Abstract
Specific-ion effects are ubiquitous in nature; however, their underlying mechanisms remain elusive. Although Hofmeister-ion effects on proteins are observed at higher (>0.3M) salt concentrations, in dilute (<0.1M) salt solutions nonspecific electrostatic screening is considered to be dominant. Here, using effective charge (Q*) measurements of hen-egg white lysozyme (HEWL) as a direct and differential measure of ion-association, we experimentally show that anions selectively and preferentially accumulate at the protein surface even at low (<100 mM) salt concentrations. At a given ion normality (50 mN), the HEWL Q* was dependent on anion, but not cation (Li+, Na+, K+, Rb+, Cs+, GdnH+, and Ca2+), identity. The Q* decreased in the order F− > Cl− > Br− > NO3− ∼ I− > SCN− > ClO4− ≫ SO42−, demonstrating progressively greater binding of the monovalent anions to HEWL and also show that the SO42− anion, despite being strongly hydrated, interacts directly with the HEWL surface. Under our experimental conditions, we observe a remarkable asymmetry between anions and cations in their interactions with the HEWL surface.
Keywords: ion–protein interactions, protein charge, Hofmeister series, electroselectivity series, specific-ion effects
Introduction
First reported by Hofmeister in the 1880s,1 specific-ion effects play an ubiquitous role in modulating myriad biological and chemical processes. However, the underlying mechanism(s) governing ion-specific effects are yet to be fully understood and remain a subject of significant interest.2 The namesake “Hofmeister effect” is observed at moderate to high (>0.3M) salt concentrations, wherein “kosmotropic” ions, classically postulated to form water structure, are observed to decrease protein solubility and increase conformational stability, whereas “chaotropic” ions, postulated to disrupt water structure, have the opposite effects.3 Ion partitioning studies with model nonpolar (e.g., benzene) and polar, peptidyl compounds provide evidence in support of the Hofmeister effect occurring by salting-out nonpolar groups through ion-specific increases in the surface tension of water4,5 and by salting-in the peptide backbone via ion binding.4,6
The effects observed at high salt concentrations, however, may not be applicable to lower, physiologically relevant, salt concentrations. Below salt concentrations of 0.1M, protein–ion interactions are thought to be predominantly governed by nonspecific, electrostatic interactions/screening.7,8 Yet, there are numerous reports in literature describing ion-specific effects on protein self-association,9 solubility,10,11 thermal stability,12 viscosity,13 and aggregation kinetics14 in this concentration range. In addition, below the protein pI, kosmotropic anions have been reported to increase protein solubility while chaotropic ions are effective salting-out or crystallizing agents, which results in an apparent inverse ranking of the Hofmeister monovalent anions.10,15
Through a series of articles, Collins15–18 proposed that ion effects on water structure are limited to only the first hydration shell and that ions may interact directly with proteins. Inner-sphere ion pairs are proposed to be formed preferentially by ions of opposite sign and similar size. Chaotropic ions, having low charge density, interact poorly with water and are able to reduce the number of ion–water interactions through ion pairing. In contrast, kosmotropic ions are able to form more energetically favorable electrostatic interactions with other kosmotropes than with water due to the short interatomic distances and high charge density of anion–cation pairs. Consistent with this qualitative description of ion interactions, simulations, and spectroscopic studies with aqueous halide solutions indicate that ion effects on water structure may be limited to the first hydration shell.2,19 Per the Collins model, the positively charged amines and backbone amide nitrogens on proteins are weakly hydrated and interact preferentially with weakly hydrated anions, whereas the strongly hydrated side-chain carboxylates and backbone carbonyls are predicted to interact preferentially with similarly strongly hydrated cations.
Anion binding to proteins was first proposed by Scatchard and Black in the 1940s on the basis of a systematic increase in pH (Cl− < I− < SCN−) on salt addition to isoionic solutions of bovine serum albumin (BSA).20 Any ion binding or association to the protein surface should accordingly modulate protein charge, and this effect was exploited to examine anion binding near the isoionic points of β-lactoglobulin and BSA by Longsworth and Jacobsen.21 In our studies, we employed protein effective charge (Q*) measurements to directly probe specific-ion interactions between hen-egg white lysozyme (HEWL) and Hofmeister salt solutions, up to concentrations of 100 mM. The Q* of a protein (in elementary charge units, e) in a given solution and temperature (T) is related to its electrophoretic mobility (μ), translational diffusion coefficient (Dt), and the Boltzmann's constant (kB), by (1).22
| (1) |
Thus, the dependence of Q* on ion-identity under equivalent conditions reflects differential interaction of the ion with the protein.
Results
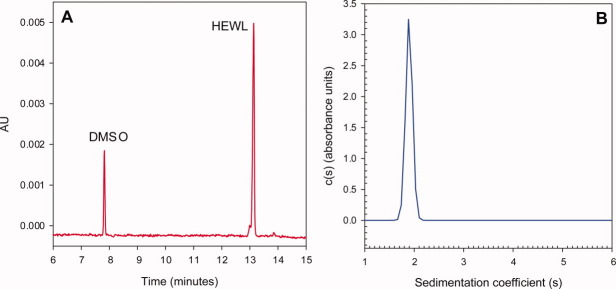
The Q* of HEWL (pI = 10.7) was measured in 10 mM acetate at pH 5.0 in the presence of two series of monovalent Hofmeister salts; an anion series (NaF, NaCl, NaBr, NaI, NaNO3, NaSCN, and NaClO4), and a cation series (LiCl, NaCl, KCl, RbCl, CsCl, and GdnHCl). The HEWL Q* was also determined in the presence of two divalent salts, CaCl2 and Na2SO4. Two comparable22 free-solution techniques, capillary zone and membrane-confined electrophoresis, have been described for measuring μ. The former was utilized with an amine-coated capillary to calculate μ from the velocity of HEWL relative to a neutral, electro-osmotic flow marker under an applied electrical field [Fig. 1(A)]. Sedimentation velocity analytical ultracentrifugation (SV-AUC) was used to obtain the Dt of HEWL from its sedimentation coefficient [Fig. 1(B)].
Figure 1.
Representative electropherogram (A) and c(s) sedimentation coefficient distribution (B) of HEWL in 10 mM acetate, 50 mM NaCl at pH 5.0. The electrophoretic mobility was calculated using the elution times of the EOF marker, (tEOF) and HEWL (tHEWL), using (2). The c(s) distribution of HEWL shows a single peak at 1.9 s with no evidence of aggregation at higher s values. The diffusion coefficient was calculated using the sedimentation coefficient (3).
The Q* decreased for all the salts tested with increasing salt concentration; however, the magnitude of the decrease was dependent on the identity of the anionic, but not the cationic, component of the salt [Fig. 2(A)]. This observation was consistent for the entire set of salts tested [Fig. 2(B)]. At a fixed ion normality of 50 mN, the HEWL Q* decreased from 3.7e to 1.7e in the order F− > Cl− > Br− > NO3− ∼ I− > SCN− > ClO4− ≫ SO42−. The trend in the Q* data shows progressively greater association of the monovalent anions and HEWL and also demonstrates that the SO42− anion, despite being strongly hydrated, interacts directly with the protein surface. In contrast, there was no effect of cation identity on the effective charge of HEWL; it remained essentially unchanged (3.4e) over the entire set of cations studied.
Figure 2.
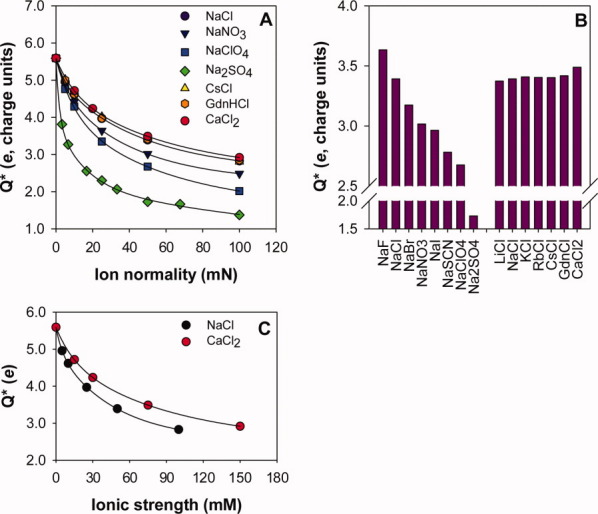
The effective charge (Q*) of hen-egg white lysozyme (HEWL) in different Hofmeister salt solutions. Panel A: The HEWL Q* in NaCl ( ), NaNO3 (
), NaNO3 ( ), NaClO4 (
), NaClO4 ( ), Na2SO4 (
), Na2SO4 ( ), CsCl (
), CsCl ( ), GdnHCl (
), GdnHCl ( ), and CaCl2 (
), and CaCl2 ( ) as a function of ion normality. Note that the NaCl, CsCl, GdnHCl, and CaCl2 data coincide. The lines through data series are presented as guides to the eye only. Panel B: At 50 mM ion normality, Q* is dependent on anion but not cation, identity. Panel C: Plotting HEWL Q* in NaCl (
) as a function of ion normality. Note that the NaCl, CsCl, GdnHCl, and CaCl2 data coincide. The lines through data series are presented as guides to the eye only. Panel B: At 50 mM ion normality, Q* is dependent on anion but not cation, identity. Panel C: Plotting HEWL Q* in NaCl ( ) and CaCl2 (
) and CaCl2 ( ) versus ionic strength (I) results in a higher Q* in CaCl2, suggestive of calcium binding to HEWL; however, the trends coincide when plotted versus ion normality (Panel A).
) versus ionic strength (I) results in a higher Q* in CaCl2, suggestive of calcium binding to HEWL; however, the trends coincide when plotted versus ion normality (Panel A).
The classical Debye-Hückel-Henry (DHH) model provides an approximation to estimate the formal protein charge (Q) by accounting for counter-ion size, bulk electrostatic screening, and the electrophoretic effect.23 The DHH model was applied in an attempt to correct for the observed anion-specific changes in Q* (Table I). However, a similar decrease was observed in HEWL charge as a function of anion identity.
Table I.
Calculated Formal Protein Charge (Q) for Hen Egg-White Lysozyme (HEWL) as a Function of Anion Identity at an Ionic Strength (I) of 0.05 M Using the Debye–Hückel–Henry (DHH) Model
| Ion | Q* (e) | Ionic radius44 Å | Q (e) |
|---|---|---|---|
| F− | 3.6 | 1.66 | 8.9 |
| Cl− | 3.4 | 1.21 | 8.2 |
| Br− | 3.2 | 1.18 | 7.6 |
| NO3− | 3.0 | 1.29 | 7.3 |
| I− | 3.0 | 1.20 | 7.1 |
| SCN− | 2.8 | 1.40 | 6.7 |
| ClO4− | 2.7 | 1.37 | 6.5 |
| SO42− | 2.1 | 2.30 | 5.1 |
We evaluated the charge heterogeneity of the HEWL surface using the Molecular Operating Environment (MOE) software and a 0.65 Å HEWL crystal structure. The solvent accessible surface area was calculated to be 6,530.5 Å2. The MOE software was used to assign partial charges at pH 5.0 using the AMBER99 force-field. A cartoon of the electrostatic surface is shown in Figure 3. Using a conservative cutoff of 1800 mV at 1.4 Å from the van der Waals surface and Poisson-Boltzmann electrostatics, 41 charged regions were identified, distributed heterogeneously over 19.0% of the molecule. Positive regions (19) covered 4.6% of the molecule, whereas negative regions (22) covered 14.3%. One large region (negative) was identified, covering 472 Å2. The remaining regions averaged 19 Å2 with a standard deviation of 18 Å2, and ranged from 6.13 Å2 to 94.6 Å2. Despite the prevalence of negative regions in terms of area, the protein was calculated to have a net-positive potential.
Figure 3.
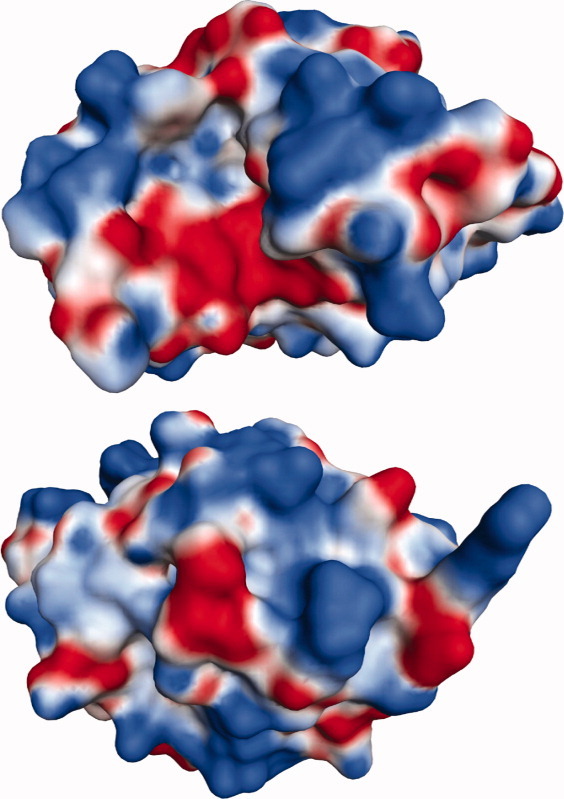
Two views of the HEWL electrostatic surface, rotated 180° along the horizontal axis. The surface is heterogeneous with respect to charge. The largest charged region (negative, red) appears as part of the cleft in the upper structure. Positively charged regions are shown in blue.
Discussion
Salts are routinely used for protein denaturation and crystallization at molar concentrations, and the literature is replete with studies of salt effects on protein solubility and stability at high salt concentrations.3,4 Our experimental approach enabled us to directly probe ion–protein interactions in the physiologically relevant, low salt concentration regime of <0.1M. A protein surface is chemically heterogeneous, composed of a complex set of charged, polar, and apolar groups, capable of interacting with ions by different mechanisms depending on the salt concentration. Ramos and Baldwin,24 in their study of salt effects on ribonuclease A (RNase A) stability, provide an elegant, systematic framework for discussing ion–protein interactions in terms of (i) electrostatic screening, (ii) specific-ion binding, and (iii) the Hofmeister effect. We follow a similar approach in the discussion of our results. The first two effects can be expected to be important under dilute salt conditions with the Hofmeister effect becoming increasingly important at higher salt concentrations.
Electrostatic screening
The HEWL charge in the presence of various sodium salts (I = 0.05M) estimated using the DHH model varies from ∼9 in the presence of F− to ∼5 in SO42− (Table I). The DHH model accounts for counter-ion screening due to the protein's charge, thus the reduction in Q from its expected value of +11 at pH 5.0 must result from anion binding to HEWL. In this context, binding includes any ions exceeding the Debye-Hückel ion-cloud that traverse with the protein (i.e., are within the Stokes radius) and does not distinguish between specific sites of anion interaction.
The positively charge HEWL can be envisioned to be surrounded by a Debye-Hückel ion-cloud predominantly composed of anions at pH 5.0. The observed independence of Q* on cation identity may thus be reconciled on the basis of a depleted cation concentration in the immediate vicinity of the molecule. However, such an explanation is simplistic given that HEWL is not uniformly charged at pH 5.0 but has a heterogeneously charged surface (Fig. 3) capable of supporting a corresponding localization of counter-ions.
The anion and CaCl2Q* data raise an important question about the appropriateness of comparing ion-specific effects of mono- and multivalent salts by normalizing to ionic strength (I). Plotting HEWL Q* versus I [Fig. 2(C)] resulted in higher Q* values in CaCl2 versus NaCl at a given I suggesting the possibility of either Ca2+ association to the HEWL surface or an inhibitory effect of Ca2+ on Cl− association. However, the Q* values in both CaCl2 and NaCl are nearly coincidental when plotted against ion normality or Cl− concentration [Fig. 2(A)], demonstrating negligible effect of the divalent Ca2+ on the Q* of HEWL at pH 5.0. Our interpretation of the calcium data is consistent with the recent calorimetric ion binding studies of HEWL under similar conditions, which show negligible binding of Ca2+ to HEWL compared with anions at pH 4.6.25
Anions selectively and directly interact with the HEWL surface
The HEWL effective charge data in sodium salts provide compelling, direct experimental evidence of preferential accumulation of anions at the protein surface even under dilute (<0.1M) salt conditions. Our work finds support in X-ray crystallographic studies of HEWL. Specific-ion binding to HEWL was proposed by Ries-Kautt and coworkers based on the ability of anions to aid in crystallization of this basic enzyme under acidic (pH 4.5) conditions.26 Specifically, larger monovalent anions were observed to be more effective in crystallizing HEWL, consistent with the Q* data, which show greater interaction of these anions with the HEWL surface. More recently, anion effects on the HEWL cloud-point in sodium salts (<0.2M) have similarly been interpreted in terms of surface charge neutralization by anion binding.11 Further, numerous cocrystal structures of HEWL-salts depict various anions namely chloride, bromide, iodide, nitrate, and thiocyanate localized on the protein surface.27–30 Our work demonstrates that direct anion–protein interactions occur in free solution even under dilute salt conditions.
Our results are also consistent with Collins'16 theoretical proposition and with recent molecular dynamics simulations that predict preferential interactions of weakly hydrated anions with the protein surface.31 However, direct anion–protein interactions may not be limited to large, weakly hydrated monovalent anions. Based on ion hydration, Collins has also proposed that the strongly hydrated, divalent SO42− anion (grouped with F− in the Hofmeister-ion series) interacts minimally with proteins.15 In contrast, the Q* values indicate significant interaction of SO42− with HEWL. Recent simulations also predict strong ion-pairing interactions of SO42− with ionized basic side-chains.32
The Hofmeister and electroselectivity series of anions
The placement of F− and SO42− on opposite ends of the sodium-salt series is an important result of this study. The sulfate ion was observed to be more effective at reducing HEWL charge than the chaotropic ClO4− ion. The distinctly different order results from differential anion binding to HEWL and is not to be misinterpreted as the “inverse” Hofmeister series. In the Hofmeister-anion series, F− and SO42− are grouped together, as kosmotropes. In addition to this grouping, the Hofmeister-ion effect has another characteristic feature: the effect varies monotonically with ion normality and continues to very high ion-normalities (>1N). At high salt concentrations, differential effects on the bulk surface tension of the solvent become increasingly important along with ion-binding to the peptide backbone (4). A good example is studies on the effect of the sulfate-ion on RNase A stability,24 which show stabilization of this enzyme by sulfate via anion-specific binding at dilute concentrations (0–0.1M) and via the Hofmeister effect at higher concentrations (0.1–1.0M). Our studies provide direct experimental evidence in support of such anion-specific binding under dilute conditions. We must point out that the anion series observed in this work (F−, Cl−, Br−, NO3−, I−, SCN−, ClO4−, SO42−) has also been termed as the electroselectivity series in the literature. The series, based on strong (SO42− > SCN− > I−) or weak (F− < Cl− < Br−) interaction of the anions with an anion-exchange resin, appears to be used to differentiate from the Hofmeister-ion series. Whenever an anion-specific dependence following the electroselectivity series in processes such as protein aggregation and fibrillation,14,33 and protein stability34 has been observed, the results have been interpreted in terms of anion-binding. Our demonstration of direct sulfate ion–protein interactions is also consistent with the sulfate ion, and other related but possibly weakly hydrated sulfated glycosaminoglycans35 accelerating protein fibril formation, attributed to direct sulfate–protein interactions.36–38
Cations do not interact with the HEWL surface
Contrary to the Collins model of strongly hydrated cations (i.e., Li+ and Na+) interacting preferentially with the protein carboxylates, the HEWL Q* was insensitive to cation identity. Although a strong anion-dependence was observed in the sodium salt series, no dependence on cation identity was observed in the chloride salt series. We also recently reported asymmetric binding of anions to three IgG2 monoclonal antibodies.12 The effective charge data for HEWL in sodium-cation salts in dilute salt solutions is consistent with our previous observations with monoclonal antibodies and a fusion protein that showed independence of cation identity in the modulation of precipitation, aggregation and apparent melting temperature,10 and oligomerization9 under similar conditions (<0.1M). In contrast, Benas et al. reported increased HEWL solubility in CsCl relative to RbCl at higher salt concentrations (0.6–1.0M).39 Solubility could be further increased with the use of MnCl2 or YbCl3; the increases could not be explained solely based on the change in chloride concentration. The observations can be rationalized by a transition in the importance of ion-binding effects versus Hofmeister-type taking place at the higher salt concentration.
No evidence of preferential GdnH+ accumulation was observed at the HEWL surface up to concentrations of 100 mM. The comparability of guanidinium (GdnH+) to other cations is also surprising. Mande and Sobhia40 have identified two GdnH+ binding sites on HEWL under acidic conditions (pH 4.6); in addition, structural perturbations caused by binding at one of these sites introduced a Na+ binding site.40 The HEWL crystal was produced in 0.1M acetate, 2M NaCl, and 1.2M GdnHCl, pH 4.6. Our data, also under acidic but dilute salt conditions, shows no response to GdnH+ (or Na+) relative to other cations. The charge measurement method we used would not necessarily be able to discern the binding of anions or cations to a small number of high affinity sites on the protein surface, because these could become saturated at <1 mM salt concentrations. Alternatively, the crystallization of HEWL in the presence of high-concentrations of GdnH+ may lead to the observation of even low affinity interactions that would not necessarily be favorable under dilute solution conditions. Our interpretation of the cation data and is that in the low-salt regime (≤0.1M), differential specific-ion binding to the protein surface is essentially restricted to anions and is supported by recent calorimetric binding studies of Hofmeister ions and HEWL.25 However, there are exceptions; cation-specific effects under dilute salt solutions have been reported in molecules bearing a high density of anionic residues (e.g., DNA and tubulin), and through the chelation of multivalent metal ions by proteins.17,41
Conclusions
Our studies with HEWL, consistent with the pioneering work of Scatchard20 and Longsworth,21 provide direct experimental evidence for anion association to the protein surface. Furthermore, the data point to a remarkable asymmetry between anions and cations for this interaction in a low-concentration salt regime considered to be dominated by nonspecific, electrostatic interactions. However, studies on a set of proteins with measurements made above and below the pI will shed further light on the relative propensity of cations and anions for interaction. Our results underscore the importance of considering ion-specific effects at low salt concentrations and have broad significance for biological function, biochemical processes, and disease.
Material and Methods
Sample preparation
Hen-egg white lysozyme (HEWL) was obtained from Sigma-Aldrich (catalog #L7651) and dissolved in a 10 mM acetic acid, pH 5.0 (adjusted with NaOH) buffer (10A5) to an approximate concentration of 70 mg/mL. The dissolved HEWL was buffer exchanged into the 10A5 buffer using gel-filtration to remove any extraneous ions. Briefly, 0.5 mL of the 70 mg/mL HEWL solution was applied to a NAP5 gel-filtration column (GE Healthcare, NJ) pre-equilibrated with the 10A5 buffer, and subsequently eluting with 0.95 mL of 10A5. The concentration was determined using an extinction coefficient of 2.65 mL/mg cm, yielding a buffer-exchanged stock HEWL solution at ∼30 mg/mL.
Stock salt solutions were prepared at 200 mM concentrations in 10A5. Analytical samples were prepared by combining the 10A5, the buffer-exchanged stock HEWL solution (∼30 mg/mL), and a stock salt solution in appropriate ratios to yield HEWL solutions with desired protein and salt concentrations.
Electrophoretic mobility measurements
The electrophoretic mobility of HEWL in the different salt solutions was measured using a Beckman Coulter PA 800 instrument and a 60 cm, 50-μm ID, eCap amine capillary. Protein samples were prepared at 0.3 mg/mL and were injected immediately after the injection of an electroosmotic flow (EOF) marker using hydrodynamic injection (0.5 psi for 3 s). For most solutions, 0.02% (v/v) DMSO was used as the EOF marker with detection at 214 nm. For solutions containing iodide or thiocyanate, the protein concentration was increased to 1 mg/mL and 0.2% (v/v) benzyl alcohol was used as the EOF marker with detection at 280 nm. The EOF was slower in higher concentration salt solutions, necessitating a higher applied field to prevent excessive, >60 min, run times. The applied voltages ranged from 8,000 to 23,000 volts in reverse polarity. The correlation between the current and applied voltage remained constant (r2 = 0.99), even at the highest voltages used. For each salt and concentration, measurements were made using four different applied fields in a 6,000 volt range (e.g., 11,000, 13,000, 15,000, and 17,000 volts). The value of μ was determined by linear regression using the relationship in (2), in which tEOF and tHEWL are the elution times of the EOF marker and HEWL, Ct is the total capillary length, Cd is the capillary length before the detector (the distance traversed by the marker/protein before detection), and P is the applied potential.
| (2) |
Diffusion coefficient measurements
The diffusion coefficient (Dt) was measured using sedimentation velocity analytical ultracentrifugation (SV-AUC) experiments. The Dt is related to the sedimentation coefficient (s), universal gas constant (R), molecular weight (Mw), partial specific volume (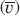 ), and solvent density (ρ) by the Svedberg equation [(3)].
), and solvent density (ρ) by the Svedberg equation [(3)].
| (3) |
Sedimentation coefficients were obtained using a Beckman Coulter (Fullerton, CA) XL-I centrifuge. Samples were loaded into two-sector sedimentation velocity centerpieces and equilibrated to 20°C before being centrifuged at 50,000 rpm. Sedimentation was monitored using 280 nm absorbance scans. The resulting data were analyzed using the program SEDFIT42 to obtain c(s) distributions to ensure sample stability with respect to aggregation and to obtain weight average sedimentation coefficients. The different solution conditions had a negligible effect on HEWL sedimentation coefficient, and the range of measured s values was within instrument variability. There was no systematic effect of ion-size on s. For this reason, an average value (1.9 ± 0.1 s), obtained from all studied conditions was used in the calculation of Dt. A value of 0.715 mL/g was used for 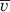 , derived from the published primary structure of HEWL (PDB 2VB1). The solvent density values used in the calculation of Dt were obtained, when possible, using the program SEDNTERP v. 1.09.43 For salts not available in SEDNTERP (NaI, NaClO4, NaSCN, Na2SO4, LiCl, KCl, and RbCl), values were obtained by measuring protein-free 10A5 salt solutions in an Anton Paar GmbH (Graz, Austria) DMA 5000 densitometer, calibrated to air and water.
, derived from the published primary structure of HEWL (PDB 2VB1). The solvent density values used in the calculation of Dt were obtained, when possible, using the program SEDNTERP v. 1.09.43 For salts not available in SEDNTERP (NaI, NaClO4, NaSCN, Na2SO4, LiCl, KCl, and RbCl), values were obtained by measuring protein-free 10A5 salt solutions in an Anton Paar GmbH (Graz, Austria) DMA 5000 densitometer, calibrated to air and water.
Application of the DHH model
The DHH model22 was applied to Q* values using (4) to correct for ionic radii and ionic strength effects. In the equation, κ is the inverse Debye length, a is the combined Stokes radii of HEWL (1.9 nm) and the salt anion, and f(κa) is Henry's function. The ionic radius was calculated based on literature diffusion values for each anion.44 The Debye length and the Henry's function were estimated using a program, developed at the University of New Hampshire, called ZUTILITIES.
| (4) |
Modeling of HEWL surface charge heterogeneity
The HEWL surface at pH 5.0 was modeled using the MOE software package produced by The Chemical Computing Group (Montreal, Canada) and a 0.65 Å HEWL crystal structure (triclinic), PDB accession number 2VB1.
Acknowledgments
The authors thank Dr. Randal R. Ketchem (Amgen, Inc.) for his help with modeling of the electrostatic surface of HEWL and construction of the related figure.
References
- 1.Kunz W, Henle J, Ninham BW. Zur Lehre von der Wirkung der Salze (about the science of the effect of salts): Franz Hofmeister's historical papers. Curr Opin Coll Inter Sci. 2004;9:19–37. [Google Scholar]
- 2.Tobias DJ, Hemminger JC. Chemistry. Getting specific about specific ion effects. Science. 2008;319:1197–1198. doi: 10.1126/science.1152799. [DOI] [PubMed] [Google Scholar]
- 3.Melander W, Horvath C. Salt effect on hydrophobic interactions in precipitation and chromatography of proteins: an interpretation of the lyotropic series. Arch Biochem Biophys. 1977;183:200–215. doi: 10.1016/0003-9861(77)90434-9. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin RL. How Hofmeister ion interactions affect protein stability. Biophys J. 1996;71:2056–2063. doi: 10.1016/S0006-3495(96)79404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nandi PK, Robinson DR. The effects of salts on the free energies of nonpolar groups in model peptides. J Am Chem Soc. 1972;94:1308–1315. doi: 10.1021/ja00759a043. [DOI] [PubMed] [Google Scholar]
- 6.Nandi PK, Robinson DR. The effects of salts on the free energy of the peptide group. J Am Chem Soc. 1972;94:1299–1308. doi: 10.1021/ja00759a042. [DOI] [PubMed] [Google Scholar]
- 7.Pegram LM, Record MT., Jr Thermodynamic origin of hofmeister ion effects. J Phys Chem. 2008;112:9428–9436. doi: 10.1021/jp800816a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kunz W. Specific ion effects in colloidal and biological systems. Curr Opin Coll Inter Sci. 2010;15:34–39. [Google Scholar]
- 9.Gokarn YR, Fesinmeyer RM, Saluja A, Cao S, Dankberg J, Goetze A, Remmele RL, Jr, Narhi LO, Brems DN. Ion-specific modulation of protein interactions: anion-induced, reversible oligomerization of a fusion protein. Protein Sci. 2009;18:169–179. doi: 10.1002/pro.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saluja A, Crampton S, Kras E, Fesinmeyer RM, Remmele RL, Jr, Narhi LO, Brems DN, Gokarn YR. Anion binding mediated precipitation of a peptibody. Pharm Res. 2009;26:152–160. doi: 10.1007/s11095-008-9722-0. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Cremer PS. The inverse and direct Hofmeister series for lysozyme. Proc Natl Acad Sci USA. 2009;106:15249–15253. doi: 10.1073/pnas.0907616106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fesinmeyer RM, Hogan S, Saluja A, Brych SR, Kras E, Narhi L, Brems DN, Gokran YR. Effect of ions on agitation- and temperature-induced aggregation reactions of antibodies. Pharm Res. 2009;26:903–913. doi: 10.1007/s11095-008-9792-z. [DOI] [PubMed] [Google Scholar]
- 13.Kanai S, Liu J, Patapoff TW, Shire S. Reversible self-association of a concentrated monoclonal antibody solution mediated by Fab-Fab interaction that impacts solution viscosity. J Pharm Sci. 2008;97:4219–4227. doi: 10.1002/jps.21322. [DOI] [PubMed] [Google Scholar]
- 14.Munishkina LA, Henriques J, Uversky VN, Fink A. Role of protein-water interactions and electrostatics in alpha-synuclein fibril formation. Biochemistry. 2004;43:3289–3300. doi: 10.1021/bi034938r. [DOI] [PubMed] [Google Scholar]
- 15.Collins KD. Ions from the Hofmeister series and osmolytes: effects on proteins in solution and in the crystallization process. Methods. 2004;34:300–311. doi: 10.1016/j.ymeth.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 16.Collins KD. Sticky ions in biological systems. Proc Natl Acad Sci USA. 1995;92:5553–5557. doi: 10.1073/pnas.92.12.5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collins KD. Charge density-dependent strength of hydration and biological structure. Biophys J. 1997;72:65–76. doi: 10.1016/S0006-3495(97)78647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins KD. Ion hydration: implications for cellular function, polyelectrolytes, and protein crystallization. Biophys Chem. 2006;119:271–281. doi: 10.1016/j.bpc.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 19.Omta AW, Kropman MF, Woutersen S, Bakker HJ. Negligible effect of ions on the hydrogen-bond structure in liquid water. Science. 2003;301:347–349. doi: 10.1126/science.1084801. [DOI] [PubMed] [Google Scholar]
- 20.Scatchard G, Black ES. The effect of salts on the isoionic and isoelectric points of proteins. J Phys Colloid Chem. 1949;53:88–99. [PubMed] [Google Scholar]
- 21.Longsworth LG, Jacobsen CF. An electrophoretic study of the binding of salt ions by beta-lactoglobulin and bovine serum albumin. J Phys Colloid Chem. 1949;53:126–135. [PubMed] [Google Scholar]
- 22.Durant JA, Chen C, Laue TM, Moody TP, Allison SA. Use of T4 lysozyme charge mutants to examine electrophoretic models. Biophys Chem. 2002;101–102:593–609. doi: 10.1016/s0301-4622(02)00168-0. [DOI] [PubMed] [Google Scholar]
- 23.Moody TP, Kingsbury JS, Durant JA, Wilson TJ, Chase SF, Laue TM. Valence and anion binding of bovine ribonuclease A between pH 6 and 8. Anal Biochem. 2005;336:243–252. doi: 10.1016/j.ab.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 24.Ramos CH, Baldwin RL. Sulfate anion stabilization of native ribonuclease A both by anion binding and by the Hofmeister effect. Protein Sci. 2002;11:1771–1778. doi: 10.1110/ps.0205902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boncina M, Lah J, Rescic J, Vlachy V. Thermodynamics of the lysozyme—salt interaction from calorimetric titrations. J Phys Chem. 114:4313–4319. doi: 10.1021/jp9071845. YEAR. [DOI] [PubMed] [Google Scholar]
- 26.Ries-Kautt MM, Ducruix AF. Relative effectiveness of various ions on the solubility and crystal growth of lysozyme. J Biol Chem. 1989;264:745–748. [PubMed] [Google Scholar]
- 27.Lim K, Nadarajah A, Forsythe EL, Pusey ML. Locations of bromide ions in tetragonal lysozyme crystals. Acta Cryst D. 1998;54:899–904. doi: 10.1107/s0907444998002844. [DOI] [PubMed] [Google Scholar]
- 28.Steinrauf LK. Structures of monoclinic lysozyme iodide at 1.6 A and of triclinic lysozyme nitrate at 1.1 A. Acta Cryst D. 1998;54:767–780. doi: 10.1107/s0907444997016922. [DOI] [PubMed] [Google Scholar]
- 29.Vaney MC, Broutin I, Retailleau P, Douangamath A, Lafont S, Hamiaux C, Prange T, Ducruix A, Ries-Kautt M. Structural effects of monovalent anions on polymorphic lysozyme crystals. Acta Cryst D. 2001;57:929–940. doi: 10.1107/s0907444901004504. [DOI] [PubMed] [Google Scholar]
- 30.Walsh MA, Schneider TR, Sieker LC, Dauter Z, Lamzin VS, Wilson KS. Refinement of triclinic hen egg-white lysozyme at atomic resolution. Acta Cryst D. 1998;54:522–546. doi: 10.1107/s0907444997013656. [DOI] [PubMed] [Google Scholar]
- 31.Lund M, Vrbka L, Jungwirth P. Specific ion binding to nonpolar surface patches of proteins. J Am Chem Soc. 2008;130:11582–11583. doi: 10.1021/ja803274p. [DOI] [PubMed] [Google Scholar]
- 32.Mason PE, Dempsey CE, Vrbka L, Heyda J, Brady JW, Jungwirth P. Specificity of ion-protein interactions: complementary and competitive effects of tetrapropylammonium, guanidinium, sulfate, and chloride ions. J Phys Chem. 2009;113:3227–3234. doi: 10.1021/jp8112232. [DOI] [PubMed] [Google Scholar]
- 33.Jain S, Udgaonkar JB. Salt-induced modulation of the pathway of amyloid fibril formation by the mouse prion protein. Biochemistry. 2010;49:7615–7624. doi: 10.1021/bi100745j. [DOI] [PubMed] [Google Scholar]
- 34.Muzammil S, Kumar Y, Tayyab S. Anion-induced stabilization of human serum albumin prevents the formation of intermediate during urea denaturation. Proteins. 2000;40:29–38. doi: 10.1002/(sici)1097-0134(20000701)40:1<29::aid-prot50>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 35.Asthagiri D, Schure MR, Lenhoff AM. Calculation of hydration effects in the binding of anionic ligands to basic proteins. J Phys Chem B. 2000;104:8753–8761. [Google Scholar]
- 36.Calamai M, Kumita JR, Mifsud J, Parrini C, Ramazzotti M, Ramponi G, Taddei N, Chiti F, Dobson CM. Nature and significance of the interactions between amyloid fibrils and biological polyelectrolytes. Biochemistry. 2006;45:12806–12815. doi: 10.1021/bi0610653. [DOI] [PubMed] [Google Scholar]
- 37.Bellotti V, Chiti F. Amyloidogenesis in its biological environment: challenging a fundamental issue in protein misfolding diseases. Curr Opin Struct Biol. 2008;18:771–779. doi: 10.1016/j.sbi.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 38.Pedersen JS, Flink JM, Dikov D, Otzen DE. Sulfates dramatically stabilize a salt-dependent type of glucagon fibrils. Biophys J. 2006;90:4181–4194. doi: 10.1529/biophysj.105.070912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benas P, Legrand L, Riess-Kautt M. Strong and specific effects of cations on lysozyme chloride solubility. Acta Cryst D. 2002;58:1582–1587. doi: 10.1107/s0907444902014518. [DOI] [PubMed] [Google Scholar]
- 40.Mande SC, Sobhia ME. Structural characterization of protein-denaturant interactions: crystal structures of hen egg-white lysozyme in complex with DMSO and guanidinium chloride. Protein Eng. 2000;13:133–141. doi: 10.1093/protein/13.2.133. [DOI] [PubMed] [Google Scholar]
- 41.Wolff J, Sackett DL, Knipling L. Cation selective promotion of tubulin polymerization by alkali metal chlorides. Protein Sci. 1996;5:2020–2028. doi: 10.1002/pro.5560051008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laue T, Shah B, Ridgeway T, Pelletier S. Computer-aided Interpretation of Analytical Sedimentation Data for Proteins. In: Harding S, Rowe A, Horton J, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Cambridge, UK: Thomas Graham House, The Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 44.Vanysek P. Thermochemistry, Electrochemistry, and Kinetics. In: Lide DR, editor. CRC Handbook of Chemistry and Physics. Boca Raton: Taylor and Francis Group LLC; 2008. pp. 5-76–75-78. [Google Scholar]