 2076 homologous sites where CAGE tags occur at syntenic nucleotide positions in annotated 3′UTRs of both species, which accounted for ∼20% of total 3′UTR CAGE tags (Supplementary Table S2). Compared to non-conserved sites, these conserved sites are enriched (2.7-fold, P < 0.01 t-test) for high-frequency CAGE tag mappings. Although we did not observe greater evolutionary conservation of the sequences at syntenic 3′UTR CAGE mapping sites, we did observe a distinct peak of conservation ∼40 nt downstream of CAGE sites consistent with a previous report (20). We were unable to detect any enriched motifs within the conserved 3′UTR CAGE mapping sites.
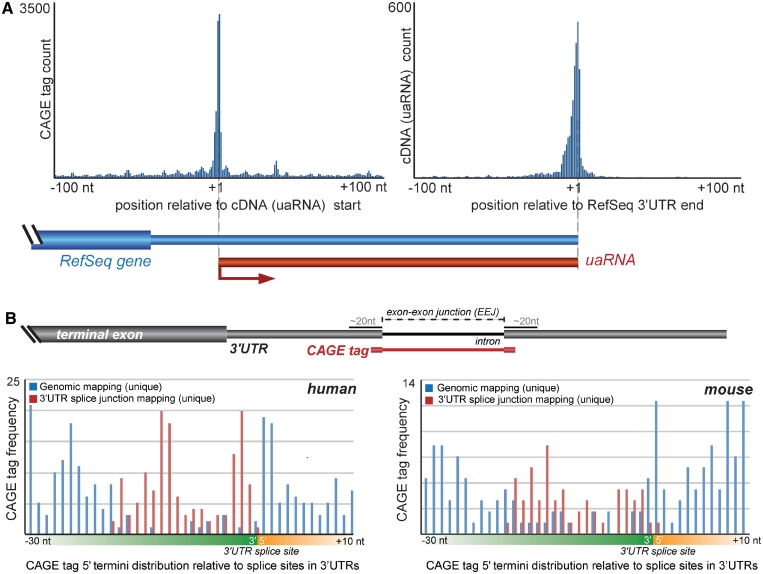
To independently verify the occurrence of transcripts derived from 3′UTRs, we also examined full-length cDNA sequences in mouse. We identified 3718 full-length mouse cDNAs (∼3.6% of total cDNAs) whose 5′ end maps within 2766 3′UTRs, and 1227 (33%) of these had 5′ start sites directly supported by a CAGE cluster in sense direction within ±50 bp (Figure 1A, Supplementary Table S3). Furthermore, 92% of these transcripts shared the polyadenylation site with the host RefSeq gene, suggesting that they are not 5′ ends of longer transcripts that extend past the end of the RefSeq gene (Figure 1A). To assess the potential for the 3′UTR transcripts to encode proteins or polypeptides, we analyzed their sequences using the CRITICA algorithm (11). CRITICA reported that only 2.9% (108/3718) of 3′UTRs were likely to encode proteins (Supplementary Table S3). In addition, an examination of the PeptideAtlas (21) revealed 22 of the transcripts intersected with peptide-mapping regions (16 of which substantiated CRITICA’s protein-coding predictions; Supplementary Table S3). Together, these data suggest that these transcripts are, as anticipated for UTRs, predominantly noncoding.
Figure 1.
Post-transcriptional processing of 3′UTRs. (A) Full-length cDNA transcripts associated with 3′UTRs. Histogram (left) shows enrichment of CAGE tags with the 5′ termini of cDNAs mapping within 3′UTRs (i.e. uaRNAs). Histogram (right) shows correlated enrichment of the terminal regions of these cDNA transcripts with the 3′ end of the host RefSeq gene. (B) The top panel shows a schematic representation of the mapping strategy employed to discern CAGE tags that span exon–exon junctions (EEJs) and therefore suggest post-transcriptional processing. The lower panels show the distribution of CAGE tags mapping proximal to EEJs in human (left) and mouse (right). The frequencies of CAGE tags that map uniquely to the genome (blue) are under-represented adjacent to EEJs. This under-representation is reconciled by considering CAGE tags that map across EEJs (red).
Putative mechanisms for uaRNA biogenesis
Next, we considered possible molecular mechanisms underlying the biogenesis of uaRNAs. Given that CAGE tags indicate 5′-capped ends, it has been previously assumed that these tags correspond to RNA polymerase II (RNAPII)-dependent transcription start sites. This conclusion was supported by evidence that the sequences upstream of a small sample of 3′UTR CAGE tags could drive expression of a reporter gene and a recent annotation of human promoters found that a considerable portion occur within 3′UTRs (22). Dynamic chromatin domains have been shown to be reliable indicators of transcription start sites (23). We identified several 3′UTRs that contained signatures of dynamic chromatin domains, such as in the 3′UTRs of the Klhl31 and Notch1 (Supplementary Figure S2). Within mouse embryonic stem cells, we identified 19 3′UTRs that contain multiple transcription factor binding sites and 17 3′UTRs that fully encompass chromatin domains indicative of transcription initiation (i.e. H3K4me3, H3K27me3) (Supplementary Table S4).
To determine whether dynamic chromatin domains were a general feature associated with uaRNAs, we examined deep sequencing tags from chromatin immunoprecipitation using antibodies against various histone modifications that mark active promoters, including H3K4me1, H3K4me2 and H3K4me3 (13,24). Despite the instances described above, we did not observe any enrichment of modified histones with 3′UTR CAGE sites in stark contrast to strong enrichment of modified histones at conventional gene promoters (Supplementary Figure S3). Furthermore, there was no enrichment for RNAPII occupancy at 3′UTR CAGE sites. Together, these results suggest that uaRNAs generally did not originate from active transcription from RNAPII promoters located within 3′UTRs, and that they are not derived via the same mechanisms as mRNAs.
In agreement with this hypothesis, it was recently found that many CAGE tags mapping across exon–exon junctions occurred in such close proximity to the splice junction that they were unlikely to be efficiently spliced, suggesting that 5′ caps on transcripts could also result from end-modification associated with post-transcriptional and post-splicing cleavage of a longer precursor (25–28). Although introns are rare in 3′UTRs, because they are thought to trigger nonsense-mediated decay (29), we were able to identify 59 mouse and 141 human CAGE tags spanning exon–exon junctions (Figure 1B). Together with the lack of signatures of active promoters, this suggests that some uaRNAs arise as a consequence of post-transcriptional cleavage rather than conventional transcription initiation.
Comparison of the expression of 3′UTRs and coding exons
We employed custom microarrays to compare directly the expression of 138 uaRNAs and their associated protein-coding sequences in two developmental systems in mouse: embryonic stem (ES) cell differentiation, and ovary and testis formation (see ‘Materials and Methods’ section). We found that for 54% (74 of 138; R2 < 0; Supplementary Table S5) of genes, the expression levels between 3′UTRs and the associated coding exons were discordant in one (67%) or more (33%) developmental systems.
The complex relationships between the expression of coding exons and the associated 3′UTRs are illustrated by transcriptional profiling during mouse ES cell differentiation (Supplementary Figure S4). Consistent with the conventional case where the 3′UTR is part of the same mRNA, 50% of the 93 genes expressed above background showed concordant expression (r ≥ 0.5) of the 3′UTR and upstream protein-coding sequence (Supplementary Table S5). However, 34% of genes showed no correlation (−0.5 < r < 0.5) and 16% showed a negative correlation (r ≤ –0.5). For example, the 3′UTR of Tmpo, a gene with roles in cell differentiation and proliferation (30), exhibits an inverse expression profile relative to the upstream coding exons (Supplementary Figure S4). Furthermore, 12 genes show increased expression of the 3′UTR compared to the CDS at specific stages during differentiation (Supplementary Table S5). There are three interpretations for these results, which are not mutually exclusive: (i) differential expression of alternatively spliced mRNAs that contain different combinations of coding exons and 3′UTRs; (ii) 3′UTRs can be transcribed independently of the coding region; or (iii) 3′UTRs are post-transcriptionally processed from the mRNA and the resulting RNAs are differentially regulated.
Developmental stage- and tissue-specific expression of uaRNAs
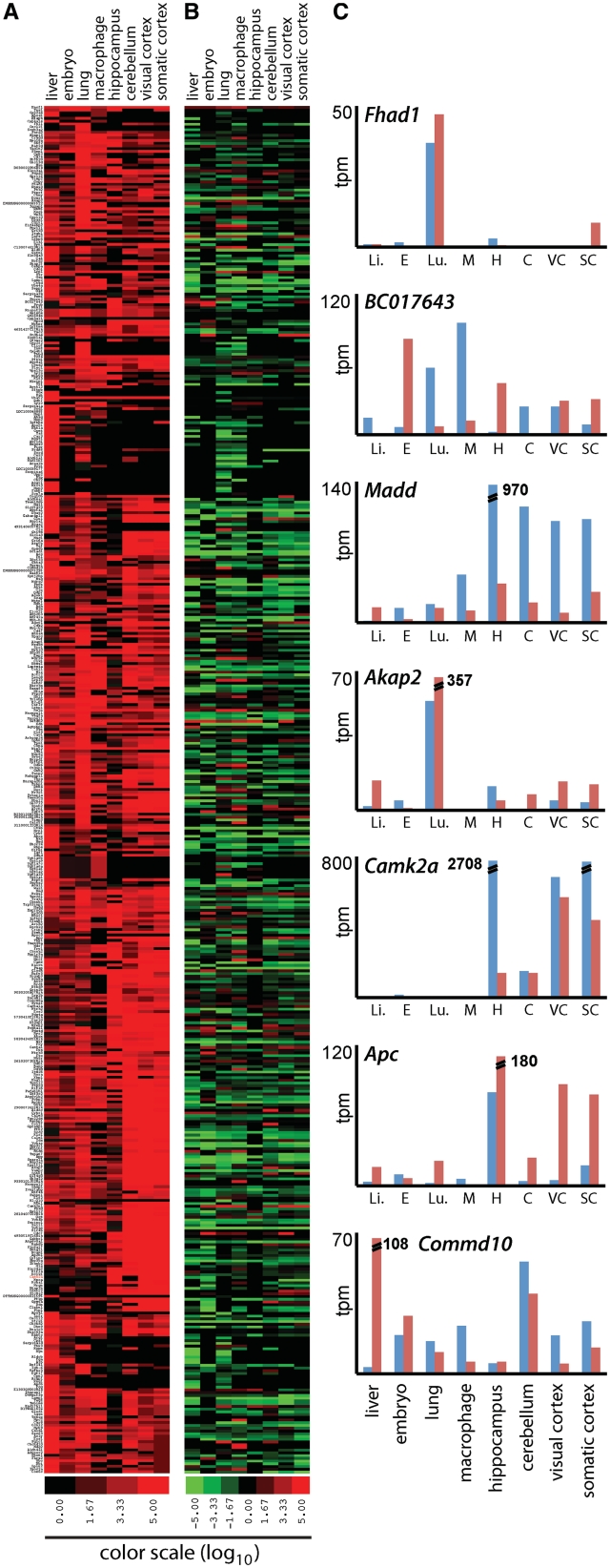
To determine whether the tissue-specific expression observed in the microarrays is the result of alternative splicing or distinct expression, we analyzed differential CAGE tag frequency in 3′UTRs. Because CAGE tags correspond to the 5′ RNA termini, their differential frequencies are unlikely to arise from alternative splicing. To compare the frequency of CAGE tags across samples, we examined CAGE libraries from eight different mouse tissues (14,20). We found 4491 genes containing CAGE tags within their 3′UTR. Clustering of the top 500 genes containing the highest frequency of 3′UTR CAGE tags clearly showed that there was a wide dynamic range of tissue-specific expression of uaRNAs (Figure 2A). We did not observe a correlation between the number of CAGE tags mapping to the 3′UTR ( of determination; r2 = 0.0064) and the number of CAGE tags mapping to the promoter of the same gene, but rather a range of ratios that varied across tissue types (Figure 2, Supplementary Table S6). This suggests uaRNA expression is a regulated process rather than a byproduct of host gene expression. In this analysis, we also observed instances where uaRNA initiation occurred preferentially at different sites in different tissues. For example, in the Camk2a gene (31), uaRNAs are preferentially initiated at a different site in the hippocampus than in the somato-sensory and visual cortex (Figure 3A).
Figure 2.
Specific and distinct expression profiles for 3′UTRs in mouse tissues. (A) Cluster analysis of the expression of the 500 genes containing the highest 3′UTR CAGE tag frequency shows the differential expression levels (no expression, black; high expression, red) of 3′UTRs in various mouse tissues. Expression level was determined as the normalized CAGE tag frequency mapping to 3′UTRs. (B) The diverse ratios of promoter to 3′UTR CAGE frequency (high, green; low, red) for each tissue indicates independent expression of mRNAs and 3′UTRs. (C) Illustrative examples indicating promoter (blue) and 3′UTR (red) CAGE frequency. tpm, tags per million; Li, liver; E, embryo; Lu, lung; M, macrophage; C, cerebellum; H, hippocampus; VC, visual cortex; SC, somatic cortex.
Figure 3.
Illustrative examples of 3′UTR transcripts in mouse and fruit fly. (A) Schematic representation of CAGE tag clusters in the 3′UTR of the mouse Camk2a gene from somatosensory cortex (green), visual cortex (orange) and hippocampus (blue). Different sites with high frequency of CAGE tags map preferentially in hippocampus (68 tags per million; tpm), the visual cortex (52 tpm) and somato-sensory cortex (56 tpm). (B) Schematic representation of the fruit fly gene oskar showing specific SAGE tags in the 3′UTR in young and old females.
We also analyzed the temporal expression pattern of uaRNAs during the differentiation of the human THP-1 myelomonocytic leukemia cell line upon stimulation with phorbol myristate acetate (PMA) at six time points between 0 and 96 h (15). We identified 2726 genes that contained CAGE tags within their 3′UTRs. Considering the expression of the top 500 genes, we again observed dynamic expression profiles that could not be accounted for by the host gene expression (Supplementary Figure S5, Supplementary Table S7). Together with the observation that several of the identified genes have important roles in macrophage activation, such as ITGB2 (15) and SRGN (32), these results raise the possibility that the regulated expression of uaRNAs has biological consequences.
Independent regulated 3′UTR expression in Drosophila melanogaster
To examine whether uaRNAs occur outside of vertebrates, we examined Drosophila melanogaster SAGE libraries, which, like CAGE, define the 5′ termini of RNA transcripts. We found 27 832 SAGE tags (1.2% of the total) in four libraries (embryo, S2 cells, young and old females) that mapped within RefSeq annotated 3′UTRs of 275 genes (Supplementary Table S1). Unlike the human and mouse 3′UTR CAGE tags, we did not identify the GGG motif or the 40-nt downstream conserved region associated in the D. melanogaster 3′UTR SAGE tags. However, we did observe elevated evolutionary conservation downstream of 3′UTR SAGE tags (Supplementary Figure S6). During this analysis, we also noted that the 3′UTR of oskar (osk), a gene whose 3′UTR had previously been indicated to function independently to the host gene (33), showed large numbers of SAGE tags specific to adult stages (Figures 3B and S6; see ‘Discussion’ section). Together, these results provide independent validation for the existence of uaRNAs in an evolutionarily distant lineage, indicating their possible widespread biological role(s).
Candidate uaRNAs are specifically expressed during mouse embryogenesis
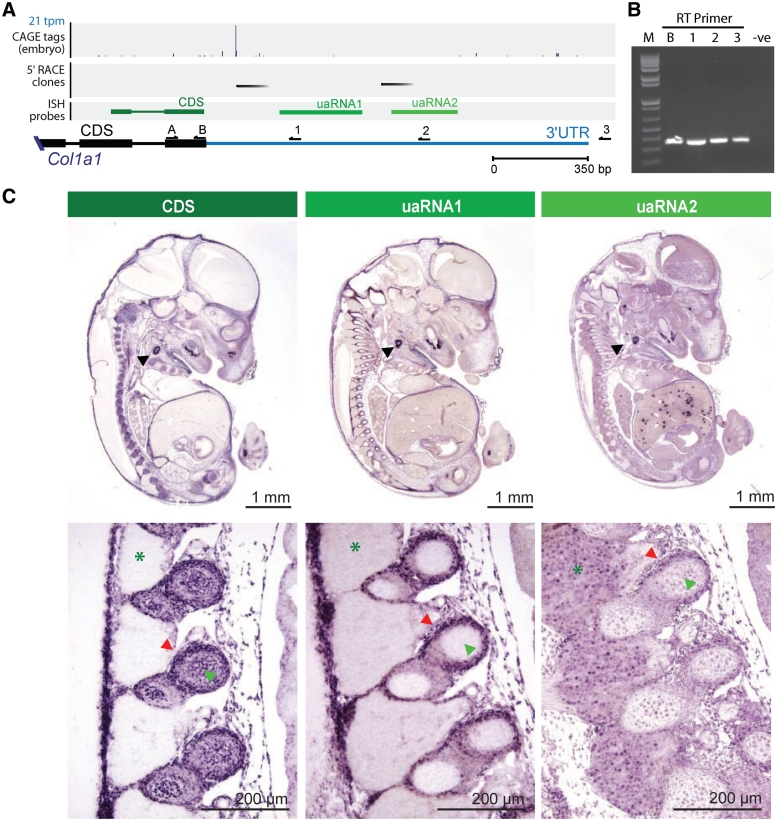
To further characterize the discordant expression observed between the coding sequence (CDS) and 3′UTR in differentiating mouse ES cells (see above), we performed 5′ rapid amplification of complementary DNA ends (RACE) on six uaRNA candidates (Col1a1, Mef2c, Mical2, Nfia, Mylk and Myadm) and compared the expression of their coding regions and corresponding 3′UTRs using ISH. These candidates were selected on the basis of their discordant expression by microarray profiling, 3′UTR conservation and presence of high-confidence 3′UTR CAGE tags. The RACE analysis identified 5′ termini within the 3′UTRs of Col1a1, Mef2c, Mical2, Nfia and Mylk (Figure 4A; Supplementary Table S8), confirming the presence of independent transcripts for these loci in whole mouse embryos. Interestingly, in most cases, several distinct 5′ RACE products were cloned at different frequencies, suggesting that multiple uaRNAs can arise from the same 3′UTR and that these can be differentially expressed (Supplementary Table S8). To confirm that these uaRNAs were indeed encompassed within the 3′UTRs of the respective full-length mRNAs, we performed gene and strand-specific reverse transcription (RT) followed by polymerase chain reaction (PCR) of the terminal exon (see Supplementary Data). Consistent with expectations based on the RefSeq annotations for these genes, in all cases examined, we confirmed that the 3′UTRs were connected to the terminal coding exon (Figures 4B and S7). ISH in whole-mouse embryos revealed discordant expression between the CDS and 3′UTR in three of the candidates, Col1a1, Nfia and Myadm, which are discussed in further detail below.
Figure 4.
uaRNAs within Col1a1 3′UTR. (A) Genome browser view of Col1a1 3′UTR showing; histogram of CAGE distribution and density (top panel; tpm, tags per million), 5′ ends inferred from high confidence 5′ RACE products (second panel, black bars); riboprobes used in in situ hybridization (third panel; green bars); Col1a1 annotated coding sequence (CDS; black bar) and 3′UTR (blue bar); reverse transcriptase (RT, 1–3) and PCR primers (A and B) used in (B) (black arrows). (B) Confirmation of 3′UTR annotation by RT using primers B, 1, 2 or 3 followed by PCR using primers A (forward) and B (reverse). Lanes contain (from left to right) 1-kb plus ladder (M), positive CDS control primer (B), RT primers (1–3) and no RT negative control (-ve). (C) ISH using one riboprobe in the terminal constitutive coding exon (CDS, dark green) of the Col1a1 gene and two probes (uaRNA1 and uaRNA2, light green) corresponding to 3′UTR sequences, one downstream of each of the two 5′ RACE clusters. All three probes exhibit expression at sites of chondrogenesis, such as the otic vesicle (upper panel, black arrowheads) and the developing ribs (lower panel, red arrowheads). Expression of the coding region is apparent during ossification of vertebrae, in contrast to both uaRNAs whose expression is absent (lower panel, green arrowheads). The uaRNA2 probe detects expression in the the dorsal root ganglia (lower panel, green asterisks); this expression is not detected by the CDS or uaRNA1 probes. Higher magnification of the dorsal root ganglia shows that the uaRNA expression is localized to the nucleus (lower panel, green asterisk).
The tissue and cellular expression of Col1a1, which encodes the procollagen type I alpha 1 chain, was detected using an RNA probe targeting the constitutive terminal coding exon and two probes targeting the 3′UTR sequences, one downstream of each of the two major 5′ RACE clusters (Figure 4A). The specificity of all three probes to Col1a1 was confirmed by northern blot (Supplementary Figure S7). Interestingly, the most distal 3′UTR probe also detected a ∼120-nt transcript, which may represent a processed uaRNA (Supplementary Figure S7). Section ISH on whole-mouse embryos at 13.5 days post coitum (dpc; Figure 4C) demonstrated that all three regions are expressed at sites of chondrogenesis, such as the otic vesicle and the developing ribs. The expression of the coding region persisted during ossification, whereas no expression of the uaRNAs was detected. Moreover, the most distal uaRNA probe revealed distinct expression of the targeted sequence in the dorsal root ganglia, i.e. expression was not detected in these areas by the coding probe or the other uaRNA probe. Higher magnification of the dorsal root ganglia indicated that the uaRNA is localized to the nucleus (Figure 4C). Together, the different expression profiles of the Col1a1 exons and the uaRNAs verified that uaRNAs are distinctly expressed in a tissue-specific manner. To confirm the discordant expression profiles observed between the 3′UTR and coding regions were not due to alternative splicing, we performed ISH with two additional independent probes targeting the coding region. In all cases, we observed the same expression pattern consistent with the first coding probe, thereby discounting alternative splicing as a cause for the discordant expression between 3′UTRs and coding exons (Supplementary Figure S8).
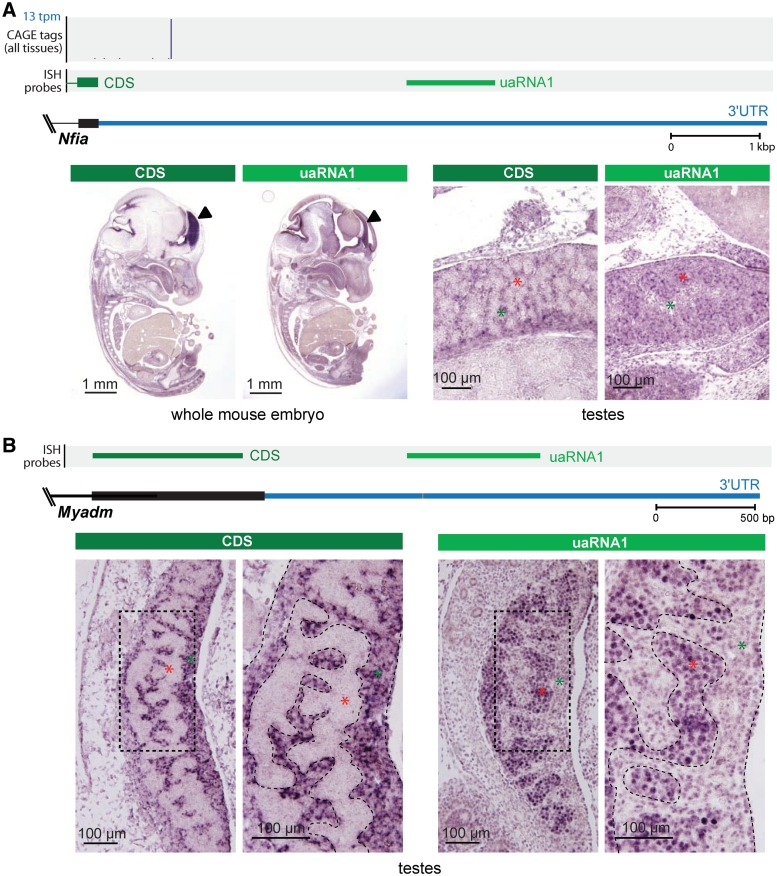
Nfia encodes a transcription factor required for brain development (34), and has a highly conserved 7.5-kb 3′UTR. Comparison of section ISH using a probe targeting the terminal exon of the CDS and a probe targeting the 3′UTR (uaRNA) showed discordant expression in the developing forebrain (Figure 5A), a region where Nfia plays important roles in regulating gene expression (35). We also detected Nfia CDS expression in interstitial cells of developing testes (Figure 5A). The function of Nfia during testis differentiation is unknown, but it has been reported that homozygous Nfia-null male mice are sterile (34). In contrast to the CDS, we detected expression of the uaRNA within the testis cords (Figure 5A), rather than the interstitium, suggesting that the Nfia uaRNA plays a role independent from the NFIA protein.
Figure 5.
uaRNAs within the Nfia and Myadm 3′UTRs. (A) (Top panel) Genomic context of Nfia 3′UTR showing annotated coding sequence (CDS, black) and 3′UTR (blue), histogram of CAGE distribution and density (tpm, tags per million), and riboprobes used in ISH that target the terminal constitutive coding exons (dark green; CDS) and 3′UTR (light green; uaRNA1). (Bottom panel) Section ISH showing the Nfia CDS probe detecting expression in interstitial cells (green asterisks) of 12.5 dpc testes (marker) and the uaRNA probe detecting expression within the testis cords (red asterisks). (B) (Top panel) Genomic context of Myadm 3′UTR showing annotated coding sequence (CDS, black) and 3′UTR (blue), riboprobes used in ISH that target the terminal constitutive coding exon (dark green; CDS) and 3′UTR (light green; uaRNA1). (Bottom panel) Section ISH showing the Myadm CDS probe detecting expression in the cytoplasm of interstitial cells (green asterisks) in the developing testis at 12.5 dpc and the uaRNA probe detecting expression in the nuclei of Sertoli cells and germ cells in the testis cords (red asterisks). High Myadm expression was not detected by either the CDS or the uaRNA probe elsewhere in the embryo (see Supplementary Figure S8).
The gene encoding myeloid-associated differentiation marker Myadm (36) showed decoupled expression of the CDS and uaRNA in the developing gonad (Figure 5B). ISH using independent CDS and uaRNA probes showed the terminal coding exon of Myadm was expressed in interstitial cells of the developing testis, whereas the uaRNA was detected in Sertoli cells and germ cells within testis cords. Moreover, high magnification examination of the ISH images revealed different subcellular localizations, with the coding exon being detected in the cytoplasm of interstitial cells, and the uaRNA localized in the nuclei of germ and Sertoli cells (Figure 5B). Interestingly, we detected only low levels of Myadm uaRNA expression in testes before 12.5 dpc or in ovaries at any stage investigated, suggesting that the expression of this uaRNA was highly tissue- and developmentally specific. Furthermore, northern analysis identified a testis-specific small uaRNA transcript of ∼140 nt (Supplementary Figure S7), suggesting the uaRNA is processed into a smaller stable RNA.
One general observation derived from the ISH data was that for each of the candidate genes, specific tissues could be identified where the 3′UTR probe detected expression, but the CDS probe did not. As already discussed above, the absence of indicators of active transcription suggest that uaRNAs are not transcribed independently of the associated mRNA, but rather are a cleaved product of the full-length transcript. Therefore, the presence of 3′UTR expression in the absence of CDS expression can be explained by rapid degradation of the upstream transcript containing the CDS. To examine this possibility, we performed quantitative RT-PCR (qRT-PCR) on the CDS and 3′UTR regions of Col1a1, Nfia and Myadm on total RNA extracted from the tissues and cell types identified by ISH as showing exclusive 3′UTR expression. Unfortunately, we were unable to extract sufficient RNA from the developing spine for analysis of Col1a1. However, for Nfia and Myadm, where the ISH showed elevated 3′UTR expression relative to the CDS, the qRT-PCR revealed a similar tissue-specific increase in 3′UTR expression in brain and testis, respectively (Supplementary Figure S7). In addition to independently validating the ISH data, this result supports a model where the 3′UTR is cleaved to give rise to a uaRNA, while the upstream coding region is degraded.
DISCUSSION
The conventional understanding of 3′UTRs is that they exclusively operate in cis, via regulatory proteins and microRNAs, to control the translation, stability and localization of mRNAs. This concept has remained unquestioned largely due to the lack of evidence to the contrary and the inherent difficulties of discerning trans-acting roles of 3′UTRs. Our data show that the distinct expression of 3′UTR sequences is a widespread phenomenon that is developmentally regulated, with stage-, tissue- and subcellular-specific expression. Together, these results suggest that 3′UTR sequences can fulfill biological roles different from their normally associated mRNA.
The independent function we propose for uaRNAs is supported by a number of published observations. As previously noted, the oskar 3′UTR in Drosophila has been shown to rescue an oogenesis defect in oskar null-mutants, independently of the oskar protein (33). Our SAGE tag analysis supported the independent expression of the oskar 3′UTR, consistent with the idea that the oskar 3′UTR is expressed and functions independently in vivo. Similarly, a number of additional reports have shown that the 3′UTRs of troponin I, tropomyosin, alpha-cardiac actin, ribonucleotide reductase, DM protein kinase and prohibitin genes can act in trans to control cell proliferation and differentiation in the absence of associated coding-regions (37–41). Such roles may also contribute toward disease etiology. For example, the 3′UTR of the DM protein kinase gene, which is involved in myotonic dystrophy, inhibits the differentiation of C2C12 myoblasts (41) and the ectopic expression of the prohibitin 3′UTR has been shown to block cell cycle progression and contains characteristic mutations in breast cancer-derived cells (42).
Despite such examples showing 3′UTRs acting in trans, the challenge remains to understand the mechanism by which they are generated and by which they act. The mechanism of post-transcriptional cleavage is unknown, but could potentially involve specific RNA-binding proteins or trans-acting RNAs that can target cleavage enzymes in a sequence-specific manner similar to the targeting of chromatin-modifying complexes by long ncRNAs and targeting of RISC enzymes by miRNAs (43). In addition, a non-transcriptionally linked capping enzyme was recently discovered in the cytoplasm of several human cell lines that may be responsible for the post-transcriptional capping of cleaved transcripts (44). The function of uaRNAs may, at least in part, be inferred from known 3′UTR function. For example, uaRNAs may act as decoys to titrate trans-acting factors and thereby fine-tune their regulatory function, similar to the ncRNA IpsPS1, which was recently shown to sequester the microRNA miR-399 in Arabidopsis thaliana (45). Alternatively, they may act as a scaffold to localize proteins into regulatory complexes that are required even in the absence of the associated mRNA, as suggested by the observation that the oskar 3′UTR is necessary for trafficking and accumulation of Staufen from nurse cells to the oocyte during oogenesis (33). Therefore, it remains to be determined what specific information uaRNAs contribute toward gene regulation and the advantage gained by having such information either linked to mRNA or decoupled in other contexts.
Regardless of novel and unexpected roles of uaRNA transcripts, the existence and distinct expression of uaRNAs should be considered in future studies that assume the 3′UTR expression to be coincident with the associated mRNAs. Within this study, we demonstrated a number of genes that exhibited discordant expression patterns between their CDSs and 3′UTRs in both microarray and ISH (46). Given these examples, results gained using probes targeted to 3′UTRs should be interpreted with care.
In summary, our findings enhance the current understanding of 3′UTR sequences and their role in regulating differentiation and developmental processes. This study and recent observations that 3′UTRs have undergone rapid expansion in eukaryotic evolution suggest more sophisticated functionality inherent in 3′UTR sequences than previously suspected. Moreover, the finding that 3′UTRs can be independently expressed as developmentally regulated ncRNAs further blurs the distinction between coding and noncoding RNAs (47) and serves as a reminder that the traditional concept of the gene is becoming increasingly outmoded (48,49), requiring a reassessment of our understanding of the genomic programming of complex organisms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Australian Research Council/University of Queensland co-sponsored Federation Fellowship (FF0561986; to J.S.M.); an Australian Research Council Discovery Project Grant (DP0879913; to D.W.); National Health and Medical Research Council of Australia Career Development Awards (CDA631542; to M.E.D., CDA519937; to D.W.); a Queensland Government Department of Employment, Economic Development and Innovation Smart Futures Fellowship (to M.E.D.); the University of Milan (to G.S.). Funding for open access charge: The University of Queensland.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kuersten S, Goodwin EB. The power of the 3′ UTR: translational control and development. Nat. Rev. Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- 2.Frith MC, Pheasant M, Mattick JS. The amazing complexity of the human transcriptome. Eur. J. Hum. Genet. 2005;13:894–897. doi: 10.1038/sj.ejhg.5201459. [DOI] [PubMed] [Google Scholar]
- 3.Mazumder B, Seshadri V, Fox PL. Translational control by the 3′-UTR: the ends specify the means. Trends Biochem. Sci. 2003;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- 4.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furuno M, Pang KC, Ninomiya N, Fukuda S, Frith MC, Bult C, Kai C, Kawai J, Carninci P, Hayashizaki Y, et al. Clusters of internally primed transcripts reveal novel long noncoding RNAs. PLoS Genet. 2006;2:e37. doi: 10.1371/journal.pgen.0020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, et al. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahsan B, Saito TL, Hashimoto S, Muramatsu K, Tsuda M, Sasaki A, Matsushima K, Aigaki T, Morishita S. MachiBase: a Drosophila melanogaster 5′-end mRNA transcription database. Nucleic Acids Res. 2009;37:D49–D53. doi: 10.1093/nar/gkn694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin H, Zhang Z, Zhang MQ, Ma B, Li M. ZOOM! Zillions of oligos mapped. Bioinformatics. 2008;24:2431–2437. doi: 10.1093/bioinformatics/btn416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Badger JH, Olsen GJ. CRITICA: coding region identification tool invoking comparative analysis. Mol. Biol. Evol. 1999;16:512–524. doi: 10.1093/oxfordjournals.molbev.a026133. [DOI] [PubMed] [Google Scholar]
- 12.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl Acad. Sci. USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valen E, Pascarella G, Chalk A, Maeda N, Kojima M, Kawazu C, Murata M, Nishiyori H, Lazarevic D, Motti D, et al. Genome-wide detection and analysis of hippocampus core promoters using DeepCAGE. Genome Res. 2009;19:255–265. doi: 10.1101/gr.084541.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki H, Forrest AR, van Nimwegen E, Daub CO, Balwierz PJ, Irvine KM, Lassmann T, Ravasi T, Hasegawa Y, de Hoon MJ, et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat. Genet. 2009;41:553–562. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilhelm D, Hiramatsu R, Mizusaki H, Widjaja L, Combes AN, Kanai Y, Koopman P. SOX9 regulates prostaglandin D synthase gene transcription in vivo to ensure testis development. J. Biol. Chem. 2007;282:10553–10560. doi: 10.1074/jbc.M609578200. [DOI] [PubMed] [Google Scholar]
- 18.Kawaji H, Kasukawa T, Fukuda S, Katayama S, Kai C, Kawai J, Carninci P, Hayashizaki Y. CAGE Basic/Analysis Databases: the CAGE resource for comprehensive promoter analysis. Nucleic Acids Res. 2006;34:D632–D636. doi: 10.1093/nar/gkj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 20.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 21.Deutsch EW, Lam H, Aebersold R. PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 2008;9:429–434. doi: 10.1038/embor.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trinklein ND, Karaoz U, Wu J, Halees A, Force Aldred S, Collins PJ, Zheng D, Zhang ZD, Gerstein MB, Snyder M, et al. Integrated analysis of experimental data sets reveals many novel promoters in 1% of the human genome. Genome Res. 2007;17:720–731. doi: 10.1101/gr.5716607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 24.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Fejes-Toth K, Sotirova V, Sachidanandam R, Assaf G, Hannon GJ, Kapranov P, Foissac S, Willingham AT, Duttagupta R, Dumais E, et al. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ni T, Corcoran DL, Rach EA, Song S, Spana EP, Gao Y, Ohler U, Zhu J. A paired-end sequencing strategy to map the complex landscape of transcription initiation. Nat. Methods. 2010;7:521–527. doi: 10.1038/nmeth.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plessy C, Bertin N, Takahashi H, Simone R, Salimullah M, Lassmann T, Vitezic M, Severin J, Olivarius S, Lazarevic D, et al. Linking promoters to functional transcripts in small samples with nanoCAGE and CAGEscan. Nat. Methods. 2010;7:528–534. doi: 10.1038/nmeth.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mercer TR, Dinger ME, Bracken CP, Kolle G, Szubert JM, Korbie DJ, Askarian-Amiri ME, Gardiner BB, Goodall GJ, Grimmond SM, et al. Regulated post-transcriptional RNA cleavage diversifies the eukaryotic transcriptome. Genome Res. 2010;20 doi: 10.1101/gr.112128.110. doi:10.1101/gr.112128.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong X, Scofield DG, Lynch M. Intron size, abundance, and distribution within untranslated regions of genes. Mol. Biol. Evol. 2006;23:2392–2404. doi: 10.1093/molbev/msl111. [DOI] [PubMed] [Google Scholar]
- 30.Dorner D, Vlcek S, Foeger N, Gajewski A, Makolm C, Gotzmann J, Hutchison CJ, Foisner R. Lamina-associated polypeptide 2alpha regulates cell cycle progression and differentiation via the retinoblastoma-E2F pathway. J. Cell. Biol. 2006;173:83–93. doi: 10.1083/jcb.200511149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wayman GA, Lee YS, Tokumitsu H, Silva A, Soderling TR. Calmodulin-kinases: modulators of neuronal development and plasticity. Neuron. 2008;59:914–931. doi: 10.1016/j.neuron.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niemann CU, Kjeldsen L, Ralfkiaer E, Jensen MK, Borregaard N. Serglycin proteoglycan in hematologic malignancies: a marker of acute myeloid leukemia. Leukemia. 2007;21:2406–2410. doi: 10.1038/sj.leu.2404975. [DOI] [PubMed] [Google Scholar]
- 33.Jenny A, Hachet O, Zavorszky P, Cyrklaff A, Weston MD, Johnston DS, Erdelyi M, Ephrussi A. A translation-independent role of oskar RNA in early Drosophila oogenesis. Development. 2006;133:2827–2833. doi: 10.1242/dev.02456. [DOI] [PubMed] [Google Scholar]
- 34.das Neves L, Duchala CS, Tolentino-Silva F, Haxhiu MA, Colmenares C, Macklin WB, Campbell CE, Butz KG, Gronostajski RM. Disruption of the murine nuclear factor I-A gene (Nfia) results in perinatal lethality, hydrocephalus, and agenesis of the corpus callosum. Proc. Natl Acad. Sci. USA. 1999;96:11946–11951. doi: 10.1073/pnas.96.21.11946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shu T, Butz KG, Plachez C, Gronostajski RM, Richards LJ. Abnormal development of forebrain midline glia and commissural projections in Nfia knock-out mice. J. Neurosci. 2003;23:203–212. doi: 10.1523/JNEUROSCI.23-01-00203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pettersson M, Dannaeus K, Nilsson K, Jonsson JI. Isolation of MYADM, a novel hematopoietic-associated marker gene expressed in multipotent progenitor cells and up-regulated during myeloid differentiation. J. Leukoc. Biol. 2000;67:423–431. doi: 10.1002/jlb.67.3.423. [DOI] [PubMed] [Google Scholar]
- 37.Fan H, Villegas C, Huang A, Wright JA. Suppression of malignancy by the 3′ untranslated regions of ribonucleotide reductase R1 and R2 messenger RNAs. Cancer Res. 1996;56:4366–4369. [PubMed] [Google Scholar]
- 38.Rastinejad F, Blau HM. Genetic complementation reveals a novel regulatory role for 3′ untranslated regions in growth and differentiation. Cell. 1993;72:903–917. doi: 10.1016/0092-8674(93)90579-f. [DOI] [PubMed] [Google Scholar]
- 39.Rastinejad F, Conboy MJ, Rando TA, Blau HM. Tumor suppression by RNA from the 3′ untranslated region of alpha-tropomyosin. Cell. 1993;75:1107–1117. doi: 10.1016/0092-8674(93)90320-p. [DOI] [PubMed] [Google Scholar]
- 40.Jupe ER, Liu XT, Kiehlbauch JL, McClung JK, Dell'Orco RT. The 3′ untranslated region of prohibitin and cellular immortalization. Exp. Cell Res. 1996;224:128–135. doi: 10.1006/excr.1996.0120. [DOI] [PubMed] [Google Scholar]
- 41.Amack JD, Paguio AP, Mahadevan MS. Cis and trans effects of the myotonic dystrophy (DM) mutation in a cell culture model. Hum. Mol. Genet. 1999;8:1975–1984. doi: 10.1093/hmg/8.11.1975. [DOI] [PubMed] [Google Scholar]
- 42.Jupe ER, Liu XT, Kiehlbauch JL, McClung JK, Dell'Orco RT. Prohibitin in breast cancer cell lines: loss of antiproliferative activity is linked to 3′ untranslated region mutations. Cell Growth Differ. 1996;7:871–878. [PubMed] [Google Scholar]
- 43.Karginov FV, Cheloufi S, Chong MM, Stark A, Smith AD, Hannon GJ. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol. Cell. 2010;38:781–788. doi: 10.1016/j.molcel.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otsuka Y, Kedersha NL, Schoenberg DR. Identification of a cytoplasmic complex that adds a cap onto 5′-monophosphate RNA. Mol. Cell. Biol. 2009;29:2155–2167. doi: 10.1128/MCB.01325-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 46.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dinger ME, Pang KC, Mercer TR, Mattick JS. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput. Biol. 2008;4:e1000176. doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dinger ME, Amaral PP, Mercer TR, Mattick JS. Pervasive transcription of the eukaryotic genome: functional indices and conceptual implications. Brief Funct. Genomic Proteomic. 2009;8:407–423. doi: 10.1093/bfgp/elp038. [DOI] [PubMed] [Google Scholar]
- 49.Gerstein MB, Bruce C, Rozowsky JS, Zheng D, Du J, Korbel JO, Emanuelsson O, Zhang ZD, Weissman S, Snyder M. What is a gene, post-ENCODE? History and updated definition. Genome Res. 2007;17:669–681. doi: 10.1101/gr.6339607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.