Abstract
In a single experiment, chromatin immunoprecipitation combined with high throughput sequencing (ChIP-seq) provides genome-wide information about a given covalent histone modification or transcription factor occupancy. However, time efficient bioinformatics resources for extracting biological meaning out of these gigabyte-scale datasets are often a limiting factor for data interpretation by biologists. We created an integrated portable ChIP-seq data interpretation platform called seqMINER, with optimized performances for efficient handling of multiple genome-wide datasets. seqMINER allows comparison and integration of multiple ChIP-seq datasets and extraction of qualitative as well as quantitative information. seqMINER can handle the biological complexity of most experimental situations and proposes methods to the user for data classification according to the analysed features. In addition, through multiple graphical representations, seqMINER allows visualization and modelling of general as well as specific patterns in a given dataset. To demonstrate the efficiency of seqMINER, we have carried out a comprehensive analysis of genome-wide chromatin modification data in mouse embryonic stem cells to understand the global epigenetic landscape and its change through cellular differentiation.
INTRODUCTION
Chromatin immunoprecipitation (ChIP) allows the quantitative measurement of protein (i.e. transcription factor) occupancy or the presence of post-translational epigenetic histone modifications at defined genomic loci. Recent technological developments combining ChIP with direct high throughput sequencing of the immunoprecipitated DNA fragments (ChIP-seq) allowed the mapping and analysis of transcription factor genomic occupancy or epigenetic chromatin marks at the genome-wide (GW) scale in a relatively unbiased manner. Over the last years, numerous studies have used ChIP-seq as a central method to create GW binding maps for a particular genomic feature (1,2), installing ChIP-seq as the gold standard in the functional genomics toolbox.
Each ChIP-seq run generates data at the gigabyte scale that requires successive steps of bioinformatics treatment prior to biological interpretation [reviewed in ref. (3)]. In a standard analysis pipeline, two major steps can currently be distinguished. First, genomic locations presenting relevant enrichment in the ChIP-seq signal are identified and annotated with respect to known genomic sequence features (genes, transcripts, repeat elements, etc). Second, by performing multiple rounds of analyses using various methods, the biological meaning of the dataset has to be extracted and often compared with other datasets. Many methods and softwares have been released over the past months in order to easily perform the initial analysis stage with good accuracy [reviewed in ref. (3,4)]. However, unlike the first step that distinguishes relevant signal from noise to provide information on a given factor or chromatin mark, the second analysis stage requires the laborious combination of various methodologies to answer complex biological questions. Thus, development of integrated complementary approaches is a prerequisite to make ChIP-seq analysis as easy and routine as possible.
Several initiatives devoted to particular biological questions have already contributed to enrich the ChIP-seq analysis toolbox. For example, numerous tools, integrated or not in larger analysis pipelines, have been proposed that allow: annotation of genomic features present in the neighbourhood of the relevant enrichment signals (5–7), detection and de novo definition of consensus DNA motifs (6), cross and compare information from distinct datasets (8) and comprehensive visualization of the obtained GW results (9–11). More recently, sophisticated statistical approaches were proposed to predict association between sets of genomic locations and numerous genomic features (12,13). Nevertheless, due to the multiplicity of biological questions that may be asked by the ChIP-seq method, many analysis issues remain un-addressed.
In particular, the definition of the genomic rules governing the function of a particular factor is a complex, but central question. The most common method for addressing this question is to investigate the frequency of co-occurrence of the analysed factor/mark with other genomic features by (i) extracting relevant signals (peaks) in the two datasets to be compared and (ii) calculating the number of events that are close enough between the datasets to be considered as overlapping. This approach, which has been previously used (8,14), presents several limitations. First, the analysis is biased towards the peak-searching step and the empirical cut-off values used to discriminate between relevant and background signals. The second drawback comes from comparisons of datasets containing either sharp (i.e. a transcription factor) or non-discrete binding sites (i.e. spread histone marks like H3K27me3), where peak detection is more difficult. Third, an increasing number of studies show that particular factors have more than a single function in the genome [i.e. (15)], challenging the direct interpretation of one to one comparisons. Finally, it becomes clear that definition of genomic regulatory elements cannot rely on a single feature, but need integration of multiple sources of information to be properly identified (16,17). Moreover, tools able to analyse multiple information sources are required to allow comprehensive qualitative and quantitative ChIP-seq analysis.
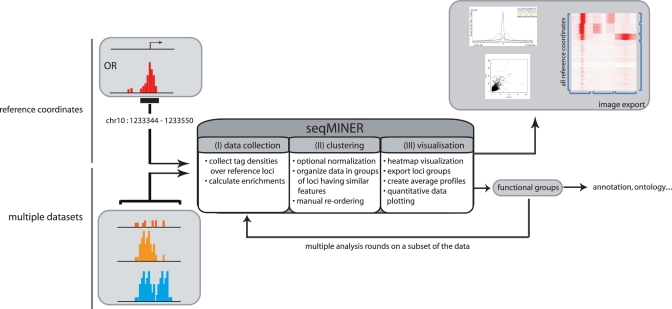
To overcome the above-described limitations, we have developed seqMINER, an integrated user friendly platform that addresses central questions in the ChIP-seq analysis workflow. seqMINER is designed in order to make it as easy as possible for the biologist to carry out in depth interpretation of the analysed datasets to answer their biological question. The purpose of seqMINER is to allow qualitative and quantitative comparisons between a reference set of genomic positions and multiple ChIP-seq datasets (workflow presented in Figure 1). Different analysis modules have been implemented to allow users to search for full characteristics of a particular feature genome-wide. Starting from a set of reference coordinates that can be a list of ChIP-seq enrichment clusters (peaks) for a particular target (i.e. a transcription factor), seqMINER proposes two complementary methods to analyse the signal enrichment status in multiple other tracks: (i) a method that computes a density array over a defined window around the reference coordinate; (ii) a method that computes a single enrichment value over a defined window around the reference coordinate. Following these steps, automated as well as manual reordering methods of the analysed loci are implemented to assist users in defining functional subgroups in their data. Finally, graphical representations of the data are proposed through heatmaps and dotplots to illustrate particular, as well as general properties of the data. We show that the different resources of seqMINER taken together allow a comprehensive analysis of genome-wide chromatin modification data to understand the global epigenetic landscape and its change through cellular differentiation.
Figure 1.
Schematic representation of the general workflow of seqMINER. seqMINER takes as an input a single set of reference loci (i.e. gene promoters or binding sites) and multiple raw sequencing datasets. seqMINER can collect tag densities or calculate enrichment values around the set of reference coordinates. Using a combination of automated clustering and manual reordering methods, seqMINER helps the user to create functional groups within the reference set. Each specific sub-group in the dataset can be visualized as heatmaps, dotplots or average plots and the groups of loci can be exported for further analysis.
MATERIALS AND METHODS
seqMINER algorithms
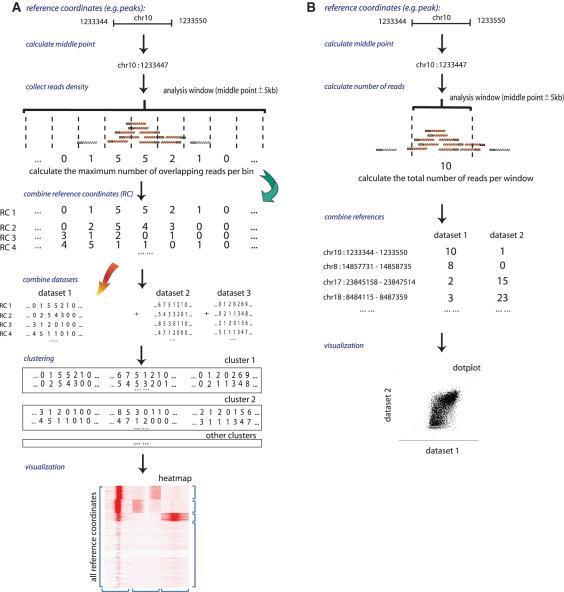
As input, seqMINER supports multiple file formats popular for storage of high throughput sequencing data (BED, SAM/BAM and Bowtie). seqMINER uses two methods to quantify ChIP-seq signal depending on the type of performed analysis (Figure 2). For both methods the middle of the reference coordinate (i.e. peak) is calculated and used as reference locus in further computation. Moreover, the strand orientation of the reference feature is (by default, if the reference locus contains strand information) taken in account in order to orientate all analysed features on the same direction. For both methods, prior to analysis, all reads are extended from a user defined size (default 200 bp). For the calculation of densities over a defined window, methods are derived from the one generally used to generate density files [i.e. (18) except that tag extension is performed only on the direction of the tag (not in both strand orientations)].
Figure 2.
Schematic representation of the algorithms for each method implemented in seqMINER. In both methods, prior to quantification, reads are extended from a user-defined value (default 200 bp). (A) Density array method: a user defined number of bins are created in a fixed window around the reference coordinate and for each bin the maximal number of overlapping reads is computed in each dataset, the collected values are pooled and submitted to clustering process and the generated clusters are visualized as heatmap. (B) Enrichment based method: the number of tags presents or overlapping a user-defined window (default 2 kb) around the reference site are counted. The values from the different datasets are computed and ploted.
For the density array method, a user defined number of bins is created over a fixed size window around the reference coordinate (sizable by the user) and for each bin the maximal number of overlapping tags is computed (Figure 2A).
To calculate a single enrichment value for a binding site, tag density is defined as the number of tags present or overlapping in user-defined window (default 2 kb) around the reference site (Figure 2B). ChIP-seq enrichments (e) are defined as e = log 2 [(foreground tags + q)/(background tags + q)]. q is defined as an empirical constant in the range of 10; foreground tags are the density value computed in the data track; background tags, the density value in the control track. q is used to lower the contribution of noise variations that is assumed to be higher at low-count levels. The use of the constant q reduces the influence of the signal variation in the noise measurement on the ratio calculation. Increasing q-value will increase the stringency of the analysis by lowering the contribution of low-density values to high-enrichment calculations.
Two normalization methods are implemented in seqMINER, namely linear and ranked-based normalization. These methods aim to lower the bias on the clustering procedure due to inter-samples intensities differences inherent to the ChIP-seq procedure. Normalized data are used only to perform the clustering step but raw intensity data are displayed in the final heatmap to recover the original patterns from the data.
Linear normalization: in each dataset, all xi values from the calculated density array are divided by the percentile P of the distribution of the all values xi > T (P is chosen by the user, default P = 75, the third quartile).
Ranked-based normalization: in each dataset, the values xi from the calculated density array are sorted in ascending order: x1 < x2 < … xi < … < xN (with N the total number of values in the density array). Then each value xi should be replaced by its rank ri, if xi > T (with T a threshold chosen by the user, default T = 10): rN = N, rN−1 = N−1,…, ri = i. All values xi ≤ T are replaced by 0, which ensures that all background values are similarly considered during the clustering process.
Data source and treatment
ChIP-seq datasets were downloaded from the public data bank Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/gds) under the accession number: GSM307618 (H3K4me3-ESC); GSM281696 (H3K4me3-brain); GSM307625 (input-ESC); GSM307620 (H3K36me3-ESC) and GSM307619 (H3K27me3-ES). Reference coordinates list is established using MACS (17).
Code repository
The source code, tutorial and wiki for seqMINER is available at: http://bips.u-strasbg.fr/seqminer/ under General Public License (GPL3).
Discussions and bug report
A google group is created to allow discussions on future developments and easy interaction of seqMINER users at: http://groups.google.com/group/seqminer?hl=en.
RESULTS
Correlative integration of multiple datasets
Most genomic studies aim to define the function of a particular regulatory factor or a given chromatin modification by understanding globally how it affects other co-occurring events in a regulatory circuit (i.e. chromatin modifications, binding of other factors) and the consequences on outputs of the regulated system (i.e. gene expression). Thus we designed seqMINER to allow the integration of multiple ChIP-seq datasets in a quick and user friendly manner.
seqMINER uses a list of genomic coordinates (i.e. loci bound by a particular factor, set of genes, set of promoters, etc.) as a reference for investigating information in other genomic datasets. Three stages can be distinguished in the analysis process (Figure 1). First, in the data collection module, seqMINER collects the read density from a reference dataset over a user-defined window around a set of coordinates and then calculates the read density in the same window in one or multiple other datasets. Second, in the clustering module, seqMINER uses a clustering procedure (k-means) to organize the identified loci presenting similar read densities within the specified window. Third, in the visualization module, seqMINER allows at glance, visualization of the entire output dataset through various graphical representations.
seqMINER proposes two related complementary tag (read) density based methods to analyse the signal enrichment status in multiple tracks (Figure 2). The first method, named ‘density array method’ defines general patterns and functional sub-groups in the dataset: densities over a window around the reference coordinates can be calculated at different resolutions in multiple tracks (Figure 2A). The created matrix can be organized by k-means clustering to isolate groups of loci having similar features as developed earlier (16,19). At this stage, clusters can optionally be reorganized manually according to the biological significance. Visualization of the whole dataset is in this case achieved through heatmaps. This method allows easy visualization of signal distribution over multiple loci and identifies general patterns over the dataset, which can be plotted as average profiles. Information on signal distribution, that can be an important biological feature, is conserved (i.e. broad, sharp enrichment peaks). However, visualization of quantitative phenomenon is more complex. The second method, named ‘enrichment based method’, allows calculation of raw tag counts as well as normalized enrichment over a control track (Figure 2B). The created matrix allows easy plotting of quantitative information and interpretation for one to one comparisons. Moreover, the method produces numerical data that are necessary to integrate sequencing data in larger mathematical models of a particular system.
All visual features as well as the list of loci can be exported for further analysis with other methodologies [i.e. gene annotation (5), ontology (20)]. Moreover, output data can be used in new clustering rounds using different sets of data or analysis parameters. This possibility facilitates multiple iterative steps of analysis inherent to the genomic data analysis.
Normalization procedure
A current limitation in ChIP-seq data inter-comparison comes from technical issues in the ChIP experiment itself. It appears to be relatively difficult to obtain quantitatively comparable data due to high variation in immunoprecipitation efficiency that is affected by a multitude of imponderable factors (antibody specificity and efficiency, natural abundance of the particular ChIPed feature, etc.) and due to the total number of reads that could differ from one sample to another. For example, if a dataset presents significantly higher enrichment compared to the others, it will likely over-contribute to the clustering and eventually lead to sub-optimal locus organization in the final clustering. A simple way to overcome this problem is to normalize data at the beginning of seqMINER analysis.
Several methods have already been proposed for normalizing high throughput sequencing datasets [i.e. (21–23)]. However, to date, no real consensus has appeared in the field on widely applicable normalization methods. seqMINER proposes two normalization methods (based on linear or ranking normalization) that interfere directly at the clustering stage but do not affect the final visualization results. Conceptually, both methods replace absolute intensity values by proportional values that allow a comparable contribution of each dataset to the clustering procedure.
In order to test the efficiency of the different normalization methods, we compared the clustering results of the analysis performed with each method using an identical input dataset (Supplementary Figure S1). We choose input datasets with various ChIP efficiencies (H3K4me3-strong signal; H3K27me3-low and H3K36me3-low signals) to test the normalization efficiency. This comparison shows that the normalization allows to better take in account the lower intensities signals (H3K36me3 and H3K27me3) in the clustering procedure and to create more biologically consistent groups in the output heatmap (Supplementary Figure S1). Use of these methods improves the cluster determination, particularly in the case of comparison of several experiments having very different efficiencies.
Performance optimization
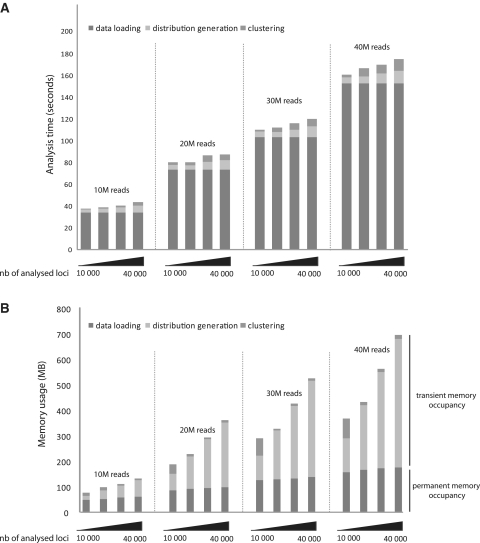
seqMINER takes advantage of the portability of the Java Virtual Machine (JVM) allowing an installation free multiplatform usage. seqMINER is designed to optimize all time limiting steps in the manipulation of the large raw sequencing datasets. For all the computational tasks, the implementation combines an optimal use of the random access memory (RAM), avoiding repetitive access to the hard drive and simultaneous multi threading (SMT) to fully use available computational resources and lower the time of analysis. SMT permits the program to execute multiple independent threads to better utilize the resources provided by modern processor architectures. Particular attention is paid to the design of the genomic data storing objects to optimize space occupancy and efficiency of data retrieval algorithms. seqMINER performances were assessed by performing the analysis using cumulatively four standard size (10 M reads) ChIP-seq datasets and using as a reference set increasing number of loci (10 000–40 000 distinct loci) (Figure 3A). All software trainings were performed on a personal computer (PC) running under windows with standard performances (CPU-3.0 GHz core-duo; RAM-4 GB). For all the tested sets, the limiting step appears to be the loading of the dataset in the memory (Figure 3A), which is usually performed only once and followed by multiple cycles of analysis. Nevertheless, the time required for a complete analysis by seqMINER for all tested datasets, regardless of the number of reference coordinates used, and including data loading is <3 min (Figure 3A).
Figure 3.
Time and memory required to complete a typical analysis with seqMINER. (A) Time required for the different stages of the analysis (namely data loading, distributions generation and clustering) by seqMINER, using raw ChIP-seq datasets of various sizes (10, 20, 30 and 40M reads) and various number of reference coordinates (10 000, 20 000, 30 000, 40 000 reference positions). The analysis was performed on a PC running under windows with standard performances (CPU-3.0 GHz core-duo; RAM-4 GB). (B) Memory required for the different stages of the analysis tested as above. Note that only the data loading step stores objects in memory, the two subsequent steps free the memory once completed allowing multiple successive analyses with limited memory attribution (<500 MB).
To bring the analysis time by seqMINER to acceptable levels we had to optimize the clustering process for the density arrays. The density array based method generates a large array of values (i.e. for a 10 kb window at a 25 bp resolution, an array of 400 values is created per dataset analysed) for each reference position. The use of existing implementation (24) of clustering algorithms to organize these large datasets appeared to slow down considerably the analysis workflow (over an hour for the 8470 reference sites and two datasets). Thus, we decided to create a novel implementation of this algorithm using recent technology developments in programming resources. We implemented the k-means algorithm, taking advantage of SMT technology and using Java Machine Learning libraries (25) for the data structure implementation. This new algorithm dramatically improves the clustering speed compared to existing implementations (Figure 3A) as <10 s are needed for clustering all the tested datasets, while more than an hour was needed for the smallest dataset tested with the former algorithm. Thus this new implementation used in seqMINER brings the efficiency of clustering routines to the requirements of ChIP-seq data analysis.
Besides time optimization, another key aspect of automated high throughput data analysis is the capacity of handling of large datasets within the available memory. Thus seqMINER is optimized to limit the memory (RAM) occupancy in order to allow simultaneous analysis of a reasonably high number of datasets on a standard PC. Using this strategy, seqMINER requires less than 50 MB for storing a standard size ChIP-seq file of 10M reads (Figure 3B). Moreover, in all the conditions tested (up to four datasets, with 40 000 reference sites), not more than 500 MB (Figure 3B) is transiently required to perform the data collection and clustering step (note that contrary to data loading, this memory is free once the task is completed).
These combined efforts allow the analysis of multiple raw sequencing datasets in a few seconds with standard computer performances, considerably speeding up the ChIP-seq analysis workflow and facilitating the testing of multiple biological assumptions in a time efficient manner.
Genome-wide analysis of the chromatin landscape of genes in embryonic stem cells
Comparative analysis of epigenetic profiles in several cell types allows a global view and better understanding of chromatin dynamics and its role in gene regulation. In the last few years, numerous genomic studies focused on embryonic stem cells (ESC) that can both self-renew indefinitely or differentiate in cell types that form the three primary germ layers. These interesting properties were broadly studied and were shown to be highly dependent on transcriptional and epigenetic regulatory networks (26–28). In order to train seqMINER and demonstrate its efficiency in addressing complex biological questions, we decided to (i) make a comprehensive description of the chromatin states at all annotated promoters of mouse ESCs, and (ii) quantitatively compare the active chromatin marking in ESCs with that observed in the differentiated brain tissue.
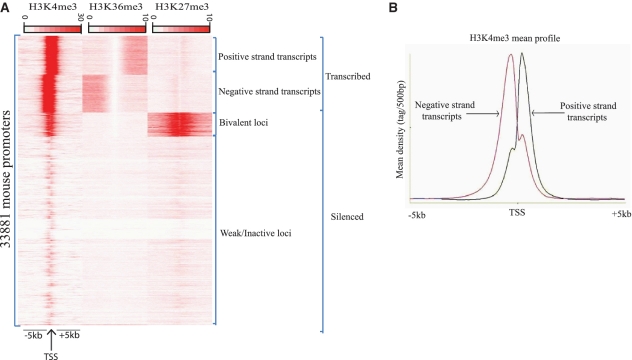
First, we used seqMINER to characterize the epigenetic profile of mouse genes in ESCs using three representative histone modifications known to mark active or inactive genes. The active histone marks H3K4me3 and H3K36me3, are enriched in the promoter regions and transcribed gene bodies, respectively. In contrast, the repressive mark, H3K27me3, is known to give a broader distribution over genes. As reference coordinates, we used transcription start sites (TSS; assumed to be at the 5′-end of annotated genes) of the 33 881 mouse genes referenced in ENSEMBL (v58) database. Tag densities from each dataset were collected in a window of 10 kb around the reference coordinates and the collected values were subjected to k-means clustering (using linear normalization method).
Out of this initial clustering, two major groups of loci could be distinguished (Figure 4A), the first group contains transcribed loci, while the second group contains silenced loci. Amongst transcribed loci we can identify two clusters of active genes transcribed on the negative strand or on the positive strand, as determined by the H3K36me3 mark (Figure 4A). Additionally, two clusters of silent loci could be identified, one marked by both H3K4me3 and H3K27me3 marks (known as bivalent promoters) and one showing no significant enrichment of the three studied marks. Interestingly, while the bivalent loci are known to correspond to transiently repressed genes required for later differentiation of ESCs, the unmarked loci likely represent strongly repressed genes that could harbor constitutive heterochromatin marks. Thus starting from heterogeneous datasets and based only on three chromatin features, we could identify consistent gene categories for which average profiles can be automatically drawn (Figure 4B), illustrating general regulatory features of these loci.
Figure 4.
Characterization of epigenetic profiles of mouse promoters in ESCs using seqMINER. (A) Read densities of regions surrounding the whole set of TSS (assumed to be the 5′-end of the annotated transcript) of mouse genes from ENSEMBL (v58). TSSs were used as reference coordinates to collect data in publicly available H3K4me3, H3K36me3 and H3K27me3 datasets. Tag densities from each ChIP-seq dataset were collected within a window of 10 kb around the reference coordinates, the collected data were subjected to k-means clustering (using linear normalization). The major groups and clusters are indicated. (B) Using seqMINER, the average profile for selected clusters was automatically calculated and plotted. The H3K4me3 mean profile for transcripts actively transcribed on the negative strand (pink) and positive (blue) strand was calculated and represented.
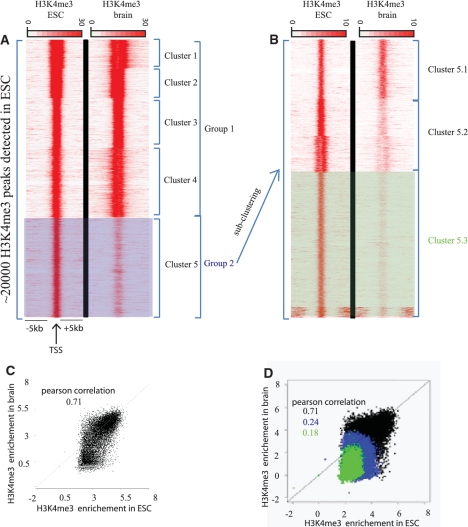
Second, we aimed to test seqMINER in a study to compare quantitative changes in the histone H3K4me3 mark in ESCs and brain tissue. In order to compare these two datasets, we generated a list of reference coordinates enriched in H3K4me3 in ESCs. We used both methods proposed in seqMINER to compare the signal in the two selected datasets over the reference loci. First, we used the density array based method to organize groups in the dataset (Figure 5A). With this method, two major groups could be identified. The first group contains loci equally enriched in H3K4me3 mark in ES and brain cells, while the second group contains loci with higher H3K4me3 signal in ESCs relative to brain tissue (group 2 in Figure 5A). This analysis suggests that group 2 comprises only ES specific genes. However, by performing an additional round of clustering on group 2, three additional clusters were identified (5.1, 5.2 and 5.3), illustrating the importance of the possibility given by seqMINER to perform iterative rounds of analysis to organize the data with precision. Analysis of the additional clusters in group 2 after this second clustering step indicates that the seemingly ES specific loci can now be reclassified as weakly/not, moderately or strongly enriched in ES cells relative to brain tissue (Figure 5B). The results now suggest for example that cluster 5.1 is also transcribed in the brain cells analysed.
Figure 5.
Quantitative changes of H3K4me3 mark in mouse brain cells relative to mESCs. Tag densities of regions surrounding the H3K4me3 enriched loci in ESCs. Publicly available ChIP-seq datasets for H3K4me3 in ESCs and in brain cells were used in this comparative analysis. (A) H3K4me3 enriched loci in ESC were detected using MACS software, these loci were used as reference coordinates. Tag densities from H3K4me3-ESC and H3K4me3-brain datasets were collected within a window of 10 kb around the reference coordinates, and then the density files were subjected to k-means clustering. Two major groups can be isolated: group1 contains loci with significant and equal enrichment of H3K4me3 in both ESC and brain; group 2 contains loci with higher enrichment of H3K4me3 in ESC relative to brain tissue. (B) As a second step of analysis, the loci in group 2 were used as reference. The densities around these loci were recollected and a second round of clustering was performed. After the second round of clustering, three clusters can be isolated: cluster 5.1, 5.2 and 5.3 corresponding to loci weakly, moderately and strongly enriched in H3K4me3 mark in ESC relative to brain, respectively. Note that the bottom of the cluster 5.3 that has H3K4me3 enrichment distant from the cluster center was not considered as a separate entity since we focused our analysis on the differential signal in the cluster center. (C) Quantification of the changes observed between the two conditions. Dot-plot representing H3K4me3 enrichments in ESC versus brain. Enrichments were calculated for H3K4me3-ESC and H3K4me3-brain datasets within a window of 2 kb around the complete set of reference coordinates (black dots), (D) and against previously isolated subsets of the reference coordinates (group2 in blue and subset 5.3 in green).
In order to quantitatively determine the differences between ESC and brain tissue, we collected enrichments over the input loci (H3K4me3 bound regions) using the enrichment based method implemented in seqMINER. Following this step, we used the visualization module of seqMINER to plot enrichment in ESC versus brain tissue (Figure 5A). Using this method, we again observe a heterogeneous pattern when using the total set of loci (Figure 5C) with a large proportion showing similar enrichments in ESC and brain tissue (Pearson correlation coefficient = 0.71), but also a subset of loci that are enriched in ESCs, but not in brain tissue. Importantly, when we collect the same data on subsets isolated with the density array method (subsets 2 and 5.3) and overlay this information on the plot (Figure 5D, subset 2 in blue and 5.3 in green), we can confirm that these subsets correspond to the loci that change their status between the two conditions. Additional information is gained by this method since we observe that most of the loci that are differentially enriched in H3K4me3 between the two cell types display low enrichment in ESC. Consequently, loci that are highly enriched in the H3K4me3 mark are invariant in both cell types and probably correspond to house-keeping genes (Figure 5C).
These analyses demonstrate how the two methods implemented in seqMINER can be combined to select populations of loci having similar features and quantitatively follow their behavior in different conditions. Importantly, we show how clusters can be used as reference coordinates for iterative rounds of analysis to detect potential sub-populations. Finally through some biological examples, we illustrate the possibilities given by seqMINER to create visual representations of the isolated genomic features.
DISCUSSION
Extracting the biological meaning from genome-wide studies requires the use of various methodologies to answer complex biological questions. With seqMINER, we attempted to develop a standalone analysis platform that allows users to make biological interpretation of their high throughput sequencing data. We implemented two methods that allow high-level correlative integration of multiple sequencing datasets to identify general as well as specific genomic characteristics that emerge from the analysed features. We provided a number of visualization methods, in order to allow rapid assessment of datasets from different analysis perspectives (i.e. general patterns, sub-groups, quantitative data comparisons). Thus, we provide a novel broad bioinformatics resource that should give analysis solutions in many ChIP-seq based biological applications. Additionally, we significantly improved the efficiency of broadly used clustering algorithms by re-implementing them using recent technology developments (i.e. SMT). Finally, we have implemented simple normalization procedures that greatly improve the efficiency of multiple ChIP-seq data comparisons. Thus seqMINER should prove to be a valuable resource for a broad range of applications in the bioinformatics community.
By using JVM for code interpretation, we aimed to avoid both operating system (OS) incompatibility and to limit users efforts for installation, making seqMINER accessible to a broad range of users. However, on most OS, by default, only a small RAM memory space is attributed to the application, limiting the number of genomic features that can be analysed simultaneously. In order to overcome this limitation, the user has to manually set the desired RAM space dedicated for seqMINER analysis. Another implementation choice regarding memory usage is to load all the necessary data in the RAM prior to analysis to avoid repetitive hard disk access and speed up the computational tasks. This can be a limitation for seqMINER usage on local computers that are commonly equipped with 2–4 GB RAM and a 32 bit operation system that limits the maximum RAM of Java virtual machine at ∼1.5 GB. Thus, to perform multi-file comparison (∼10< sequencing raw files), the use of seqMINER on a server is recommended.
In the present version of seqMINER, we aimed to address numerous biological questions commonly raised in classical ChIP-seq analysis pipelines. As users may have specific questions they wish to ask and as the arrival of future technologies extends the scope of the questions that can be asked, we designed seqMINER in a flexible and open mode. We implemented seqMINER in an organized architecture (following Model-View-Controller guidelines) and making the source code available open source (GPL3). For example, dedicated methods for integrating ChIP-seq with RNA-seq will be the next developments needed to improve seqMINER functionality.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
INSERM-Région Alsace (to A.R.K.); Association pour la Recherche sur le Cancer (ARC to A.R.K.); French Ministry of Education, Research and Technology (MNERT to M-A.C.); ARC (to M-A.C.); ANR (GenomATAC; ANR-09-BLAN-0266) (to L.T.); the EU EPIDIACAN and the Ligue National contre le cancer (to L.T.); the EU (EUTRACC, LSHG-CT-2007-037445) (to L.T. and I.D.); the Institut National du Cancer; Ligue National contre le cancer, ‘équipe labélisée’ (to I.D.). Funding for open access charge: by the EU EUTRACC.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to M. Stadler for technical suggestions, M. De Dieuleveult, S. Le Gras, T. Strub, N. Paul for useful comments on the software and the article; and K. Karmodiya for critical reading of the article.
REFERENCES
- 1.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genome-wide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 4.Laajala TD, Raghav S, Tuomela S, Lahesmaa R, Aittokallio T, Elo LL. A practical comparison of methods for detecting transcription factor binding sites in ChIP-seq experiments. BMC Genomics. 2009;10:618. doi: 10.1186/1471-2164-10-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krebs A, Frontini M, Tora L. GPAT: retrieval of genomic annotation from large genomic position datasets. BMC Bioinformatics. 2008;9:533. doi: 10.1186/1471-2105-9-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shin H, Liu T, Manrai AK, Liu XS. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25:2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- 8.Blankenberg D, Taylor J, Schenck I, He J, Zhang Y, Ghent M, Veeraraghavan N, Albert I, Miller W, Makova KD, et al. A framework for collaborative analysis of ENCODE data: making large-scale analyses biologist-friendly. Genome Res. 2007;17:960–964. doi: 10.1101/gr.5578007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH. JBrowse: a next-generation genome browser. Genome Res. 2009;19:1630–1638. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manske HM, Kwiatkowski DP. LookSeq: a browser-based viewer for deep sequencing data. Genome Res. 2009;19:2125–2132. doi: 10.1101/gr.093443.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bock C, Halachev K, Buch J, Lengauer T. EpiGRAPH: user-friendly software for statistical analysis and prediction of (epi)genomic data. Genome Biol. 2009;10:R14. doi: 10.1186/gb-2009-10-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hon G, Ren B, Wang W. ChromaSig: a probabilistic approach to finding common chromatin signatures in the human genome. PLoS Comput. Biol. 2008;4:e1000201. doi: 10.1371/journal.pcbi.1000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blahnik KR, Dou L, O'Geen H, McPhillips T, Xu X, Cao AR, Iyengar S, Nicolet CM, Ludascher B, Korf I, et al. Sole-Search: an integrated analysis program for peak detection and functional annotation using ChIP-seq data. Nucleic Acids Res. 38:e13. doi: 10.1093/nar/gkp1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 2009;23:2484–2489. doi: 10.1101/gad.1837309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 20.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 21.Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, Stunnenberg HG. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28:1418–1428. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taslim C, Wu J, Yan P, Singer G, Parvin J, Huang T, Lin S, Huang K. Comparative study on ChIP-seq data: normalization and binding pattern characterization. Bioinformatics. 2009;25:2334–2340. doi: 10.1093/bioinformatics/btp384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, Bjornson R, Carriero N, Snyder M, Gerstein MB. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat. Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 25.Abeel T, de Peer YV, Saeys Y. Java-ML: a Machine Learning Library. J. Machine Learn. Res. 2009;2009:931–934. [Google Scholar]
- 26.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 27.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 28.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.