Abstract
Human γD-crystallin (HγD-Crys) is a highly stable protein that remains folded in the eye lens for the majority of an individual's lifetime. HγD-Crys exhibits two homologous crystallin domains, each containing two Greek key motifs with eight β-strands. Six aromatic pairs (four Tyr/Tyr, one Tyr/Phe and one Phe/Phe) are present in the β-hairpin sequences of the Greek keys. Ultraviolet damage to the aromatic residues in lens crystallins may contribute to the genesis of cataract. Mutant proteins with these aromatic residues substituted with alanines were constructed and expressed in E. coli. All mutant proteins except F115A and F117A had lower thermal stability than the WT protein. In equilibrium experiments in guanidine hydrochloride (GuHCl), all mutant proteins had lower thermodynamic stability than the WT protein. N-terminal domain (N-td) substitutions shifted the N-td transition to lower GuHCl concentration, but the C-terminal domain (C-td) transition remained unaffected. C-td substitutions led to a more cooperative unfolding/refolding process, with both the N-td and C-td transitions shifted to lower GuHCl concentration. The aromatic pairs conserved for each Greek key motif (Greek key pairs) had larger contributions to both thermal stability and thermodynamic stability than the other pairs. Aromatic-aromatic interaction was estimated as 1.5–2.0 kcal/mol. In kinetic experiments, N-td substitutions accelerated the early phase of unfolding, while C-td substitutions accelerated the late phase, suggesting independent domain unfolding. Only substitutions of the second Greek key pair of each crystallin domain slowed refolding. The second Greek keys may provide nucleation sites during the folding of the double-Greek-key crystallin domains.
Keywords: crystallin, aromatic residues, Greek key, stability, folding, kinetics, nucleation site
Introduction
Crystallins make up over 90% of the eye lens proteins.1 α-Crystallins are small heat shock proteins (sHSP) with chaperone functions,2,3 while βγ-crystallins are the major structural proteins.4 βγ-crystallins are a family of proteins exhibiting two homologous crystallin domains. Each domain contains two Greek key motifs forming a β-sandwich of eight intercalated β-strands5 (Fig. 1). β-Crystallins form dimers and higher-order oligomers, while γ-crystallins are monomeric.4 βγ-Crystallin-like proteins are also found in a number of lower organisms lacking the lens tissue.6–8 Lens βγ-crystallins together with these βγ-crystallin-like proteins form the βγ-crystallin superfamily.
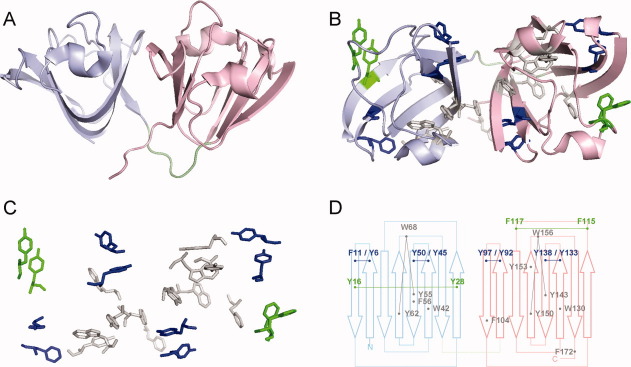
Figure 1.
Crystal structure of HγD-Crys (1HK0)5. Three-dimensional structures were prepared by PyMOL (DeLano Scientific). (A) “Side view” of HγD-Crys, showing the two crystallin domains. Pale blue: N-td; pink: C-td; light green: linker. (B) “Top view” of HγD-Crys. All aromatic residues are shown in stick representation. Blue: four β-hairpin aromatic pairs (the “Greek key pairs”); green: two additional β-hairpin aromatic pairs (the “non-Greek-key pairs”); gray: all other aromatic residues. (C) The aromatic residues in HγD-Crys in the same orientation and color scheme as in panel B. the cartoon representation of the nonaromatic residues are removed for a clearer view. (D) Topology diagram of HγD-Crys and the locations of the aromatic residues. Color scheme is the same as in B. Block arrows represent β-strands. The six aromatic pairs and the two Tyr-Trp-Tyr clusters are indicated by thin lines connecting dots.
Lens epithelial cells lose their organelles upon differentiation into the mature lens fiber cells, and cannot synthesize crystallins afterwards.9 The central region of the eye lens, the nucleus, is formed in utero. Human γ-crystallins, particularly γC- and γD-crystallins, are enriched in the lens nucleus, representing some of the oldest molecules in the human body.10–12 To maintain lens transparency, the crystallins must remain soluble and folded throughout the majority of an individual's lifetime. In fact, human γD-crystallin (HγD-Crys) is resistant to denaturation by up to 8 M urea.13 The half time of the initial unfolding step was about 19 years,14 and the thermal denaturation midpoint was 83.8°C.15 The complex topology of the Greek key motifs may contribute to the high stability of βγ-crystallins.16
Like all βγ-crystallins, the 173-amino-acid HγD-Crys has two homologous crystallin domains [Fig. 1(A)]: N-terminal domain (N-td, residues 1–82), and C-terminal domain (C-td, residues 88–173) and four Greek key motifs (named Greek key 1 through 4 from N- to C-terminus).5 Each crystallin domain has two highly conserved tryptophans. The fluorescence spectrum of denatured HγD-Crys becomes higher and red shifted compared to that of the native state.17–20 This unusual native-state fluorescence quenching property of HγD-Crys provides a sensitive reporter of the transition between the native and unfolded states.
During protein expression in E. coli cytoplasm, a substantial proportion of recombinant HγD-Crys successfully folded into the soluble native state. Purified HγD-Crys also readily refolds into a native-like state in vitro at 37°C when diluted from 5 M guanidine hydrochloride (GuHCl) to ∼1 M, without any assistance of chaperones. Below 1 M, however, partially folded HγD-Crys forms high-molecular-weight aggregates.13 In equilibrium experiments in GuHCl, an intermediate with the N-td unfolded and C-td folded was identified in the unfolding/refolding pathway. Mutations of the hydrophobic core, polar pairs interacting across the domain interface, and N-td congenital mutations of HγD-Crys affected the thermodynamic stability and the refolding rate of the N-td but not the C-td.15,21–23 Kinetic experiments revealed a sequential refolding pathway where the C-td folds first, followed by the N-td.17,24 Computational studies using molecular dynamics supported this pathway.25 A more detailed pathway was proposed by Flaugh et al., where the four Greek keys unfold successively from the N- to the C-terminus, and refold successively in the opposite direction.15 However, experimental evidence regarding the unfolding/refolding pathway within each double-Greek-key crystallin domain of HγD-Crys remains scarce.
HγD-Crys contains a high percentage of aromatic residues distributed throughout the protein: 14 Tyr, 6 Phe, and 4 Trp. 18 of these 24 residues have aromatic partners within 5 Å [Fig. 1(B–D]. These aromatic residues cluster into several structural elements. The “tyrosine corner” is a classic feature of the Greek key motifs, bridging two β-strands by a hydrogen bond between the tyrosine hydroxyl group and a backbone carboxyl group.26 The hydroxyl groups were suggested to be essential for stability, but not for nucleating the folding of Greek key proteins.27 HγD-Crys has two homologous tyrosine corners, Y62 in the N-td and Y150 in the C-td.
Besides the tyrosine corners, four homologous aromatic pairs with partners five residues apart in the primary sequence are present (three Tyr/Tyr and one Tyr/Phe, N-td: Y6/F11, Y45/Y50, C-td: Y92/Y97, Y133/Y138, termed “Greek key pairs”). They are conserved at the homologous β-hairpins of the four Greek key motifs. Two additional aromatic pairs (one Tyr/Tyr and one Phe/Phe; N-td: Y16/Y28, C-td: F115/F117, termed “non-Greek-key pairs”) are found in other β-hairpins. Two three-residue Tyr-Trp-Tyr clusters are also present, one in each crystallin domain. Epidemiological studies of humans and laboratory experiments on animals supported ultraviolet (UV) radiation being a causative factor for cataract, a major cause of blindness in the world.28 Trp, Tyr, and Phe are the main UV absorbers in proteins and are susceptible to direct and indirect UV damage.29,30 UV damage to these aromatic residues may impair the folding and stability of lens crystallins, such as HγD-Crys, and contribute to the genesis of cataract.
Aromatic molecules can interact through partial charges on the δ(–) ring faces and δ(+) ring edges.31 Aromatic residues tend to cluster in proteins, and prefer a distance of ∼5.5 Å between phenyl ring centroids.32 Two frequently observed orientations are perpendicular32 and “parallel displaced stacking.”33 Aromatic-aromatic interaction energy was estimated to be about −0.6 to −1.3 kcal/mol.32 A rich body of experimental evidence suggests that aromatic-aromatic interactions play important roles in stability and/or folding of diverse proteins,34–38 especially β-sheet proteins.16,39–42 Aromatic interactions have also been studied in peptides, and employed in de novo peptide design.43 As in proteins, β-hairpin peptides could be stabilized by aromatic pairs and clusters.44–46
The β-hairpin aromatic pairs are a prominent feature of the Greek keys in βγ-crystallins. Assessing the contributions of these aromatic pairs to the folding and stability of HγD-Crys is important in advancing our understanding of the folding and stability of this and other Greek key and β-sheet proteins, as well as other proteins containing aromatic pairs. In this study, we used site-directed mutagenesis, equilibrium and kinetic experiments, and both thermal and GuHCl denaturation to dissect the roles of the six β-hairpin aromatic pairs in the folding and stability of HγD-Crys.
Results
Conservation of the aromatic pairs in βγ-crystallins
Sequence comparisons between the lens βγ-crystallins and the βγ-crystallin-like relatives in lower organisms were previously conducted for protein S,6 spherulin 3a,7 and Ciona crystallin.8 In these three βγ-crystallin-like proteins, when Phe or Tyr was considered to be conserved, the Greek key aromatic pairs were maintained, while the non-Greek-key pairs were not. The Greek key pair consensus Y/FXXXXY/FXG, was used for defining the βγ-crystallin superfamily.47 To evaluate the conservation of the aromatic pairs in lens βγ-crystallins, we performed a sequence alignment with 43 β-crystallins and 36 γ-crystallins from human and seven other vertebrates, using ClustalW248 (Supporting Information Fig. S1). Residues homologous to the six aromatic pairs in HγD-Crys were examined for their conservation.
When either Phe or Tyr was considered to be conserved in each position, conservation are >80% for the Greek key pairs, and ∼20–70% for the non-Greek-key pairs in the 79 βγ-crystallins selected (Table I). For the two less conserved non-Greek-key pairs, most γ-crystallins, but none of the β-crystallins, maintain the pair Y16/Y28; all of the γD-, γE-, γF-crystallins, but almost none of the other γ-, or β-crystallins, maintain both residues of the pair F115/F117 (Supporting Information Fig. S1). For the positions not occupied by Tyr or Phe, there was no obvious pattern for amino acid usage. The conservation of the Greek key pairs in the lens βγ-crystallins and βγ-crystallin-like relatives, in addition to the well-defined primary sequence and three-dimensional structure, suggest that for the functions these aromatic pairs serve in βγ-crystallins, the Greek key pairs are much more important than the non-Greek-key pairs.
Table I.
Conservation of Aromatic Pairs in Selected βγ-Crystallin Sequences
| Positiona | Usage of Tyr or Pheb |
|---|---|
| Y6 | 82% |
| F11 | 100% |
| Y16 | 44% |
| Y28 | 44% |
| Y45 | 100% |
| Y50 | 91% |
| Y92 | 100% |
| Y97 | 90% |
| F115 | 68% |
| F117 | 19% |
| Y133 | 91% |
| Y138 | 99% |
Amino acid positions are as in HγD-Cyrs.
Percentages were calculated from the sequence alignment result shown in Supporting Information Figure S1.
Mutant protein construction, expression, and purification
To assess experimentally the roles of the six aromatic pairs in HγD-Crys, 12 alanine single mutants of each of the nine Tyr and three Phe residues were constructed by site-directed mutagenesis. Each mutant coding sequence had an N-terminal 6xHis tag. The recombinant mutant proteins were expressed in E. coli at 37°C and purified using a Ni-NTA column. The purification protocol was originally optimized for the wildtype (WT) HγD-Crys, but was reasonably efficient with the mutant proteins. Soluble protein yields for 10 out of the 12 single mutant proteins were comparable to the WT protein. Both the soluble and insoluble fractions of the lysate contained considerable amounts of the recombinant proteins by SDS-PAGE. However, two single mutant proteins Y133A and Y138A of Greek key 4 remained largely in the insoluble inclusion body fractions, and the soluble fractions yielded less than 1/10 of the WT amount. Expression at 18°C for longer periods of time failed to improve the soluble yields of these two mutant proteins. Nonetheless, sufficient amounts of these two single mutant proteins were recovered in soluble form for the subsequent experiments. Three double mutants were also constructed, and used for double mutant cycle analysis described in a later section. All other experiments involved only the 12 single mutant proteins.
Structural assessment
Far-UV circular dichroism (CD) was used for gross secondary structural assessment of the purified WT and mutant HγD-Crys. Overall, the spectra of the mutant proteins were very similar to that of the WT protein, displaying a major peak at 218 nm with similar intensity (Fig. 2). This indicated that the mutant proteins maintained the folded β-sheet structures very similar to the WT protein. A small inflection at 208 nm may reflect a minimal amount of helical content for the WT protein. This feature was also retained for all the mutant proteins, although it appeared less obvious for some of the mutant proteins (Fig. 2).
Figure 2.
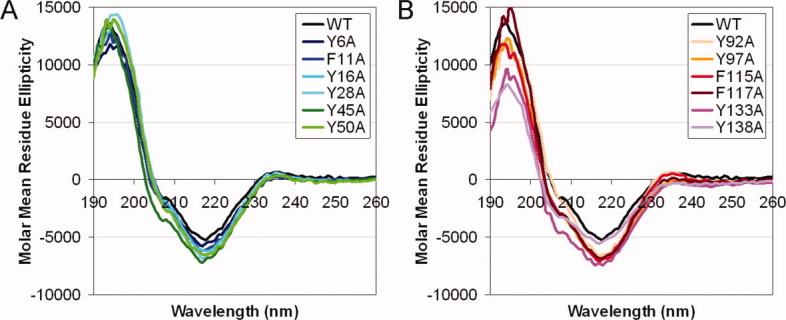
Circular dichroism for WT and mutant HγD-Crys. The samples were prepared as: 100 μg/mL protein in buffer containing 10 mM NaH2PO4/Na2HPO4, pH 7.0. Each protein was scanned from 190–260 nm, at 25°C. Molar mean residue ellipticity was calculated from the raw signal. Averages of independent triplicates for each protein are shown. (A) N-td mutant proteins. (B) C-td mutant proteins.
Thermal denaturation
As an initial test for stability, thermal denaturation experiments were performed on the WT and mutant HγD-Crys. Ellipticity at 218 nm was monitored while the samples were heated slowly from 25 to 95°C. Wavelength 218 nm is the characteristic wavelength of antiparallel β-sheet structure, and also the major peak wavelength found in the initial structural assessment.
The results showed that for the WT and all mutant proteins, ellipticity at 218 nm became less negative as temperature increased (Fig. 3). At the end of each experiment, the protein solution became turbid from particulate aggregates. The decrease in ellipticity signal was therefore likely due to both the loss of secondary structure and aggregate formation that increased light scattering. All WT and mutant HγD-Crys had similar two-state thermal denaturation traces and similar degrees of cooperativity, with no obvious intermediates. The transition midpoint for the WT protein was 80.9°C. For the mutant proteins, the transition midpoints varied from 71.3 to 81.1°C (Fig. 3; Table II). Mutant proteins F115A and F117A had transition midpoints indistinguishable from the WT protein. The other 10 mutant proteins had transition midpoints lower than the WT protein to different degrees, indicating that the corresponding five aromatic pairs but not F115/F117 contributed to the thermal stability of HγD-Crys.
Figure 3.
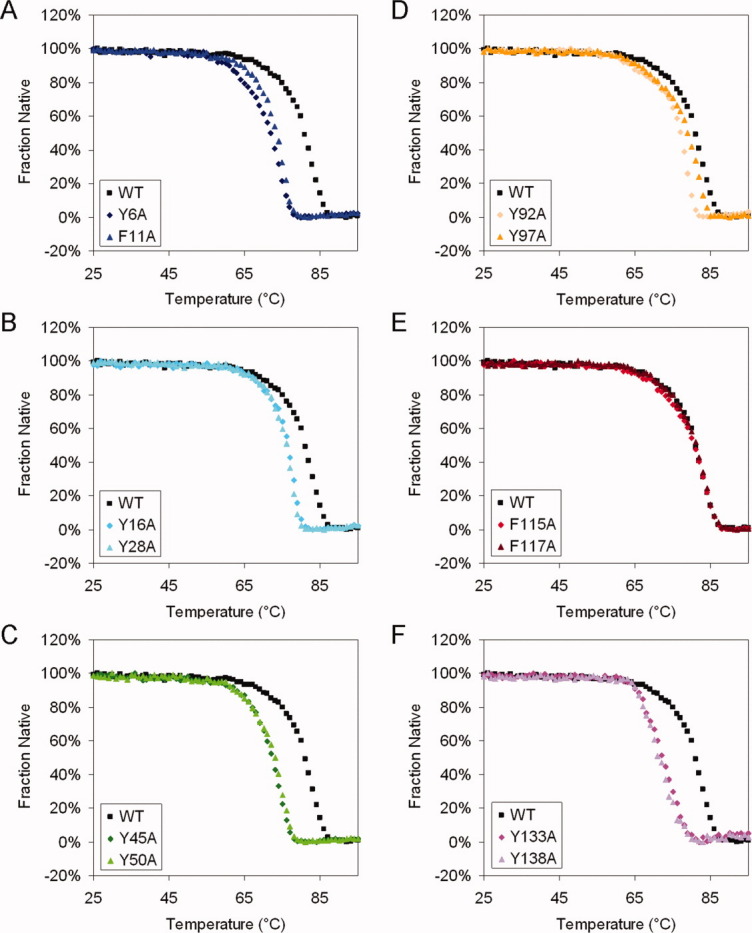
Thermal denaturation for WT and mutant HγD-Crys. The samples were prepared as: 100 μg/mL purified protein in 10 mM NaH2PO4/Na2HPO4, pH 7.0. Temperature was slowly raised from 25 to 90°C in 1°C steps, and equilibrated for 1 min at each temperature; CD at 218 nm was measured for every °C. Averages of at least two independent repeats for each protein are shown. The data were normalized to the highest and lowest data points. (A) Y6A, F11A. (B) Y16A, Y28A. (C) Y45A, Y50A. (D) Y92A, Y97A. (E) F115A, F117A. (F) Y133A, Y138A. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Table II.
Thermal Denaturation and Equilibrium Unfolding Parameters for WT and Mutant HγD-Crys
| Thermal Denaturation | Equilibrium Transition 1 | Equilibrium Transition 2 | |||||
|---|---|---|---|---|---|---|---|
| Protein | Tma | Cmb | Apparent m valuec | Apparent ΔΔG0d | Cmb | Apparent m valuec | Apparent ΔΔG0d |
| N-td | |||||||
| WT | 80.9 ± 0.3 | 2.21 ± 0.05 | 4.6 ± 0.4 | 0.0 ± 0.8 | 2.99 ± 0.03 | 2.9 ± 0.3 | 0.0 ± 1.3 |
| Y6A | 72.0 ± 0.1 | 1.12 ± 0.03 | 4.5 ± 0.4 | −4.8 ± 0.3 | 3.02 ± 0.03 | 2.8 ± 0.1 | −0.3 ± 0.5 |
| F11A | 73.4 ± 0.2 | 1.47 ± 0.05 | 4.2 ± 0.3 | −3.7 ± 0.3 | 3.09 ± 0.09 | 2.6 ± 0.8 | −1.1 ± 2.2 |
| Y16A | 76.4 ± 0.1 | 1.85 ± 0.03 | 4.4 ± 0.1 | −1.7 ± 0.2 | 3.00 ± 0.06 | 2.8 ± 0.3 | −0.8 ± 0.7 |
| Y28A | 76.0 ± 0.0 | 1.77 ± 0.05 | 4.1 ± 0.8 | −2.5 ± 1.3 | 2.91 ± 0.09 | 3.0 ± 0.4 | −0.2 ± 1.1 |
| Y45A | 72.2 ± 0.1 | 1.13 ± 0.03 | 3.8 ± 0.2 | −5.5 ± 0.2 | 2.99 ± 0.02 | 2.9 ± 0.3 | −0.3 ± 0.8 |
| Y50A | 72.9 ± 0.1 | 1.20 ± 0.02 | 4.4 ± 0.5 | −4.6 ± 0.6 | 2.94 ± 0.06 | 2.8 ± 0.5 | −0.9 ± 1.3 |
| C-td | |||||||
| WT | 80.9 ± 0.3 | 2.81 ± 0.02 | 0.6 ± 0.0 | 0.0 ± 0.0 | − | – | – |
| Y92A | 77.0 ± 0.2 | 1.80 ± 0.04 | 3.0 ± 0.5 | 3.6 ± 0.7 | – | – | – |
| Y97A | 79.0 ± 0.2 | 2.21 ± 0.04 | 1.6 ± 0.4 | 1.7 ± 0.9 | – | – | – |
| F115A | 80.6 ± 0.1 | 2.50 ± 0.02 | 1.1 ± 0.1 | 1.0 ± 0.2 | – | – | – |
| F117A | 81.1 ± 0.0 | 2.58 ± 0.07 | 1.0 ± 0.1 | 0.9 ± 0.2 | – | – | – |
| Y133A | 72.1 ± 0.2 | 1.93 ± 0.01 | 1.7 ± 0.1 | 1.4 ± 0.3 | – | – | – |
| Y138A | 71.3 ± 0.6 | 1.98 ± 0.01 | 2.2 ± 0.1 | 2.6 ± 0.2 | – | – | – |
Thermal denaturation transition midpoints in units of °C.
Equilibrium unfolding transition midpoints in units of M GuHCl.
Apparent m values in units of kcal mol−1 M−1.
Difference of free energy change extrapolated to 0 M GuHCl, as compared to the WT protein, in units of kcal mol−1. A negative number means that the mutant is less stable than the WT.
“–” not applicable. Errors are standard deviations of three trials.
Substitutions of the pairs Y6/F11, Y45/Y50, and Y133/Y138 had the largest effects on thermal stability: the transition midpoints ranged from 71.3 to 73.4°C, substantially lower than 80.9°C of the WT protein (Table II). Within each crystallin domain, substitutions of the Greek key pairs (N-td: Y6/F11, Y45/Y50; C-td: Y92/Y97, Y133/Y138) had larger effects than substitutions of the non-Greek-key pairs (N-td: Y16/Y28; C-td: F115/F117), indicating that the Greek key pairs are especially important for the thermal stability of the proteins. For the first Greek key pair in each crystallin domain, substitution of the first residue (N-td: Y6; C-td: Y92) had a larger effect than substitution of the second residue (N-td: F11; C-td: Y97).
Equilibrium unfolding/refolding
To assess the effects of the Tyr or Phe substitutions on thermodynamic stability of HγD-Crys, equilibrium unfolding/refolding experiments were performed on the WT and mutant proteins. The samples were prepared in GuHCl at pH 7.0, and incubated at 37°C for 24 hr. The fluorescence ratio 360/320 was used to assess the unfolded/native state. Upon refolding at very low GuHCl concentration, WT HγD-Crys aggregation competes with refolding, and interferes with the fluorescence signal.13 This behavior was also observed for all 12 single mutant proteins. At 1.0 M or higher GuHCl concentration, the equilibrium refolding traces overlapped very well with the unfolding traces. Therefore, the refolding traces were used mainly to ensure that the samples were fully equilibrated. The unfolding traces are used for the following analysis.
For WT HγD-Crys, a three-state equilibrium model best fit the equilibrium trace (Fig. 4): black traces in each panel. The bottom baseline at low GuHCl concentration corresponded to the native state, while the top baseline at high GuHCl concentration corresponded to the unfolded state. The intermediate at fluorescence ratio ∼1.0 (a small kink in the trace) was previously identified as a single-domain-folded species with the N-td unfolded and C-td folded. The N-td transition midpoint was 2.21 M GuHCl, and the C-td transition midpoint was 2.99 M (Table II).
Figure 4.
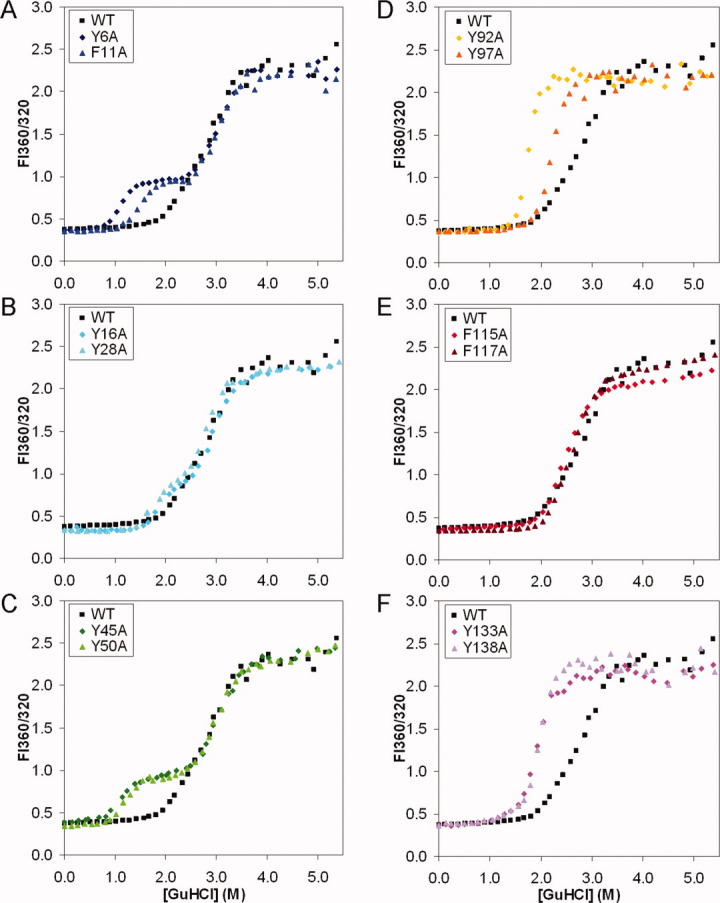
Equilibrium unfolding for WT and mutant HγD-Crys. The samples were prepared as: 10 μg/mL protein, in 100 mM NaH2PO4/Na2HPO4, 5 mM DTT, 1 mM EDTA, in varying [GuHCl] at pH 7.0, 37°C, incubated for 24 hr. Fluorescence spectra were measured at excitation wavelength 295 nm. Ratios of fluorescence intensity of 360/320 are extracted and plotted against [GuHCl]. Representative traces of independent triplicates for each protein are shown. (A) Y6A, F11A. (B) Y16A, Y28A. (C) Y45A, Y50A. (D) Y92A, Y97A. (E) F115A, F117A. (F) Y133A, Y138A. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Each of the mutant proteins had a different equilibrium profile compared to the WT (Fig. 4). From the equilibrium traces, parameters including transition midpoints, apparent m values and ΔΔG's compared to the WT protein were calculated (Table II). Figure 4(A–C) shows that compared to the WT protein, each of the N-td mutant proteins had the N-td transition shifted to lower GuHCl concentration, indicating a destabilized N-td. The C-td transitions remained unaffected. Thus, an increased population of the single-domain-folded intermediate was revealed. A three-state equilibrium model was best fit to these data. Two transition midpoints were calculated: the N-td transition midpoints ranged from 1.12 to 1.85 M, significantly lower than 2.21 M for the WT protein. The C-td transition midpoints ranged from 2.91 to 3.09 M, closely centered around 2.99 M for the WT protein. Figure 4(D–F) showed that compared to the WT protein, C-td mutant proteins had both the N-td and C-td destabilized. Also, the N-td and C-td transitions were closer and inseparable, indicating a more cooperative unfolding/refolding process. A two-state equilibrium model was best fit to these data. One transition midpoint was calculated. This ranged from 1.80 to 2.58 M, significantly lower than 2.81 M of the WT protein, if the WT transition was also considered two-state. These results clearly showed that all the aromatic pairs were important for the thermodynamic stability of HγD-Crys.
Comparing ΔΔG's and transition midpoints yielded similar results for the N-td mutant proteins. However, for the C-td mutant proteins, because the N-td and C-td transitions partially overlap, the apparent ΔΔG values probably reflect the level of separation of the two transitions, rather than the genuine thermodynamic stability of the mutant proteins. Therefore, our analysis focuses on the transition midpoints. Comparing the mutant proteins carrying substitutions within the same crystallin domain, two patterns regarding the extents of destabilization emerged, and they were consistent with the patterns of thermal denaturation. (1) The Greek key pairs (N-td: Y6/F11, Y45/Y50; C-td: Y92/Y97, Y133/Y138) had larger contributions to the thermodynamic stability than the non-Greek-key pairs (N-td: Y16/Y28, C-td: F115/F117). (2) For the first Greek key pair in each crystallin domain, the first residue (N-td: Y6, C-td: Y92) had a larger contribution than the second residue (N-td: F11, C-td: Y97).
Kinetic unfolding/refolding at 37°C
Decreased thermodynamic stability of the mutant HγD-Crys pointed to changes in unfolding and/or refolding rates. To further distinguish the effects on unfolding vs. refolding, kinetic unfolding and refolding experiments were performed on the WT and mutant proteins. Initial kinetic experiments were performed at 37°C, with one final GuHCl concentration each for unfolding and refolding. For unfolding, native protein was mixed into 5.5 M GuHCl. For refolding, fully denatured protein in 5.5 M GuHCl was diluted to a final concentration of 1.0 M GuHCl. One molar was chosen because at concentration below 1.0 M, refolding HγD-Crys species formed aggregates that interfered with the fluorescence signal. Fluorescence at 350 nm was monitored for 1 hr. The results showed that the WT protein exhibited an early/fast and a late/slow phase, corresponding to the two crystallin domains as previously suggested [Fig. 5(A) and 6(A)].15,17 The fast phase was not resolved by the instrument at 37°C, so our initial analysis focuses on the slow phase.
Figure 5.
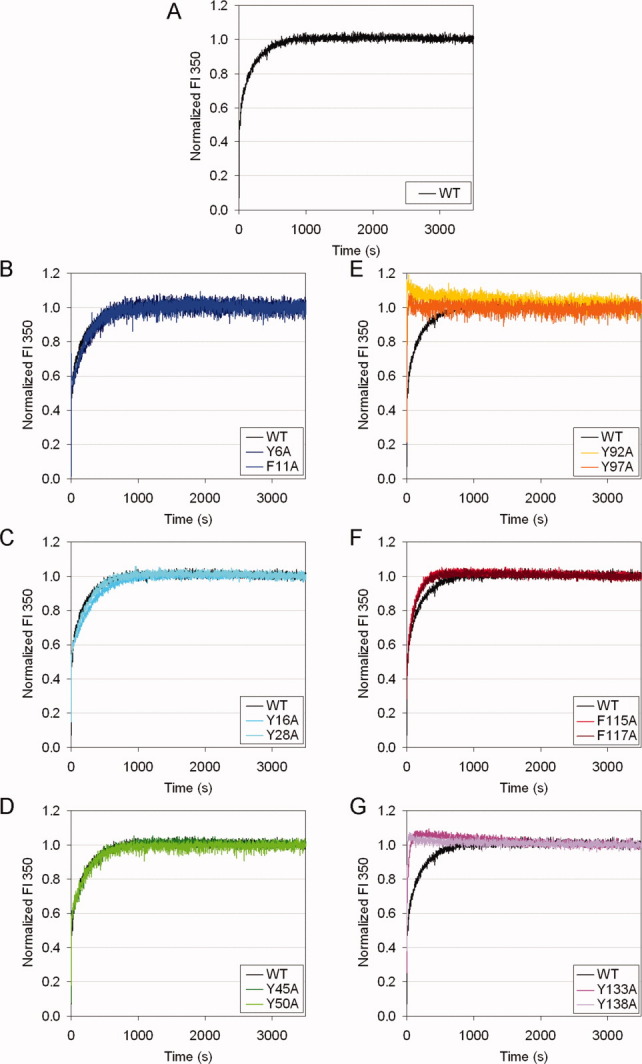
Kinetic unfolding for WT and mutant HγD-Crys at 37°C. The samples were prepared in 100 mM NaH2PO4/Na2HPO4, 5 mM DTT, 1 mM EDTA, pH 7.0, at 37°C. Unfolding was initiated by mixing native protein into 5.5 M GuHCl. Fluorescence at 350 nm was monitored. Fluorescence intensity was normalized to the native and unfolded protein controls. Representative traces of independent triplicates for each protein are shown. (A) WT. (B) Y6A, F11A. (C) Y16A, Y28A. (D) Y45A, Y50A. (E) Y92A, Y97A. (F) F115A, F117A. (G) Y133A, Y138A.
For unfolding, the N-td mutant proteins had no observable difference compared to the WT protein [Fig. 5(B–D)]. In contrast, C-td mutant proteins unfolded significantly faster than the WT protein [Fig. 5(E–G)]. In particular, mutant proteins of the Greek key pairs unfolded extremely fast, with the fluorescence signals reaching their final levels in less than 1 min, compared to about 15 min for the WT protein [Fig. 5(E,G)]. Mutant proteins of the non-Greek-key pair unfolded slightly faster than the WT protein [Fig. 5(F)]. These results showed that the C-td aromatic pairs made important contributions to the kinetic stability of HγD-Crys, while the N-td aromatic pairs were less important.
For refolding, mutant proteins of the second Greek key pair in each crystallin domain refolded significantly slower than the WT protein [Fig. 6(D,G)], but affected different phases. For Y45A, Y50A proteins, the initial 70% of the fluorescence changes occurred at similar rates as for the WT protein, but the later 30% occurred at slower rates. This result was similar to the refolding kinetics of the interface mutant HγD-Crys.15,21,22 Unlike Y45A and Y50A proteins, the refolding of Y133A and Y138A proteins were slower than the WT protein from the beginning of the reaction. From these results, it appeared that the pair Y133/Y138 was important for the early phase in the refolding process, while the pair Y45/Y50 was important for the late phase. Except for these two pairs, all other mutant proteins had no observable differences compared to the WT protein [Fig. 6(B,C,E,F)]. These results indicated that the second Greek key pair in each crystallin domain contributed to the refolding of HγD-Crys, while the other pairs were less important.
Figure 6.
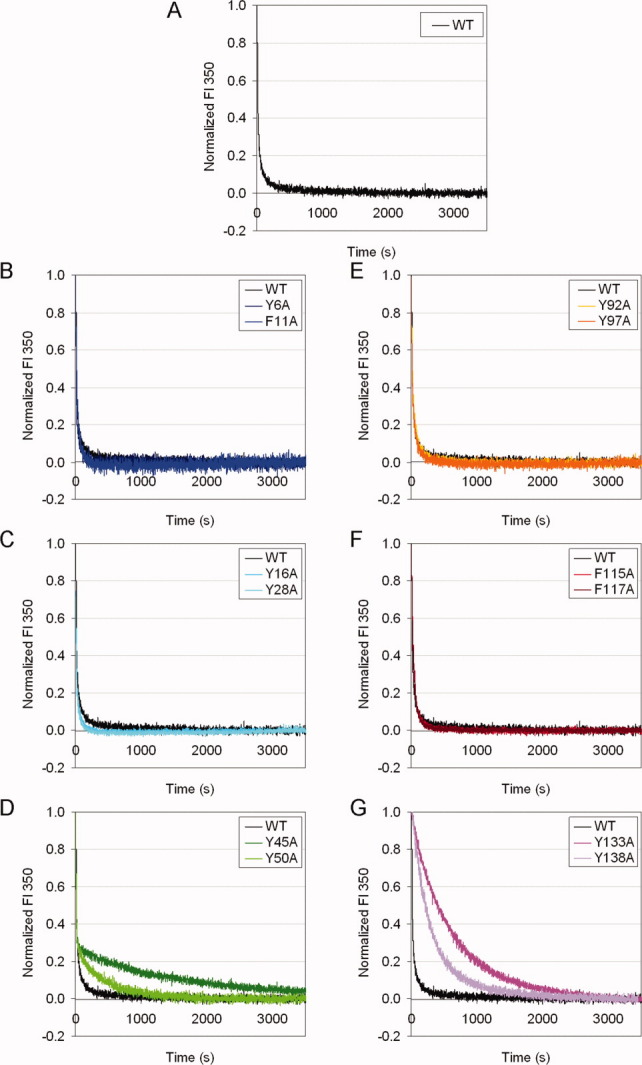
Kinetic refolding for WT and mutant HγD-Crys at 37°C. The samples were prepared in 100 mM NaH2PO4/Na2HPO4, 5 mM DTT, 1 mM EDTA, pH 7.0, at 37°C. Refolding was done by diluting fully unfolded protein in 5.5 M GuHCl into a final GuHCl concentration of 1.0 M. Fluorescence at 350 nm was monitored. Fluorescence intensity was normalized to the native and unfolded protein controls. Representative traces of independent triplicates for each protein are shown. (A) WT. (B) Y6A, F11A. (C) Y16A, Y28A. (D) Y45A, Y50A. (E) Y92A, Y97A. (F) F115A, F117A. (G) Y133A, Y138A.
Kinetic unfolding/refolding at 18°C
In the kinetic experiments at 37°C reported above, it was puzzling to see that four mutant HγD-Crys, Y6A, F11A, Y16A, and Y28A, had no obvious differences compared to the WT protein in either unfolding or refolding. To slow down the fast phase of the kinetic traces that were not resolved by the instrument at 37°C, the kinetic experiments were repeated under identical conditions to those described above, only at 18°C instead of 37°C.
The unfolding process of the WT and the four mutant proteins tested was much slower at 18°C than at 37°C, lasting for about 3 hr before reaching the final level, compared to about 15 min at 37°C [Fig. 7(A–C insets)]. As a whole, the unfolding traces of the WT and the mutant proteins overlapped very well. However, a close examination of the beginning of the traces revealed a “burst” for the mutant proteins, where the mutant proteins unfolded slightly but significantly faster than the WT protein [Fig. 7(A–C main panels)]. The mutant proteins were then caught up by the WT protein about 10 min into the experiment and at about 50% of the final fluorescence level. All four mutant proteins behaved similarly. This result indicated that these N-td aromatic pairs determined the unfolding rate of the early phase; however, the late phase was independent of these N-td aromatic pairs.
Figure 7.
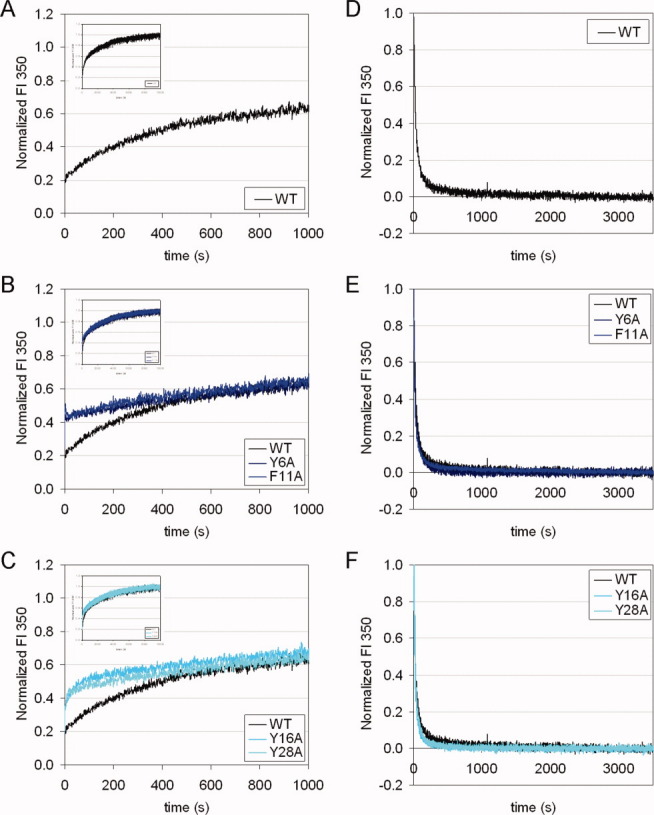
Kinetic unfolding/refolding for WT and mutant HγD-Crys at 18°C. Experiments were performed under identical conditions to those shown in Figures 5 and 6, except that temperature was maintained at 18°C, instead of 37°C. Left panels (A)–(C): unfolding. The main panels show the first 20 min, and the insets show the full-length experiments for 3 hr (x-axis maximum: 10,000 sec). Right panels (D)–(F): refolding. Top panels (A) and (D): WT. Middle panels (B) and (E): Y6A, F11A. Bottom panels (C) and (F): Y16A, Y28A.
For refolding, lowering the temperature from 37 to 18°C did not slow down the refolding process for neither the WT nor mutant proteins [Fig. 7(D–F)]. The refolding processes reached the final level in about 10 min at both temperatures. The refolding traces of the WT and mutant proteins still overlapped very well at 18°C, like at 37°C [Fig. 7(E,F)]. This result further supported our conclusion that the refolding kinetics of each crystallin domain depended on only the second Greek key pair but not any other pairs.
Double mutant cycle
Aromatic-aromatic interaction was estimated to be ∼1.3 kcal/mol in the case of barnase by double mutant cycle analysis.35 We employed this method for the three N-td aromatic pairs, because the N-td mutant proteins had separated, well-defined N-td and C-td equilibrium transitions. Three additional mutants of HγD-Crys: double mutants Y6A/F11A, Y16A/Y28A, and Y45A/Y50A were constructed, expressed and subjected to equilibrium unfolding/refolding experiments described above. ΔG's were calculated and compared with those of the corresponding pair of single mutant proteins. Aromatic-aromatic interaction energies for pairs Y6/F11, Y16/Y28, Y45/Y50 were −1.6, −1.5, and −2.0 kcal/mol, respectively. These numbers were within reasonable range of the literature values,32,35 consistent with the notion that aromatic interaction is a “weak force.” The results also suggested that the contributions of these aromatic residues to thermodynamic stability originated partially from the pair-wise aromatic-aromatic interaction, and partially from the interactions of these residues with other neighboring residues.
Discussion
The stability of crystallins is critical for maintenance of the transparency and appropriate refractive index of the vertebrate eye lens. The amino acid determinants of the stability of the βγ-crystallins probably reflect the folding/unfolding of the intertwined β-strands of the double Greek keys. The UV-damage model of cataractogenesis suggests that the aromatic residues could be the key to the folding and stability of lens crystallins. HγD-Crys contains six Tyr/Tyr, Tyr/Phe, or Phe/Phe pairs: four highly conserved Greek key pairs, and two moderately conserved non-Greek-key pairs. They are all located at structurally crucial β-hairpins, bridging two neighboring β-strands. Five of these six pairs (except for Y16/Y28) have partners five or fewer residues apart in the primary sequence. Burly and Petsko in their pioneering work suggested that aromatic-aromatic interactions may be nucleation sites for protein folding.32 The proximity of the partners in the primary sequence makes these aromatic pairs in HγD-Crys very likely to be such nucleation sites during the early steps of the folding process. After the protein matures into the native structure, these aromatic pairs may also act as “clasps” to stabilize local conformations, and thus contribute to the overall stability of the protein. Using site-directed mutagenesis followed by thermal, equilibrium, and kinetic experiments, we performed initial tests of these hypotheses. The results allowed us to confirm and expand upon the previously proposed HγD-Crys unfolding/refolding pathway.
Soluble protein yields and far-UV CD spectra showed that loss of one or two aromatic residues did not prevent most of the mutant HγD-Crys from folding into near-native structures. However, pair Y133/Y138 seemed essential for folding, since newly synthesized HγD-Crys was largely insoluble without either of these two residues. This was presumably due to intracellular aggregation into inclusion bodies that overwhelmed a defective folding process. For the small fractions of Y133A and Y138A proteins that did fold up and were soluble, they were able to maintain the near-native structure in buffer. These results indicated that these six aromatic pairs were not required for dictating the Greek key fold. However, the subsequent results indicated that the stability and unfolding/refolding kinetics were compromised by substitutions of specific subsets of these aromatic pairs.
During thermal denaturation, substitutions at various positions throughout HγD-Crys generated very similar two-state transition traces without obvious intermediates, but with transition midpoints shifted to lower temperatures. This result suggested that thermal denaturation of HγD-Crys was probably a highly cooperative process. All the stabilizing factors within the protein collectively determine a single “melting point,” at which different parts of the protein unfold simultaneously. However, aggregation occurred during the thermal denaturation process, and it was not clear when aggregation started to dominate during thermal denaturation. It is possible that the observed two-state process simply reflected the nature of the aggregation process, but not the unfolding process. In this case, more complicated unfolding features such as a three-state transition might be masked. The aggregation may be a result of the heating procedure, higher protein concentration, buffer with low ionic strength, or a combination of these. Nonetheless, the transition midpoints should be valid indicators of the general stability. The results indicated that five out of the six pairs (except F115/F117) clearly contributed to the thermal stability of HγD-Crys. However, the lack of an identifiable intermediate prevented the inference of a sequential pathway of HγD-Crys thermal unfolding.
β-Crystallins generally lack the two non-Greek-key pairs and can therefore be viewed as “natural” aromatic mutants of the γ-crystallins. Human βB1-crystallin lacks three of the four non-Greek-key paired residues while maintaining all of the Greek key pairs. In experiments similar to those in this study, Lampi et al. showed that the thermal transition temperature of βB1-crystallin was about 67°C.49 This transition temperature was lower than any of the single aromatic mutant HγD-Crys, despite the possible dimeric state that could enhance the stability of βB1-crystallin. The aromatic residues may be the key to the different thermal stabilities of β- and γ-crystallins.
Comparing the relative stabilities of the WT and mutant HγD-Crys carrying substitutions within the same crystallin domain, we observed two patterns common in both thermal and equilibrium experiments. (1) Substitutions of the Greek key pairs had larger effects than substitutions of the non-Greek-key pairs. This result was not surprising considering the higher level of conservation of the Greek key pairs, across the four Greek keys in HγD-Crys and across the βγ-crystallin superfamily, in terms of both primary sequence and three-dimensional structure. It can be concluded that one of the functions of these Greek key pairs is maintenance of protein stability. (2) For the first Greek key pair in each crystallin domain, substitution of the first residue had larger effects than substitution of the second residue. This may be because the first residue is slightly more buried, and thus has slightly more interaction with the core of the motif.
The result that all mutant HγD-Crys had destabilized equilibrium profiles indicated that all six aromatic pairs must contribute to some aspect(s) of unfolding and/or refolding kinetics. Surprisingly, for the four N-td mutant proteins Y6A, F11A, Y16A, and Y28A, neither kinetic unfolding nor refolding experiment revealed any obvious difference compared to the WT protein at 37°C. The mixing system used in all the kinetic experiments in this study had a dead time on the order of seconds. Therefore, we suspected that there might be differences in the fast phase that were not detected due to this limitation. By repeating the kinetic experiments at 18°C, we successfully captured a subtle but significant “burst” for the four N-td mutant proteins Y6A, F11A, Y16A, and Y28A compared to the WT at the beginning of the unfolding traces. These four N-td substitutions accelerated the early phase, but did not subsequently accelerate the late phase. This indicated that the fast phase, although occurred before the slow phase, was not a prerequisite for the slow phase to occur. At 37°C, only C-td but not N-td substitutions accelerated the late phase. Combining these results at both temperatures, we propose that the unfolding of the two crystallin domains of HγD-Crys are independent, but appear to be sequential due to intrinsic stability difference [Fig. 8(A)]. All β-hairpin aromatic pairs contribute to the kinetic stability of each crystallin domain. Double mutant cycle analysis showed that aromatic-aromatic interactions of the pairs was one source of stability. However, our results did not show evidence regarding the sequential unfolding of the two Greek key motifs within a crystallin domain.
Figure 8.
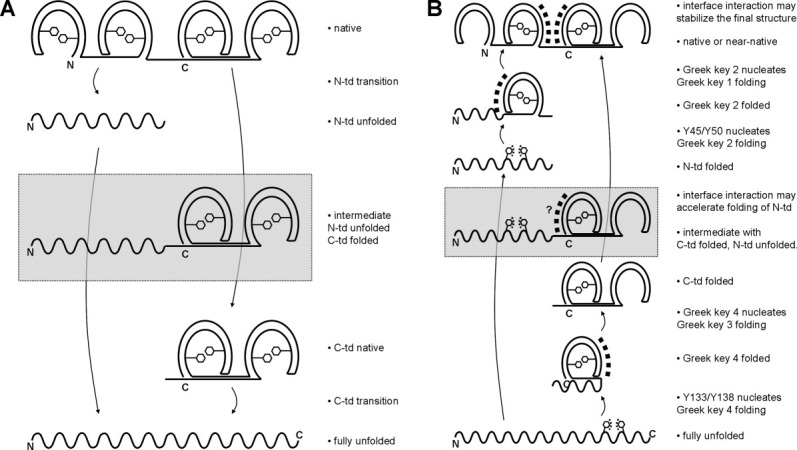
Schematic diagram of the proposed HγD-Crys unfolding and refolding model. The four Greek keys are colored blue, green, yellow, and red from N- to C-terminus. Horseshoe-shape symbols represent folded Greek keys, while the wavy lines represent unfolded chains. Hexagonal symbols represent Greek key aromatic pairs. Square dots indicate molecular events in action. See text next to each panel for more explanation. The species in gray boxes are the observed, but probably not obligate intermediates, due to independent domain unfolding/refolding. (A) Unfolding. (B) Refolding.
The refolding rate of HγD-Crys was only significantly slowed down by substitutions of the second Greek key pair in each crystallin domain but not any other pair, even though the two Greek key pairs in a crystallin domain are homologous in sequence and structure. Lowering the temperature to 18°C neither slowed down the refolding process of the WT and mutant proteins Y6A, F11A, Y16A, Y28A, nor revealed any differences in refolding among the WT and these mutant proteins. These results indicated that unlike unfolding, refolding had limited temperature dependency, and also confirmed the lesser role of the corresponding residues in refolding. Flaugh et al. proposed that unfolding/refolding of a crystallin domain is reversible through an intermediate with the first Greek key unfolded and the second at Greek key folded.15 This proposal with details at the Greek key level was largely based on data that showed that the second Greek key had stabilizing effects,16,50 but might overlook other structural elements in the first Greek key that are equally stabilizing50 [Fig. 4(A,D)]. Our experimental evidence of unequal contributions to the refolding kinetics from the two homologous Greek key pairs in the same crystallin domain strongly argues that the second Greek keys are involved in the rate-determining step of crystallin domain folding. Based on previous studies suggesting that aromatic-aromatic interactions may nucleate protein folding,32,50 we propose that this rate-determining step is a nucleation event from the aromatic-aromatic interaction of the second Greek key pair, and then the first Greek key folds using the folded second Greek key as a template [Fig. 8(B)]. However, we cannot rule out the possibility that this rate-determining step is a late step that is vulnerable to amino acid substitution. Yet another alternative is that the apparent unequal contributions may be due to the asymmetric positions of tryptophans within the crystallin domain, leading to different sensitivity of the fluorescence signal to structural changes in different parts of the crystallin domain.
At the crystallin domain level, although it is well-established that the C-td folds first, followed by the N-td, during HγD-Crys refolding [Fig. 6(D,G)],15,17,24 the role of the domain interface in folding remains unclear. While Flaugh et al. showed that substitutions of interface residues slowed down the refolding of the N-td,15,21,22 Mills et al. found that isolated N-td was able to refold with a comparable rate to full-length. HγD-Crys, although it may have slight structural alterations from the N-td in the full-length protein.24 Independent domain folding was also shown to be a property of bovine γB-crystallin.51 In HγD-Crys, the interface interactions may not be required for the initial refolding process of the N-td, but could be important for stabilizing the final conformation of the N-td after it folds up [Fig. 8(B)].
A number of computational studies offered insights on the HγD-Crys folding pathway. Using a structure-based model, Itoh and Sasai recaptured the sequential folding model of Flaugh et al.15,52 Their results also showed that the connectivity of HγD-Crys determined the folding sequence of the two crystallin domains. Among the six aromatic pairs, the Greek key pair Y133/Y138 is particularly interesting because substitutions of this pair affected all the parameters tested, including thermal stability, thermodynamic stability, unfolding, and refolding kinetics. These results helped to explain the insolubility of mutant proteins Y133A and Y138A during expression. Interestingly, Das et al. identified a salt bridge, E134-R141, as a nucleation site for folding by molecular dynamics simulations,25 and the location coincided with the pair Y133/Y138, our proposed nucleation site of the initial folding of HγD-Crys. The region around the β-hairpin at Greek key 4 seems to be highly crucial for the folding and structural integrity of HγD-Crys.
Materials and Methods
Site-specific mutagenesis
N-terminally 6xHis tagged WT HγD-Crys pQE.1 plasmid (Qiagen) was amplified by PCR-based mutagenesis using primers encoding the desired mutations. The primers were designed from PrimerX http://www.bioinformatics.org/primerx/ and ordered from Integrated DNA Technologies. The mutant plasmids were amplified in E. coli Nova Blue, purified by miniprep and sequenced at Massachusetts General Hospital to confirm the correct mutations. Double mutants were constructed using the confirmed single mutant plasmids as templates in a two-step procedure.
Expression and purification
E. coli M15(pREP4) were transformed with the purified plasmids, cultured on shakers at 37°C until OD600 reached ∼1.0, then induced with 1 mM IPTG, and allowed for expression for 3 hr. The cells were pelleted and resuspended in 300 mM NaCl, 50 mM NaH2PO4/Na2HPO4, 18 mM imidazole, pH 8.0. The cells were lysed by sonication and centrifuged at 20,000g for 30 min. The supernatant was applied to a Ni-NTA column (Qiagen) by an FPLC system (GE Healthcare), eluted with a linear gradient of imidazole from 18 to 250 mM at 4°C. Purity was ensured by SDS-PAGE. Fractions exceeding about 95% purity were pooled, dialyzed with 10 mM ammonium acetate, pH 7.0, at 4°C, and concentrated with Amicon centrifuge filters with 10 kDa MWCO. Protein concentrations were measured by A280 in 5.5 M GuHCl, using extinction coefficient calculated from Expasy http://ca.expasy.org/tools/protparam.html.
Structural assessment by CD
Far-UV CD spectra were obtained with an AVIV Model 202 CD spectrophotometer. The samples were prepared as follows: 100 μg/mL protein in 10 mM NaH2PO4/Na2HPO4, pH 7.0, 0.2 μm filtered, degassed for >1 hr. Each protein was scanned at 190–260 nm, 5 sec averaging time of signal, at 25°C. Triplicates of independent experiments were done for each protein.
Thermal denaturation
Sample preparation and the instrument were the same as in the Structural Assessment section above. Temperature was raised from 25 to 90°C in 1°C steps, equilibrated for 1 min at each temperature, CD signal at 218 nm was measured for every °C, 10 sec averaging time. Triplicates of independent experiments were done for each protein. The data were normalized to the maxima and minima, transition midpoints were determined by the intersections of the traces and the 50% lines.
Equilibrium unfolding/refolding
The buffer system was as follows: 100 mM NaH2PO4/Na2HPO4, 5 mM DTT, 1 mM EDTA, pH 7.0, 0.2 μm filtered. Unfolding samples were prepared by diluting native protein into the buffer above, with final protein concentration 10 μg/mL, as well as 38 different concentrations of GuHCl in the range of 0–5.5 M. The samples were incubated at 37°C for 24 hr. Refolding samples were prepared similarly, except that the native protein was first denatured at protein concentration of 100 μg/mL, in 5.5 M GuHCl at 37°C for 5 hr, before being diluted into a 0.55–5.5 M GuHCl gradient.
Fluorescence spectra of the samples were obtained using a Hitachi F-4500 fluorometer with the following specifications: excitation wavelength 295 nm, emission wavelength 310–400 nm, scan speed 60 nm/min, excitation and emission slit band width 10 nm. Temperature was maintained at 37°C. GuHCl concentration was calculated from the refractive index of each sample. The ratio of fluorescence intensity at 360/320 nm was calculated for analysis. The data were fit to two- or three-state equilibrium equations using KaleidaGraph (Synergy software). Triplicates of independent experiments were done for each protein.
Kinetic unfolding/refolding
The buffer system used was the same as in the Equilibrium Unfolding/Refolding section above. For unfolding, buffer with GuHCl at a final concentration of 5.5 M was incubated in a cuvette at 37°C (or 18°C), and stirred with a mini spin bar. Fluorescence was monitored by a Hitachi F-4500 fluorometer with excitation wavelength 295 nm and emission wavelength 350 nm. Native protein at 100 μg/mL (10×) was syringe injected through a small injection port into the cuvette. The final protein concentration was 10 μg/mL. Fluorescence was monitored until a plateau was reached. Triplicates of independent experiments were done for each protein. Refolding experiments were similar to unfolding, except that the 10× protein was incubated with 5.5 M GuHCl at 37°C for 5 hr before being injected into the buffer containing 0.5 M GuHCl. Final GuHCl concentration was 1.0 M.
Acknowledgments
The Biophysical Instrumentation Facility for the Study of Complex Macromolecular Systems at MIT is gratefully acknowledged. We thank Dr. Kate Moreau for providing mutant plasmids, Brian Sosa and Jeannie Chew for preliminary data, and Dr. Jiejin Chen, Dr. Kate Moreau, Dr. Ligia Acosta-Sampson, and Dr. Kelly Knee for insightful discussions.
Glossary
Abbreviations:
- C-td
C-terminal domain
- CD
circular dichroism
- GuHCl
guanidine hydrochloride
- HγD-Crys
Human γD-crystallin
- N-td
N-terminal domain
- sHSP
small heat shock protein
- UV
ultraviolet
- WT
wildtype.
References
- 1.Bron AJ, Vrensen GF, Koretz J, Maraini G, Harding JJ. The ageing lens. Ophthalmologica. 2000;214:86–104. doi: 10.1159/000027475. [DOI] [PubMed] [Google Scholar]
- 2.Horwitz J. The function of alpha-crystallin in vision. Semin Cell Dev Biol. 2000;11:53–60. doi: 10.1006/scdb.1999.0351. [DOI] [PubMed] [Google Scholar]
- 3.Horwitz J. Alpha-crystallin. Exp Eye Res. 2003;76:145–153. doi: 10.1016/s0014-4835(02)00278-6. [DOI] [PubMed] [Google Scholar]
- 4.Bloemendal H, de Jong W, Jaenicke R, Lubsen NH, Slingsby C, Tardieu A. Ageing and vision: structure, stability and function of lens crystallins. Prog Biophys Mol Biol. 2004;86:407–485. doi: 10.1016/j.pbiomolbio.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Basak A, Bateman O, Slingsby C, Pande A, Asherie N, Ogun O, Benedek GB, Pande J. High-resolution X-ray crystal structures of human gammaD crystallin (1.25 A) and the R58H mutant (1.15 A) associated with aculeiform cataract. J Mol Biol. 2003;328:1137–1147. doi: 10.1016/s0022-2836(03)00375-9. [DOI] [PubMed] [Google Scholar]
- 6.Wistow G, Summers L, Blundell T. Myxococcus xanthus spore coat protein S may have a similar structure to vertebrate lens beta gamma-crystallins. Nature. 1985;315:771–773. doi: 10.1038/315771a0. [DOI] [PubMed] [Google Scholar]
- 7.Wistow G. Evolution of a protein superfamily: relationships between vertebrate lens crystallins and microorganism dormancy proteins. J Mol Evol. 1990;30:140–145. doi: 10.1007/BF02099940. [DOI] [PubMed] [Google Scholar]
- 8.Shimeld SM, Purkiss AG, Dirks RP, Bateman OA, Slingsby C, Lubsen NH. Urochordate betagamma-crystallin and the evolutionary origin of the vertebrate eye lens. Curr Biol. 2005;15:1684–1689. doi: 10.1016/j.cub.2005.08.046. [DOI] [PubMed] [Google Scholar]
- 9.Bassnett S. Lens organelle degradation. Exp Eye Res. 2002;74:1–6. doi: 10.1006/exer.2001.1111. [DOI] [PubMed] [Google Scholar]
- 10.Siezen RJ, Thomson JA, Kaplan ED, Benedek GB. Human lens gamma-crystallins: isolation, identification, and characterization of the expressed gene products. Proc Natl Acad Sci U S A. 1987;84:6088–6092. doi: 10.1073/pnas.84.17.6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brakenhoff RH, Aarts HJ, Reek FH, Lubsen NH, Schoenmakers JG. Human gamma-crystallin genes. A gene family on its way to extinction. J Mol Biol. 1990;216:519–532. doi: 10.1016/0022-2836(90)90380-5. [DOI] [PubMed] [Google Scholar]
- 12.Lampi KJ, Ma Z, Shih M, Shearer TR, Smith JB, Smith DL, David LL. Sequence analysis of betaA3, betaB3, and betaA4 crystallins completes the identification of the major proteins in young human lens. J Biol Chem. 1997;272:2268–2275. doi: 10.1074/jbc.272.4.2268. [DOI] [PubMed] [Google Scholar]
- 13.Kosinski-Collins MS, King J. Protein Sci. 2003;12:480–490. doi: 10.1110/ps.0225503. In vitro unfolding, refolding, and polymerization of human gammaD crystallin, a protein involved in cataract formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mills-Henry I. Stability, unfolding, and aggregation of the gamma D and gamma S human eye lens crystallins. Department of Biology. Massachusetts Institute of Technology; 2007. [Google Scholar]
- 15.Flaugh SL, Mills IA, King J. Glutamine deamidation destabilizes human gammaD-crystallin and lowers the kinetic barrier to unfolding. J Biol Chem. 2006;281:30782–30793. doi: 10.1074/jbc.M603882200. [DOI] [PubMed] [Google Scholar]
- 16.MacDonald JT, Purkiss AG, Smith MA, Evans P, Goodfellow JM, Slingsby C. Unfolding crystallins: the destabilizing role of a beta-hairpin cysteine in betaB2-crystallin by simulation and experiment. Protein Sci. 2005;14:1282–1292. doi: 10.1110/ps.041227805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kosinski-Collins MS, Flaugh SL, King J. Probing folding and fluorescence quenching in human gammaD crystallin Greek key domains using triple tryptophan mutant proteins. Protein Sci. 2004;13:2223–2235. doi: 10.1110/ps.04627004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Flaugh SL, Callis PR, King J. Mechanism of the highly efficient quenching of tryptophan fluorescence in human gammaD-crystallin. Biochemistry. 2006;45:11552–11563. doi: 10.1021/bi060988v. [DOI] [PubMed] [Google Scholar]
- 19.Chen J, Toptygin D, Brand L, King J. Mechanism of the efficient tryptophan fluorescence quenching in human gammaD-crystallin studied by time-resolved fluorescence. Biochemistry. 2008;47:10705–10721. doi: 10.1021/bi800499k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, Callis PR, King J. Mechanism of the very efficient quenching of tryptophan fluorescence in human gammaD- and gammaS-crystallins: the gamma-crystallin fold may have evolved to protect tryptophan residues from ultraviolet photodamage. Biochemistry. 2009;48:3708–3716. doi: 10.1021/bi802177g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flaugh SL, Kosinski-Collins MS, King J. Contributions of hydrophobic domain interface interactions to the folding and stability of human gammaD-crystallin. Protein Sci. 2005;14:569–581. doi: 10.1110/ps.041111405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flaugh SL, Kosinski-Collins MS, King J. Interdomain side-chain interactions in human gammaD crystallin influencing folding and stability. Protein Sci. 2005;14:2030–2043. doi: 10.1110/ps.051460505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreau KL, King J. Hydrophobic core mutations associated with cataract development in mice destabilize human gammaD-crystallin. J Biol Chem. 2009;284:33285–33295. doi: 10.1074/jbc.M109.031344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mills IA, Flaugh SL, Kosinski-Collins MS, King JA. Folding and stability of the isolated Greek key domains of the long-lived human lens proteins gammaD-crystallin and gammaS-crystallin. Protein Sci. 2007;16:2427–2444. doi: 10.1110/ps.072970207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das P, King JA, Zhou R. beta-strand interactions at the domain interface critical for the stability of human lens gammaD-crystallin. Protein Sci. 2009;19:131–140. doi: 10.1002/pro.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hemmingsen JM, Gernert KM, Richardson JS, Richardson DC. The tyrosine corner: a feature of most Greek key beta-barrel proteins. Protein Sci. 1994;3:1927–1937. doi: 10.1002/pro.5560031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamill SJ, Cota E, Chothia C, Clarke J. Conservation of folding and stability within a protein family: the tyrosine corner as an evolutionary cul-de-sac. J Mol Biol. 2000;295:641–649. doi: 10.1006/jmbi.1999.3360. [DOI] [PubMed] [Google Scholar]
- 28.McCarty CA, Taylor HR. A review of the epidemiologic evidence linking ultraviolet radiation and cataracts. Dev Ophthalmol. 2002;35:21–31. doi: 10.1159/000060807. [DOI] [PubMed] [Google Scholar]
- 29.Grossweiner LI. Photochemistry of proteins: a review. Curr Eye Res. 1984;3:137–144. doi: 10.3109/02713688408997195. [DOI] [PubMed] [Google Scholar]
- 30.Davies MJ, Truscott RJ. Photo-oxidation of proteins and its role in cataractogenesis. J Photochem Photobiol B. 2001;63:114–125. doi: 10.1016/s1011-1344(01)00208-1. [DOI] [PubMed] [Google Scholar]
- 31.Meyer EA, Castellano RK, Diederich F. Interactions with aromatic rings in chemical and biological recognition. Angew Chem Int Ed Engl. 2003;42:1210–1250. doi: 10.1002/anie.200390319. [DOI] [PubMed] [Google Scholar]
- 32.Burley SK, Petsko GA. Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science. 1985;229:23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- 33.McGaughey GB, Gagne M, Rappe AK. pi-Stacking interactions. Alive and well in proteins. J Biol Chem. 1998;273:15458–15463. doi: 10.1074/jbc.273.25.15458. [DOI] [PubMed] [Google Scholar]
- 34.Hecht MH, Sturtevant JM, Sauer RT. Effect of single amino acid replacements on the thermal stability of the NH2-terminal domain of phage lambda repressor. Proc Natl Acad Sci U S A. 1984;81:5685–5689. doi: 10.1073/pnas.81.18.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serrano L, Bycroft M, Fersht AR. Aromatic-aromatic interactions and protein stability. Investigation by double-mutant cycles. J Mol Biol. 1991;218:465–475. doi: 10.1016/0022-2836(91)90725-l. [DOI] [PubMed] [Google Scholar]
- 36.Betts S, Haase-Pettingell C, Cook K, King J. Buried hydrophobic side-chains essential for the folding of the parallel beta-helix domains of the P22 tailspike. Protein Sci. 2004;13:2291–2303. doi: 10.1110/ps.04676704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simkovsky R, King J. An elongated spine of buried core residues necessary for in vivo folding of the parallel beta-helix of P22 tailspike adhesin. Proc Natl Acad Sci U S A. 2006;103:3575–3580. doi: 10.1073/pnas.0509087103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kannan N, Vishveshwara S. Aromatic clusters: a determinant of thermal stability of thermophilic proteins. Protein Eng. 2000;13:753–761. doi: 10.1093/protein/13.11.753. [DOI] [PubMed] [Google Scholar]
- 39.Hillier BJ, Rodriguez HM, Gregoret LM. Coupling protein stability and protein function in Escherichia coli CspA. Fold Des. 1998;3:87–93. doi: 10.1016/S1359-0278(98)00014-5. [DOI] [PubMed] [Google Scholar]
- 40.Rodriguez HM, Vu DM, Gregoret LM. Role of a solvent-exposed aromatic cluster in the folding of Escherichia coli CspA. Protein Sci. 2000;9:1993–2000. doi: 10.1110/ps.9.10.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith CK, Regan L. Guidelines for protein design: the energetics of beta sheet side chain interactions. Science. 1995;270:980–982. doi: 10.1126/science.270.5238.980. [DOI] [PubMed] [Google Scholar]
- 42.Rea AM, Simpson ER, Meldrum JK, Williams HE, Searle MS. Aromatic residues engineered into the beta-turn nucleation site of ubiquitin lead to a complex folding landscape, non-native side-chain interactions, and kinetic traps. Biochemistry. 2008;47:12910–12922. doi: 10.1021/bi801330r. [DOI] [PubMed] [Google Scholar]
- 43.Waters ML. Aromatic interactions in peptides: impact on structure and function. Biopolymers. 2004;76:435–445. doi: 10.1002/bip.20144. [DOI] [PubMed] [Google Scholar]
- 44.Yao J, Dyson HJ, Wright PE. Three-dimensional structure of a type VI turn in a linear peptide in water solution. Evidence for stacking of aromatic rings as a major stabilizing factor. J Mol Biol. 1994;243:754–766. doi: 10.1016/0022-2836(94)90045-0. [DOI] [PubMed] [Google Scholar]
- 45.Wu L, McElheny D, Takekiyo T, Keiderling TA. Geometry and efficacy of cross-strand Trp/Trp, Trp/Tyr, and Tyr/Tyr aromatic interaction in a beta-hairpin peptide. Biochemistry. 2010;49:4705–4714. doi: 10.1021/bi100491s. [DOI] [PubMed] [Google Scholar]
- 46.Griffiths-Jones SR, Searle MS. Structure, Folding, and Energetics of Cooperative Interactions between the β-Strands of a de Novo Designed Three-Stranded Antiparallel β-Sheet Peptide. J Am Chem Soc. 2000;122:8350–8356. [Google Scholar]
- 47.Aravind P, Mishra A, Suman SK, Jobby MK, Sankaranarayanan R, Sharma Y. The betagamma-crystallin superfamily contains a universal motif for binding calcium. Biochemistry. 2009;48:12180–12190. doi: 10.1021/bi9017076. [DOI] [PubMed] [Google Scholar]
- 48.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 49.Lampi KJ, Kim YH, Bachinger HP, Boswell BA, Lindner RA, Carver JA, Shearer TR, David LL, Kapfer DM. Decreased heat stability and increased chaperone requirement of modified human betaB1-crystallins. Mol Vis. 2002;8:359–366. [PubMed] [Google Scholar]
- 50.Bagby S, Go S, Inouye S, Ikura M, Chakrabartty A. Equilibrium folding intermediates of a Greek key beta-barrel protein. J Mol Biol. 1998;276:669–681. doi: 10.1006/jmbi.1997.1563. [DOI] [PubMed] [Google Scholar]
- 51.Rudolph R, Siebendritt R, Nesslauer G, Sharma AK, Jaenicke R. Folding of an all-beta proteindependent domain folding in gamma II-crystallin from calf eye lens. Proc Natl Acad Sci U S A. 1990;87:4625–4629. doi: 10.1073/pnas.87.12.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Itoh K, Sasai M. Cooperativity, connectivity, and folding pathways of multidomain proteins. Proc Natl Acad Sci U S A. 2008;105:13865–13870. doi: 10.1073/pnas.0804512105. [DOI] [PMC free article] [PubMed] [Google Scholar]