Abstract
The bifunctional Escherichia coli glutathionylspermidine synthetase/amidase (GspSA) catalyzes both the synthesis and hydrolysis of Gsp. Its amidase domain (GspA), which catalyzes the hydrolysis of Gsp into glutathione and spermidine, plays an important role in redox sensing and protein S-thiolation. To gain insight of the regulation and catalytic mechanism of and further understand the recycling of the Gsp dimer and Gsp-S-protein adducts, we solved two crystal structures of GspA and GspSA both with the C59A mutation and bound with the substrate, Gsp. In both structures, Cys59, His131, and Glu147 form the catalytic triad, which is similar to other cysteine proteases. Comparison of the GspA_Gsp complex and apo GspSA structures indicates that on binding with Gsp, the side chains of Asn149 and Gln58 of the amidase domain are induced to move closer to the carbonyl oxygen of the cleaved amide bond of Gsp, thereby participating in catalysis. In addition, the helix-loop region of GspA, corresponding to the sequence 30YSSLDPQEYEDDA42, involves in regulating the substrate binding. Our previous study indicated that the thiol of Cys59 of GspA is only oxidized to sulfenic acid by H2O2. When comparing the active site of GspA with those of other cysteine proteases, we found that limited space and hydrophobicity of the environment around Cys59 play an important role to inhibit its further oxidation. The structural results presented here not only elucidate the catalytic mechanism and regulation of GspA but also help us to design small molecules to inhibit or probe for the activity of GspA.
Keywords: glutathionylspermidine; amidase; structure; mechanism; regulation; cysteine,; histidine-dependent amidohydrolases/peptidases
Introduction
The bifunctional glutathionylspermidine synthetase/amidase (GspSA) from Escherichia coli catalyzes both the ATP-dependent formation of an amide bond between N1 of spermidine (N-(3-amino)propyl-1,4-diaminobutane) and the glycine carboxylate of glutathione (GSH, γ-Glu-Cys-Gly) and the opposing hydrolysis of this amide bond.1 These two activities are separate in two protein domains: the N-terminal amidase domain and the C-terminal synthetase domain.2 Both spermidine and GSH are important metabolites and are present at high concentrations (0.1−10 mM) in most cells.3,4 Spermidine is a polycationic molecule that interacts with proteins, phospholipids, and nucleic acids,5 affecting primarily cell proliferation and differentiation.3,6,7 GSH, a primary antioxidant, is important in maintaining the redox balance and in reductively scavenging reactive oxygen species.4 Alternatively, Gsp is required for the synthesis of trypanothione (bis(glutathionyl)spermidine), which is used to defend against oxidative stress in protozoal parasites of genera Trypanosoma and Leishmania.8,9 Although GspSA was first identified in E. coli more than three decades ago,10 the precise physiological role of GspSA was not clear until recently.
Previously, we solved the X-ray crystal structure of GspSA and its complex structures with the substrate, product, and inhibitor to clarify the mechanistic details of the synthetase reaction.11 Furthermore, we proved recently that Gsp amidase (GspA) plays a significant role in the reduction of Gsp-S-S-Gsp and Gsp-S-protein adducts of E. coli.12 Gsp S-thiolated proteins (GspSSPs) in E. coli have mixed disulfides of Gsp and protein thiols, standing for a new type of post-translational modification and the amounts of these Gsp derivatives increased when E. coli was treated with H2O2.12 The accumulation of GspSSPs probably occurred because of inactivation of GspA by H2O2 with Gsp synthetase (GspS) remains unaffected.12 H2O2 inactivated the activity of GspA by oxidizing the thiol of the amidase active-site nucleophile, Cys59, to a sulfenic acid.12 In addition, GspA can hydrolyze a variety of Gsp-derived substrates yielding Spd and GSH S-thiolated proteins/peptides.12 With elimination of the oxidative threat, GspA, GSH reductase, and glutaredoxin act in concert to convert oxidized Gsp (as the disulfide of Gsp, mixed disulfides of Gsp and other small thiol-containing compounds, and/or GspSSPs) to GSH.12 Such oxidative stress defense is likely the principal biological role of E. coli GspA, and it is the first time an essential role of the amidase domain was identified in the hydrolysis of Gsp that ultimately enables the reduction of Gsp-S-S-Gsp and Gsp-S-protein adducts, as E. coli does not have an enzyme to reduce Gsp-S-S-Gsp.
Therefore, it is important to gain more insight into the activity of the enzyme on these larger substrates and further understand the regulation of the two opposite activities because it plays a significant role in avoiding a futile GSH-dependent ATPase cycle. In this study, we present the structures of the amidase domain alone (GspA) and the full length of E. coli GspSA both with a mutation of C59A and in complex with the amidase substrate, Gsp, to further understand the regulation, the catalytic mechanism and S-thiolation of GspA, which have not been previously discussed fully in details.
Results
Overall structure
We obtained two crystal complex structures, (1) GspA (residues 1–197 of GspSA) with the C59A mutation containing Gsp (GspA(C59A)_Gsp) and (2) GspSA with the C59A mutation containing ADP and Gsp (GspSA(C59A)_ADP_Gsp) [Fig. 1(A,B), respectively]. The complex structure of GspSA(C59A)_ADP_Gsp is a homodimer, and each monomer contains the N-terminal amidase and C-terminal synthetase domains connected by a linker in between. The overall structure shares nearly identical folding with the previously reported GspSA (wild-type) structure (with the RMSD value of 2.0 Å),11 and the relative orientation of the two separate domains does not show obvious difference. Interestingly, the GspSA_Gsp complex was formed during crystal growth by addition of the substrates for the synthetase domain, that is, GSH, spermidine, ATP, and Mg2+. The resulting structure contained GSP in the GspA domain active site and the product ADP in the GspS domain active site [Fig. 1(B)]. On the other hand, the amidase domain structure, GspA(C59A)_Gsp, is a monomer showing similar folding to the amidase domain of the GspSA(C59A)_ADP_Gsp structure with the RMSD value of 0.36 Å (Supporting Information Fig. S1) and contains the substrate, Gsp, bound in the active site [Fig. 1(A)]. All of the following structural descriptions are based on the engineered GspA domain structure because it is of higher resolution (1.95 Å; Table I).
Figure 1.
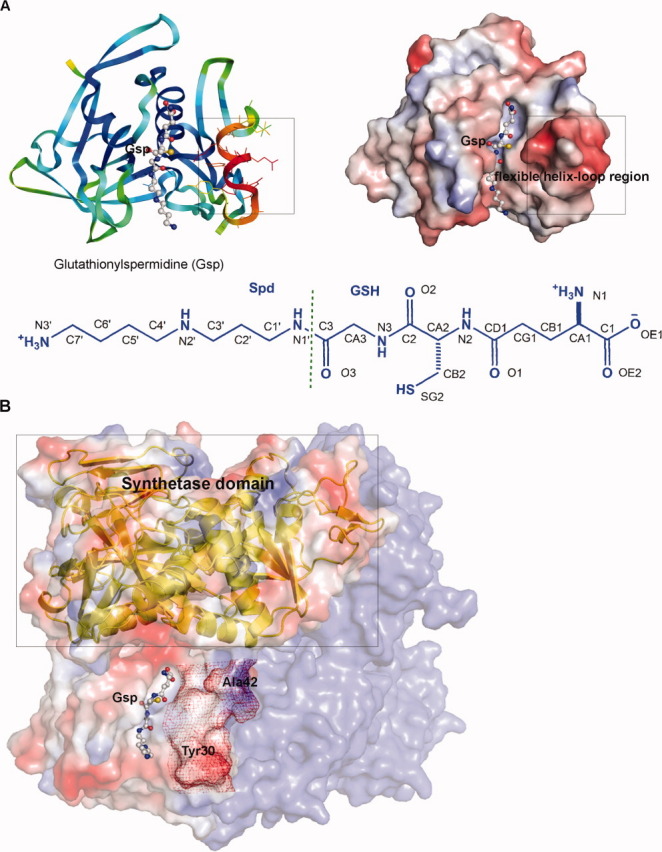
(A) The overall fold of the GspA(C59A)_Gsp structure (left) and the electrostatic surface representation of the GspA(C59A)_Gsp complex structure (right). The Gsp molecule bound at the active site is shown as a ball-and-stick model. The boxes indicate the flexible helix-loop region of the amino acids 30∼42. Left: The colors of the ribbon diagrams are based on different B factors with red representing high B factor, while deep blue representing the lower one. Right: The colors, red, white, and blue, of electrostatic surfaces indicate negative, neutral and positive charges, respectively. The Gsp atom nomenclature is shown below. (B) Electrostatic surface representation of the GspSA(C59A)_ADP_Gsp complex structure. The Gsp molecule is shown in the binding cavity as a ball-and-stick model. The corresponding C-terminal synthetase domain is presented by a ribbon diagram and indicated by a box. The second monomer of the dimer is colored blue. The position of the helix-loop region of GspA, corresponding to the sequence 30YSSLDPQEYEDDA42, is highlighted with red mesh.
Table I.
Data Collection and Refinement Statistics
| GspA(C59A)_Gsp | GspSA(C59A)_ADP_Gsp | |
|---|---|---|
| Space group | P6422 | P1 |
| No. of molecules per asymmetric unit | 1 | 2 |
| Unit cell parameters (Å, °) | 83.96, 83.96, 104.94 | 60.38, 76.20, 84.22, 70.81 74.37, 78.64 |
| Resolution, Å | 30–1.95 (2.02–1.95) | 30–2.8 (2.9–2.8) |
| Total no. of reflections | 222956 | 120938 |
| No. of unique reflections | 16593 | 31316 |
| Completeness, % | 99.8 (100) | 96.3 (94.7) |
| Redundancy | 13.4 (13.4) | 3.9 (3.6) |
| I/sigmaI | 61.9 (3.9) | 15.9 (2.4) |
| Rmergea | 5.6 (70.0) | 9.6 (49.2) |
| Refinement statistics | ||
| Resolution, Å b | 30–1.95 (2.02–1.95) | 30–2.8 (2.9–2.8) |
| Rwork | 20.6 (25.2) | 22.0 (28.1) |
| Rfreec | 23.0 (30.1) | 26.2 (32.8) |
| rmsd bond length, Å | 0.006 | 0.010 |
| rmsd angle, degree | 1.26 | 1.33 |
| Ramachandran plot, % | ||
| Most favored/allowed regions | 88/100 | 83.1/99.8 |
| Additional allowed | 10.8 | 16.1 |
| Generously allowed | 1.3 | 0.7 |
| Average B factor (Å2)/atoms | 43.4/1641 | 43.5/9998 |
| Waters/ atoms | 57.8/132 | 32.7/223 |
| Ligands/ atoms | 57.6/29 | 34.4/95 |
Rmerge= SUM (ABS (I − <I>))/SUM (I).
Values in parentheses are for the highest resolution shell.
Rfree = R factor calculated using 5% of the reflection data chosen randomly and omitted from the start of refinement.
Protein structure accession number
The atomic coordinates and structure factors for GspA(C59A)_Gsp and GspSA(C59A)_ADP_Gsp have been deposited in the PDB with the accession numbers of 3A2Y and 3O98, respectively.
Active site
The Gsp molecule bound to the enzyme in a similar binding mode in both full-length GspSA(C59A)_ADP_Gsp and the domain GspA(C59A)_Gsp structures (Fig. 1). Backbone atoms of Val78, Gly79, and Ala81 form hydrogen bonds with the tripeptide moiety of Gsp, and the terminal guanidinium group of Arg64 forms a salt-bridge with the bidentate carboxylate (OE1 and OE2) of the GSH moiety. Main chain nitrogen of Ala59 (substitution of Cys59) and side chains of Asn149 and Gln58 are hydrogen-bonded to O3 of Gsp [Fig. 2(A), left, Supporting Information Fig. S2 and Table II; see Fig. 1(A) for the Gsp numbering]. The main chain oxygen of Gly130 interacts with N1 of the spermidine moiety of Gsp. Val60, Thr129, and Phe126 are involved in the hydrophobic contacts (with the C2, C3, CG1, and C2′ atoms of Gsp). Lack of electron density is observed toward the end of the spermidine moiety, supporting the idea that the atoms that participate in the interactions are flexible [Fig. 2(A), left] and thus do not make significant contribution.
Figure 2.
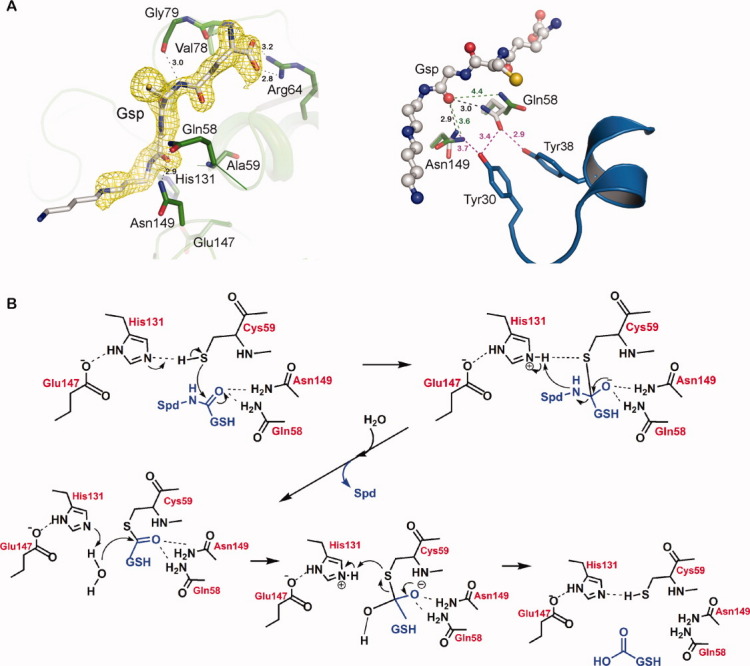
(A) The active site of GspA(C59A) complexed with the substrate, Gsp. Left: The Gsp molecule is displayed in the stick diagram. The omit map of the Gsp molecule contoured at 1σ is shown in yellow. The residues interacting with the ligand are labeled. Hydrogen bonds are depicted as dashed lines with the distances given nearby in Å. Right: GspA(C59A)_Gsp (gray) is superimposed with apo GspSA (PDB: 2IOB, green). The Gsp molecule is shown as a ball-and-stick model. The Gln58 and Asn149 side chains showing conformational changes are shown as sticks and their distances to the O3 atom of Gsp (in Å) are labeled. The distances to Tyr30 and Tyr38 located at the flexible helix-loop region are also shown. (B) The proposed mechanism of GspA. Cys59, His131, and Glu147 serve as the catalytic triad to lower the pKa of the thiol of Cys59 and thus to enhance its nucleophilicity. Gln58 and Asn149 form hydrogen bond interactions with the O3 of Gsp to either enhance the electrophilicity or to stabilize the developed oxyanion intermediate.
Table II.
H-bonds between Gsp and the GspA(C59A) Residues
| GspS residues | H-bond distance of Gsp (Å) |
|---|---|
| Asn149-Nδ2 | GSH O3 (2.9) |
| Gln58-Nɛ2 | GSH O3 (3.0) |
| Gly130-O | Spermidine N1 (3.3) |
| Ala81-N | GSH O2 (2.9) |
| Gly79-O | GSH N2 (3.0) |
| Val78-N | GSH OE1 (3.1) |
| Arg64-NH1 | GSH OE1 (3.2) |
| Arg64-NH2 | GSH OE2 (2.8) |
| Ala59-N | GSH O3 (3.2) |
The arrangement of the active site environment and the bound substrate is consistent with the catalytic mechanism13 in which Cys59, His131, and Glu147 function as the catalytic triad (Fig. 2). These residues of catalytic triad interact with each other, that is, NE2 of His131 forms the hydrogen bond with OE1 of Glu147 and ND1 of His131 is hydrogen bonded to the sulfhydryl group of Cys59 (Fig. 2).
Revealed by comparing the structure of GspA(C59A)_Gsp with that of GspSA apo protein (PDB ID: 2IOB) [Fig. 2(A), right], the side chains of Asn149 and Gln58, which is thought to flip during catalysis, are induced to move closer to the carbonyl oxygen of the cleaved amide bond of Gsp on the substrate binding to stabilize the developing oxyanion. Moreover, both Asn149 and Gln58 interact with Tyr30 and Tyr38 resided in the mobile loop-helix [Fig. 2(A), right, see below].
Conformational change on substrate binding
In the GspSA apo structure (PDB ID: 2IOB), there is a conformational change in the helix-loop region of the Gsp amidase domain corresponding to the sequence 30YSSLDPQEYEDDA42 between the chains A and B (closed and open forms, respectively; Fig. 3 or Supporting Information Figs. S3A and S3B), which is not far from the substrate binding cavity [Fig. 1(A)]. This region is somewhat disordered and appears with high B factors, suggesting its flexibility and the possibility of regulating the substrate entry. In the substrate binding structure (GspA(C59A)_Gsp), this helix-loop region appears to adopt a “more closed” form than the closed form of the apo GspSA structure (Fig. 3), and additional interactions were thus observed. Hydroxyl group of Tyr30 interacts with the Nδ atom of Asn149 and the Nɛ atom of Gln58, and a water molecule also interacts with the Oɛ atom of Gln58. In addition, the OH group of Tyr38 forms a hydrogen bond with the Nɛ atom of Gln58 [Fig. 2(A), right].
Figure 3.
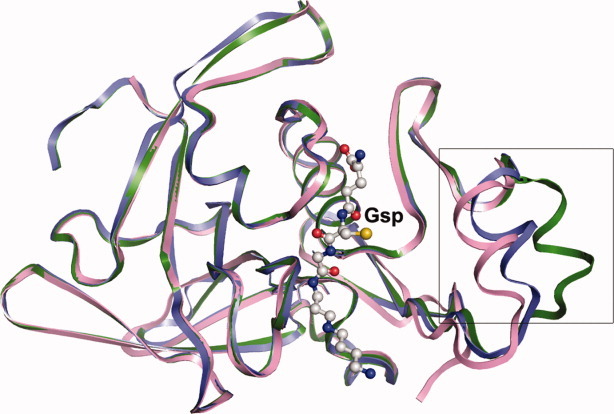
Superimposition of the Apo_GspSA amidase domain chain A (blue), Apo_GspSA chain B (green) and GspA(C59A)_Gsp (pink). The Gsp molecule is displayed as the ball-and-stick model.
Because E. coli GspSA and Leishmania major trypanothione synthetase (TryS, PDB ID: 2VOB,14) are 32% identical in amino acid sequences, and structural alignment of the amidase domains of E. coli GspSA and TryS was performed on the combinatorial extension server (http://cl.sdsc.edu/ce.html). When 168 residues of the two amidase domains were aligned, the RMSD value is 1.5 Å (Fig. 4). Intriguingly, the aforementioned flexible helix-loop region was found missing in the TryS amidase domain (Fig. 4), indicating the unique regulation role in E. coli GspA.
Figure 4.
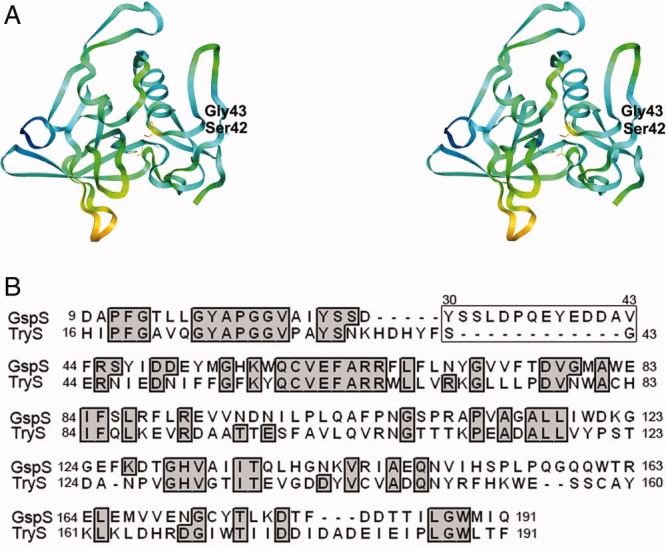
(A) Stereo view of the L. major TryS N-terminal amidase domain. Different colors represent different B factors. (B) Structural alignment of E. coli GspS amidase and L. major TryS amidase. The box indicates the flexible helix-loop region around residues 30–43. This part is missing in the TryS structure.
Comparison with other cysteine proteases
To disclose the conserved and distinct features of GspA, its crystal structure was compared with those of other cysteine proteases. When comparing GspA with 3CL protease of severe acute respiratory syndrome, N-carbamoylsarcosine amidase (CSHase) Ta0454 of Thermoplasma acidophilum and ubiquitin-specific processing protease (UBP) family of deubiquitinating enzymes, they have similar active sites, despite their very different overall structures. GspA contains the cysteine, histidine-dependent amidohydrolases/peptidases (CHAP) domain, which shows the papain-like cysteine proteinase fold (Fig. 3), whereas 3CL protease adopts a chymotrypsin protein fold15 and CSHase Ta0454 forms a Rossmann-type fold.16 CSHase Ta0454 contains the highly conserved residues, Cys123, Asp9, and Lys90, which serve as the catalytic triad.16 Similarly, UBP contains the conserved catalytic triad, that is, Cys, His, Asn, or Asp.17 3CL protease forms a catalytic dyad, Cys145 and His41 (Supporting Information Fig. S4B). Even though GspA and 3CL protease utilize similar catalytic residues, the shape of their binding pockets appears to be different, likely reflecting their substrate specificities (Supporting Information Figs. S4A vs. S4B).
Oxidation state of reactive cysteine
We previously determined a partially oxidized form of the GspA structure, whose active-site Cys59 was oxidized by H2O2 into sulfenic acid and this reaction is reversible.12 In contrast, the reactive Cys (Cys25) of papain (cysteine protease) was reported to convert to an irreversible higher oxidation state of sulfonic acid.18 Comparison of the active site of GspA (both Cys-SH and Cys-SOH forms of GspA) with that of papain cysteine protease (PDB ID: 9PAP) shows that the entrance of GspA active site, formed by the hydrophobic residues, Val132, Val60, and Ala81, is narrower than that of the papain active site (Supporting Information Fig. S5A, B vs. S5C). The distances between the Sγ atom of Cys59 and the side chains of Val132, Val60, and Ala81 are 3.8, 3.9, and 4.2 Å, respectively. Like papain, the catalytic Cys, Cys580, of small ubiquitin-like modifier (SUMO) protease Ulp1 can be oxidized to sulfenic, sulfinic, and sulfonic acids.19 According to the structural comparison of GspA-sulfenic acid and Ulp1-sulfenic acid, obviously the space around the sulfenic acid of Ulp1 is wider than that of GspA, and there is no hydrophobic patch near Cys580 of Ulp1 unlike GspA (Supporting Information Figs. S5B vs. S5D). This evidence accounts for the limited oxidation occurring in the Cys59 of GspA. Superimposing the Ulp1 structures with cysteine-sulfenic and cysteine-sulfonic acids reveals that both Trp448 and His514 shift closer to the cysteine-sulfonic acid than to the cysteine-sulfenic acid (Supporting Information Figs. S5D vs. S5E).
Discussion
The bifunctional E. coli GspSA, which catalyzes both the synthesis and hydrolysis of Gsp, has been of interest for its implications in human disease and bacterial cell maintenance and metabolism.1 Importantly, we have identified an essential role of the amidase domain in the hydrolysis of Gsp that ultimately enables the reduction of Gsp-S-S-Gsp and Gsp-S-protein adducts,12 as E. coli does not have an enzyme to reduce Gsp-S-S-Gsp. In our current structures, we can gain more insight into the possible binding mode of GspA to Gsp-S-S-Gsp and Gsp-S-protein adducts. The O3 atom of the scissile peptide bond of Gsp faces outward in the GspA_Gsp complex structure (Figs. 1 and 2); therefore, larger substrates, that is, Gsp-S-S-Gsp and Gsp-S-protein adducts would probably bind in similar mode to Gsp with the other Gsp (in the Gsp disulfide) or the non-Gsp part (in the Gsp-mixed disulfide) hanging outside the Gsp-binding cavity. In this way, any larger Gsp-S-protein adducts would be able to fit into the GspA binding cavity without major disruption to the rest of its structure.
In E. coli, the two activity domains of GspSA are regulated to avoid the futile consumption of ATP. The structures of GspSA and GspA clearly indicate that the entrance of the amidase active site, which is near the synthetase domain, is more accessible when the synthetase domain is absent [Fig. 1(A) vs. 1(B)]. Therefore, this observation suggests the synthetase domain plays a role in negatively regulating the amidase activity and supports the previous study by Lin et al.20
In contrast, a significant conformational change was noticed in the helix-loop region of GspA at sequence 30–42 in comparison with the apo form of GspSA (Fig. 3). This helix-loop region of GspA is near the substrate binding cavity and thus narrows the substrate entry upon binding to the substrate, that is, Gsp; this “more closed form” would probably increase binding affinity to Gsp. In particular, the residues of this helix-loop are mostly negatively charged [Fig. 1(A), right), which attract and facilitate binding with positively charged spermidine that holds the helix-loop and substrate tunnel in the arrangement observed [Fig. 1(A)]. These indicate that this helix-loop plays a role in regulating the substrate entry and thus possibly affects the binding affinity to the substrate and finally its activity. Furthermore, Tyr30 and Tyr38, both in the helix-loop, participate in hydrogen bonding with Asn149 and Gln58 [Fig. 2(A), right]. It appears that on binding of the substrate the helix-loop region moves in to interact with the substrate; most likely facilitating catalysis via the additional interactions. Because of the functional role of Asn149 and Gln58 in the catalysis (see below), these observations suggest that the mobile helix-loop region is likely in association with the regulation of the amidase activity, in addition to the aforementioned regulation caused by the synthetase domain. As the helix-loop plays a role in regulation of the amidase activity, mutating Tyr30 and/or Tyr38 or removing the flexible helix-loop would probably abolish the interaction with the substrate then decrease binding affinity to the substrate and finally decrease the amidase's catalysis. Certainly, this analysis would be a great experimental support to our hypotheses. Surprisingly, this helix is absent in the closely related structure of L. major TryS amidase domain [Figs. 3, Supporting Information Fig. S3 vs. 4(A) and Fig. 4(B)], explaining that such regulation by a helix of the amidase activity is unique to E. coli GspA. In contrast, in L. major trypanothione synthetase-amidase (TSA) the C-terminus, which blocks the amidase active site, provides the mechanism to regulate its activity whereby a conformational change is required to vacate the active site for substrate binding.14
Our complex structures provide further insight explaining how the catalytic machinery operates to hydrolytically cleave the amide bond. Cys59, His131, and Glu147 form the catalytic triad; the resulting Cys59-His131 thiolate-imidazolium pair is oriented by a hydrogen bond to the Glu147 carboxylate (Fig. 2). Such interactions enhance the nucleophilicity of Cys59 to attack the isopeptide bond between GSH and spermidine to yield a tetrahedral intermediate, followed by formation of the acyl-S-enzyme intermediate (covalent catalysis) as do serine and cysteine proteases [Fig. 2(B)]. The high reactivity of Cys59 is further supported by its low pKa value of 3.05 that is calculated and predicted by using the PROPKA (web interface 2.0, http://propka.ki.ku.dk/).21,22 Spermidine is liberated upon formation of the acyl-enzyme intermediate. The subsequent hydrolysis of the acyl-enzyme intermediate releases GSH from the enzyme and, thus, completes the catalytic reaction [Fig. 2(B)]. The catalytic-related residues, Gln58, Cys59, and Asn149, are highly conserved in the CHAP family.23 In this aspect, both Asn149 and Gln58 have dual roles in the catalysis. First, they form H-bonds with the carbonyl oxygen of the cleaved amide bond to make the amide a better electrophile, that is, more vulnerable to the nucleophilic attack by the thiol of Cys59. Second, Asn149 and Gln58 also form an oxyanion hole, which refers to the accommodation of the negative potential formed on the carbonyl oxygen atom at the scissile bond on binding to the substrate to stabilize the tetrahedral intermediate before the formation of an acyl-enzyme adduct.
As aforementioned, GspA hydrolyzes the isopeptide bond between GSH and spermidine [Fig. 2(B)]. Amide bonds can be hydrolyzed by a variety of unrelated or distantly related enzymes, such as amidases and autolysins or cell-wall hydrolases.24 It was shown that these proteins from bacteria, bacteriophages, archaea, and eukaryotes of the Trypanosomidae family contain a common domain, CHAP domain.23,24 Similarly, GspA has a typical CHAP domain. It thus has many features shared by the CHAP domain-containing proteins. The positioning of the invariant cysteine and histidine residues close to the start of predicted helices and sheets, respectively, is consistent with a distant relationship to the papain-like cysteine proteinase fold.24,25 The proposed nucleophilic-attack mechanism presented here involving the conserved cysteine residue as the catalytic nucleophile is probably the mechanism for all members of the CHAP superfamily.
When comparing GspA with the cysteine proteases not belonging to the CHAP superfamily, it is clear that they have similar catalytic residues and mechanisms, although they have very different overall structures. 3CL protease has a Cys-His catalytic dyad and hydrolyses the peptide bond like GspA.15 However, the binding pocket of 3CL protease has more space for the side chains of peptides, whereas GspA only has a restricted pocket for the peptide without large side chains, i.e., Gsp (Supporting Information Fig. S4). This explains how the substrate specificity defines in spite of similar active sites. Taken together, a variety of cysteine proteases share a common catalytic mechanism that involves the conserved catalytic triad, indicating a convergent evolution of their active sites, but with development of their own substrate specificity.
We reported previously that a sulfenic acid forms in the thiol of Cys59 of GspA by H2O2 oxidation.12 Because sulfenic acid is labile and easily undergoes further oxidation to generate irreversible higher oxidation states, including sulfinic and sulfonic acids, it is interesting to identify what distinct features of GspA are correlated with the stability of Cys59-sulfenic acid. The comparison of the active site of GspA with those of papain cysteine protease (PDB: 9PAP) and SUMO protease Ulp1 shows how GspA functions to operate during oxidative stress. The electrostatic surface shows that the entry of substrate channel of GspA formed by the hydrophobic residues is narrower than those of papain and Ulp1 (Supporting Information Figs. S5A, S5B vs. S5C, S5D, and S5E). Interestingly, the space around the sulfonic acid of Cys580 of Ulp1 is more limited than that of the sulfenic acid of Cys580 (Supporting Information Figs. S5D vs. S5E), indicating that the environment of Cys59-sulfenic acid mimics that of Cys508-sulfonic acid. Therefore, the hydrophobicity and the crowded space likely make it difficult for the excessive access of H2O2, revealing a correlation between the surrounding environment of cysteine and the corresponding oxidation state. Meanwhile, the life of Cys59-sulfenic acid is sustained by these steric and intramolecular-bonding factors,26 which impede further oxidation on the Sγ atom of Cys59. More importantly, the unusually short hydrogen bonding interaction that is contributed by an adjacent water molecule stabilizes the Cys59-sulfenic acid in GspA and makes it stable enough to resist further oxidation.12
The structural data demonstrated that GspA has more interactions with the GSH moiety of Gsp than it does with the spermidine moiety [Fig. 2(A) and Table II]; as a result, it would be better to mimic the former interactions when developing GspA inhibitors. This is supported by the previous study that γ-Glu-Ala-Gly-aldehyde is a potent time-dependent specific inhibitor of the amidase activity.27 Intriguingly, the sulfhydryl group of Gsp faces outward and does not form any interactions with GspA [Figs. 1(A) and 2(A), left]. Therefore, we can modify this group (e.g., replacement of Cys with Ala) without disrupting any interaction with the enzyme. The chromogenic substrate γ-Glu-Ala-Gly p-nitroanilide, for instance, has been used for activity assay.20 Such binding interactions can be useful for designing activity-based probes for specific protein tagging or in vivo visualization.
In conclusion, this study provides more structural evidence for the negative regulation and catalytic mechanism of Gsp amidase. Such negative regulation is necessary for this bifunctional enzyme, otherwise the net reaction will be a total waste of energy (simultaneous operation of the synthesis and hydrolysis). The mechanism presented here provides more insight into our knowledge of the enzymatic hydrolysis akin to the unique amidase. Importantly, this structural information provides clues for structure-based design of inhibitors or activity-based probes of amidase.
Materials and Methods
Protein expression and purification
The construction of the plasmids containing the E. coli GspSA (619 amino acids) and Gsp amidase (GspA, i.e., residues 1–197 of GspSA) genes were described before.20 The gene of GspSA was cloned into the pET22b vector (Novagen) behind the T7 promoter, whereas the gene of GspA was cloned into the pET28a vector (Novagen) behind the T7 promoter that encodes an N-terminal 6x His tag. The C59A mutants of both GspSA and GspA were prepared using the QuickChange site-directed mutagenesis kit (Stratagene). The constructs were transformed into the E. coli expression strain BL21 (DE3). Luria broth (LB) liquid cultures were grown aerobically with shaking, and protein expression was induced by addition of 500 μM IPTG. The GspSA protein was purified as previously described.1,2 The 6x His-tagged GspA(C59A) protein was purified by nickel affinity chromatography with the His Excellose Spin Kit (Yeastern Biotech). Protein concentration was determined in accordance with the Bradford method (Bio-Rad Protein Assay kit) with bovine serum albumin as the standard.
In the typical preparation, 30–40 mg of purified protein was obtained per liter of cell culture. Preparations were judged to be 90–95% pure by SDS-PAGE. Purified protein was concentrated to 20 mg/mL and stored at −80°C.
Crystallization and data collection
Crystal of the GspSA(C59A)_ADP_Gsp complex was obtained in the same crystallization condition as previously.11 The cocrystallization solution contains the GspSA(C59A) protein (20 mg/mL), Mg2+, ATP, GSH, and spermidine. For the GspA(C59A)_Gsp complex crystal, it was obtained by soaking GspA(C59A) crystal into the crystallization solution containing 2 mM Gsp (Calbiochem) for 2 days. Data were collected using ADSC Quantum 315 or Quantum 210 CCD detectors at the Synchrotron Protein Crystallography Facility BL13B1 and BL13C1 beamlines, National Synchrotron Radiation Research Center, Hsinchu, Taiwan. Data were processed and integrated using the program, HKL2000.28
Structure determination and refinement
Structures were determined by molecular replacement using the program CNS,29 which was also used in computational refinements. The known GspSA structure was used as the model.11 The coordinates, the topology, and the parameter files for ADP and Gsp, which were required for CNS calculations were taken from the HIC-UP server (http://xray.bmc.uu.se/hicup/). All manual modifications of the models were performed by XtalView30 under the guidance of (2Fo-Fc) sum difference maps. Data collection and refinement statistics are listed in Table I. The figures were produced by PyMOL.31
Acknowledgments
The authors thank National Synchrotron Radiation Research Center of Taiwan for beam time allocations. C. H. Pai and H.-J. Wu are grateful for the Postdoctoral Fellowships from Academia Sinica, Taiwan.
Glossary
Abbreviations:
- CHAP
cysteine, histidine-dependent amidohydrolases/peptidases
- CSHase
N-carbamoylsarcosine amidase
- GSH
glutathione
- Gsp
glutathionylspermidine
- GspSA
glutathionylspermidine syntetase/amidase
- GspA
Gsp amidase
- spd
spermidine
- SUMO
small ubiquitin-like modifier
- UBP
ubiquitin-specific processing protease
References
- 1.Bollinger JM, Jr, Kwon DS, Huisman GW, Kolter R, Walsh CT. Glutathionylspermidine metabolism in Escherichia coli. Purification, cloning, overproduction, and characterization of a bifunctional glutathionylspermidine synthetase/amidase. J Biol Chem. 1995;270:14031–14041. doi: 10.1074/jbc.270.23.14031. [DOI] [PubMed] [Google Scholar]
- 2.Kwon DS, Lin CH, Chen S, Coward JK, Walsh CT, Bollinger JM., Jr Dissection of glutathionylspermidine synthetase/amidase from Escherichia coli into autonomously folding and functional synthetase and amidase domains. J Biol Chem. 1997;272:2429–2436. doi: 10.1074/jbc.272.4.2429. [DOI] [PubMed] [Google Scholar]
- 3.Tabor CW, Tabor H. Polyamines. Annu Rev Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- 4.Meister A, Anderson ME. Glutathione. Annu Rev Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 5.Marton LJ, Pegg AE. Polyamines as targets for therapeutic intervention. Annu Rev Pharmacol Toxicol. 1995;35:55–91. doi: 10.1146/annurev.pa.35.040195.000415. [DOI] [PubMed] [Google Scholar]
- 6.Pegg AE. Recent advances in the biochemistry of polyamines in eukaryotes. Biochem J. 1986;234:249–262. doi: 10.1042/bj2340249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang CC. Molecular mechanisms and therapeutic approaches to the treatment of African trypanosomiasis. Annu Rev Pharmacol Toxicol. 1995;35:93–127. doi: 10.1146/annurev.pa.35.040195.000521. [DOI] [PubMed] [Google Scholar]
- 8.Fairlamb AH, Blackburn P, Ulrich P, Chait BT, Cerami A. Trypanothione: a novel bis(glutathionyl)spermidine cofactor for glutathione reductase in trypanosomatids. Science. 1985;227:1485–1487. doi: 10.1126/science.3883489. [DOI] [PubMed] [Google Scholar]
- 9.Fairlamb AH, Henderson GB, Cerami A. The biosynthesis of trypanothione and N1-glutathionylspermidine in Crithidia fasciculata. Mol Biochem Parasitol. 1986;21:247–257. doi: 10.1016/0166-6851(86)90130-1. [DOI] [PubMed] [Google Scholar]
- 10.Tabor H, Tabor CW. Isolation, characterization, and turnover of glutathionylspermidine from Escherichia coli. J Biol Chem. 1975;250:2648–2654. [PubMed] [Google Scholar]
- 11.Pai CH, Chiang BY, Ko TP, Chou CC, Chong CM, Yen FJ, Chen S, Coward JK, Wang AH, Lin CH. Dual binding sites for translocation catalysis by Escherichia coli glutathionylspermidine synthetase. EMBO J. 2006;25:5970–5982. doi: 10.1038/sj.emboj.7601440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiang BY, Chen TC, Pai CH, Chou CC, Chen HH, Ko TP, Hsu WH, Chang CY, Wu WF, Wang AH, Lin CH. Protein S-thiolation by glutathionylspermidine (GSP): the role of Escherichia coli gsp synthetase/amidase in redox regulation. J Biol Chem. 2010;285:25345–25353. doi: 10.1074/jbc.M110.133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Storer AC, Menard R. Catalytic mechanism in papain family of cysteine peptidases. Methods Enzymol. 1994;244:486–500. doi: 10.1016/0076-6879(94)44035-2. [DOI] [PubMed] [Google Scholar]
- 14.Fyfe PK, Oza SL, Fairlamb AH, Hunter WN. Leishmania trypanothione synthetase-amidase structure reveals a basis for regulation of conflicting synthetic and hydrolytic activities. J Biol Chem. 2008;283:17672–17680. doi: 10.1074/jbc.M801850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee CC, Kuo CJ, Ko TP, Hsu MF, Tsui YC, Chang SC, Yang S, Chen SJ, Chen HC, Hsu MC, Shih SR, Liang PH, Wang AH. Structural basis of inhibition specificities of 3C and 3C-like proteases by zinc-coordinating and peptidomimetic compounds. J Biol Chem. 2009;284:7646–7655. doi: 10.1074/jbc.M807947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo HB, Zheng H, Zimmerman MD, Chruszcz M, Skarina T, Egorova O, Savchenko A, Edwards AM, Minor W. Crystal structure and molecular modeling study of N-carbamoylsarcosine amidase Ta0454 from Thermoplasma acidophilum. J Struct Biol. 2010;169:304–311. doi: 10.1016/j.jsb.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu M, Li P, Li M, Li W, Yao T, Wu JW, Gu W, Cohen RE, Shi Y. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 18.Kamphuis IG, Kalk KH, Swarte MB, Drenth J. Structure of papain refined at 1.65 A resolution. J Mol Biol. 1984;179:233–256. doi: 10.1016/0022-2836(84)90467-4. [DOI] [PubMed] [Google Scholar]
- 19.Xu Z, Lam LS, Lam LH, Chau SF, Ng TB, Au SW. Molecular basis of the redox regulation of SUMO proteases: a protective mechanism of intermolecular disulfide linkage against irreversible sulfhydryl oxidation. FASEB J. 2008;22:127–137. doi: 10.1096/fj.06-7871com. [DOI] [PubMed] [Google Scholar]
- 20.Lin CH, Kwon DS, Bollinger JM, Jr, Walsh CT. Evidence for a glutathionyl-enzyme intermediate in the amidase activity of the bifunctional glutathionylspermidine synthetase/amidase from Escherichia coli. Biochemistry. 1997;36:14930–14938. doi: 10.1021/bi9714464. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 22.Bas DC, Rogers DM, Jensen JH. Very fast prediction and rationalization of pKa values for protein-ligand complexes. Proteins. 2008;73:765–783. doi: 10.1002/prot.22102. [DOI] [PubMed] [Google Scholar]
- 23.Bateman A, Rawlings ND. The CHAP domaa large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem Sci. 2003;28:234–237. doi: 10.1016/S0968-0004(03)00061-6. [DOI] [PubMed] [Google Scholar]
- 24.Rigden DJ, Jedrzejas MJ, Galperin MY. Amidase domains from bacterial and phage autolysins define a family of gamma-D,L-glutamate-specific amidohydrolases. Trends Biochem Sci. 2003;28:230–234. doi: 10.1016/s0968-0004(03)00062-8. [DOI] [PubMed] [Google Scholar]
- 25.Anantharaman V, Aravind L. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 2003;4:R11.1–R11.12. doi: 10.1186/gb-2003-4-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Claiborne A, Miller H, Parsonage D, Ross RP. Protein-sulfenic acid stabilization and function in enzyme catalysis and gene regulation. FASEB J. 1993;7:1483–1490. doi: 10.1096/fasebj.7.15.8262333. [DOI] [PubMed] [Google Scholar]
- 27.Lin CH, Chen S, Kwon DS, Coward JK, Walsh CT. Aldehyde and phosphinate analogs of glutathione and glutathionylspermidine: potent, selective binding inhibitors of the E. coli bifunctional glutathionylspermidine synthetase/amidase. Chem Biol. 1997;4:859–866. doi: 10.1016/s1074-5521(97)90118-6. [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.McRee DE. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 31.DeLano WL. The PyMOL molecular graphics system. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]


