Abstract
The effect of glycosylation on structure and stability of glycoproteins has been a topic of considerable interest. In this work, we have investigated the solution conformation of the oligosaccharide and its effect on the structure and stability of the glycoprotein by carrying out a series of long Molecular dynamics (MD) simulations on glycosylated Erythrina corallodendron lectin (EcorL) and nonglycosylated recombinant Erythrina corallodendron lectin (rEcorL). Our results indicate that, despite the similarity in overall three dimensional structures, glycosylated EcorL has lesser nonpolar solvent accessible surface area compared to nonglycosylated EcorL. This might explain the experimental observation of higher thermodynamic stability for glycosylated EcorL compared to nonglycosylated EcorL. Analysis of the simulation results indicates that, dynamic view of interactions between protein residues and oligosaccharide is entirely different from the static picture seen in the crystal structure. The oligosaccharide moiety had dynamically stable interactions with Lys 55 and Tyr 53, both of which are separated in sequence from the site of glycosylation, Asn 17. It is possible that glycosylation helps in forming long-range contacts between amino acids, which are separated in sequence and thus provides a folding nucleus. Thus our simulations not only reveal the conformations sampled by the oligosaccharide, but also provide novel insights into possible molecular mechanisms by which glycosylation can help in folding of the glycoprotein by formation of folding nucleus involving specific contacts with the oligosaccharide moiety.
Keywords: molecular dynamics of glycoprotein, protein glycosylation, unfolding simulations, lectin, oligosaccharide conformation
Introduction
Proteins are known for the diversity of functions they display within and around the cells. They are the drivers, effectors, as well as modulators of processes that drive the cellular machinery. The versatility of proteins stems from their ability to generate additional diversity by co- and post-translational modifications. Of these, glycosylation is by far the most complex and profuse form of modification. It confers structural complexity, variety as well as differential physicochemical properties to proteins. The sugar moiety decorating the protein may range from a small monosaccharide to large branched polysaccharides. Broadly speaking, glycosylation can be classified as N-linked (nitrogen of asparagine or arginine) or O-linked (hydroxy oxygen of serine, threonine, tyrosine, hydroxylysine, or hydroxyproline). Of these, the asparagine linked N-glycosylation is the most recurrent form observed in eukaryotes.1,2 It was discovered recently that glycosylation occurs even at prokaryotic level and shares several similarities with the eukaryotic glycosylation.3,4
Glycosylated proteins are involved in various phenomena like host-pathogen interactions, symbiotic associations, cell adhesion, cell-cell recognition, embryonic development, and differentiation.5,6 The specificity of signaling and receptor binding by these proteins can be attributed to their glycosylation pattern. Additionally, glycosylation can also potentially alleviate several physical instabilities of proteins that are a result of exposure to elements such as extreme temperatures, pH, amphipathic interfaces or hydrophobic surfaces, and chemical denaturants.7 Based on studies over several years on a number of glycoproteins, it has been proposed that glycosylation leads to a decrease in the overall structural dynamics of the protein and sometimes the effects can even be observed on regions far from the site of glycosylation.8–11
Recently, studies on antibodies have been carried out to understand the functional consequences of variation in glycan moieties attached to the protein.12,13 The work by Kaneko et al.12 suggests that antibody function depends greatly on the specific sequence of oligosaccharides attached to the Fc chain. It has been observed that one class of Fc-FcγR interactions generate pro-inflammatory effects while another class has anti-inflammatory effects. The distinction arises due to differential sialylation of the Fc core glycan, which provides a switch from innate anti-inflammatory activity to a pro-inflammatory one upon antigenic challenge. However, in certain cases the glycans may have no function at all and might be completely replaceable.2,14 There also have been contradictory reports, for example, in one study the carbohydrate moiety was found to enhance the folding and stability of the covalently attached protein15 whereas in another study on the same protein no increase in thermostability could be seen.16 The secondary or tertiary structures of the glycoproteins, for example, RNAse9,17 and “pars intercerebralis” major peptide-C (PMPC)11 were found unaffected by glycosylation though an enhanced thermostability was observed for both. The deglycosylation of several natural glycoproteins did not lead to substantial changes in their conformations although the glycoproteins experienced decreased thermostability.14 In the case of soybean agglutinin, it was observed that glycosylation not only affects the overall stability of the protein, but also brings about a change in its unfolding kinetics.18 In case of asialoglycoprotein receptor, deglycosylation was found to have no significant effect on cell-surface targeting, function, or turnover.19 The vesicular stomatitis virus producing nonglycosylated viral glycoprotein under the presence or effect of tunicamycin was found to have comparable specific infectivity.20 Deglycosylation of hCG had no effect of on the structure and binding to its receptor, but its function was found to be compromised.21 For horseradish peroxidase isoenzyme C (HRP) which has eight N-linked glycans, it was found that glycosylation enhanced the kinetic stability significantly but not the thermodynamic stability.22 The importance of glycosylation is highlighted by the existence of a quality control system, the calreticulin-calnexin cycle, which prevents the release of misfolded proteins. Glycans are crucial for correct folding of proteins, though a few proteins do fold in their absence. It prompts local conformational changes in the protein structure at the site of glycosylation, which in turn influences not just the stability, but also the oligomerization status of proteins in many cases.6 These studies suggest that, the effect of glycosylation on structure, stability and function of the protein depends on several factors like sequence of the protein, the type of glycan motif attached and the site of attachment and so forth. Therefore, it would be interesting to understand the effect of glycosylation on structure, stability, and function at molecular level.
One such glycoprotein whose crystal structure is available both in glycosylated and nonglycosylated form is a lectin from Erythrina corallodendron (EcorL). Figure 1(a) shows the crystal structure of the dimeric form of EcorL.23 The glycan moiety was found to be a heptasaccharide [Fig. 1(b)] Manα3(Manα6)(Xylβ2)-Manβ4-GlcNAcβ4-(Fucα3)GlcNAc24 predominantly, with Asn-17 being the site of glycosylation.25 Even though another glycosylation site have been proposed at Asn-113,26 none of the EcorL crystal structures have shown the presence of an oligosaccharide on this location. One of the surprising aspects of the structure of EcorL was that in spite of having a typical legume-lectin fold,23,27 it showed a completely novel mode of quaternary association not seen before in other legume lectins. Initially this new mode of quaternary association was attributed to the presence of the oligosaccharide.23 However, subsequent elucidation of the crystal structure of the recombinant form of EcorL (rEcorL)28,29 lacking glycosylation revealed that, it had identical quaternary association as the native glycosylated form. This indicated that glycosylation per se could not have been the cause for a new mode of quaternary association. Instead, it was found that nonglycosylated EcorL had decreased thermal stability and a drastically changed refolding pathway compared to the native glycosylated form of EcorL30 indicating the role of glycosylation in the folding and thermostability.
Figure 1.
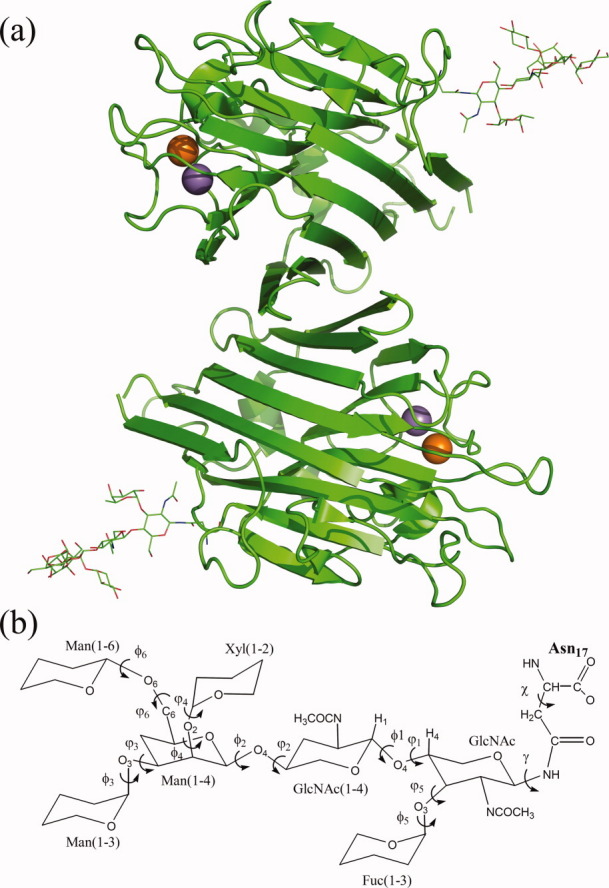
EcorL structure. (a) The cartoon depiction of the crystal structure of EcorL (PDB entry 1AX0). The orange and purple spheres represent the Ca2+ and Mn2+ metal ions. The covalently attached heptasaccharide (below) is shown as lines. (b) The chemical structure of the heptasaccharide depicting various dihedral angles (both φ and ψ) along with the glycosidic dihedral angle (γ) and the chi (χ) angle of the Asn-17 side chain. The monosaccharides have been labeled along with all the glycosidic dihedral angles.
During the last decade, major progress have been made in development of accurate forcefield parameters for the simulations of sugar molecules31,32 and easy availability of high performance computing resources have permitted long nano second scale simulations for large oligomeric glycoproteins. Recently, we were able to successfully simulate the effects of demetalation on saccharide binding and the conformational changes in the ion-binding loop in Concanavalin A.33 Similar studies involving molecular dynamics (MD) simulations on ConA, jacalin, galectin, and other plant lectins have also provided novel insights into their structure and substrate specificity.34–38 This encouraged us to explore in the current study, the effects of glycosylation on the structure and stability of EcorL by comparing the glycosylated and nonglycosylated forms of EcorL using MD simulations. Molecular dynamics simulations have been used earlier to study the structure of the EcorL oligosaccharide successfully.39 However, due to lack of adequate computational resources the protein was kept rigid and only the dynamics of the oligosaccharide was studied. MD simulations have also been used to study the retardation of unfolding of bovine pancreatic RNaseA due to glycosylation.40 In this work, we report a series of long MD simulations in explicit solvent environment for glycosylated as well as nonglycosylated dimers of EcorL for understanding effect of glycosylation on the structure and stability of this important glycoprotein.
Result and Discussion
Structural features and dynamic behavior of glycosylated and nonglycosylated EcorL: Similar or different?
The structural changes in glycosylated and nonglycosylated EcorL were analyzed by root mean square deviation (RMSD) plots. Figure 2 shows the RMSD with respect to the starting structures for each of the four simulations for glycosylated and nonglycosylated EcorL as a function of time. As can be seen, both glycosylated and nonglycosylated EcorL remain close to their respective starting structures throughout the simulations with a final RMSD close to 1.5 Å. However, nonglycosylated EcorL had a peak RMSD of ∼1.7 Å, whereas glycosylated EcorL showed slightly higher RMSD values intermittently with a peak RMSD value of 2.6 Å. The 1AX0 dimer was simulated for 10 ns in a single trajectory and its structural deviation has been compared to other 5 ns glycosylated and nonglycosylated EcorL simulations in Supporting Information Figure S2 where it is clear that the RMSD for 1AX0 dimer over 10 ns is similar to other EcorL simulations. Therefore, we used the 5 ns trajectories for further analysis as each of them had been repeated four times with different random number seeds. Figure 3 shows the residue-wise breakup of RMSD (rRMSD) between the starting structure and the final structures at the end of the 5 ns simulations for both glycosylated and nonglycosylated EcorL. It was also found that there were no regions with large differences in rRMSD values (Fig. 3) between the starting and the final structures that is, rRMSD was found to be similar across all the residues. This indicated that there were no significant structural differences between the final conformations of glycosylated and nonglycosylated EcorL neither in any local region nor in terms of the overall structure. Analysis of radius of gyration (Rg) values of the sampled conformations in all eight room temperature trajectories indicated that it was close to 24.3 Å for both glycosylated and nonglycosylated EcorL across all simulations (Supporting Information Fig. S3). We also computed the theoretical B-factor values from the fluctuations for various residues over the entire 5 ns trajectories. The comparative analysis of B-factors (Supporting Information Fig. S4) for glycosylated and nonglycosylated EcorL revealed that during the simulations all the regions of both the proteins behaved in a very similar manner except for the loop regions. Most nonloop regions had almost identical B-factor values and showed very minor variation if at all. The loop regions had sharp peaks in the B-factor plot. The average value of B-factor over all the residues including the chain ends were found to be 19, 18, 19, 23 Å2 for nonglycosylated EcorL in four different simulations at 300 k, whereas for glycosylated EcorL the corresponding values were 22, 23, 29, 21 Å2. Thus, glycosylated EcorL had slightly higher average B-factor values compared to nonglycosylated EcorL. However, there was no region in the protein, which showed any significantly different mobility, due to the presence of the glycosyl moiety.
Figure 2.
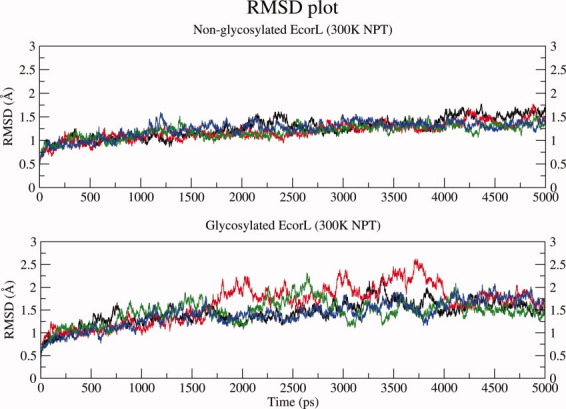
RMSD Plot. The RMSD plot for glycosylated and nonglycosylated EcorL at 300 K (NPT) simulations. The RMSD values from the four independent simulations have been shown in different colors.
Figure 3.
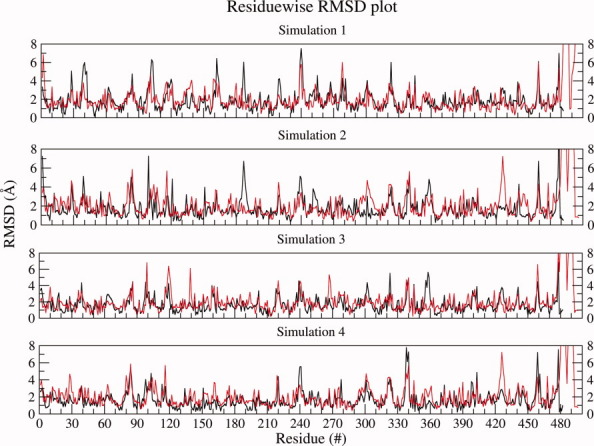
Residue wise RMSD Plot. The residue-wise RMSD values for all the residues of the final structure of nonglycosylated EcorL (black) and glycosylated EcorL (red) simulations have been plotted. The rRMSD values from all the four simulations have been plotted.
We next proceeded to analyze differences in the MD trajectories of glycosylated and nonglycosylated EcorL in terms of accessibility of side chains, and also hydrogen bonds involving backbone as well as side chains. Figure 4 shows the variation of percentage of nonpolar solvent accessible surface area (ASA) over the different MD trajectories for both glycosylated and nonglycosylated EcorL. Supporting Information Table S1 shows the mean and standard deviation values of nonpolar ASA for various simulations. The nonpolar ASA for both forms of EcorL remained around 52–54% indicating that both the forms of EcorL have stable structures at 300 K. As can be seen from Figure 4 and Supporting Information Table S1, glycosylated EcorL has lower nonpolar ASA compared to nonglycosylated EcorL in three out of the four trajectories and this is most prominent in third simulation (S3). This suggests marginally higher stability of glycosylated EcorL over its nonglycosylated counterpart. We have also tried to identify if some specific region of nonglycosylated EcorL was contributing to the higher nonpolar ASA. We first calculated the average nonpolar ASA (ASAmean) for each residue and then subtracted the average nonpolar ASA observed in nonglycosylated EcorL from the average nonpolar ASA found in glycosylated EcorL. From simulation 3 (S3; which showed maximum nonpolar ASA difference) we identified residues, which showed a difference of more than 10% in ASAmean and they are 16–17, 25, 55, 64, 97–98, 161–163, 187, and 237 (Supporting Information Fig. S5). Visualization of these residues on the structure indicated that only some of them were present adjacent to residues, which interact with sugar moiety while many were present far from sugar moiety.
Figure 4.
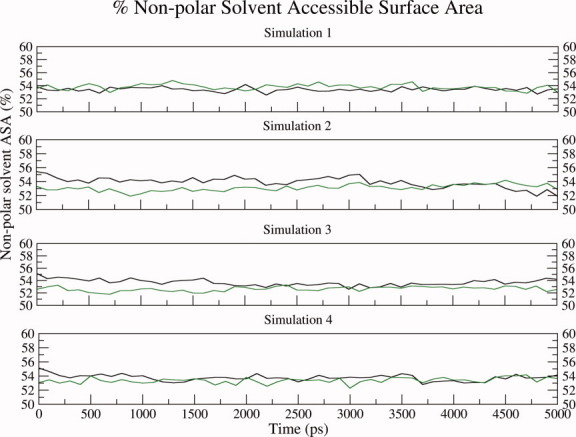
Nonpolar solvent accessible surface area plot. The nonpolar solvent accessible surface area has been plotted for both nonglycosylated EcorL (black) and glycosylated EcorL (green). The ASA values from all the four simulations have been plotted in different panels. For simulation 1 (S1) the average and standard deviation values for the nonpolar ASA of nonglycosylated and glycosylated EcorL were found to be 53.4 ± 0.3 and 53.9 ± 0.4, respectively. Similarly, for S2 these values were 53.9 ± 0.7 and 53.1 ± 0.5, for S3 they were 53.7 ± 0.5 and 52.6 ± 0.4, and for S4 they were found to be 53.7 ± 0.4 and 53.4 ± 0.4, respectively. As can be seen in three out of the four trajectories the glycosylated EcorL has lower nonpolar ASA compared to nonglycosylated EcorL and this is most prominent in simulation S3.
During the molecular dynamics simulations many of the noncovalent interactions like hydrogen bonds keep breaking and forming. Interactions that have very short resident times or occupancy are termed as “transient interactions,” while interactions having relatively longer resident times are termed “dynamically stable interactions.” We have analyzed hydrogen bonds in each window of 100 ps in the MD trajectory and hydrogen bonds, which are persistent for 10% of the simulation time have been termed “dynamically stable interactions.” Figure 5 shows the number of dynamically stable hydrogen bonds involving main chain–main chain (M–M), main chain–side chain (M–S), and side chain–side chain (S–S) for the glycosylated EcorL [Fig. 5(b)] and nonglycosylated EcorL [Fig. 5(a)] in the various 5 ns trajectories sampled at 300 K. As can be seen, the M–M count is similar in both forms of EcorL indicating that the intrabackbone interactions are similar among the two forms, though glycosylated EcorL seemed to have a more consistent and stable count. This stability might be considered as a signature of the structural stability at the fold level. Figure 5 also shows the number of hydrogen bonds involving side chains (the second panel in both Figure 5(a,b) shows total hydrogen bonds formed by side-chain atoms), which are likely to reflect the packing interactions present in the two forms. The total side-chain hydrogen bonds in nonglycosylated EcorL were observed to be more than 225 [Fig. 5(a); middle panel] whereas in glycosylated EcorL such bonds were observed to be about 200 [Fig. 5(b); middle panel]. Further, at the end of 5 ns, both of the forms had a similar average count for hydrogen bonds involving side chain interactions, but glycosylated EcorL showed a relatively larger variation in the count of side chain interactions among different simulations. It may be noted that, even though for individual trajectories there was a difference in the number of dynamically stable S–M/S and S–S hydrogen bonds between glycosylated and nonglycosylated EcorL, the differences in the numbers were of the same order as the differences, which arise in multiple MD runs for the same structure. This suggests that the overall packing interactions in glycosylated and nonglycosylated EcorL are also very similar.
Figure 5.
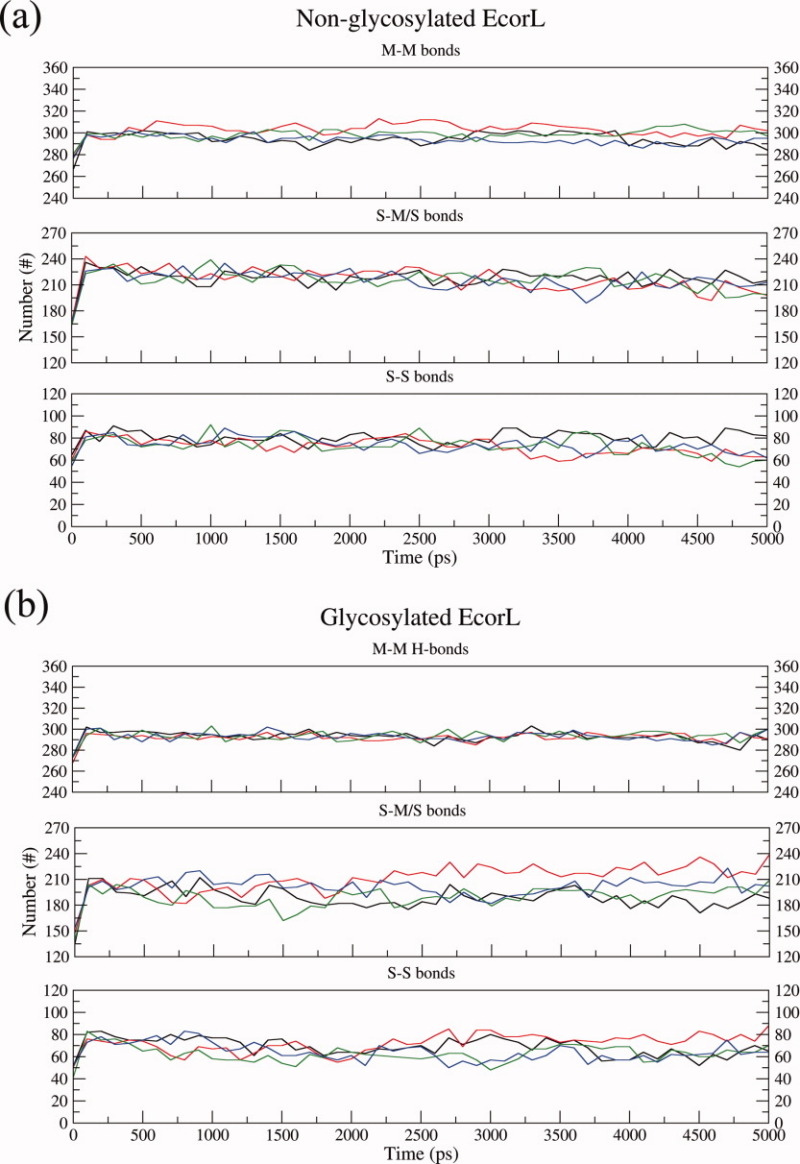
Intramolecular hydrogen bonds analysis. The intramolecular hydrogen bonds have been plotted for both nonglycosylated EcorL (a) and glycosylated EcorL (b). The hydrogen bonds have been classified as main chain-main chain (M-M) hydrogen bonds, total side chain (S-M/S) bonds and side chain-side chain (S-S) interactions. The values from all the four simulations have been plotted in different colors.
Similar final RMSD and almost identical number of backbone interactions strongly suggested that at the fold level there is no difference among the two forms. These observations supported the previous studies that the tertiary structure of a glycoprotein was unaffected by glycosylation.9,11 However, some differences were still observed among the two forms, for example intermittent high RMSD value, slightly increased Rg and B-factors of the loops in glycosylated EcorL as compared to nonglycosylated EcorL suggested an increased structural flexibility of the loops, possibly due to glycosylation. A decreased and varying count of side chain interactions also appeared as a constant feature in all four simulations. The decreased side chain interactions might have led to increased Rg values and B-factors of some residues or vice versa without affecting the stability of the molecule. Instead, it was found that glycosylated EcorL had lesser exposure of nonpolar regions to solvent compared to nonglycosylated EcorL indicating an extra stability advantage of glycosylated EcorL over nonglycosylated EcorL. These observations from our MD simulations indicate that effects of glycosylation on EcorL manifests mainly in the rearrangement of the side chains so as to decrease the ASA. This could be the reason for the greater thermal stability of glycosylated EcorL over nonglycosylated EcorL. Thus the results of our long MD simulation studies are in agreement with the reported biophysical studies regarding stability differences between glycosylated and nonglycosylated EcorL. Our simulation results are also in agreement with observations from crystallographic studies29 that, even though glycosylation does not lead to a change in the overall structure of EcorL, it affects the side chain packing.
Conformation of the oligosaccharide moiety at 300 K
We also analyzed the conformational changes occurring in the oligosaccharide moiety of glycosylated EcorL during the 5 ns of simulations. The flexibility of the heptasaccharide has already been investigated in earlier studies39 using MD simulations. However, in those studies the majority of the EcorL protein was restrained and the simulations were for short durations. Since our current study involved multiple numbers of long simulations without any restraints, it might give a more realistic picture of the conformational dynamics of the oligosaccharide moiety. Table I lists the average values of the various dihedral angles in the oligosaccharide along with the standard deviations over the 5 ns simulations for each of the four trajectories for glycosylated EcorL at 300 K. For the purpose of comparison we have also listed in Table I, the average and standard deviation values of the corresponding dihedral angles in the N- and O-linkage conformations in various crystal structures. These values have been reported in literature by Petrescu et al.41 based on statistical analysis of crystallographic data. The most dramatic effect of glycosylation was seen in the χ angle of Asn-17 residue (Table II). For nonglycosylated EcorL the average value of the χ dihedral angle of the Asn-17 side chain was found to vary between −76° and −124° in four different simulations and the standard deviations were as high as 65°. However, in glycosylated EcorL the average value of χ ranged between −47° and −70° with a maximum standard deviation of only 13°. Thus glycosylation seems to have restricted the rotation of the Asn-17 side chain to a relatively smaller range. Similarly, the glycosidic dihedral angle (γ) connecting the oligosaccharide to the protein was found to be rigid with values close to 73° (Table II) in both the chains of glycosylated EcorL with a standard deviation of only about 10°. The φ and ψ dihedral angles [Fig. 1(b)] for each of the six disaccharide linkages were not only compared with their statistical averages in the crystallographic data,41 but also with the values reported in the earlier work by Naidoo et al.39 which involved short simulation of the oligosaccharide. It is interesting to note that, most of the φ and ψ values seen in our simulations (Table I) were in good agreement with the values obtained from statistical analysis of crystal structures.41 For example, the average values for φ1 (GlcNAc(1–4)GlcNAc linkage) in our simulations ranged between −68° and −74°, whereas φ1 value of −73.7 ± 8.4° has been reported by Petrescu et al.41 Similarly, the ψ1 value was found to vary between 128° and 131° in our simulations and was 116.8 ± 15.6° in the work of Petrescu et al.41 Thus, most φ and ψ values obtained from our simulations were in close agreement with the values reported by Petrescu et al. However, comparison of the corresponding φ and ψ values reported by Naidoo et al with the φ and ψ values reported by Petrescu et al indicate that only φ values were in agreement. The ψ values showed no direct correlation except for ψ1 and ψ5. The reason for such a disagreement could be that, the length of simulations carried out by Naidoo et al.39 was very short limiting the conformational sampling for the oligosaccharide. However, longer simulations carried out in the current work permitted efficient sampling of the conformational space, thus leading to better agreement with experimental values.
Table I.
φ and Ψ Angles of the Individual Disaccharides from the EcorL Heptasaccharide
| φ = O5–C1–Ox–Cx | Ψ = C1–Ox–Cx–Cx-1 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| X-ray | S1 | S2 | S3 | S4 | X-ray | S1 | S2 | S3 | S4 | |||
| GlcNAc-β(1-4)-GlcNAc (x = 4) | φ1 | −73.7 ± 8.4 | −74 ± 9 | −74 ±10 | −73±9 | −68 ± 9 | Ψ1 | 116.8 ± 15.6 | 129 ± 7 | 130 ± 8 | 131 ± 8 | 128 ± 7 |
| Man-α(1-4)-GlcNAc (x = 4) | φ2 | −88 ± 10.8 | −90 ± 12 | −84 ± 15 | −86 ± 19 | −80 ± 14 | Ψ2 | 107.9 ± 20.3 | 103 ± 19 | 104 ± 17 | 110 ± 16 | 115 ± 22 |
| Man-α(1-3)-Man (x = 3) | φ3 | 72.5 ± 11.0 | 68 ± 9 | 69 ± 10 | 69 ± 10 | 70 ± 11 | Ψ3 | −112.3 ± 22.5 | −121 ± 16 | −96 ± 9 | −98 ± 9 | −91 ± 11 |
| Xyl-β(1-2)-Man (x = 2) | φ4 | −91.5 ± 6.6 | −89 ± 7 | −85 ± 17 | −88 ± 16 | −90 ± 16 | Ψ4 | −105.8 ± 3.9 | −117 ± 17 | −110 ± 18 | −114 ± 17 | −112 ± 24 |
| Fuc-β(1-3)-GlcNAc (x = 3) | φ5 | −68.2 ± 9.6 | −74 ± 11 | −67 ± 13 | −74 ± 12 | −67 ± 12 | Ψ5 | −101.7 ± 8.1 | −99 ± 9 | −96 ± 9 | −98 ± 9 | −91 ± 11 |
| Man-α(1-6)-Man (x = 6) | φ6 | 65.4 ± 9.0 | 70 ± 13 | 71 ± 16 | 67 ± 14 | 70 ± 18 | Ψ6 | 182.6 ± 5.1 | 177 ± 32 | 180 ± 22 | 157 ± 45 | 160 ± 45 |
The dihedral angles for individual disaccharides are given as phi (φ) and psi (ψ) values, which refer to the O5–C1–Ox–Cx and C1–Ox–Cx–Cx-1 angles, respectively. The columns S1, S2, S3, and S4 correspond to dihedral angles obtained from the four independent simulations carried out with different initial velocities.
Table II.
Glycosidic Dihedral Angle (γ) and Chi (χ) Angle for Asn17 of EcorL
| Nonglycosylated EcorL | Glycosylated EcorL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Chain | S1 | S2 | S3 | S4 | S1 | S2 | S3 | S4 | |
| γ (Cγ-N-C1-O5) | A | – | – | – | – | 74 ± 9 | 73 ± 10 | 73 ± 9 | 72 ± 9 |
| B | – | – | – | – | 73 ± 10 | 73 ± 13 | 75 ± 10 | 73 ± 9 | |
| χ (N-Cα-Cβ-Cγ) | A | −96 ± 49 | −97 ± 49 | −103 ± 62 | −124 ± 57 | −68 ± 12 | −68 ± 13 | −63 ± 11 | −70 ± 11 |
| B | −102 ± 52 | −76 ± 32 | −107 ± 52 | −111 ± 65 | −59 ± 8 | −47 ± 8 | −60 ± 11 | −70 ± 11 | |
The glycosidic dihedral angle (γ) measured as Cγ-N-C1-O5 (between Cγ-N of Asn-17 and C1-O5 of GlcNAc) and the chi (χ) angle measured as N-Cα-Cβ-Cγ for the side chain of Asn-17 have been calculated for both chains of the EcorL dimer. The columns S1, S2, S3, and S4 correspond to dihedral angles obtained from the four independent simulations carried out with different initial velocities.
Even though most dihedral angles of the oligosaccharide moiety showed only limited flexibility, due to the combinatorial effect of the six disaccharide linkages, the oligosaccharide was highly mobile and sampled a large volume of space around the location of its tethering [Fig. 6(a)] during the 5 ns simulation time. So, we analyzed if during its motion the oligosaccharide made contacts with the amino acid residues in protein. We searched for any oligosaccharide atoms that were closer than 5 Å to the protein atoms. It was found that most of the oligosaccharide residues, except the ones which are sequential neighbors due to covalent connectivity, were interacting more with the solvent than the protein [Fig. 6(a)]. The interactions, whenever occurred, were of transient nature and no stable noncovalent contact could be located between the protein and oligosaccharide atoms. Figure 6(b) shows for one trajectory, the residues that were found uniformly closer (≤5 Å) to the oligosaccharide, the oligosaccharide contacting residues for all the four simulations are listed in Supporting Information Table S2. As can be seen from Figure 6(c) all these contacting residues 4–8, 15–17, 53, and 55 were in spatial proximity of the Asn 17, which is covalently connected to the sugar moiety. Hence, our analysis suggests that the oligosaccharide shows high degree of flexibility and it interacts predominantly with the solvent. It may be noted that Naidoo et al.39 had also made the similar observations about the oligosaccharide's preferential location being the solvent.
Figure 6.
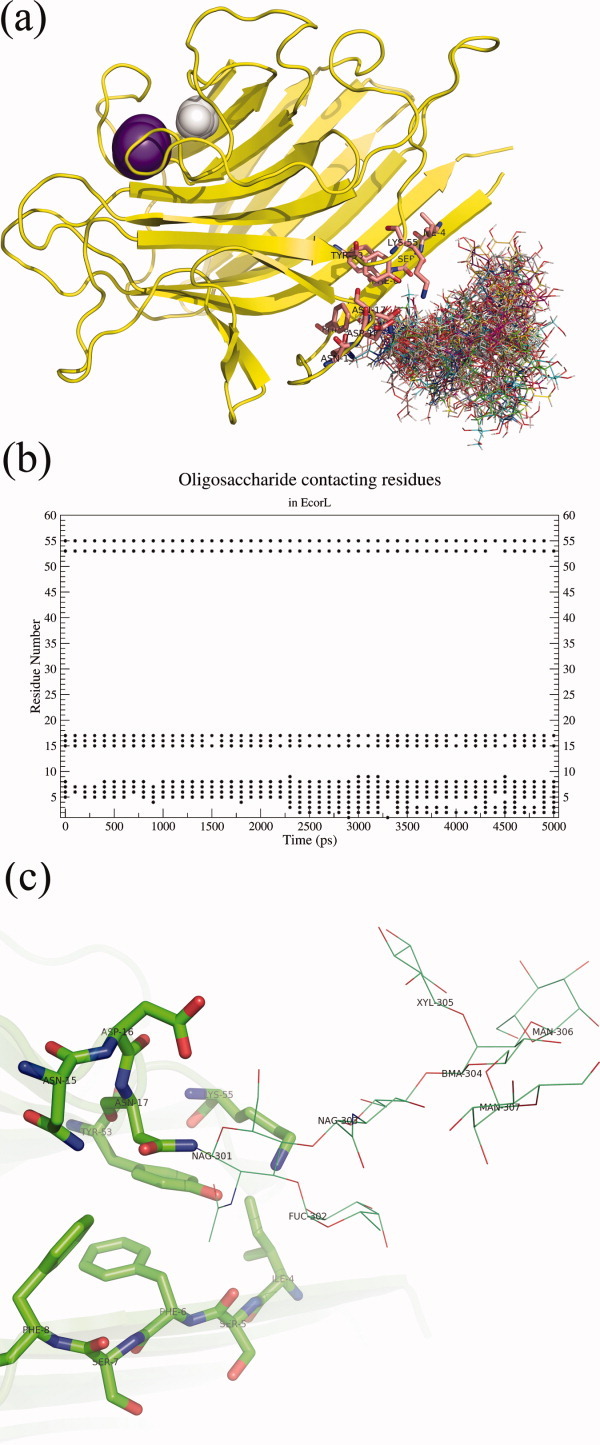
Oligosaccharide solution conformation and interacting residues. (a) A cartoon depiction of glycosylated EcorL and the various conformations of the oligosaccharide sampled during the 5 ns simulation. The structures were extracted at an interval of 100 ps and aligned with each other using the protein co-ordinates. However, only one conformation of the protein has been shown for the purpose of clarity. The Mn2+ and Ca2+ ions are depicted as purple and white spheres, respectively. The residues at a distance closer than 5 Å to oligosaccharide have been shown as sticks. (b) A graph showing residues which had contact (closer than 5 Å) with the oligosaccharide. Residues, for example, 5, 6, 7, 15, 17, 53, and 55, were seen to be in 5 Å distance of oligosaccharide throughout the simulations. (c) The residues found to be in contact with the oligosaccharide were also located in the crystal structure (1AX0). The pink transparent cartoon depicts the protein and the residues contacting oligosaccharide are shown in sticks. The oligosaccharide itself is shown in lines.
Interactions of Lys55 and Tyr53 with oligosaccharide
Our main objective has been to understand how the oligosaccharide moiety can help in the folding process. Analysis of the glycosylated EcorL indicated that a total of 13 residues showed interactions with the oligosaccharide. However, out of these 13 residues 11 residues are proximal in sequence to the site of glycosylation, that is, Asn17. Proximity in one dimensional sequence will automatically result in proximity in terms of spatial distance. Thus these 11 residues are likely to be proximal to the oligosaccharide both in folded as well as unfolded state irrespective of whether they preferentially interact with oligosaccharide or not. Therefore, interaction of the oligosaccharide with these 11 residues is unlikely to help much in the folding process. However, Lys55 and Tyr53 are sequentially far from Asn17, the site of glycosylation and preferential interaction between oligosaccharide and these two residues would result in the formation of tertiary native contacts, thus it might function as a folding nucleus. Therefore, we carried out detailed analysis of interactions between glycan and these two residues.
Earlier studies have reported hydrogen bonding interactions between oligosaccharide and Tyr53 as well as Lys55, based on the analysis of crystal structure of glycosylated EcorL.30 In view of the known flexibility of the oligosaccharide group, it was not known whether these interactions seen in the static crystal structure will be stable under dynamic conditions, which are of significance in solution. We analyzed the dynamic behavior of these oligosaccharide-protein interactions in various MD trajectories. One of the novel observation from our multiple trajectory MD simulation studies is that, hydrogen bonds seen in the crystal structure do not persist during the dynamic simulations, they constantly form and break during the simulation. Analysis of hydrogen bonds involving sugar residues which persist for at least 10 ps in each 100 ps window indicated that, apart from the proximal oligosaccharide residues like GlcNac 301 and Fuc 302 which form hydrogen bonds with Lys55 in the crystal structure, even distal sugar residues like GlcNac 303, Xyl 305, and Man 307 can also form hydrogen bonds with Lys55. Figure 7 shows interactions of these oligosaccharide residues with Lys55 from a single trajectory. For the sake of clarity results have been shown from a single trajectory only instead of all four trajectories. The top five panels of Figure 7 actually correspond to interactions of Lys55 with five different sugar residues, namely Fuc 302, Man 307, Xyl 305, GlcNac 303, and GlcNac 301. Each of these panels depicts the occupancy of various hydrogen bonds formed between a particular oligo-saccharide residue and Lys55. For example, the topmost panel shows the occupancy of hydrogen bonds formed between Lys55 and Fucose residue (Fuc 302) of the oligosaccharide. It can be seen clearly that hydrogen bonds between O2 of Fucose and Lys55 form during the first nanosecond only. Similarly, the second panel shows that hydrogen bonds between O6 of Man 307 and Lys55 form only after 2 ns during the simulation. These results indicate that different hydrogen bonds form at different time points in the trajectory and persist for different lengths of time. The other three trajectories also show similar results, but differ in the type of hydrogen bonds, their occupancy and the time line of their formation and disappearance on the trajectory (data not shown). Even though the trajectory from simulation 4 (top 5 panels in Fig. 7) shows lack of hydrogen bonds between oligosaccharide and Lys55 for considerable period of time, averaging of results from all four simulations indicated presence of two hydrogen bonds between oligosaccharide and Lys55 during most of the 5 ns simulation (Fig. 7, bottom most panel). It is also clear from the bottom panel of Figure 7 that on average two hydrogen bonds existed during most of the 5 ns simulation, thus indicating that the bonds between the oligosaccharide residues and Lys55 are not transient. This dynamic view of interactions between protein residues and oligosaccharide is entirely different from the static picture seen in the crystal structure.30 The hydroxyl oxygen of Y53 was observed to interact, almost exclusively, with the carboxyl oxygen of acetyl group of the first NAcGlc (GlcNAc-301) unit of the oligosaccharide.
Figure 7.
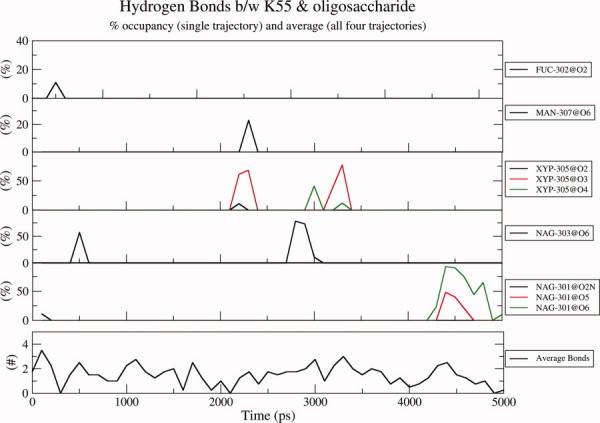
Hydrogen bonds between the oligosaccharide residues and Lys55. The hydrogen bonds between various oligosaccharide residues and Lys55 have been shown in different panels except for the last (detached) panel which provides the average hydrogen bonds between the oligosaccharide and K55 during one of the four simulations. For the sake of clarity results from a single trajectory instead of all four trajectories are shown. Top five panels correspond to interactions of Lys55 with five different sugar residues, namely Fuc 302, Man 307, Xyl 305, GlcNac 303, and GlcNac 301. Each of these panels depicts the occupancy of various hydrogen bonds formed between a particular oligo-saccharide residue and Lys55 during a single trajectory. For example, the topmost panel shows the occupancy of hydrogen bonds formed between Lys55 and Fucose residue (Fuc 302) of the oligosaccharide. It can be seen clearly that hydrogen bonds between O2 of Fucose and Lys55 form during the first nanosecond only. Similarly, the second panel shows that hydrogen bonds between O6 of Man 307 and Lys55 form only after 2 ns during the simulation. The bottommost panel shows the averaging of results from all four simulations and indicates presence of two hydrogen bonds between oligosaccharide and Lys55 during most of part of the 5 ns simulation.
Figure 8(a) shows that Tyr53 of nonglycosylated EcorL (glycan-free enzyme) interacts with only two other residues, namely, Asn17 and Lys55 and that the average number of such hydrogen bonds over the four trajectories was around 0.25, that is, no hydrogen bond was present during 75% of the simulation time. On the contrary, in Figure 7, on average more than 1.5 hydrogen bonds exist between K55 and the oligosaccharide during the simulations, that is, at least two hydrogen bonds are present 75% of the simulation time. And, despite similar spatial proximity the average number of hydrogen bonds between Tyr53 and Asn17 in the nonglycosylated structure is only 0.25 [Fig. 8(a)]. Comparison of Figure 8 a with Figure 7 indicates that, there is a preferential interaction between oligosaccharide and Lys55, which does not arise entirely from spatial proximity. This also suggests that even though Y53 and K55 could interact intermittently with N17 in nonglycosylated EcorL, glycosylation led to formation of stronger and dynamically stable interactions of Y53 and Lys55 with the oligosaccharide in glycosylated EcorL. The higher stability and number of these interactions clearly show that the hydrogen bonds between K55, Y53 and oligosaccharide residues in glycosylated EcorL are not a mere replacement of weak intraprotein hydrogen bonds in nonglycosylated EcorL. This is further supported by results from simulations on glycosylated EcorL mutants as discussed later.
Figure 8.
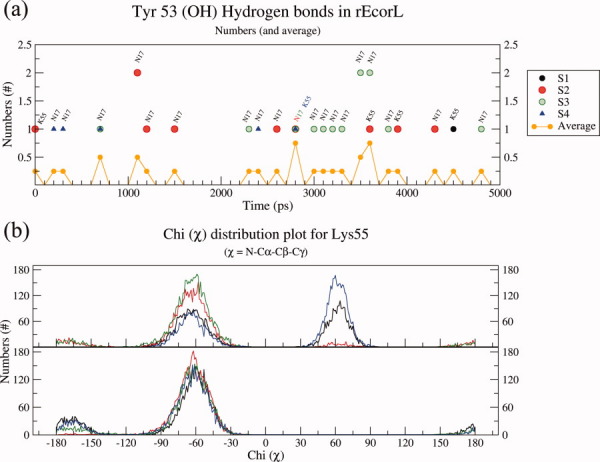
Effects of glycosylation on orientations of Tyr53 and Lys55. (a) Hydrogen bonding interactions of Y53 in nonglycosylated EcorL have been plotted from all the four simulations and indicated with different colors. Residues with which the hydrogen bonds were observed have been labeled. Also shown is the average number of hydrogen bonds involving Y53 from all four nonglycosylated EcorL simulations at 300 K. (b) Dihedral angle value distribution for χ of K55 of nonglycosylated EcorL and glycosylated EcorL has been plotted. The χ values from all the four simulations have been plotted in different colors.
The interactions with oligosaccharide led to no conspicuous effects on the orientation of Y53 side-chain with respect to the backbone. However, K55 side chain seemed to have been constrained as could be seen from the range of values sampled by the χ angle. In nonglycosylated EcorL, the χ of K55 was found to exist in two rotameric states, one corresponding to χ value 65° and the other one with a χ value close to −65° [Fig. 8(b)]. In glycosylated EcorL, only the peak corresponding to the χ value of −65° was observed. These results suggested that due to interactions with the oligosaccharide atoms the K55 side chain gets locked in a specific conformation in contrast to the two rotameric states seen in the nonglycosylated EcorL.
Effect of Y53A and K55A mutation on oligosaccharide interactions
Since the analysis of the simulations on glycosylated EcorL indicated preferential interactions between oligosaccharide moiety, and the residues Y53 and K55, we wanted to analyze the effect of mutations at these sites on the interactions between protein and oligosaccharide. Simulations were carried out on a double-mutant of glycosylated EcorL (DM-EcorL; Y53A, K55A) and conformations and orientations of the oligosaccharide were analyzed. This hypothetical double mutant of native glycosylated EcorL (1AX0 dimer) was created by replacing both Y53 and K55 side chains with Ala side chain. DM-EcorL and native glycosylated EcorL were simulated for 5 and 10 ns, respectively. The side chain of Ala lacks hydrogen bond donor or acceptor atoms therefore hydrogen bonds cannot be used to analyze these interactions. The interactions between residues 53 (Tyr or Ala) and 55 (Lys or Ala) with oligosaccharide residues were analyzed by measuring the distance between center of mass of each oligosaccharide residue from Cβ atom (CB) of residue 53 and 55. As can be seen from Supporting Information Table S3 and Supporting Information Figure S6, compared to the wild type EcorL the distance between oligosaccharide residues and residues 53, 55 in DM-EcorL was higher by at least 5 Å. Additionally, the B-factors of oligosaccharide residues were found to be higher compared to the corresponding values in the wild type EcorL (Supporting Information Table S4). These results further demonstrate that the observed interactions between oligosaccharide and residues at position 53 and 55 arise from hydrogen bonding and not from their spatial proximity.
Thus, our simulations indicated that glycosylation at N17 leads to formation of dynamically stable interactions between sites which are separated in sequence and the glycosylation helps in locking long flexible side chains in specific conformations. The stable interactions between the oligosaccharide and the residue 53 and 55 could act as a folding nucleus and help in enhanced folding as seen in earlier experimental studies.30 The folding nucleus proposed here involved hydrogen bonding interactions, but generally folding nucleus involves hydrophobic residues. The observations about hydrophobic interactions driving the process of protein folding are mostly based on folding studies on nonglycosylated proteins. In case of glycoproteins hydrogen bonding interactions might initiate the process of folding. However, a caveat may be noted before we infer the formation of a folding nucleus from our simulation results. Since we have not carried out folding simulations, our results do not unambiguously demonstrate that these interactions are indeed a folding nucleus. This is a hypothesis based on the results of our simulations.
Thermal unfolding of the glycosylated and nonglycosylated EcorL
As discussed earlier the analysis of the results of various simulations at room temperature indicated that both glycosylated and nonglycosylated EcorL had similar structural features, except for a decreased nonpolar ASA and altered side chain-backbone interactions in glycosylated EcorL. Some additional destabilizing factors, for example, low pH,42 might have improved the chances of observing the differences in stability among the two forms of EcorL. However, to avoid generating complications in the results we have not tried to test the effects of other parameters on the structure of EcorL. The lower nonpolar ASA would suggest higher stability for glycosylated EcorL compared to nonglycosylated EcorL. Earlier experimental studies30 have also shown that deglycosylation leads to lower stability of nonglycosylated EcorL compared to glycosylated EcorL. We wanted to further investigate the differences in the stability of glycosylated and nonglycosylated EcorL using in silico thermal unfolding studies by carrying out MD simulations at elevated temperatures. MD simulations were carried out for glycosylated and nonglycosylated EcorL at 400, 500, and 600 K. Unfolding of the structures at various elevated temperatures was monitored by RMSD plots over the trajectories (Fig. 9) and compared with the RMSD plot from 300 K simulation. As can be seen, at 300 K the RMSD increased to ∼1Å within first 500 ps and for the rest of the 4.5 ns simulation it only increased another 0.5 Å. However, at 400 K RMSD for both glycosylated and nonglycosylated EcorL had increased to ∼3 Å at about 1 ns and for initial 3.5 ns the RMSD of glycosylated EcorL was less compared to nonglycosylated EcorL. At 500 K the RMSD for both the forms rose to ∼5 Å in the initial 3 ns of the simulation, but in the subsequent 2 ns run the final RMSD for glycosylated EcorL increased to ∼6 Å, whereas nonglycosylated EcorL had RMSD value of ∼7.5 Å rising at the end of 5 ns MD run. The lower RMSD of glycosylated EcorL at 400 K for a major period and also a lower final RMSD at 500 K suggest differences in the stability of glycosylated and nonglycosylated EcorL. The relatively lower deviation from the native structure in the glycosylated EcorL compared to the nonglycosylated form was also seen in regions which specifically interacted with the oligosaccharide. Supporting Information Figure S7 shows the conformation of the β strand containing the amino acids Tyr 53 and Lys 55, which preferentially interact with the oligosaccharide and could possibly form the folding nucleus. As can be seen, at elevated temperatures this β strand has relatively lower deviation from its native structure in the presence of glycosylation. This suggests that the dynamically stable contacts between the oligosaccharide and the residues Tyr 53 and Lys 55, can not only act as potential folding nucleus, they can also impart additional stability to the glycosylated form. Additionally, the analysis of nonpolar ASA of the structures glycosylated and nonglycosylated EcorL sampled at elevated temperature simulations showed that glycosylated EcorL had relatively lower nonpolar ASA compared to nonglycosylated EcorL even at higher temperatures, which was prominent in 500 and 600 K simulations (Fig. 10).
Figure 9.
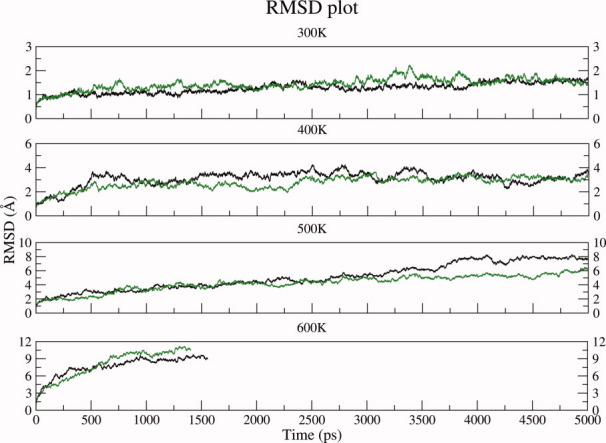
RMSD plot for conformations sampled during high temperature simulations. RMSD plot for nonglycosylated EcorL (black) and glycosylated EcorL (green) at 300 K and higher temperatures (400, 500, and 600 K) from simulations.
Figure 10.
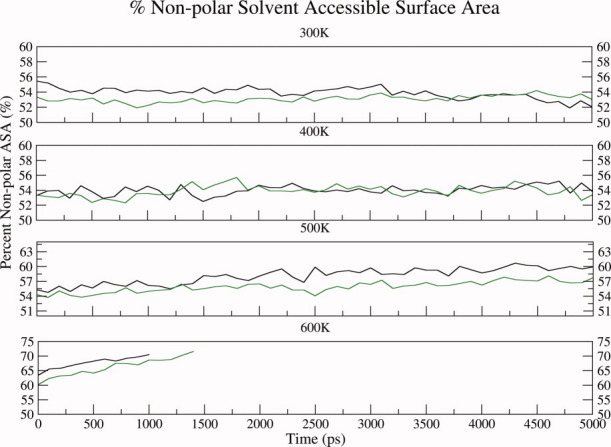
Nonpolar ASA plot for conformations sampled during high temperature simulations. The nonpolar ASA plot for nonglycosylated EcorL (black) and glycosylated EcorL (green) at 300 K and higher temperatures (400, 500, and 600 K) from simulations. The nonpolar ASA was calculated every 100 ps from the trajectory.
Thus, the results from these elevated temperature simulations were also in agreement with results from simulations at 300 K in terms of stability differences between glycosylated and nonglycosylated EcorL. At 600 K the simulations could not be continued beyond 1.5 ns and within this period the RMSD for nonglycosylated and glycosylated EcorL had steeply increased to ∼9 Å and >10.5 Å, respectively. This indicated that both the forms now had a hugely unfolded structure compared to the native structure at 300 K. At such high temperatures the contributions from glycosylations to stability was not adequate to compete with temperature induced unfolding. Thus the high temperature simulations were also in qualitative agreement with the earlier experimental studies which suggested lower stability of nonglycosylated EcorL compared to glycosylated EcorL. We also carried out detailed analysis of the rRMSD to understand the mechanistic details of temperature induced unfolding of glycosylated and nonglycosylated EcorL. It was found that, in all the high temperature simulations only the loop regions of the protein showed higher RMSD from native structures while the primary hydrophobic core of the proteins consisting of the β-sheet including the dimer interface remained unperturbed even at 600 K.
Conclusion
A series of long MD simulations (5 ns each) in explicit solvent environment for glycosylated as well as nonglycosylated dimers of EcorL were carried out at various temperatures for understanding the effect of glycosylation on the structure and stability of this glycoprotein. Detailed analysis of these multiple MD trajectories indicated that, glycosylation led to neither local nor overall change in EcorL structure. These results have interesting implications for efforts towards expression of glycoproteins in heterologous systems for therapeutic purposes. Even though the structure of the glycosylated EcorL was found to be very similar to its nonglycosylated counterpart, the percent nonpolar solvent ASA for glycosylated EcorL was found to be less compared to nonglycosylated EcorL for most of the simulation time, which might indicate higher stability of glycosylated EcorL compared to nonglycosylated EcorL. To understand the effect of glycosylation on thermal unfolding of EcorL, a series of MD simulations were also carried out on glycosylated as well as nonglycosylated EcorL at elevated temperatures of 400, 500, and 600 K. The unfolding of the structures at elevated temperatures was measured by RMSD from native structure and nonpolar accessible surface areas. Interestingly, in many of the high temperature simulations the glycosylated EcorL showed lower RMSD and nonpolar ASA compared to nonglycosylated EcorL. Thus these high temperature simulations are also in qualitative agreement with earlier experimental studies, which suggested slightly higher stability for glycosylated EcorL compared to the nonglycosylated form.
Analysis of the various conformations sampled by the oligosaccharide moiety indicated that, during the simulations the oligosaccharide moiety was highly flexible. However, the average values of various dihedral angles was in close agreement with average values of the corresponding dihedrals reported in literature based on analysis of various crystal structures. In various trajectories the oligosaccharide moiety preferentially interacted with Lys 55 and Tyr 53 which are separated in sequence from the site of glycosylation Asn 17. It may be noted that, hydrogen bonds between oligosaccharide and these residues as seen in the crystal structure do not persist during the dynamics, they constantly form and break during the simulation. Apart from the proximal oligosaccharide residues like GlcNac 301 and Fuc 302, which form hydrogen bonds with Lys55 in the crystal structure, even distal sugar residues like GlcNac 303, Xyl 305, and Man 307 can also form hydrogen bonds with Lys55. This dynamic view of interactions between protein residues and oligosaccharide is entirely different from the static picture seen in the crystal structure. We believe that it constitutes a novel observation from the current studies. In nonglycosylated EcorL the Lys 55 side chain sampled two rotameric states, whereas in glycosylated EcorL Lys 55 remained predominantly in one rotameric state due to interactions with the sugar groups. It is possible that glycosylation helps in forming long range contacts between amino acids, which are separated in sequence and thus provides a folding nucleus. Interestingly, the β strand containing Tyr 53 and Lys 55, which might work as folding nucleus also shows relatively lower deviations from native structure in glycosylated EcorL compared to the nonglycosylated counterpart. Thus, our simulations are not only in agreement with experimental observations about higher stability of glycosylated EcorL compared to nonglycosylated EcorL, but also provide novel insights into possible molecular mechanisms by which glycosylation can help in enhanced folding of the glycoprotein by formation of folding nucleus involving specific contacts with the oligosaccharide moiety. However, it must be noted that for unambiguous demonstration of involvement of oligosaccharide moiety in folding nucleus would require folding simulations, which are currently impossible for such large proteins. Nevertheless our simulations provide valuable clues, which can help in design of experiments for better understanding the folding and stability of glycoproteins.
Materials and Methods
The coordinates for the dimeric nonglycosylated form of EcorL were obtained from C and D chains of the PDB entry 1SFY. The coordinates for the dimeric glycosylated EcorL were obtained by superposing the monomer coordinates from the PDB entry 1AX0 on the nonglycosylated EcorL dimer. The glycosylation has been retained at Asn-17 only, as none of crystal structures of EcorL have coordinates for the glycan at Asn-113. The recombinant and glycosylated forms of EcorL have almost identical structure and all atoms RMSD between monomeric chains of 1SFY and 1AX0 was only 0.8Å. Therefore, to generate a glycosylated structure with protein coordinates exactly identical to the nonglycosylated counterpart, a glycosylated dimer was generated by using protein coordinates from 1SFY and oligosaccharide coordinates from 1AX0. Metal ions were included for both the dimers for the simulations but ligand coordinates were removed as it has been known that the ligand binding induces conformational changes in the protein although minor they may be. Thus, we obtained the metal-bound and ligand-free coordinates of both glycosylated and nonglycosylated EcorL dimers. MD simulations were carried out using AMBER9 package.43 The ff0344 and glycam0632 force-fields were used for the polypeptide and heptasaccharide segments respectively of the glycoprotein. The oligomeric EcorL was solvated inside a TIP3P45 water box extending 10 Å from the protein surface along all three axes. Energy minimization was carried out for the solvated protein to remove steric clashes if any between the protein and the solvent molecules. To remove bad contacts in an experimental structure of a protein an energy minimization of the protein alone might be warranted before solvating it. However, significant proportion of EcorL structure is composed of loops and minimization in vacuum might have altered the conformation of these loops. Besides, water molecules have been known to participate in structuring certain residues and the EcorL structures do not have steric clashes, therefore we have minimized the solvated protein. The temperature of the system was raised to 300 K over a 20 ps constant volume simulation and then equilibrated at 300 K for 200 ps by carrying out constant pressure (NPT) simulations at 1 atmospheric pressure. Subsequently, the production dynamics was run for a period of 5 ns under NPT conditions. A time step of 1 fs and a nonbonded cutoff radius of 8 Å were used in all the simulations. The periodic boundary conditions were used to mimic the infinite solvent system and the electrostatic potential was calculated using particle-mesh Ewald summation method.46 The coordinates were stored every ps during the dynamics so that the 5 ns trajectory consisted of a set of 5000 structures. The nonglycosylated form of EcorL was also simulated for 5 ns under identical conditions at 300 K and 1 atm pressure. Each of the simulations for glycosylated and nonglycosylated EcorL were repeated four times using different random number seeds for assigning different starting velocities. These simulations have been called S1, S2, S3, and S4, respectively. MD simulations for both glycosylated and nonglycosylated EcorL were also carried out at elevated temperatures of 400, 500, and 600 K. For these high temperature simulations at 300 and 400 K, the temperature of the system was gradually raised to the desired value and the system was equilibrated for 200 ps at constant pressure. During the constant pressure equilibration at higher temperatures, it was seen that the density of the system dropped from ∼1.05 g/cm3 at 300 K to ∼0.96 and 0.83 g/cm3 at 400 and 500 K, respectively. At 600 K the density of the system kept dropping to lower values. Hence, the production dynamics was initiated after a 400 ps of equilibration at 600 K. At 400, 500, and 600 K, only a single MD run was carried out for glycosylated and nonglycosylated EcorL. Thus, the high temperature simulations gave a total of six trajectories for glycosylated and nonglycosylated EcorL.
As mentioned earlier, the glycosylated EcorL simulations were carried out on a hybrid structure using protein coordinates from nonglycosylated form and sugar coordinates from the glycosylated EcorL crystal structure. In view of the very high structural similarity (backbone atoms RMSD < 0.4 Å) between the glycosylated (1AX0) and nonglycosylated (1SFY) EcorL structures, simulations of 1AX0 dimer or 1SFY-oligosaccharide hybrid dimer would give very similar results for glycosylated EcorL. However, for the purpose of comparison, we also carried out a simulation on the glycosylated EcorL crystal structure, that is, 1AX0. The 1AX0 dimer was simulated for 10ns in a single run. To examine whether increasing the equilibration for longer periods would affect the system properties, namely, temperature, pressure, or density any further, we ran this simulation with thermalization period increased to 500 ps and NPT equilibration period extended to 1ns. The temperature, pressure, and density values from this simulation were very similar to the corresponding values obtained from simulations with 20 ps of thermalization and 200 ps of equilibrations (Supporting Information Fig. S1).
The trajectories were analyzed using the Ptraj module of AMBER943 package. The various structural and dynamic parameters, which were calculated from the trajectories were RMSD from the starting structure, Rg, B-factors, and hydrogen bonds were performed using ptraj. RMSD and B-factors were calculated for the backbone atoms only. Rg was calculated for protein residues only. For calculation of the persistence of hydrogen bonds, the distance, angle and percentage occupancy cutoff for interacting atoms were 3.5Å, 120°, and 10%, respectively, in a window of 100 ps of each trajectory. The hydrogen bonds were divided into two categories, the backbone interactions (M-M), which exist between the main chain atoms (M) only, and the side-chain interaction (M/S-S) which exist between a side chain atom (S) and either a backbone atom (M) or another side chain atom (S). NACCESS47 was used to calculate the solvent accessible surface area (ASA). ProFit48 was used for the calculation of residue-wise RMSDs (rRMSD). PyMol49 was used for depiction of structural models.
Acknowledgments
S. K. is a recipient of Senior Research Fellowship from CSIR, India. Computational resources provided under BTIS project of DBT, India, are gratefully acknowledged.
References
- 1.Nalivaeva NN, Turner AJ. Post-translational modifications of proteins: acetylcholinesterase as a model system. Proteomics. 2001;1:735–747. doi: 10.1002/1615-9861(200106)1:6<735::AID-PROT735>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 2.Wyss DF, Wagner G. The structural role of sugars in glycoproteins. Curr Opin Biotechnol. 1996;7:409–416. doi: 10.1016/s0958-1669(96)80116-9. [DOI] [PubMed] [Google Scholar]
- 3.Upreti RK, Kumar M, Shankar V. Bacterial glycoproteins: functions, biosynthesis and applications. Proteomics. 2003;3:363–379. doi: 10.1002/pmic.200390052. [DOI] [PubMed] [Google Scholar]
- 4.Weerapana E, Imperiali B. Asparagine-linked protein glycosylation: from eukaryotic to prokaryotic systems. Glycobiology. 2006;16:91R–101R. doi: 10.1093/glycob/cwj099. [DOI] [PubMed] [Google Scholar]
- 5.Varki A. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitra N, Sinha S, Ramya TN, Surolia A. N-linked oligosaccharides as outfitters for glycoprotein folding, form and function. Trends Biochem Sci. 2006;31:156–163. doi: 10.1016/j.tibs.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Sola RJ, Griebenow K. Effects of glycosylation on the stability of protein pharmaceuticals. J Pharm Sci. 2009;98:1223–1245. doi: 10.1002/jps.21504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wormald MR, Dwek RA. Glycoproteins: glycan presentation and protein-fold stability. Structure. 1999;7:R155–160. doi: 10.1016/s0969-2126(99)80095-1. [DOI] [PubMed] [Google Scholar]
- 9.Joao HC, Scragg IG, Dwek RA. Effects of glycosylation on protein conformation and amide proton exchange rates in RNase B. FEBS Lett. 1992;307:343–346. doi: 10.1016/0014-5793(92)80709-p. [DOI] [PubMed] [Google Scholar]
- 10.Gervais V, Zerial A, Oschkinat H. NMR investigations of the role of the sugar moiety in glycosylated recombinant human granulocyte-colony-stimulating factor. Eur J Biochem. 1997;247:386–395. doi: 10.1111/j.1432-1033.1997.00386.x. [DOI] [PubMed] [Google Scholar]
- 11.Mer G, Hietter H, Lefevre JF. Stabilization of proteins by glycosylation examined by NMR analysis of a fucosylated proteinase inhibitor. Nat Struct Biol. 1996;3:45–53. doi: 10.1038/nsb0196-45. [DOI] [PubMed] [Google Scholar]
- 12.Kaneko Y, Nimmerjahn F, Ravetch JV. Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science. 2006;313:670–673. doi: 10.1126/science.1129594. [DOI] [PubMed] [Google Scholar]
- 13.Burton DR, Dwek RA. Immunology. Sugar determines antibody activity. Science. 2006;313:627–628. doi: 10.1126/science.1131712. [DOI] [PubMed] [Google Scholar]
- 14.Wang C, Eufemi M, Turano C, Giartosio A. Influence of the carbohydrate moiety on the stability of glycoproteins. Biochemistry. 1996;35:7299–7307. doi: 10.1021/bi9517704. [DOI] [PubMed] [Google Scholar]
- 15.Chu FK, Trimble RB, Maley F. The effect of carbohydrate depletion on the properties of yeast external invertase. J Biol Chem. 1978;253:8691–8693. [PubMed] [Google Scholar]
- 16.Schulke N, Schmid FX. The stability of yeast invertase is not significantly influenced by glycosylation. J Biol Chem. 1988;263:8827–8831. [PubMed] [Google Scholar]
- 17.Williams RL, Greene SM, McPherson A. The crystal structure of ribonuclease B at 2.5-A resolution. J Biol Chem. 1987;262:16020–16031. doi: 10.2210/pdb1rbb/pdb. [DOI] [PubMed] [Google Scholar]
- 18.Sinha S, Surolia A. Attributes of glycosylation in the establishment of the unfolding pathway of soybean agglutinin. Biophys J. 2007;92:208–216. doi: 10.1529/biophysj.106.092668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breitfeld PP, Rup D, Schwartz AL. Influence of the N-linked oligosaccharides on the biosynthesis, intracellular routing, and function of the human asialoglycoprotein receptor. J Biol Chem. 1984;259:10414–10421. [PubMed] [Google Scholar]
- 20.Gibson R, Leavitt R, Kornfeld S, Schlesinger S. Synthesis and infectivity of vesicular stomatitis virus containing nonglycosylated G protein. Cell. 1978;13:671–679. doi: 10.1016/0092-8674(78)90217-9. [DOI] [PubMed] [Google Scholar]
- 21.Kalyan NK, Bahl OP. Role of carbohydrate in human chorionic gonadotropin. Effect of deglycosylation on the subunit interaction and on its in vitro and in vivo biological properties. J Biol Chem. 1983;258:67–74. [PubMed] [Google Scholar]
- 22.Tams JW, Welinder KG. Glycosylation and thermodynamic versus kinetic stability of horseradish peroxidase. FEBS Lett. 1998;421:234–236. doi: 10.1016/s0014-5793(97)01573-1. [DOI] [PubMed] [Google Scholar]
- 23.Shaanan B, Lis H, Sharon N. Structure of a legume lectin with an ordered N-linked carbohydrate in complex with lactose. Science. 1991;254:862–866. doi: 10.1126/science.1948067. [DOI] [PubMed] [Google Scholar]
- 24.Ashford D, Dwek RA, Welply JK, Amatayakul S, Homans SW, Lis H, Taylor GN, Sharon N, Rademacher TW. The beta 1—2-D-xylose and alpha 1—3-L-fucose substituted N-linked oligosaccharides from Erythrina cristagalli lectin. Isolation, characterisation and comparison with other legume lectins. Eur J Biochem. 1987;166:311–320. doi: 10.1111/j.1432-1033.1987.tb13516.x. [DOI] [PubMed] [Google Scholar]
- 25.Adar R, Richardson M, Lis H, Sharon N. The amino acid sequence of Erythrina corallodendron lectin and its homology with other legume lectins. FEBS Lett. 1989;257:81–85. doi: 10.1016/0014-5793(89)81791-0. [DOI] [PubMed] [Google Scholar]
- 26.Young NM, Watson DC, Yaguchi M, Adar R, Arango R, Rodriguez-Arango E, Sharon N, Blay PK, Thibault P. C-terminal post-translational proteolysis of plant lectins and their recombinant forms expressed in Escherichia coli. Characterization of “ragged ends” by mass spectrometry. J Biol Chem. 1995;270:2563–2570. doi: 10.1074/jbc.270.6.2563. [DOI] [PubMed] [Google Scholar]
- 27.Chandra NR, Prabu MM, Suguna K, Vijayan M. Structural similarity and functional diversity in proteins containing the legume lectin fold. Protein Eng. 2001;14:857–866. doi: 10.1093/protein/14.11.857. [DOI] [PubMed] [Google Scholar]
- 28.Arango R, Adar R, Rozenblatt S, Sharon N. Expression of Erythrina corallodendron lectin in Escherichia coli. Eur J Biochem. 1992;205:575–581. doi: 10.1111/j.1432-1033.1992.tb16815.x. [DOI] [PubMed] [Google Scholar]
- 29.Kulkarni KA, Srivastava A, Mitra N, Sharon N, Surolia A, Vijayan M, Suguna K. Effect of glycosylation on the structure of Erythrina corallodendron lectin. Proteins. 2004;56:821–827. doi: 10.1002/prot.20168. [DOI] [PubMed] [Google Scholar]
- 30.Mitra N, Sharon N, Surolia A. Role of N-linked glycan in the unfolding pathway of Erythrina corallodendron lectin. Biochemistry. 2003;42:12208–12216. doi: 10.1021/bi035169e. [DOI] [PubMed] [Google Scholar]
- 31.Guvench O, Greene SN, Kamath G, Brady JW, Venable RM, Pastor RW, Mackerell AD., Jr Additive empirical force field for hexopyranose monosaccharides. J Comput Chem. 2008;29:2543–2564. doi: 10.1002/jcc.21004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woods RJ, Dwek RA, Edge CJ, Fraser-Reid B. Molecular mechanical and molecular dynamic simulations of glycoproteins and oligosaccharides. 1. GLYCAM_93 parameter development. J Phys Chem. 1995;99:3832–3846. [Google Scholar]
- 33.Kaushik S, Mohanty D, Surolia A. The role of metal ions in substrate recognition and stability of concanavalin A: a molecular dynamics study. Biophys J. 2009;96:21–34. doi: 10.1529/biophysj.108.134601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansia P, Dev S, Surolia A, Vishveshwara S. Insight into the early stages of thermal unfolding of peanut agglutinin by molecular dynamics simulations. Proteins. 2007;69:32–42. doi: 10.1002/prot.21512. [DOI] [PubMed] [Google Scholar]
- 35.Sharma A, Sekar K, Vijayan M. Structure, dynamics, and interactions of jacalin. Insights from molecular dynamics simulations examined in conjunction with results of X-ray studies. Proteins. 2009;77:760–777. doi: 10.1002/prot.22486. [DOI] [PubMed] [Google Scholar]
- 36.Gupta G, Vishveshwara S, Surolia A. Stability of dimeric interface in banana lectInsight from molecular dynamics simulations. IUBMB Life. 2009;61:252–260. doi: 10.1002/iub.162. [DOI] [PubMed] [Google Scholar]
- 37.Meynier C, Feracci M, Espeli M, Chaspoul F, Gallice P, Schiff C, Guerlesquin F, Roche P. NMR and MD investigations of human galectin-1/oligosaccharide complexes. Biophys J. 2009;97:3168–3177. doi: 10.1016/j.bpj.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pratap JV, Bradbrook GM, Reddy GB, Surolia A, Raftery J, Helliwell JR, Vijayan M. The combination of molecular dynamics with crystallography for elucidating protein-ligand interactions: a case study involving peanut lectin complexes with T-antigen and lactose. Acta Cryst D. 2001;57:1584–1594. doi: 10.1107/s0907444901011957. [DOI] [PubMed] [Google Scholar]
- 39.Naidoo KJ, Denysyk D, Brady JW. Molecular dynamics simulations of the N-linked oligosaccharide of the lectin from Erythrina corallodendron. Protein Eng. 1997;10:1249–1261. doi: 10.1093/protein/10.11.1249. [DOI] [PubMed] [Google Scholar]
- 40.Choi Y, Lee JH, Hwang S, Kim JK, Jeong K, Jung S. Retardation of the unfolding process by single N-glycosylation of ribonuclease A based on molecular dynamics simulations. Biopolymers. 2008;89:114–123. doi: 10.1002/bip.20867. [DOI] [PubMed] [Google Scholar]
- 41.Petrescu AJ, Petrescu SM, Dwek RA, Wormald MR. A statistical analysis of N- and O-glycan linkage conformations from crystallographic data. Glycobiology. 1999;9:343–352. doi: 10.1093/glycob/9.4.343. [DOI] [PubMed] [Google Scholar]
- 42.Esposito L, Daggett V. Insight into ribonuclease A domain swapping by molecular dynamics unfolding simulations. Biochemistry. 2005;44:3358–3368. doi: 10.1021/bi0488350. [DOI] [PubMed] [Google Scholar]
- 43.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Rossa WS, Kollman PA. AMBER 9. San Francisco: University of California; 2006. [Google Scholar]
- 44.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 45.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 46.Darden T, York D, Pedersen L. Particle mesh Ewald: An N [center-dot] log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 47.Hubbard SJ, Thornton JM. NACCESS version 2.1.1, Computer Program, Department of Biochemistry and Molecular Biology. University College London; 1993. [Google Scholar]
- 48.Martin ACR, Porter CT. ProFit v2.6, Computer Program. University College London: 2008. [Google Scholar]
- 49.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, California: DeLano Scientific LLC; 2002. [Google Scholar]


