Abstract
Many mutation events in microsatellite DNA sequences were traced to the first embryonic divisions. It was not known what makes the first replication cycles of embryonic DNA different from subsequent replication cycles. Here we demonstrate that an unusual replication mode is involved in the first cycle of replication of DNA introduced in mammalian cells. This alternative replication starts at random positions, and occurs before the chromatin is fully assembled. It is detected in various cell lines and primary cells. The presence of single-stranded regions increases the efficiency of this alternative replication mode. The alternative replication cannot progress through the A/T-rich FRA16B fragile site, while the regular replication mode is not affected by it. A/T-rich microsatellites are associated with the majority of chromosomal breakpoints in cancer. We suggest that the alternative replication mode may be initiated at the regions with immature chromatin structure in embryonic and cancer cells resulting in increased genomic instability. This work demonstrates, for the first time, differences in the replication progression during the first and subsequent replication cycles in mammalian cells.
INTRODUCTION
The first replication cycle of the fertilized mammalian egg remains understudied, for apparent reasons. However, some information is available from studying amphibian embryogenesis (1). During fertilization the sperm DNA enters the egg in complex with protamines, which are degraded inside the zygote and eventually replaced with histones (2). The chromatin of an egg also goes through intensive remodeling (3). The chromatin structure of a fertilized egg takes a long time to form, and is not completed even after the first division of the zygote (4).
An intriguing feature of the first zygotic cycle is increased genomic instability. The first replication cycle evokes many more mutations than the subsequent somatic cells’ divisions (5). In Friedreich’s ataxia the expansion of the GAA repeat responsible for the disease can be traced to early embryogenesis (6); for review see (7). The origins of this increased mutability of the first replication cycle remain undetermined. However, there are some known differences between the first replication cycle and the subsequent ones.
One of the characteristics of the first replication cycle, which changes later on, is the high number and apparent randomness of initiation points (8,9). In the first divisions of Xenopus laevis and Drosophila melanogaster embryos, replication origins are closely spaced, random sequences (8,9). Similarly, replication initiates randomly in exogenous DNA added to Xenopus egg extract (10). Later in the development, a switch to less frequent replication initiation occurs (8,9). This switch is most likely associated with epigenetic changes, which restrict replication initiation to specific areas of the genome.
Mammalian replication initiation in its essence is a fairly random process (11). The origin recognition complex (ORC) does not have a consensus recognition site and would initiate replication of any kind of DNA fragment in vitro (12), while its binding is limited to specific areas in vivo (13). An in vitro oligonucleotide-binding assay showed that the ORC complex preferred synthetic A/T-rich polydeoxynucleotides (12), but no consensus sequence was revealed.
It appears that many more sequences have the potential to serve as replication origins than are actually used in somatic cells (14). In early G1, the binding of Cdc6 and Cdt1 to ORC facilitate the loading of minichromosome maintenance (MCM) complexes onto DNA (13). MCM complexes possess the helicase activity necessary for unwinding the double helix for DNA polymerase α/primase loading (15). A single ORC can initiate the loading of more than 20 MCM complexes (16); however, only a few of them initiate replication (17). Additionally, random fragments of mammalian chromosome of more than several thousand base pairs that normally do not initiate genomic replication support the autonomous replication of plasmids in mammalian cells (18).
An unknown mechanism restricts initiation to certain positions. The limitations most likely result from the chromatin structure, which is different in the first replication cycle (3). Moreover, the absence of mature chromatin structure in the first replication cycle may lead to the initiation of the non-canonical replication mode that is more prone to stalling at obstacles.
This alternative replication mode could also take place in cancer cells that often have an abnormal chromatin structure (19). Chromosomal instability is common in cancer cells (20,21); more than half of the breakpoints in cancer-causing chromosomal translocations were found to be associated with A/T-rich microsatellites (22).
One of the A/T-reach microsatellites, a rare fragile site FRA16B, can cause spontaneous chromosomal breakage by an unknown mechanism (23). FRA16B is composed of a 33-bp A/T-rich repeat (24). While most people have around 7–12 units in their FRA16B locus, it can sometimes expand to >2000 repeated units (25). FRA16B is characterized by increased flexibility and instability of the double helix (23). The cause of FRA16B fragility is still unknown. We hypothesized that the fragility of FRA16B is caused by an alternative replication mode that might be initiated due to an aberrant chromatin structure.
In this study, we analyzed the first replication cycle of an SV40-based plasmid in fibroblasts. Similar to the first replication round of the zygote, the chromatin structure of this plasmid was still in the formation stage when the first replication cycle took place. Interestingly, we discovered that an alternative replication mode was involved in this plasmid’s first replication cycle. This mode initiated randomly all over the plasmid, similar to the replication observed in Xenopus eggs (26). Furthermore, the FRA16B sequence caused its complete blockage, which could lead to fragility.
MATERIALS AND METHODS
Plasmids and strains
The plasmid pUCneo was obtained by replacing the EcoRI–NdeI fragment of pSV2Neo (Stratagene) with the PvuII fragment of pUC19 in an orientation that would lead to the inversion of the ampicillin resistance gene. pUCneo was used in place of the original pSV2Neo to improve the stability of cloned repeats, which are sometimes unstable in transcribed areas. The 1300-bp FRA16B fragment containing 26- to 33-bp-long A/T-rich repeats was amplified by PCR out of genomic DNA isolated from an EBV-transformed line of human lymphoblasts, using primers JS27 5′-CATAGTATATACTATAGTATATAG-3′ and JS29 5′-GTACTATACTATACTATATTATACAG-3′ as previously described (27). The amplified fragment was cloned into a pGEM-T cloning vector. The FRA16B-containing fragment was isolated by AatII-PstI digest of pGEM-FRA16B, and inserted in two orientations in the AatII site of the pUCneo located between the SV40 origin and the ampicillin resistance gene. The plasmid Ori(-) was obtained by replacing the HindIII-AccI fragment of pUCneo with the annealed oligonucleotides 5′-ATGGTGTGGAAAGTCCCCAGGCTCCCCAGCAGCGCAGAAGTATGCAAAGCATGCATCTCAATTAGTCAGCAACCA-3′ and 5′-AGCTTGGTTGCTGACTAATTGAGATGCATGCTTTGCATACTTCTG CGCTGCTGGGGAGCCTGGGGACTTTCCACACC-3′. The sequence of oligonucleotides contained the 72-bp repeat from the SV40 origin, which can also serve as a nuclear localization sequence (NLS) for improved nuclear delivery. The plasmid ΔAT was obtained from Ori(-) by deletion of the Bsp119I-EcoRI fragment.
pUC19-BsmI plasmid with a unique site for nicking endonuclease Nb. BsmI was created by replacing a EcoRI-PstI polylinker fragment of pUC19 with BsmI-containing EcoRI-PstI fragment of Ori(-).
All plasmid samples were isolated from the XL1-Blue strain (Stratagene).
Obtaining a highly supercoiled plasmid
A highly supercoiled plasmid was obtained as described in (28). In brief, pUC19-BsmI plasmid was nicked at a single site using the nicking endonuclease BsmI, incubated at 65°C to inactivate BsmI, purified with phenol/chlorophorm extractions, precipitated with ethanol and then resuspended in TE buffer (10 mM Tris–HCl pH 8.0, 1 mM EDTA).
The nicked pUC19-BsmI was re-ligated with T4 DNA ligase in the presence of 6 µg/ml ethidium bromide for 3 h at room temperature. Then ethidium bromide was removed by three rounds of extraction with 1-butanol. The DNA was ethanol precipitated, and resuspended in TE buffer.
Cell culture and transient transfections for replication analysis
All of the cell lines (COS-1 monkey fibroblasts, 293A embryonic kidney fibroblast, and HeLa cells) were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% newborn calf serum. Primary fibroblasts, kindly provided by Dr Andrea Mastro (Penn State University), were grown in DMEM supplemented with 10% fetal bovine serum.
All transfections of immortalized cell lines were performed with lipofectin transfection reagent (Invitrogen) according to the manufacturer’s instructions. Primary fibroblasts were transfected with Express-In transfection reagent (Open Biosystems) according to the manufacturer’s instructions.
Mimosine treatment of cultured mammalian cells
Cells were grown in normal growth media until they reached 50% confluency. Then the growth media was supplemented with 0.5 mM mimosine and the cells were incubated for another 13 h. After that the cells were transfected with 10 µg of plasmid with lipofectin (Invitrogen) according to the manufacturer’s protocol except that all the media was supplemented with 0.5 mM mimosine. Cells were lysed 12 h after transfection and replication intermediates were isolated and analyzed by 2D electrophoresis as described below.
Flow cytometry analysis
Cells were washed with PBS, harvested, fixed with 70% ethanol, and stained in PBS buffer (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.47 mM KH2PO4) supplemented with 50 µg/ml propidium iodide, 01% triton X-100, and 0.1mg/ml RNAse A. Samples were analyzed at the Penn State Flow Cytometry Facility.
Isolation of replication intermediates
The isolation of the intermediate products of replication was performed by modified Hirt extraction protocol (29). Briefly, the transfected cells growing on one 100 cm2 plate were washed with TBS (50 mM Tris–HCl pH 7.0, 150 mM NaCl), and lysed by incubation for 15–25 min at room temperature in 0.75 ml of lysis solution (50 mM Tris–HCl pH 7.0, 20 mM EDTA, 10 mM NaCl, 10% SDS, 0.2 mg/ml proteinase K). Then chromosomal DNA was precipitated by the addition of 0.187 ml of 5M NaCl followed by incubation for 8–24 h at 4°C. Then the chromosomal DNA was precipitated by centrifugation for 50 min at 27 200g at 4°C. The supernatant was collected, supplemented with 5 µl of proteinase K (20 mg/ml), and incubated at 55°C for 2 h. DNA was further purified by phenol/chloroform and chloroform extraction followed by precipitation with isopropanol. The pellets were washed with 70% ethanol, vacuum-dried, and dissolved in TE buffer.
Analysis of replication progression by 2D neutral–neutral agarose gel electrophoresis
Separation of replication intermediates was performed as described in reference (30). Each sample was separated on first dimension 0.4% agarose gel at 1 V/cm for 13 h followed by second dimension electrophoresis at 4°C, 5 V/cm for 8–13 h.
The gel was transferred to Zeta-probe nylon membrane (BioRad) according to manufacturer’s instructions and hybridized with a 32P-labeled radioactive probe corresponding to the fragment of interest.
Detection of semimethylated DNA
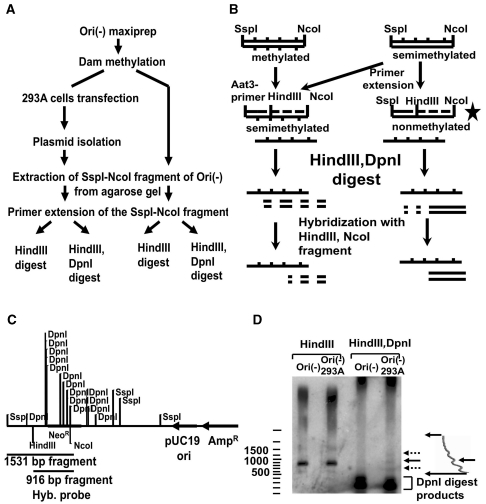
We developed the following method for the detection of small amounts of semimethylated plasmid (outlined in Figure 5A). Ori(-) plasmid isolated from E. coli was methylated with dam methylase according to the manufacturer’s instructions. Then the 293A cells were transfected with the methylated Ori(-) plasmid, and lysed in 12 h. The plasmid DNA was isolated from the 293A cells by the same protocol we used for the analysis of replication intermediates. The methylated plasmid and the plasmid isolated from the 293A cells were digested with SspI and NcoI, and the 1531-bp fragment was extracted from the gel (Figure 5C). From each of the extracted fragments, 0.1µg was mixed with 0.3 µM of the Aat5 primer, which anneals to the 5′ part of the fragment (Figure 5B), and supplemented with 0.3 mM dNTPs, 1× Pfx amplification buffer, 1× PCRx Enhancer solution, 1 mM MgSO4, and 1 unit of Platinum Pfx DNA polymerase (Invitrogen). The reaction mixture was denatured for 4 min at 94°C, and primer was extended for 2 min at 68°C. The reactions were purified by phenol/chlorophorm extractions followed by ethanol precipitation, and digested overnight with either 10 units of HindIII or 10 units of HindIII together with 10 units of DpnI. The digests were analyzed by 1% agarose gel electrophoresis followed by Southern hybridization with a radiolabeled HindIII-NcoI fragment.
Figure 5.
(A) The scheme of experiment on detection of semimethylated replication intermediates of Ori(-) isolated from 293A cells. (B) The two different fates of methylated and semimethylated fragments in our experiment. The DpnI resistant fragment is shown with a star. (C) The restriction map of Ori(-) plasmid. The DpnI sites are only shown inside the 1531-bp SspI-NcoI fragment. (D) The presence of DpnI-resistant fraction in the plasmid isolated from mammalian cells. The Ori(-) plasmid isolated from 293A cells is partially DpnI-resistant (see ‘Materials and methods’ section for the detailed description of the assay). The samples in lanes 1 and 3 were obtained from the plasmid isolated from bacteria. The samples in lanes 2 and 4 were isolated from transiently transfected 293A cells. The fragments were separated on 1% agarose gel and analyzed by Southern-blot hybridization with HindIII-NcoI fragment of Ori(-) plasmid. The 916-bp HindIII-NcoI fragment is shown by a solid arrow. The DpnI digest products are shown by the bracket. The fragments shown by the dotted arrows correspond to the partially denatured fragment and the primer extension product of the self-annealed single-stranded fragments. The densitogram of the region of lane 4 is shown on the right.
Depletion of RAD51 in HeLa cells
shRNAs against human RAD51, expressed from the lentiviral vector pLKO.1, were purchased from Open Biosystems as bacterial clones. Vectors were isolated using endotoxin-free kit (Sigma) and packaged as viruses by co-transfection into 293T cells with the packaging plasmid pCMV-dR8.2 dvpr and envelope plasmid pCMV-VSVG, as previously described (31). Viral supernatants were collected, filtered through 45-micron filters and stored at −80°C until use. The ability of each shRNA to deplete its target protein was determined by transducing human fibroblasts or HeLa cells with a specific shRNA or control lentivirus over 24 h, followed by puromycin selection (1.5 μg/ml) for 4–5 days. The resulting puromycin-resistant cells were then screened for protein depletion by western blotting with anti-RAD51 antibodies purchased from Millipore (clone 3C10) and Calbiochem (Ab-1). RAD51-depleted cells were further characterized (J.M. Sidorova et al. submitted for publication). Human CHK1 protein was visualized on the same western blots (using an antibody purchased from Santa Cruz, clone G-4) as an internal control for loading. Depletion levels were quantified on PhosphoImager scans of western blots using ImageQuant software. We chose the representative depletor shRNA clone TRCN0000018877 for further experiments.
To determine whether the first replication cycle was affected by RAD51, HeLa cells were infected with shRNA-expressing pLKO.1, puromycin selected and then lipofectin-transfected with the pUCneo plasmid as described above.
Micrococcal nuclease digestion
Digestion was performed by the following protocol. The 50% confluent cells from a single 100-mm-diameter plate were washed with 1× PBS three times, collected, and centrifuged at 250 g for 5 min. The pellets were resuspended in 2 ml HB buffer (10 mM Tris–HCl pH 7.6, 10 mM KCl, 1.5 mM MgCl2, 1 mM DTT, and 1% Triton X-100), incubated on ice for 10 min, and centrifuged at 2000g for 10 min. The pellets were resuspended in 1.5 ml N2 buffer (10 mM HEPES pH 7.4, 5 mM MgCl2, 1 mM CaCl2, 10% glycerol, 0.1 mM PMSF, and 0.5 mM DTT), and CaCl2 was added up to 5 mM. The sample was divided into 200 µl aliquots to be used in Micrococcal nuclease (MNase) reactions. Different amounts of MNase were added to each aliquot and incubated at 37°C for 10 min before the digestion reaction was stopped with 200 µl of MNase stop buffer (20 mM EDTA, 200 mM NaCl, and 1% SDS). Then the samples were treated with 10 µl of 10 mg/ml RNase A for 1 h followed by incubation with 0.25 mg/ml proteinase K for 2 h at 37°C. The samples were purified by phenol/chloroform extraction, precipitated with ethanol, and dissolved in 20 µl of TE buffer. The samples were separated on 1% agarose gel, transferred to the nylon membrane and hybridized with the AflIII fragment of the pUCneo plasmid containing the ampicillin resistance gene.
RESULTS
Bimodal replication during the first 2–3 days after transfection
To study the first replication cycle in mammalian cells and compare it to subsequent cycles, we used an SV40-based plasmid, pUCneo (Figure 1A), that contains the SV40 replication origin for the initiation of plasmid replication in mammalian cells expressing T-antigen (32). This plasmid is comprised of bacterial and SV40 sequence elements (see ‘Materials and methods’ section for cloning details). We studied the first and subsequent replication cycles of this plasmid by isolating replication intermediates at different time points after transfection and resolving them by 2D gel electrophoresis. This approach has been applied to SV40-based plasmid replication before (33); however, researchers never focused on the first replication round after transfection. The subsequent rounds of SV40-based replication were shown to originate at the SV40 origin through its interaction with T-antigen (33).
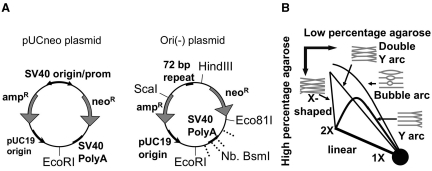
Figure 1.
(A) The restriction maps of pUCneo and Ori(-) plasmids. The sites of the single-stranded breaks generated by Nb. BsmI are shown by dashed lines. (B) The schematic presentation of the possible arcs, which can be generated by 2D-gel electrophoresis of the digested plasmids.
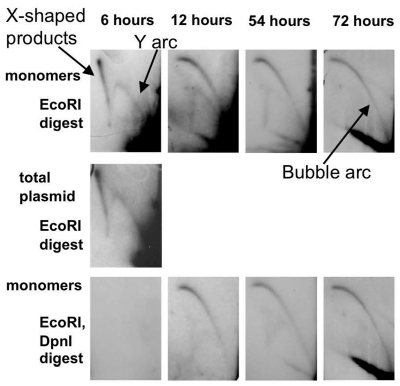
The pUCneo plasmid replication intermediates were digested with EcoRI restriction endonuclease at a unique site opposite the origin (Figure 1A), and separated by 2D electrophoresis. The replication intermediates initiated at the SV40 origin were expected to form a bubble arc on the 2D gel image (Figure 1B and Supplementary Figure 1C, left). However, during the first 2 days after transfection we observed an unexpected pattern, corresponding to replication initiated outside of the SV40 origin (Figure 2, upper panels). In 6 h after transfection, we observed a Y arc, while a bubble arc was not visible. At 12–54 h, a composite pattern consisting of both a Y and a bubble arc was observed. After 72 h transfection, we could detect a bubble arc, but not the Y arc. The presence of the Y arcs on the gel indicated that replication initiated close to the ends of the linearized plasmid fragment and not at the SV40 origin (30) (Supplementary Figure 1C, right). Additionally, we observed an arc formed by X-shaped intermediates (Figures 1B and 2). These could correspond to the digested products of intermolecular recombination or hemicatenation, and were most abundant 6 h after transfection when the concentration of the plasmid in the cell was at its highest.
Figure 2.
The 2D separation of replication intermediates of pUCneo plasmid isolated at 6, 12, 54 and 72 h after Cos-1 cells transfection. Upper panels: replication intermediates digested with EcoRI opposite to the origin. Purified monomers of pUCneo were used for transfection. Middle panel: replication intermediates isolated 6 h after transfection and digested with EcoRI opposite to the origin. Total plasmid sample that consisted of monomers, dimers, and other multimers, was used in this transfection. Lower panel: replication intermediates digested with EcoRI and DpnI. Replication pattern gradually switches from random (Y arc) to origin-based (bubble arc). The Y, bubble and X-shaped recombination products arcs are shown with arrows.
It was possible that the Y arc was caused by the presence of multimerized plasmids (Supplementary Figure 1C, middle). If the SV40 origin fired in one part of a dimer, or a multimer, the replication forks could enter the EcoRI fragment from the end resulting in a Y arc (Supplementary Figure 1C, middle). To avoid this, we used gel-purified plasmid monomers for this and other experiments.
To additionally confirm that the Y arc we observed was not caused by the trace amount of dimers left in the plasmid sample after gel extraction, we compared the replication of the gel-purified monomer plasmid (Figure 2, left-upper panel) with the replication of total plasmid DNA, purified from E. coli (Figure 2, middle panel). The total plasmid DNA sample contained ∼20% dimers and other multimers (data not shown), which greatly exceeded the trace amount that could be present in a gel-purified monomer plasmid sample. The intensity of the Y arc seen in the total plasmid sample was very close to that in the monomer-only sample, hence the replication of plasmid dimers could not account for the prevalence of Y arc shortly after transfection.
We next asked whether the observed Y arc resulted from the first or subsequent replication round by digesting replication intermediates with restriction endonuclease DpnI, which cleaves DNA at the methylated GATC sequence (Figure 2, lower panels). GATC methylation occurs in E. coli, but not in mammalian cells (34). Plasmid DNA that went through two or more replication rounds becomes DpnI-resistant when both of its strands are synthesized in mammalian cells. The plasmid that went through a single-replication round is semimethylated since only one of its strands was synthesized in mammalian cells. Since methylated and semimethylated plasmids are cleaved by DpnI, non-replicated plasmid and the first replication cycle’s intermediates are digested by DpnI, while the second and further replication cycles’ products persist.
When we digested replication intermediates from different time points after transfection with DpnI, the Y arc mostly disappeared, but the bubble arc persisted (Figure 2, lower panels). Thus, Y arc intermediates either preexisted in plasmid DNA isolated from bacteria, or originated from the first round of replication in mammalian cells. The following considerations make it unlikely that the Y arc intermediates pre-existed in plasmid DNA used in transfections. First, no Y arc intermediates were detected by 2D-gel analysis in plasmid DNA samples prior to transfection (data not shown), and it is unlikely that any plasmid replication intermediates can survive the alkali lysis procedure used to generate plasmid DNA. Second, the unusual replication that we observed was not initiated at the plasmid ColE1 origin, as will be discussed below.
In summary, the 2D data suggested that upon transfection, the pUCneo plasmid was initially replicated by an alternative replication mode that was not initiated at the SV40 origin. This alternative initiation starts before the regular, SV40-initiated mode, coexists with it for a few days, and subsides 3 days after transfection.
The SV40 replication origin and T-antigen are not necessary for the first replication cycle
Since the first replication cycle did not initiate at the SV40 origin, we decided to confirm that the SV40 replication origin and T-antigen were not required for it. To this end, we analyzed the replication of pUCneo derivative, Ori(-) plasmid, in which all of the SV40 origin except for the 72-bp repeat element was deleted (Figure 1A). The 72-bp repeat contains a nuclear localization sequence (NLS), which increases the delivery of plasmids into the cell nucleus (35) but is insufficient for T-antigen-initiated replication (36).
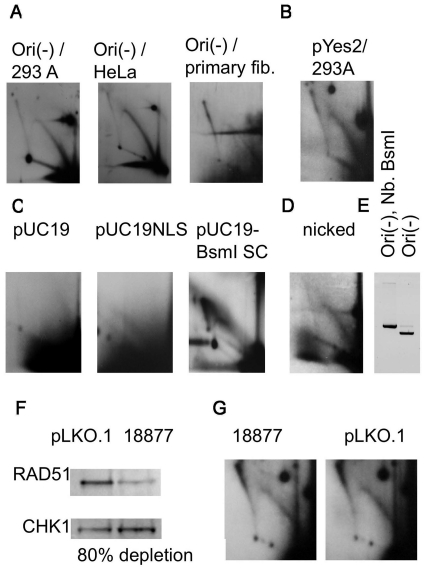
We transfected 293A and HeLa cell lines that did not contain T-antigen with Ori(-) plasmid and still detected Y arc intermediates in the 2D-replication patterns (Figure 3A). Hence, the first replication cycle relies on mammalian cell components and is independent of the SV40 virus replication origin or T-antigen. Consistent with this, we also detected Y arcs in primary fibroblasts (Figure 3A, right), which confirmed that this mode of replication was not an artifact of cell transformation.
Figure 3.
(A) The first round of replication can take place in different cell lines. The 2D electrophoresis of the replication intermediates isolated from 293A, HeLa, and primary fibroblasts transfected with Ori(-) plasmid and digested with EcoRI. The horizontal smear in the primary fibroblast picture is not a part of a pattern; it resulted from the overexposure due to the low amount of intermediates. (B) The 2D electrophoresis of the replication intermediates isolated from 293A cells transfected with pYES2 plasmid. The intermediates were digested with NdeI, which cuts pYES2 at a unique site located in the middle of the 2-µ replication origin. (C) pUC19 plasmid is only capable of replicating in mammalian cells when it is highly supercoiled. The 2D electrophoresis of the replication intermediates isolated from 293A cells transfected with pUC19 or highly supercoiled pUC19-BsmI plasmids. The plasmids were digested at a unique HindIII site. (D) Non-canonical replication is not caused by single-stranded breaks in the plasmid. The 2D electrophoresis of the replication intermediates isolated from 293A cells transfected with Ori(-) plasmid, treated with nicking endonuclease Nb.BsmI. Replication intermediates were digested with HindIII, which cuts the plasmid opposite to the single stranded breaks introduced by Nb.BsmI. (E) Ori(-) was completely converted into an open circular form by Nb. BsmI. (F) Western blot confirms depletion of RAD51. CHK1 was used as an internal control for loading (see ‘Materials and methods’ section). pLKO.1, parental vector containing no shRNA. 18877, abbreviated ID number for the RAD51-depleting shRNA. Protein levels were quantified using ImageQuant software. RAD51 abundance was normalized to CHK1 abundance. Depletion level (below the image) was calculated as a difference between normalized RAD51 levels in pLKO.1 and 18877 cell extracts. (G) Alternative replication mode persists in RAD51-depleted HeLa cells. Replication intermediates of pUCneo were isolated from RAD51-depleted and control cell lines 6 h after transfection and digested with HindIII. Designations are as in (F).
The hypothesis that the Y arc originated from the first cycle of replication was consistent with our observation that the replication arc of Ori(-) plasmid in 293A cells was only visible for the first 24 h (Figure 4A). We did not observe any DpnI-resistant fraction in the Ori(-) samples isolated from mammalian cells, confirming that the Ori(-) plasmid does not contain a replication origin for stable maintenance in the mammalian cell (Figure 4C).
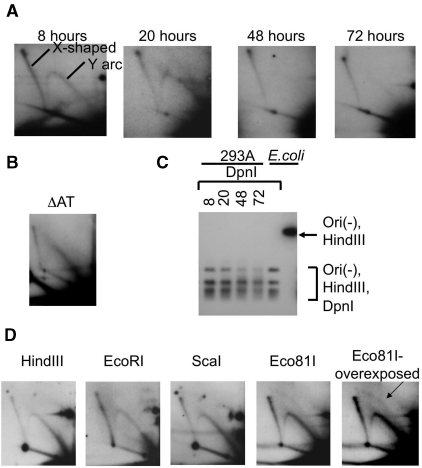
Figure 4.
(A) The 2D electrophoresis of the replication intermediates isolated from 293A cells transfected with Ori(-) plasmid. Replication intermediates were isolated at different time points after transfcetion (8, 20, 48 and 72 h). Replication was only visible during the first 24 h. (B) The A/T-rich region in the plasmid is not responsible for non-canonical replication. Replication intermediates of ΔAT plasmid plasmid were isolated from HeLa cells 6 h after transfection, and digested with HindIII. (C) Ori(-) plasmid does not go through more than one replication round. Lanes 1–4. Replication intermediates of Ori(-) plasmid isolated from 293A cells in 8, 20, 48 and 72 h after transfection (also analyzed in Figure 4A) were digested with HindIII and DpnI enzymes. Lanes 5 and 6. Ori(-) plasmid isolated from E. coli digested with HindIII and DpnI, or with HindIII alone. Ori(-) plasmid isolated from mammalian cells did not show any DpnI-resistance. (D) Random initiation of the alternative replication mode. The 2D electrophoresis of the replication intermediates isolated from 293A cells transfected with Ori(-) plasmid and digested with different restriction endonucleases. A small amount of bubble arc is visible in the overexposed picture for Eco81I digest (indicated by an arrow).
We analyzed other unrelated plasmids for their ability to go through the first replication cycle. The yeast shuttle plasmid pYES2 also replicated in HeLa cells (Figure 3B). This plasmid does not contain a mammalian replication origin, and is not capable of long-term replication in mammalian cells (37).
However, we were not able to detect the replication arc when we transfected HeLa cells with pUC19 plasmid (Figure 3C, pUC19). pUC19 is a short (2.8 kb) plasmid comprised entirely of bacterial DNA; 87% of its sequence is also a part of the Ori(-) plasmid. Cloning the NLS-containing 72-bp repeat in pUC19 still did not allow us to observe a complete Y arc (Figure 3C, pUC19NLS plasmid); however, the ascending part of the Y arc could be detected at very long exposures. The absence of the characteristic Y arc for pUC19 can serve as independent confirmation that the arc is indeed generated by replication in mammalian cells, and not by bacterial replication.
The efficiency of the alternative replication is increased by superhelicity
The plasmid pUC19 is about twice as short as the Ori(-) plasmid (5.6 kb), and characterized by the same superhelical density; therefore it has twice fewer supercoils than Ori(-). Since local unwinding of the double helix is facilitated by negative supercoils, we speculated that increased superhelicity could promote the random replication of pUC19. To this end, we generated a highly negatively supercoiled sample of pUC19. First, we generated a pUC19-BsmI plasmid with a unique site for nicking endonuclease BsmI (see ‘Materials and methods’ section). Then we increased superhelical density of the pUC19-BsmI by nicking it with BsmI and re-ligating the nick in the presence of 6 µg/ml of ethidium bromide. This results in highly supercoiled DNA with a superhelical density of ∼−0.6 (28), which can promote the melting of the DNA duplex and the generation of permanently single-stranded stretches of DNA (38). For this highly supercoiled plasmid, we were able to observe a Y arc consisting of both ascending and descending parts (Figure 3C, pUC19-BsmI SC). We concluded that the efficiency of the alternative replication was strongly increased by the single-stranded regions in template DNA.
The first replication round is not stimulated by damaged DNA and does not depend on homologous recombination
It is possible that the Y arc that we observed is produced by repair replication initiated at sites of DNA damage, such as single or double-strand breaks. If single-strand breaks were a requirement, the first replication round would be greatly enhanced at nicked DNA. To test this possibility, we treated our Ori(-) plasmid with the nicking endonuclease Nb. BsmI and confirmed by agarose gel that the superhelical form was no longer visible (Figure 3E). BsmI generates three single-stranded breaks in Ori(-) located within 1500 bp. We transfected HeLa cells with the nicked plasmid sample and analyzed its replication intermediates digested with HindIII on a 2D gel. The Y arc was visible for this open circle form (Figure 3D), but it was somewhat more diffuse than the arc generated by transfection with the supercoiled plasmid (Figure 3A). We did not observe an increase in the efficiency of the first replication cycle in the presence of single-stranded breaks; thus we concluded that single-stranded breaks in the plasmid could not account for the random-replication mode.
Another explanation for the Y-arc intermediates observed within 6 h of transfection could be that these represent replication that takes place during the repair of double strand breaks by homologous recombination. Specifically, they might represent break-induced replication, which is thought to enable the synthesis of long (>Kb) stretches of DNA in yeast (39). If so, the presence of Y-arc would depend on the DNA strand invasion carried out by the RAD51 protein (40). We have established depletion of RAD51 in human cells using lentiviral delivery of short hairpin (sh) inhibitory RNAs (see ‘Materials and methods’ section). We achieved 80% depletion (Figure 3F), which correlates with slow growth, prolonged G2 phase, and other phenotypes previously described for RAD51 deficiency (J.M. Sidorova et al., submitted for publication). Higher depletion levels are likely to result in cell death since RAD51 is an essential protein.
We transfected HeLa cells depleted of Rad51 and mock-depleted controls with pUCneo and isolated the plasmids 6 h later. We detected comparable levels of Y intermediates in RAD51-depleted and control samples (Figure 3G). Thus, it is unlikely that the first replication round is a manifestation of homologous recombination repair.
A large A/T-rich region of Ori(-) is not required for the non-canonical replication
It is conceivable that long A/T-rich regions present in pUCneo and Ori(-) promoted the first replication cycle. For example, this region is not present in pUC19 that does not go through the first replication cycle. We therefore deleted the highest A/T-content region in the Ori(-) plasmid (from Bsp119I to EcoRI; 60% A/T versus 40–50% in the rest of the sequence) (Supplementary Figure 4E), and analyzed the replication of the resulting deletion derivative, ΔAT. We found that the first round of replication still took place in ΔAT (Figure 4B). We concluded that A/T content was not critical for the non-canonical first replication cycle.
An alternative replication mode initiates throughout transfected DNA
We next searched for specific replication initiation points on the Ori(-) plasmid. To this end, we transfected 293A cells with the Ori(-) plasmid, which did not contain the SV40 replication origin. Ori(-) replication intermediates were digested at either of four unique restriction sites (HindIII, EcoRI, ScaI or Eco81I) more or less evenly distributed along the plasmid (Figure 1A), and analyzed for the presence of the bubble arc (Supplementary Figure 1C, left). If replication preferentially initiated at a particular site, we would detect a strong bubble arc for one of the digests. However, all of the digests resulted in similar patterns on 2D gels that mostly consisted of Y arc (Figure 4D). A small amount of bubble arc could be observed in overexposed pictures of each of the digests (only shown for Eco81I, right panel). Since each of the sites produced a similar pattern, we concluded that the plasmid’s first replication round does not initiate at a particular origin site at the plasmid but rather at positions scattered throughout its sequence.
Ori(-) DNA isolated from 293A cells is partially semimethylated at GATC sequence
We used an alternative approach to confirm that the Ori(-) plasmid isolated from the mammalian cells contains some molecules that went through at least one replication round. We took advantage of the fact that DpnI restriction endonuclease digests the DNA that has been methylated at GATC by E. coli dam methylase, but does not digest non-methylated DNA. DpnI also digests semimethylated DNA, although with lower efficiency. The DNA that went through a single-replication round in a mammalian cell is semimethylated, and therefore is not resistant to DpnI.
Since DpnI digests the products of the first replication cycle, we developed a method to detect a small amount of semimethylated DNA. We used primer extension to synthesize non-methylated DNA, which is resistant to DpnI (Figure 5A). The DNA isolated from E. coli contains some fraction of semimethylated plasmid, which can be explained by the delay of dam methylation relative to replication in E. coli. This delay is utilized by the bacterial cell to distinguish between the maternal and daughter strands for the purpose of mismatch repair (41). Thus, to ensure that we started out with a completely methylated DNA sample, we treated the Ori(-) plasmid with dam methylase before transfections.
To detect plasmid DNA that replicated in mammalian cells, the 293A cells were transfected with dam-methylated Ori(-) (initial sample), and plasmid DNA was isolated (293A sample). We next digested the initial and 293A samples with NcoI and SspI (Figure 5A and B) and extracted the 1531-bp fragment from the agarose gel (Figure 5C). We expected that the NcoI-SspI fragment from the initial sample was fully methylated and the same fragment from the 293A sample contained a small fraction of semimethylated DNA, generated by replication in mammalian cells. Next, both fragments were mixed with the Aat5 primer, which annealed close to the SspI side of the fragment, and the primer was extended (Figure 5B). Primer extension is expected to convert any semimethylated DNA present in the sample to a non-methylated form, which would be resistant to DpnI (Figure 5B, indicated by a star), and the bulk of methylated DNA into semimethylated form, still susceptible to DpnI.
Other products that can result in DpnI resistant species in our assay are the single-stranded NcoI-SspI fragment since the DpnI does not digest single-stranded DNA (Figure 5B), and a misannealed fragment (data not shown). In order to discriminate between double-stranded DpnI-resistant DNA and single-stranded or misannealed molecules, we digested our DNA samples with HindIIII along with the DpnI enzyme. Only HindIII-digested, DpnI-resistant species correspond to DNA molecules that underwent a round of replication in mammalian cells.
We separated the digested samples on agarose gel followed by Sothern hybridization with HindIII-NcoI radiolabeled fragment (Figure 5C). The plasmid sample isolated from mammalian cells (Figure 5D) showed a DpnI-resistant band at the position of the 916-bp HindIII-NcoI fragment (Figure 5D, lanes 1 and 2, shown by a solid arrow). The two other bands shown with the dotted arrows most likely corresponded to the single-stranded and misannealed DNA. The presence of the band at the position of the HindIII-NcoI fragment in the sample isolated from mammalian cells (Figure 5D, lane 4), but not in the control (Figure 5D, lane 3), confirmed that the plasmid went through at least one replication round in mammalian cells.
Origin-independent replication can take place outside of the S phase
Since the initiation of the random replication mode preceded the T-antigen-driven initiation event at the SV40 replication origin, we suspected that random replication might take place even before the S phase. We used mimosine, a replication initiation inhibitor that is known to arrest cells at the G1/S border (42). Mimosine addition to the cells does not allow the initiation of new replication forks while the existent replication forks still complete replication (43). Mimosine prevents the initiation of both genomic and SV40-based replication initiation (44), thus the absence of the bubble arc from the SV40 origin can be used as a control that the mimosine indeed blocked the initiation. To confirm that genomic replication was severely inhibited in the presence of mimosine, we analyzed the cell-cycle distribution of mimosine-treated and control cultures by flow cytometry. The mimosine-treated culture was mostly in G0/G1, and the fraction of cells in S-phase was considerably lower than in the control culture.
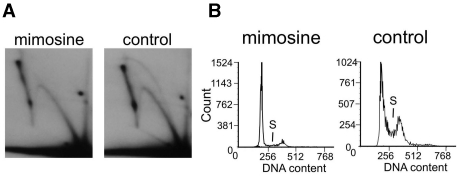
We next investigated the initiation of replication in the SV40-based pUCneo plasmid 12 h after transfection when both the random and the SV40 origin-initiated replication modes take place. We transfected Cos-1 cells incubated with mimosine for 13 h with pUCneo. A control culture of Cos-1 cells that was not treated with mimosine was transfected in parallel. Replication intermediates from the mimosine and control samples were isolated and digested with EcoRI at the site opposite the SV40 origin. The separated intermediates from the control sample (Figure 6, right) formed a Y and a bubble arc, while only a Y arc was present upon mimosine treatment (Figure 6, left). The SV40-based replication was limited to S phase (45) and was not able to initiate under mimosine treatment. However, the Y arc resulting from the random-replication mode was not affected by mimosine (Figure 6, left). The initiation of the random-replication mode in the presence of mimosine is indicative of an alternative mechanism of initiation, which does not involve some of the factors disabled by mimosine treatment. Mimosine is known to chelate zinc and other metals, which may be important for some of the factors that participate in replication initiation (46). The presence of this alternative initiation in G1/G0 phases is also intriguing since it somehow overrides the usual constraints for origin firing until the beginning of S phase. The above observations are indicative of the replication mechanism missing some of the key players of the regular S phase replication fork.
Figure 6.
Unusual replication also takes place in the cells synchronized at G1/S border with mimosine. (A) The 2D electrophoresis of the replication intermediates isolated from Cos-1 cells, which were incubated in the media containing 0.5 mM mimosine and transfected with pUCneo plasmid. Replication intermediates were digested with EcoR1 opposite to the replication origin. In the presence of mimosine only the Y arc corresponding to random replication is visible, while in the absence of mimosine both Y arc and bubble arc is present. (B) Cell cycle analysis by flow cytometry shows that mimosine-treated culture mostly contains cells in G0/G1 phase, and the number of cells in S-phase is significantly less than that in the control.
The timing of the first replication cycle coincides with the nucleosome structure formation
The first replication round of the zygote occurs in the presence of immature chromatin. We analyzed the chromatin structure of the pUCneo plasmid to confirm that in the timeframe of the first replication cycle the chromatin structure of the pUCneo was still at the formation stage. We compared the chromatin structure of the pUCneo plasmid 8 h after transfection when alternative replication occurred, and 72 h after transfection when we could no longer detect an alternative replication.
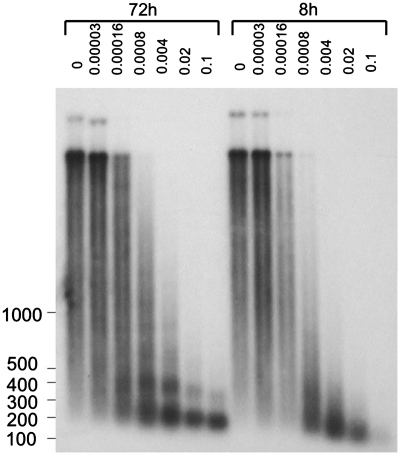
The chromatin structure was analyzed by a micrococcal nuclease (MNase) digest of intact chromatin in situ. MNase induces double-stranded breaks in inter-nucleosomal linker regions, but only single-stranded breaks in DNA within a nucleosome. Cos-1 cells were treated with varying amounts of MNase 8 and 72 h after transfection, and samples were analyzed by agarose-gel electrophoresis followed by Southern hybridization (Figure 7). The treatment of cells 72 h after transfection with 1.6 × 10−4 to 4 × 10−3 units of MNase resulted in a ladder of bands corresponding to two, three or more nucleosomes. This ladder could be interpreted as a regular nucleosome structure of the plasmid. At 8 h, we could only observe some protection by single nucleosomes. We concluded that 8 h after transfection, chromatin started to assemble in nucleosomes but did not yet form a regular structure. The timing of chromatin formation roughly coincided with the timing of the random-replication mode. We suggest that the first replication cycle takes place on naked DNA and is not initiated once the plasmid is covered by nucleosomes.
Figure 7.
Chromosomal structure of the plasmid is more regular in 72 than in 8 h after transfection. Cos-1 cells were transfected with pUCneo plasmid and digested with different amounts (0.000032, 0.00016, 0.0008, 0.004, 0.02 and 0.1 units) of micrococcal nuclease in 8 and 72 h. The plasmid samples were isolated and resolved on 1% agarose gel followed by Southern hybridization with the AflIII fragment of pUCneo. Undigested plasmid sample: lane 1 (72 h), lane 8 (8 h). The ladder fragments’ sizes are shown on the left.
The first replication cycle, but not subsequent cycles, is affected by the presence of the fragile site FRA16B
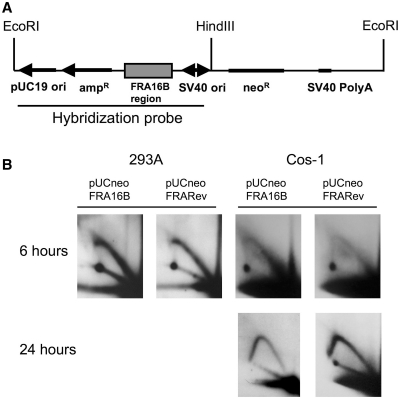
We analyzed how replication was affected by the FRA16B sequence of a rare fragile site. We cloned the FRA16B sequence in the pUCneo plasmid in direct and reverse orientation relative to the SV40 origin to obtain the pUCneoFRA16B and pUCneoFRARev plasmids. Cos-1 and 293A cells were transfected with these plasmids and replication progression through FRA16B was analyzed by 2D-gel electrophoresis. To observe how replication progressed through FRA16B, we digested replication intermediates isolated from Cos-1 cells with HindIII and EcoRI enzymes. This digest resulted in a fragment with the FRA16B spanning a 1.0-kb region in its second quarter (Figure 8A). The first replication cycle was analyzed in both 293A and Cos-1 cells that were lysed 6 h after transfection. Subsequent replication cycles were analyzed in Cos-1 cells lysed 24 h after transfection. In order to concentrate exclusively on the subsequent cycles, replication intermediates isolated from Cos-1 cells were also digested with DpnI, which eliminated the products of the first replication cycle.
Figure 8.
Random replication cannot progress through FRA16B region. (A) The scheme of pUCneoFRA16B plasmid. (B) The 2D electrophoresis of the replication intermediates isolated from 293A or Cos-1 cells transfected with pUCneoFRA16B or pUCneoFRARev plasmid. The pUCneoFRARev plasmid contains the FRA16B repeat in the reverse orientation. The intermediates isolated from 293A cells were isolated 6 h after transfection, and digested with EcoRI and HindIII. Replication intermediates isolated from Cos-1 cells 6 h after transfection were digested with EcoRI and HindIII; those isolated 24 h after transfection were digested with EcoRI, HindIII and DpnI.
We observed that FRA16B had a very strong effect on the first round of replication in both orientations and in both cell lines. The presence of the FRA16B sequence leads to the disappearance of a subset of the large Y replication intermediates, suggestive of a decreased ability of forks to traverse the whole EcoRI-HindIII segment. Interestingly, however, FRA16B had no effect on replication initiated at the SV40 origin (Figure 8B). We expect that the Y arc in Figure 8 is a result of the superposition of the two Y-arcs from two replication forks entering the fragment from the EcoRI and HindIII ends. We did not observe a distinct replication-stalling site, but rather a termination of the arc within the FRA16B region. Since termination of the leftward replication fork occurred within the second half of the fragment, the interruption of the Y arc was observed at the beginning of its descending part.
The initiation of the alternative replication mode in the proximity of FRA16B would result in its termination, which could lead to chromosome breakage at this region. However, this only happens when the alternative replication mode is involved in the replication of FRA16B. This finding indicates that alternative replication through A/T-rich microsatellite repeats may promote chromosomal abnormalities.
DISCUSSION
This work demonstrated for the first time that SV40 origin-based plasmids widely used as a model system for replication studies exhibit bimodal replication in mammalian cells. Along with the SV40 origin-initiated replication driven by T-antigen, there is another mode, which starts shortly after transfection. This mode is independent of the SV40 origin or T-antigen, and takes place in all cell lines studied here, including the skin primary fibroblasts. This origin-independent mode is capable of completing only one full round of plasmid replication. Two, three days after transfection, the efficiency of the random-replication mode significantly decreases.
A possible explanation for the decline of random replication over time is the gradual formation of chromatin on plasmid DNA. It was shown that episomal plasmids in mammalian cells were covered with histones (47). The formation of chromatin was speculated to cause an initially high level of transgene expression to drop 2 days after transfection (48,49). Our experiments confirmed the presence of chromatin on the transfected plasmids. The timing of the first replication cycle of the plasmid coincided with the time when chromatin was still in the process of formation. The fact that the replication process itself can promote the deposition of chromatin (1) can account for the observation that the alternative replication only goes through one cycle on plasmid DNA.
Examples of the random initiation of replication were previously reported for both plasmids and chromosomes (26,50). The most studied case is the random-replication initiation in Xenopus eggs and egg extracts (26). Plasmid DNA of any origin can be replicated in eggs extract, and specific initiation and termination sites were not observed (10,51,52). However, only one initiation site per plasmid was active in each replication round (26). Replication of salmon sperm DNA also initiated randomly in Xenopus extracts and eggs (10). Some preference to initiation at A/T-rich symmetrical sequences was detected in the replication of phage λ DNA in Xenopus extracts (53). However, this preference was not observed in the initiation of replication in Xenopus eggs (52). In our experiments the efficiency of the random-replication mode was not significantly affected by the deletion of a long-A/T-rich region from the plasmid.
Another example of random-replication initiation came from experiments selecting for the sequences that can support replication of plasmids in mammalian cells (54). It turned out that, combined with an element that provides the proper segregation of the plasmid, sufficiently long stretches of mammalian genome can support long-term maintenance of the plasmid inside the cell (18). It also turned out that these plasmids do not possess a specific replication origin, but rather that replication is initiated at random positions along the plasmid (55). The plasmids with the deleted SV40 origin that we studied here did not contain any mammalian genome sequences, and were not maintained in mammalian cells. Some of these plasmids contained strictly bacterial or yeast sequences. Nevertheless, they supported the randomly initiated first replication round.
Recent data revealed that the initiation of mammalian genomic DNA replication also involves some random origin selection. Replication in each consecutive S phase is initiated in the same areas, but the selection of specific initiation positions in these defined areas is random and never exactly the same (11). The switch in the initiation pattern to the less frequent has been suggested to happen due to changes in chromatin structure (8,9). Chromosome packaging and histone modifications were shown to affect origin selection (56). It appears that acetylation of histones favors the selection of an area as a potential origin (13). We propose that chromatin structure also inhibits the promiscuous replication of our plasmids.
Other factor that increased the efficiency of random replication was the superhelicity of the plasmid. We also noticed that the presence of the regions capable of adopting non-B DNA structures can promote the random-replication mode (data not shown). Both of these factors increase the likelihood of the existence of single-stranded DNA areas. We concluded that random initiation mostly happens on nucleosome-free DNA that contains single-stranded regions. Both of the above can take place during the first replication cycle of the zygote. The formation of non-B DNA structures could also be more plausible in the absence of mature chromatin in the zygote.
It is interesting that the first replication cycle seems to be much more sensitive to the obstacles in DNA than subsequent replication cycles of the plasmid. We showed that randomly initiated replication forks terminated at the FRA16B spontaneous fragile site, and were severely stalled at the GAA repeat (M. M. Krasilnikova, unpublished data). Both of these sequences are capable of forming non-canonical DNA structures. The next replication cycles of the plasmid are much less affected by the above structures, possibly because the formation of nucleosomes makes alternative structures more difficult to form. We speculate that the first replication cycle of the zygote, which happens in the presence of immature chromatin structure, can also be different from subsequent replication cycles, and encounter more frequent obstacles in the form of alternative DNA structures. The first mitosis is characterized by a high level of DNA damage, which might be a consequence of stalled replication forks (5). Since DNA polymerases stall at non-canonical DNA structures such as hairpins (57) and triplexes (58), causing an increase in their mutability, the first replication cycle of the zygote can be a major source of genomic instability. It can account for the observation that the changes in the number of repeats in the microsatellite DNA can be traced to the first divisions of the egg, when the cells are not yet specialized. After this initial jump in the repeat numbers, the subsequent changes during the following cell divisions can also take place, but they are much less drastic (6,59).
It is intriguing that origin-independent replication can occur in the cells outside of the S phase. This is indicative of the initiation mechanism being different from initiation at the mammalian chromosomal origins. The latest data point out the importance of microhomology-stimulated replication initiated at single-stranded regions (60). The sequences with the potential to support single-stranded DNA conformation are frequently found close to spots of increased mutability and chromosomal rearrangements, which often lead to diseases (60). For instance, many rearrangements found in cancers are in proximity of the A/T-rich sequences, similar to FRA16B, which caused a disruption in the first replication cycle of our plasmids (22).
What could be the mechanism of the alternative initiation we observed? It is clear that it is different from that involved in the regular duplication of the mammalian genome. We do not know which components of the replication fork are present at random initiation sites. Apparently, the absence of some of them can lead to a loss of control over the origin firing. For example, a mutation of Cdc6 in yeast promotes a promiscuous initiation pattern and overreplication of the genome (50,61).
There is also a possibility of origin-independent loading of the primase-polymerase α complex. The assembly of the primase-polymerase α complex on single-stranded DNA was previously reported; however, efficient replication of double-stranded plasmids was not observed (62). We speculate that superhelicity or alternative DNA conformations support a local unwinding of DNA, which results in the initial replication by primase-polymerase α. The recruitment of DNA polymerase α/primase can be facilitated by topoisomerase I, which binds to negatively supercoiled DNA (63). DNA polymerase α/primase is present in its active form at the end of the G1 phase (64), which can explain the presence of the random-replication mode in late G1. A switch to polymerase δ or possibly some other polymerase can follow the initial synthesis and bring the plasmid replication to completion. It is possible that Y-family DNA polymerases that carry out translesion synthesis are involved in alternative replication (65,66).
The initiation of promiscuous replication is limited to one round per plasmid, and is inhibited after this round is completed. This replication activity, which is extremely strong on the plasmid template, should be inhibited in genomic DNA. Promiscuous origin firing in the G1 phase can lead to overreplication of the genome, which can result in genomic instability and tumorigenesis (67). More research is necessary to determine the role of promiscuous replication in the instability of the genome, but this potentially damaging ready-to-go mechanism should certainly deserve further attention.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Penn State University Huck Institute (to M.K.); Summer Discovery grant (to G.C.). Funding for open access charge: Waived by Oxford University Press.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Jean Brenchley, Kristin Eckert, Kateryna Makova, Sergei Mirkin, Frank Pugh, Robert Schlegel and Yanming Wang for valuable suggestions.
REFERENCES
- 1.Bell S, Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 2.McLay D, Clarke H. Remodelling the paternal chromatin at fertilization in mammals. Reproduction. 2003;125:625–633. doi: 10.1530/rep.0.1250625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spinaci M, Seren E, Mattioli M. Maternal chromatin remodeling during maturation and after fertilization in mouse oocytes. Mol. Reprod. Dev. 2004;69:215–221. doi: 10.1002/mrd.20117. [DOI] [PubMed] [Google Scholar]
- 4.Imschenetzky M, Puchi M, Gutierrez S, Montecino M. Sea urchin zygote chromatin exhibit an unfolded nucleosomal array during the first S phase. J. Cell Biochem. 1995;59:161–167. doi: 10.1002/jcb.240590205. [DOI] [PubMed] [Google Scholar]
- 5.Derijck A, van der Heijden G, Giele M, Philippens M, de Boer P. DNA double-strand break repair in parental chromatin of mouse zygotes, the first cell cycle as an origin of de novo mutation. Hum. Mol. Genet. 2008;17:1922–1937. doi: 10.1093/hmg/ddn090. [DOI] [PubMed] [Google Scholar]
- 6.De Michele G, Cavalcanti F, Criscuolo C, Pianese L, Monticelli A, Filla A, Cocozza S. Parental gender, age at birth and expansion length influence GAA repeat intergenerational instability in the X25 gene: pedigree studies and analysis of sperm from patients with Friedreich's ataxia. Hum. Mol. Genet. 1998;7:1901–1906. doi: 10.1093/hmg/7.12.1901. [DOI] [PubMed] [Google Scholar]
- 7.Mirkin S. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 8.Hyrien O, Maric C, Mechali M. Transition in specification of embryonic metazoan DNA replication origins. Science. 1995;270:994–997. doi: 10.1126/science.270.5238.994. [DOI] [PubMed] [Google Scholar]
- 9.Sasaki T, Sawado T, Yamaguchi M, Shinomiya T. Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolalpha-dE2F locus of Drosophila melanogaster. Mol. Cell Biol. 1999;19:547–555. doi: 10.1128/mcb.19.1.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blow J, Laskey R. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- 11.Maya-Mendoza A, Tang C, Pombo A, Jackson D. Mechanisms regulating S phase progression in mammalian cells. Front Biosci. 2009;14:4199–4213. doi: 10.2741/3523. [DOI] [PubMed] [Google Scholar]
- 12.Vashee S, Cvetic C, Lu W, Simancek P, Kelly T, Walter J. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 2003;17:1894–1908. doi: 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sclafani R, Holzen T. Cell cycle regulation of DNA replication. Annu. Rev. Genet. 2007;41:237–280. doi: 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DePamphilis M. Replication origins in metazoan chromosomes: fact or fiction? Bioessays. 1999;21:5–16. doi: 10.1002/(SICI)1521-1878(199901)21:1<5::AID-BIES2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 15.Maiorano D, Lutzmann M, Mechali M. MCM proteins and DNA replication. Curr. Opin. Cell Biol. 2006;18:130–136. doi: 10.1016/j.ceb.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Ibarra A, Schwob E, Mendez J. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc. Natl Acad. Sci. USA. 2008;105:8956–8961. doi: 10.1073/pnas.0803978105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinoshita Y, Johnson E. Site-specific loading of an MCM protein complex in a DNA replication initiation zone upstream of the c-MYC gene in the HeLa cell cycle. J. Biol. Chem. 2004;279:35879–35889. doi: 10.1074/jbc.M401640200. [DOI] [PubMed] [Google Scholar]
- 18.Heinzel S, Krysan P, Tran C, Calos M. Autonomous DNA replication in human cells is affected by the size and the source of the DNA. Mol. Cell Biol. 1991;11:2263–2272. doi: 10.1128/mcb.11.4.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellis L, Atadja P, Johnstone R. Epigenetics in cancer: targeting chromatin modifications. Mol. Cancer Ther. 2009;8:1409–1420. doi: 10.1158/1535-7163.MCT-08-0860. [DOI] [PubMed] [Google Scholar]
- 20.Duensing A, Spardy N, Chatterjee P, Zheng L, Parry J, Cuevas R, Korzeniewski N, Duensing S. Centrosome overduplication, chromosomal instability, and human papillomavirus oncoproteins. Environ. Mol. Mutagen. 2009;50:741–747. doi: 10.1002/em.20478. [DOI] [PubMed] [Google Scholar]
- 21.McGivern D, Lemon S. Tumor suppressors, chromosomal instability, and hepatitis C virus-associated liver cancer. Annu. Rev. Pathol. 2009;4:399–415. doi: 10.1146/annurev.pathol.4.110807.092202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burrow A, Williams L, Pierce L, Wang Y. Over half of breakpoints in gene pairs involved in cancer-specific recurrent translocations are mapped to human chromosomal fragile sites. BMC Genomics. 2009;10:59. doi: 10.1186/1471-2164-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukusa T, Fryns J. Human chromosome fragility. Biochim. Biophys. Acta. 2008;1779:3–16. doi: 10.1016/j.bbagrm.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Hsu Y, Wang Y. Human fragile site FRA16B DNA excludes nucleosomes in the presence of distamycin. J. Biol. Chem. 2002;277:17315–17319. doi: 10.1074/jbc.M200901200. [DOI] [PubMed] [Google Scholar]
- 25.Felbor U, Feichtinger W, Schmid M. The rare human fragile site 16B. Cytogenet. Genome Res. 2003;100:85–88. doi: 10.1159/000072841. [DOI] [PubMed] [Google Scholar]
- 26.Hyrien O, Mechali M. Plasmid replication in Xenopus eggs and egg extracts: a 2D gel electrophoretic analysis. Nucleic Acids Res. 1992;20:1463–1469. doi: 10.1093/nar/20.7.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu S, Mangelsdorf M, Hewett D, Hobson L, Baker E, Eyre H, Lapsys N, Le Paslier D, Doggett N, Sutherland G, et al. Human chromosomal fragile site FRA16B is an amplified AT-rich minisatellite repeat. Cell. 1997;88:367–374. doi: 10.1016/s0092-8674(00)81875-9. [DOI] [PubMed] [Google Scholar]
- 28.Fogg J, Kolmakova N, Rees I, Magonov S, Hansma H, Perona J, Zechiedrich E. Exploring writhe in supercoiled minicircle DNA. J. Phys. Condens Matter. 2006;18:S145–S159. doi: 10.1088/0953-8984/18/14/S01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 30.Friedman K, Brewer B. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 1995;262:613–627. doi: 10.1016/0076-6879(95)62048-6. [DOI] [PubMed] [Google Scholar]
- 31.Sidorova JM, Li N, Folch A, Monnat RJ., Jr The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle. 2008;7:796–807. doi: 10.4161/cc.7.6.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bullock P. The initiation of simian virus 40 DNA replication in vitro. Crit. Rev. Biochem. Mol. Biol. 1997;32:503–568. doi: 10.3109/10409239709082001. [DOI] [PubMed] [Google Scholar]
- 33.Tack L, Proctor G. Two major replicating simian virus 40 chromosome classes. Synchronous replication fork movement is associated with bound large T antigen during elongation. J. Biol. Chem. 1987;262:6339–6349. [PubMed] [Google Scholar]
- 34.Barras F, Marinus M. The great GATC: DNA methylation in E. coli. Trends Genet. 1989;5:139–143. doi: 10.1016/0168-9525(89)90054-1. [DOI] [PubMed] [Google Scholar]
- 35.Dean D, Dean B, Muller S, Smith L. Sequence requirements for plasmid nuclear import. Exp. Cell Res. 1999;253:713–722. doi: 10.1006/excr.1999.4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartzell S, Byrne B, Subramanian K. The simian virus 40 minimal origin and the 72-base-pair repeat are required simultaneously for efficient induction of late gene expression with large tumor antigen. Proc. Natl Acad. Sci. USA. 1984;81:6335–6339. doi: 10.1073/pnas.81.20.6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tran C, Caddle M, Calos M. The replication behavior of Saccharomyces cerevisiae DNA in human cells. Chromosoma. 1993;102:129–136. doi: 10.1007/BF00356030. [DOI] [PubMed] [Google Scholar]
- 38.Vologodskii A, Lukashin A, Anshelevich V, Frank-Kamenetskii M. Fluctuations in superhelical DNA. Nucleic Acids Res. 1979;6:967–982. doi: 10.1093/nar/6.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Llorente B, Smith CE, Symington LS. Break-induced replication: what is it and what is it for? Cell Cycle. 2008;7:859–864. doi: 10.4161/cc.7.7.5613. [DOI] [PubMed] [Google Scholar]
- 40.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 41.Collier J. Epigenetic regulation of the bacterial cell cycle. Curr. Opin. Microbiol. 2009;12:722–729. doi: 10.1016/j.mib.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 42.Watson P, Hanauske-Abel H, Flint A, Lalande M. Mimosine reversibly arrests cell cycle progression at the G1-S phase border. Cytometry. 1991;12:242–246. doi: 10.1002/cyto.990120306. [DOI] [PubMed] [Google Scholar]
- 43.Kalejta R, Hamlin J. The dual effect of mimosine on DNA replication. Exp. Cell Res. 1997;231:173–183. doi: 10.1006/excr.1996.3444. [DOI] [PubMed] [Google Scholar]
- 44.Kalejta R, Li X, Mesner L, Dijkwel P, Lin H, Hamlin J. Distal sequences, but not ori-beta/OBR-1, are essential for initiation of DNA replication in the Chinese hamster DHFR origin. Mol. Cell. 1998;2:797–806. doi: 10.1016/s1097-2765(00)80294-4. [DOI] [PubMed] [Google Scholar]
- 45.Roberts J, D’Urso G. An origin unwinding activity regulates initiation of DNA replication during mammalian cell cycle. Science. 1988;241:1486–1489. doi: 10.1126/science.2843984. [DOI] [PubMed] [Google Scholar]
- 46.Perry C, Sastry R, Nasrallah I, Stover P. Mimosine attenuates serine hydroxymethyltransferase transcription by chelating zinc. Implications for inhibition of DNA replication. J. Biol. Chem. 2005;280:396–400. doi: 10.1074/jbc.M410467200. [DOI] [PubMed] [Google Scholar]
- 47.Chen Z, Riu E, He C, Xu H, Kay M. Silencing of episomal transgene expression in liver by plasmid bacterial backbone DNA is independent of CpG methylation. Mol. Ther. 2008;16:548–556. doi: 10.1038/sj.mt.6300399. [DOI] [PubMed] [Google Scholar]
- 48.Riu E, Chen Z, Xu H, He C, Kay M. Histone modifications are associated with the persistence or silencing of vector-mediated transgene expression in vivo. Mol. Ther. 2007;15:1348–1355. doi: 10.1038/sj.mt.6300177. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki M, Kasai K, Saeki Y. Plasmid DNA sequences present in conventional herpes simplex virus amplicon vectors cause rapid transgene silencing by forming inactive chromatin. J. Virol. 2006;80:3293–3300. doi: 10.1128/JVI.80.7.3293-3300.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang C, Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harland R, Laskey R. Maternal chromatin remodeling during maturation and after fertilization in mouse oocytes. Cell. 1980;21:761–771. [Google Scholar]
- 52.Mechali M, Kearsey S. Lack of specific sequence requirement for DNA replication in Xenopus eggs compared with high sequence specificity in yeast. Cell. 1984;38:55–64. doi: 10.1016/0092-8674(84)90526-9. [DOI] [PubMed] [Google Scholar]
- 53.Stanojcic S, Lemaitre J, Brodolin K, Danis E, Mechali M. In Xenopus egg extracts, DNA replication initiates preferentially at or near asymmetric AT sequences. Mol. Reprod. Dev. 2008;28:5265–5274. doi: 10.1128/MCB.00181-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krysan P, Haase S, Calos M. Isolation of human sequences that replicate autonomously in human cells. Mol. Cell Biol. 1989;9:1026–1033. doi: 10.1128/mcb.9.3.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krysan P, Calos M. Replication initiates at multiple locations on an autonomously replicating plasmid in human cells. Mol. Cell Biol. 1991;11:1464–1472. doi: 10.1128/mcb.11.3.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aggarwal B, Calvi B. Chromatin regulates origin activity in Drosophila follicle cells. Nature. 2004;430:372–376. doi: 10.1038/nature02694. [DOI] [PubMed] [Google Scholar]
- 57.Voineagu I, Narayanan V, Lobachev K, Mirkin S. Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins. Proc. Natl Acad. Sci. USA. 2008;105:993619941. doi: 10.1073/pnas.0804510105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krasilnikova MM, Mirkin SM. Replication stalling at Friedreich's ataxia (GAA)n repeats in vivo. Mol. Cell. Biol. 2004;24:2286–2295. doi: 10.1128/MCB.24.6.2286-2295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kovtun I, Thornhill A, McMurray C. Somatic deletion events occur during early embryonic development and modify the extent of CAG expansion in subsequent generations. Hum. Mol. Genet. 2004;13:3057–3068. doi: 10.1093/hmg/ddh325. [DOI] [PubMed] [Google Scholar]
- 60.Hastings P, Ira G, Lupski J. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu W, Abbas T, Dutta A. DNA replication and genomic instability. Adv. Exp. Med. Biol. 2005;570:249–279. doi: 10.1007/1-4020-3764-3_9. [DOI] [PubMed] [Google Scholar]
- 62.Mechali M, Harland R. DNA synthesis in a cell-free system from Xenopus eggs: priming and elongation on single-stranded DNA in vitro. Cell. 1982;30:93–101. doi: 10.1016/0092-8674(82)90015-0. [DOI] [PubMed] [Google Scholar]
- 63.Khopde S, Roy R, Simmons D. The binding of topoisomerase I to T antigen enhances the synthesis of RNA-DNA primers during simian virus 40 DNA replication. Biochemistry. 2008;47:9653–9660. doi: 10.1021/bi800825r. [DOI] [PubMed] [Google Scholar]
- 64.Voitenleitner C, Rehfuess C, Hilmes M, O'Rear L, Liao P, Gage D, Ott R, Nasheuer H, Fanning E. Cell cycle-dependent regulation of human DNA polymerase alpha-primase activity by phosphorylation. Mol. Cell Biol. 1999;19:646–656. doi: 10.1128/mcb.19.1.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lehmann A, Fuchs R. Gaps and forks in DNA replication: Rediscovering old models. DNA Repair (Amst) 2006;5:1495–1498. doi: 10.1016/j.dnarep.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 66.Lopes M, Foiani M, Sogo J. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 67.Hook S, Lin J, Dutta A. Mechanisms to control rereplication and implications for cancer. Curr. Opin. Cell Biol. 2007;19:663–671. doi: 10.1016/j.ceb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]