Preface
Responding to genotoxic stress is a prerequisite for development of the nervous system. Mutations in a variety of DNA repair factors can lead to human diseases that are characterized by pronounced neuropathology. In many of these syndromes the neurological component is amongst the most deleterious aspects of the disease. The nervous system poses a particular challenge in terms of clinical intervention, as the neuropathology often arises during nervous system development, and can be fully penetrant by childhood. Understanding how DNA repair deficiency impacts the nervous system will provide a rationale basis for therapies targeted at ameliorating the neurological problems in these syndromes.
[1] Introduction
Maintaining genomic integrity by DNA repair is fundamental to the function and survival of an organism. Compromising genomic DNA can lead to a spectrum of human disorders that exhibit developmental defects, immune deficiency and cancer. In particular, the nervous system is often profoundly affected by DNA repair deficiency, which can result in neurodegeneration, microcephaly or brain tumors. More broadly, defective DNA repair in mature neural tissues has also been linked to aging and recently to common neurodegenerative syndromes such as Alzheimer's disease and Parkinson disease1-4.
The developmental dynamics of the nervous system requires vigilant DNA repair processes capable of repairing multiple types of damage. DNA repair is especially critical during the early period of rapid proliferation as progenitors expand and differentiate to generate the nervous system. In the mature nervous system, DNA repair is also essential to prevent DNA damage, arising from sources such as oxidative metabolism, from blocking active transcription. Understanding the interplay between signaling networks that function to maintain genomic stability in the nervous system will be of paramount importance for the treatment of diseases associated with DNA repair deficiency. This review will discuss the role of DNA repair during the genesis of the nervous system, and how this process maintains neural homeostasis.
[2] The importance of DNA integrity during neural development
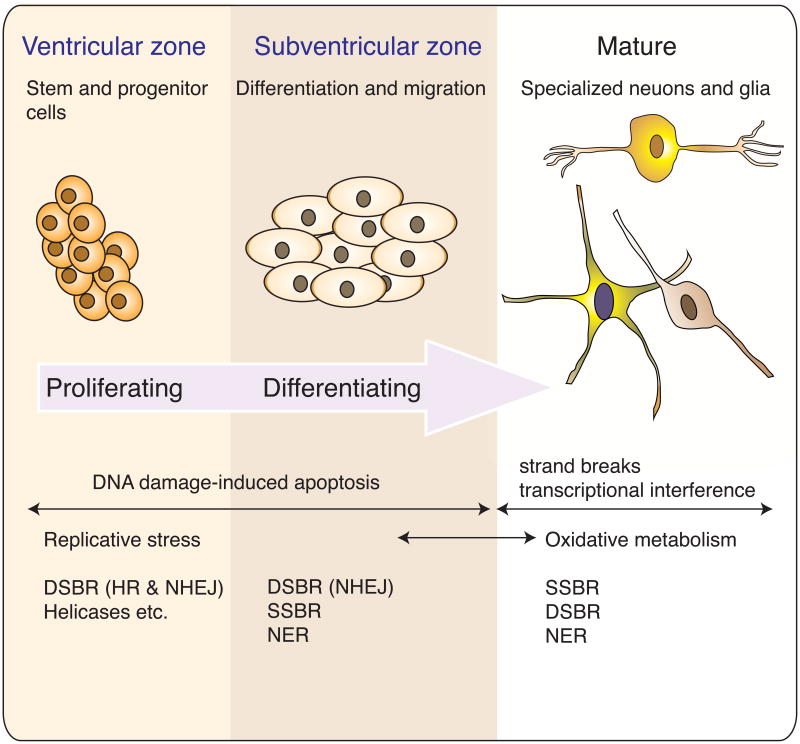
Many human DNA repair syndromes are congenital and so it is clear that the developing nervous system can be markedly affected by DNA repair deficiency. Therefore, understanding neural development is important for determining how DNA repair deficiency impacts the nervous system. Detailed reviews that summarize neural development are available5-9. In humans, the nervous system begins to form during the first trimester of gestation and continues until after birth. The complexity of the mature nervous system results from a relatively simple strategy of proliferation, differentiation and migration (Fig. 1). The engine driving neural development is rapid cellular proliferation within the ventricular and subventricular zones that line the four ventricles present in the nervous system. These regions contain neural stem and proliferating progenitor cells5, 6, 9, and DNA replication in these cells is of paramount importance since they will populate the nervous system and remain in place for the life of the organism. If these stem/progenitor cells incur mutation the expansion of these damaged cells may subsequently lead to disease. Thus, during this phase of proliferative expansion DNA repair is of the utmost importance. Proliferation of neural progenitors also occurs within the subventricular zone, the lateral ventricle and the dentate gyrus throughout adulthood9. Although it still unclear exactly how these contribute to the form and function of the mature human nervous system, evidence from mice indicates that ongoing adult neurogenesis is functionally important, suggesting that DNA repair will remain important in these areas throughout life10, 11.
Figure 1. DNA damage and repair during nervous system development.
During development the formation of the nervous system occurs via widespread proliferation, migration and differentiation. The diversity of the nervous system begins as stem/progenitors cells that divide in the ventricular zone and also, albeit to a lesser extent, the subventricular zone and then undergo differentiation, migration and maturation to give rise to the neurons and glia of the adult nervous system. A wide variety of functionally specialized cells with unique properties all originate from proliferative cells in one of four ventricles present in the nervous system. At different stages of development the nervous system is susceptible to different types of DNA damage. During proliferation, replication associated DNA strand breaks can occur that may require DNA double strand break repair (DSBR), involving homologous recombination (HR) or non-homologous end-joining (NHEJ) and associated functions of helicases and various other replication components that interface with DNA repair. In differentiating cells repair options are more limited as HR is not available after the cells exit the cell cycle. At this stage, NHEJ repairs DNA DSBs while other types of DNA damage require nucleotide excision repair (NER) or single strand break repair (SSBR). Because the nervous system can easily replace cells during development, DNA damage-induced apoptosis is also a frequent outcome at these stages of neural development. In the mature nervous system, strand breaks and DNA modification from oxidative damage engage SSBR, NER and DSBR and DNA damage that is not repaired can disrupt transcription leading to cell death.
While the bulk of cells in the nervous system are either neurons or glia, within these broad classes are a myriad of specialized cell types, which are characterized by specific functional roles that support the divergent functions of neural tissue. These cellular subgroups are characterized by their morphology, the neurotransmitters that they use for cellular communications and the connections they form with other cells. This diversity is reiterated through out the nervous system, and therefore further impacts the specific requirements for DNA repair pathways.
[3] Endogenous sources of DNA damage
Because of its relative complexity, the nervous system presents a substantial challenge for delineating the spatiotemporal requirements for DNA repair. One of the main questions associated with inherited human DNA repair deficiency syndromes is the nature and source of endogenous DNA damage in the nervous system.
The rapid cellular proliferation during development is associated with replication-induced damage and therefore an intact DNA repair system is needed at this stage. However, DNA repair remains important after replication ends and maturation commences12-14. The high oxidative load of the brain, and the free radicals produced by cellular metabolism can lead to many different types of DNA damage, and so it is possible that a limited number of etiological agents are responsible for generating a diversity of lesions. There is an increase in DNA breaks after high oxidative load such as that induced by overexpression of superoxide dismutase or increased oxygen tension15, and depending on the developmental timing, these strand breaks can lead to apoptosis14. Additionally, oxidative stress-induced strand breaks in DNA can also affect transcription16, 17.
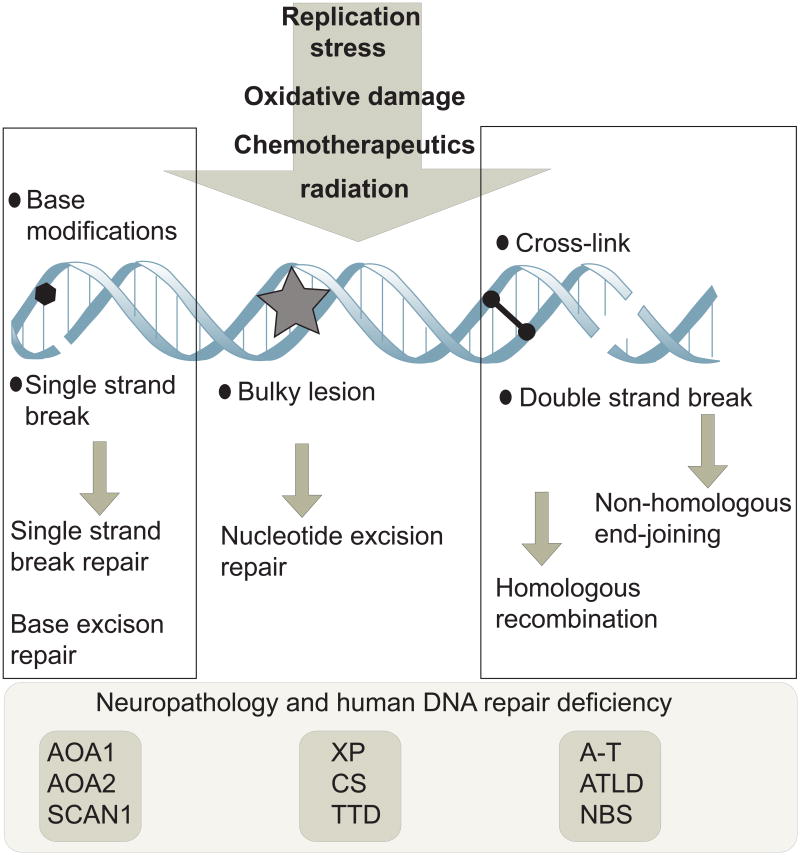
In addition to DNA strand breaks, oxidative stress may also generate specific types of lesions that can only be repaired by the excision of sections of DNA (see below)18. DNA damage may also be a by-product of normal neural functioning as ionotropic glutamate receptor activation leads to the formation of γH2AX (a phosphorylated form of the histone variant H2AX), a marker associated with DNA damage and repair19. Whatever the exact nature of the endogenous lesions, the vast repertoire of DNA damage-response factors and the many human DNA repair syndromes underscore the need to respond to a variety of insults (Fig. 2).
Figure 2. Types of DNA damage and repair.
A variety of different types of DNA damage can occur in neural cells as a result of endogenous agents such as replication stress or free radicals from oxidative metabolism. Exogenous insults such as ionizing or ultraviolet radiation and chemotherapeutics can also cause different types of DNA damage. These agents can cause single or double strand breaks in the DNA, base modifications, helix-distorting bulky lesions or cross-links of DNA strands. Biochemically distinct DNA repair pathways are available to repair each class of DNA damage. DNA repair pathways that are particularly important for nervous system function comprise pathways that repair DNA single and double strand breaks and nucleotide excision repair. When any of these pathways are defective, diseases can result that impact the nervous system; representative examples of human syndromes linked to defects in the particular DNA repair pathways are listed. Defective repair of DNA single strand breaks can lead to ataxia with oculomotor apraxia 1 or 2 (AOA1 and AOA2) or spinocerebellar ataxia with axonal neuropathy 1 (SCAN1), while defects in nucleotide excision repair can lead to xeroderma pigmentosum (XP), Cockayne Syndrome (CS) or trichothyrodystrophy (TTD). Defective responses to DNA double strand breaks can lead to ataxia telangiectasia (A-T), A-T like disease (ATLD) or Nijmegen breakage syndrome (NBS).
Although the focus of this review is the link between DNA repair deficiency and neurological disease, normal DNA repair processes can also be involved in neuropathology. In the triplet repeat expansion class of neurodegenerative diseases, such as those involving polyglutamine tracts within encoded proteins, oxidative stress in the brain has been shown to cause DNA glycosylase-mediated expansion of triplet repeats towards diseases levels20, 21. Thus, while oxidative stress may be causal in DNA repair deficient syndromes, this etiological agent could also be responsible for activating repair systems that inadvertently expand triplet sequences leading to disease.
[4] DNA damage responses
DNA can undergo a variety of modifications, including strand breaks, base damage, helical distortions and strand cross-links. Biochemically distinct DNA repair pathways have evolved to repair each of these specific lesions (Fig. 2). Basic strategies for DNA repair involve excision of the damage by removing small numbers of nucleotides via the base excision repair (BER) pathway or longer stretches via the nucleotide excision repair (NER) pathway. In the case of DNA strand breaks, either single strand break repair (SSBR) or double strand break repair (DSBR) pathways address each type of damaged DNA. Defects in these pathways can lead to human neurological disease, with the resultant neuropathology often linked to the type of damage and the developmental stage at which it occurs (Table 1). Several recent reviews discuss the detailed biochemistry of DNA repair pathways linked to human diseases22-28. Here, we will consider the main DNA damage pathways and examine the impact of defects in these pathways and how they lead to neuropathology.
Table 1. Human DNA repair deficient syndromes.
| Disease/Syndrome | Gene | Neurological | Extra-neurological |
|---|---|---|---|
| DNA DSB deficiency | |||
| Ataxia-telangiectasia | ATM | ataxia, neurodegeneration telangiectasia, dysarthria, | immunological defects, malignancy, sterility |
| Ataxia-telangiectasia-like disorder | MRE11 | ataxia, neurodegeneration dysarthria, oculomotor apraxia | mild immunological defects |
| Nijmegen breakage syndrome | NBS1 | microcephaly | immunological defects, lymphoid malignancy |
| ATR Seckel syndrome | ATR | microcephaly, mental retardation | growth defects |
| LIG4 syndrome | LIG4 | Microcephaly | developmental/growth delay, immunodeficiency, lymphoma |
| Human immunodeficiency with microcephaly | XLF/Cernunos | Microcephaly | Immunodeficiency |
| Fanconi anemia | BRCA2 | medulloblastoma | bone marrow and congenital defects |
| DNA SSB deficiency | |||
| Spinocerebellar ataxia with axonal neuropathy | TDP1 | ataxia, neurodegeneration peripheral axonal motor and sensory neuropathy muscle weakness | hypercholesterolemia, hypoalbuminemia |
| Ataxia with oculomotor apraxia 1 | APTX | ataxia, neurodegeneration oculomotor apraxia, peripheral neuropathy | hypercholesterolemia, hypoalbuminemia |
| NER deficiency | |||
| Xeroderma pigmentosum | XP (A-G) | Neurodegeneration microcephaly, | UV sensitivity, skin cancer |
| Cockayne syndrome | CS(A & B), XPB,XPD, XPG | Microcephaly Dysmyelination, | Progeria, variable presentation |
| Trichothyrodystrophy | XPD, XPB, TTD-A | Neurodevelopmental defects, Dysmyelination | Brittle hair, variable presentation |
| DNA X-link repair | |||
| Fanconi Anemia | FA(A-L) | Microcephaly, medulloblastoma (FANCD2, FANCN) | Anemia, developmental defects, cancer |
| Helicase deficiency | |||
| Werner Syndrome | WRN | ? | Severe progeria, cancer |
| Rothmund Thomson syndrome | RTS | ? | cancer |
| Bloom Syndrome | BLM | ? | Proportional dwarfism, cancer |
| Ataxia with oculomotor apraxia 2 | SETX | ataxia, neurodegeneration oculomotor apraxia | Absent/minimal |
[5] DNA DSB repair pathways and related diseases
One of the earliest recognized syndromes linking DNA damage and neurodegeneration was ataxia telangiectasia (A-T) (discussed below). Children with A-T are characterized by severe neurodegeneration with an extreme sensitivity to ionizing radiation29-31. A-T established a compelling link between faulty responses to DNA DSBs and neurodegeneration. Although this connection was recognized in the mid 70's32, it was not until 1995 that A-T was found to result from a mutation of a single gene encoding ATM (Ataxia Telangiectasia, Mutated)33, and subsequently the DNA double strand break (DSB) signaling pathway controlled by this protein kinase was elucidated34, 35.
DNA DSB repair pathways
DNA DSBs initiate a signaling cascade that leads to the repair and resolution of the break, or to apoptosis. The repair of DNA DSBs occurs by either non-homologous end joining (NHEJ) or homologous recombination (HR) (Fig. 3). These two major repair pathways maintain the integrity of DNA after DSBs from endogenous or exogenous agents, and inactivation of either NHEJ or HR results in marked defects in nervous system development14. Although these two pathways repair DSBs, they are biochemically distinct: HR operates in proliferating cells while NHEJ can function in both proliferating and differentiating cells.
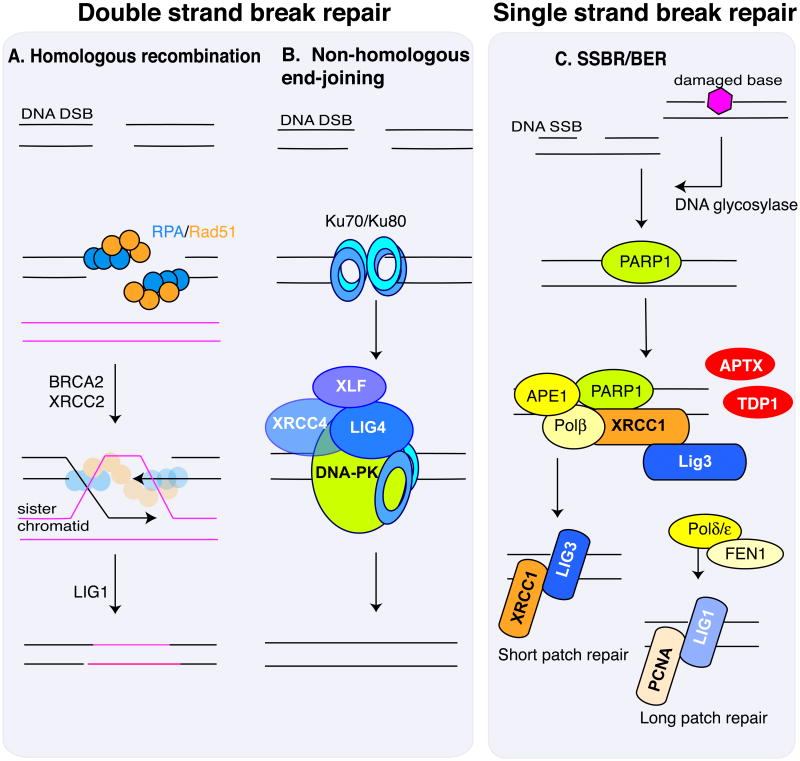
Figure 3. Repairing DNA strand breaks.
The repair of double strand breaks and single strand breaks occur by distinct biochemical pathways. Two different repair pathways can deal with a DSB, and the choice depends on the proliferative state of the cell. A. Homologous recombination can be utilized in proliferating cells. Replication Protein A (RPA) coats the resected single stranded DNA and recruits RAD51 recombinase, which together with a multitude of other factors36, 37, 41, including BRCA2 and XRCC236, 37, 41 repairs the DNA via a processes whereby template DNA from the sister-chromatid is inserted into the damaged chromatid and acts as an error-free template. Ligation of the break requires DNA ligase I (LIG1). B. Non-homologous end-joining (NHEJ) can function in proliferating and non-proliferating cells. Heterodimeric KU70 and KU80 bind DNA ends and recruit DNA-PK (catalytic subunit of the DNA-dependent kinase) which, together with XRCC4, XLF and DNA ligase IV (LIG4), reseals the DNA ends after suitable end processing from various nucleases42-44. Because some nucleotides may be lost in this process, NHEJ is often referred to as error-prone repair. However, DNA breaks are repaired, and so these ‘errors’ may generally be of little consequence in non-dividing cells like neurons. C. Strand breaks can arise from damaged bases being removed via specific DNA glycosylases (referred to as base excision repair) or direct backbone damage that severs the DNA strand. In both cases the stand break leads to Poly ADP-Ribose Polymerase (PARP) accumulation, which facilitates recruitment of the XRCC1 scaffolding protein that is important for promoting repair. XRCC1 recruits repair factors that modify the DNA ends for re-ligation. Depending upon the nature of the DNA ends present at the SSB, TDP1 (Tyrosyl DNA Phosphodiesterase 1; 3′ modified ends or topoisomerase I DNA adducts) or APTX (Aprataxin; 5′ends resulting from adenylated DNA from abortive ligation events) may be required to modify the damaged DNA for repair. Repair can involve removal of a single nucleotide (short patch repair) or a longer patch of nucleotide (long patch repair) by PCNA and Ligase 1.
HR is an error-free process that uses a sister chromatid as template DNA to achieve precise repair36, 37. Homology-directed repair initially involves DSB processing by the Mre11-Rad50-Nbs1 (MRN) complex in collaboration with CtIP (BRCA1 C-terminal Interacting Protein). This generates 3′ single stranded DNA that becomes bound by RPA (replication protein A) and Rad51 recombinase, which facilitates homology search and strand invasion, a process whereby template DNA from a sister-chromatid is inserted into the damaged chromatid and acts as an error-free template38-40, and effects recombinational repair in collaboration with a multitude of accessory factors (Fig. 3)36, 37, 41. During development, this pathway maintains the genomic integrity of neural progenitor cells.
In non-replicating cells, such as those in the mature nervous system, DNA DSBs are repaired by NHEJ (Fig. 3)42. In contrast to HR, NHEJ modifies the two DNA ends so that they are compatible for direct ligation42-44. Ligation of the DNA ends is catalyzed by DNA ligase IV (LIG4) in conjunction with its binding partner XRCC4 after end modification by the DNA-PK complex (DNA-PKCS and Ku70/80) and associated end-processing factors44. An additional factor, Cernunnos/XLF, has been identified as a binding partner of the LIG4-XRCC4 complex and is also necessary for efficient ligation via NHEJ45, 46. Hypomorphic mutations in LIG4 that lead to attenuated ligase activity have been associated with a disorder termed LIG4 syndrome47, characterized by radiosensitivity, immunodeficiency, and microcephaly48; defective XLF also leads to microcephaly46 (Table 1). The severity of LIG4 syndrome is related to the amount of residual LIG4 activity49.
DSB signaling
Coincident with DNA DSB repair is the activation of specific signaling events that initiate cell cycle arrest to stop proliferation and allow DNA repair to proceed. ATM is a key protein kinase that functions at the apex of a signaling pathway that coordinates cell cycle arrest after DNA damage34. ATM-dependent phosphorylation of cell-cycle checkpoint effectors such as p53, Chk2, SMC1 (Structural Maintenance of Chromosomes 1), BRCA1 (BReast Cancer Associated 1), and NBS1 (Nijmegen Breakage Syndrome 1) occurs to activate G1, intra-S and G2/M checkpoints (Fig. 4). DNA DSBs also induces rapid phosphorylation of histone H2AX, which facilitates the retention of numerous proteins that assemble at the break including NBS1, MDC1 (Mediator of DNA Damage Checkpoint 1) and 53BP1 (p53 Binding Protein 1)50-52. Detailed biochemical descriptions of ATM-signaling after DNA damage are available35, 52-57.
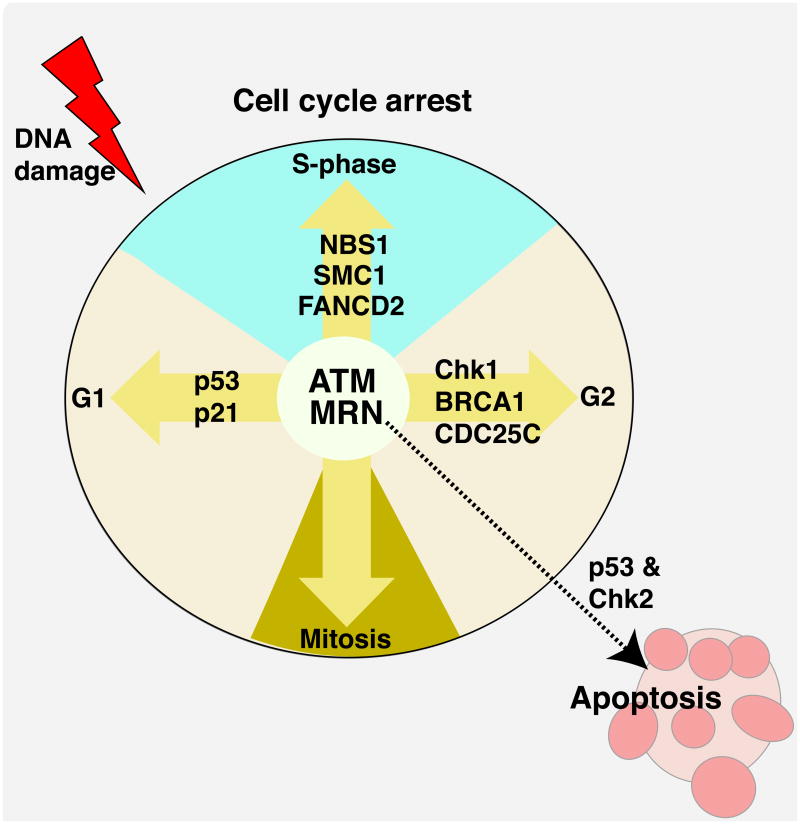
Figure 4. ATM signalling in response to DNA damage.
ATM (Ataxia-Telangiectasia, Mutated) is a serine/threonine protein kinase modulated by the MRN (Mre11-Rad50-NBS1) complex that is critically important for responding to DNA DSBs, and loss of these factors can lead to profound neuropathology. Many ATM substrates are important cell cycle and apoptotic regulators. A primary function of ATM after DNA damage is checkpoint activation. At each phase of the cell cycle ATM activates checkpoint proteins, amongst which are FANCD2 (Fanconi Anemia group D2), SMC1 (Structural Maintenance of Chromosomes 1), NBS1 (Nijmegen Breakage Syndrome 1), CDC25C (cell division cycle 25C) and BRCA1 (Breast Cancer Associated 1); an inclusive list of ATM substrates is available34, 35. Key ATM substrates are p53 and Chk2 (checkpoint kinase 2), which are responsible for activation of the G1 checkpoint and apoptosis. A main function of ATM in the nervous system may be to modulate DNA damage induced apoptosis. This is because in the nervous system, the prolific cell division that occurs allows damaged cells to be easily replaced, and so ATM fulfils an important genome monitoring function as immature cells exit the cell cycle, whereby cells with DNA damage are eliminated in an ATM dependent manner14.
In the developing nervous system, apoptosis is a common outcome after DNA DSBs and may in fact be preferred to DNA repair, as cells can easily be replaced from the abundant progenitor population present during neural development14. ATM is an important determinant of DSB-induced apoptosis in neural progenitor cells as they exit the cell cycle58, as it signals to the apoptosis machinery via Chk2 and p53 to activate proapoptotic gene expression and apoptosis59-61. Thus, the absence of ATM can result in the viability of DNA-damaged cells that would normally have been eliminated via apoptosis. This feature of ATM loss may be linked to the neuropathology found in A-T, as a failure to eliminate these damaged cells could lead to cell loss at later times.
An ATM-related protein kinase, ATR (Ataxia-Telangiectasia and Rad3-related) also participates in DNA damage signaling62. ATM and ATR are both critical for nervous system function and phosphorylate many common substrates including p53, NBS1, BRCA1 and Chk1 in response to DNA damage34, 35, 52, 55, and recent estimates suggest that these kinases can phosphorylate 700 or more proteins in response to ionizing radiation63. However, physiologically ATM and ATR have non-redundant roles. In the mouse, inactivation of ATR is lethal very early during development64, 65, while mice lacking ATM are viable, and although tumor-prone can often have a relatively normal lifespan. Hypomorphic mutations of ATR can lead to ATR-Seckel syndrome that presents with developmental defects and microcephaly, compared with progressive neurodegeneration that is characteristic of A-T25, 30, 48. Nonetheless, both kinases can cooperatively respond to DSBs during S-phase62, 66. In addition to ATM and ATR another related kinase, the catalytic subunit of DNA-PK, a central factor in NHEJ, is also important for signaling after DNA damage67. While, NHEJ is clearly important in the nervous system, roles for DNA-PKCS outside of NHEJ are not clear, although it may have some functional redundancy to ATM, as dual inactivation of ATM and DNA-PKCS is lethal early during embryogenesis68.
Disorders related to defective DSB repair and signaling
The hallmark feature of A-T is neurodegeneration and individuals with A-T are typically wheelchair-bound by their early teen years. Other features of A-T include immune dysfunction, sterility, extreme radiosensitivity and cancer predisposition29-31. Patients with A-T manifest muscle hypotonia, truncal swaying while sitting or standing and abnormalities of eye movement, features typical of cerebellar dysfunction69. Atrophy of the cerebellar folia, widespread loss of Purkinje cells, granule cell loss and significant thinning of the molecular layer are characteristic defects found in the A-T cerebellum70-72. MRI and CT studies also show cerebellar atrophy that is especially prevalent in vermal regions73-75. A-T patients also exhibit marked alterations in deep tendon reflexes, loss of the ability to sense vibration, reduction in sensory conduction velocity and axonal degeneration of peripheral nerves69, 76, 77. Widespread neurodegeneration also occurs in other regions of the nervous system, but these are variable and usually late stage phenomena69.
Two syndromes with phenotypic overlap to A-T are associated with mutations of components of the MRN complex (Fig. 4). These are ataxia telangiectasia-like disease (A-TLD) and Nijmegen Breakage syndrome (NBS), which result from mutation of Mre11 and NBS1, respectively78-80. In contrast to A-T, the disease-causing mutations in A-TLD and NBS are hypomorphic (as the MRN complex is indispensable for DNA replication and cellular survival) and while they disable some important MRN functions, the protein nonetheless retains the essential activity that support DNA replication.
Some individuals originally diagnosed with A-T, were later shown to carry hypomorphic mutations in Mre1179, 80, and therefore to have A-TLD. This finding demonstrated the interrelationship between ATM and the MRN complex, and the role of DNA damage in the aetiology of A-T. The clinical features of A-TLD are similar to those of A-T, although A-TLD patients do not have telangiectasia and have a later onset ataxia phenotype with a slower progression of neurodegeneration80. NBS was described as an A-T variant because of its close similarity to A-T81, 82. However, NBS is characterized by microcephaly rather than neurodegeneration83, 84, with occasional medulloblastoma brain tumors85, 86. Mutations of the NBS1 gene in NBS lead to synthesis of a truncated C-terminal protein87 that can interact with MRE11 and RAD50 and is sufficient for most of the functionality of NBS1 required for embryonic development.
The interrelationship of A-T, A-TLD and NBS results from the requirement of the MRN complex for normal ATM activation after DNA DSBs88, 89 (Fig. 4). While the exact details of ATM activation by the MRN complex are not fully understood, the function of MRN as a sensor of DNA damage provides insight. MRN binds to a DNA break and undergoes a conformational change leading to the formation of an inter-complex tether between MRN complexes (via Rad50) bound to opposing DNA strands to facilitate DNA repair90-92. The ability of the MRN complex to tether broken DNA ends provides a physical basis for the recruitment of critical signaling kinases such as ATM. The association of ATM with the MRN complex involves interaction of ATM with the NBS1 carboxyl terminal region93-95, and after DNA damage NBS1 is required for localization of activated ATM at sites of DSBs96.
[6] SSB repair pathways and related disease
A mechanistic counterpoint to the repair of DSBs is the utilization of distinct biochemical factors that deal with damage to only one DNA strand (Fig. 3). DNA SSBs are common DNA lesions, and can occur due to the direct effects of reactive oxygen species (ROS) or indirectly as an intermediate during BER22, 97, as each of these lesions utilize similar core machinery for repair98. The repair of a damaged base involves the action of a lesion-specific DNA glycosylase, an endonuclease that initiates base excision, followed by apurinic site incision by the APE1 endonuclease, recruitment of repair factors via XRCC1 and PARP (Poly ADP-Ribose Polymerase), end modification and gap-filling by DNA polymerase and ligation via Ligase III (short patch repair) or Ligase I in collaboration with PCNA (long patch repair)22, 98-100.
Defects in DNA SSBR are associated with ataxia with oculomotor apraxia 1 (AOA1) and spinocerebellar ataxia with axonal neuropathy (SCAN1), neurodegenerative syndromes that are similar to A-T (Table 1)26, 101-105. The neurological presentation of AOA1 is also almost identical to that of A-T, leading to AOA1 initially being classified as an A-T variant106. Notably, SSBR defects lead almost exclusively to neuropathology without the extra-neurological phenotypes associated with DSB repair deficiency. This is probably due to the availability of back-up repair pathways in proliferating and other tissues107.
The genes that are mutated in AOA1 and SCAN1 are aprataxin (APTX) and tyrosyl-DNA phosphodiesterase-1 (TDP1) respectively. TDP1 repairs altered 3′ DNA ends arising from oxidative damage and topoisomerase 1-DNA covalent complexes22, 107-112. Neural cells from a Tdp1-/- mouse have a pronounced defect in the repair of camptothecin-induced Top1-DNA cleavage complexes and hydrogen peroxide-induced SSBs113, 114. APTX is a member of the histidine triad superfamily of nucleotide hydrolases and possesses an AMP-lysine hydrolase activity, required for the repair of 5′-AMP intermediates that arise from failed DNA ligation reactions26, 115, 116. While APTX could in principal be involved in DSB repair117, the lack of extra-neurological features in AOA1 strongly suggest a primary defect in SSB repair26, 101, 103 as syndromes associated with DSB repair deficiency such as A-T or LIG4 syndrome present with multi-systemic defects.
Although rare, AOA1 is the most common recessive ataxia in Japan and second of all autosomal recessive ataxias in Portugal103. While the phenotypes associated with AOA1 and SCAN1 are similar, oculomotor apraxia is absent from SCAN1104, 118. These SSBR deficiency syndromes highlight the unique susceptibility of the nervous system to DNA damage, and despite a lack of extra-neurological features, neuropathology in these diseases is severe106, 119. However, the interrelationship between DNA repair deficiency and neuropathology is intricate and the full details remain to be elucidated (Text Box 1).
Studies of neuropathology in DNA repair syndromes aim to understand the cause and consequence of DNA damage thereby providing a rational basis for therapies to ameliorate the neurological deficits. In general, loss of DNA repair capacity impacts development and neurological issues are often apparent early in childhood. One brain region that is particularly susceptible in many DNA repair syndromes is the cerebellum. Notably, the cerebellum continues its development for around a year after birth. During this period cerebellar neurogenesis is substantial, and the granule neurons generated become the most numerous neuronal type in the brain7. Diseases such as ataxia telangiectasia (A-T), ataxia with oculomotor apraxia 1 and 2 (AOA1 and AOA2) and spinocerebellar ataxia with axonal neuropathy 1 (SCAN1) all profoundly affect the cerebellum. The underlying mutations in each disease affect the repair of DNA single or double strand breaks (see main text for details). Although the close parallel between the neuropathology of these diseases identifies the need for DNA repair, the phenotypes indicate relative differences in the timing, or types of DNA damage, that presumably reflect the specific activities of the affected enzyme. Analysis of A-T reveals cerebellar atrophy with widespread loss of purkinje and granule cells30. In AOA1, cerebellar atrophy is present, but the loss of purkinje cells is restricted to the cerebellar flocculus115. A-T and AOA1 feature oculomotor apraxia and a very early disease onset age, while in SCAN1 and AOA2 disease onset is later, and in SCAN1 oculomotor apraxia is absent. Understanding how specific types of DNA damage spatiotemporally affect different neural cell types will be required to understand the molecular basis of the phenotypic spectrum of the DNA repair syndromes.
[7] Repairing other types of DNA damage
The NER pathway
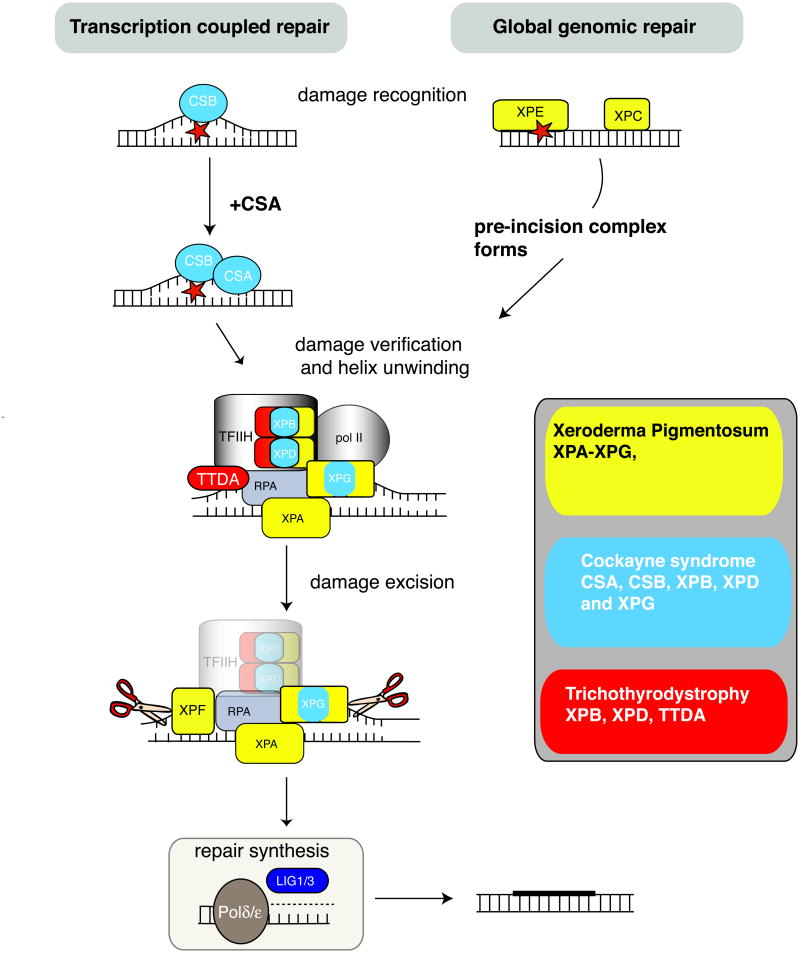
In contrast to the DNA strand break repair mechanisms discussed above, the NER pathway resolves DNA-distorting lesions typified by those generated by UV radiation. This repair pathway is functionally linked to transcription, as the basal TFIIH transcription/repair complex, which is required for RNA polymerase II-dependent transcription involves proteins also required for NER23, 120-122. NER repair of DNA during transcription is termed transcription-coupled repair (TCR), while repair that occurs independently of transcription is termed global genomic repair (GGR) (Fig. 5). These pathways utilize common repair factors, and differ only in the mode of DNA damage recognition. During GGR, it is the XPC or XPE complexes that initially recognize DNA damage28, 122-124. If the lesion occurs in a transcribed gene, it is sensed as a blockage to RNA pol II and requires CSB (and CSA) to initiate repair23. For both pathways, DNA damage verification and DNA unwinding occurs via TFIIH (complexed with XPB and XPD helicases) together with XPG, in a pre-incision complex also involving XPA125-129. The damaged DNA strand then undergoes a dual incision either side of the damage via the nuclease action of XPF and XPG130, 131 followed by gap filling by DNA polymerase δ or ε and strand rejoining by Ligase 1 or III. Detailed reviews outlining the biochemistry of this pathway are available23, 28, 122, 132, 133. The distinction between these two branches of NER is important because neuropathology in NER syndromes is associated with disabled TCR132, 134.
Figure 5. Nucleotide excision repair and related diseases.
Defects in nucleotide excision repair (NER) lead to at least three human syndromes characterized by neurodegeneration and DNA repair; xeroderma pigmentosum, Cockayne syndrome and Trichothiodystrophy (also see Table 1). NER can involve transcription-coupled repair (TCR), which occurs when the transcription complex encounters a damaged template and the more general repair pathway, global genomic repair (GGR), which repairs lesions present in non-transcribed DNA. The difference between these pathways is at the step of damage detection. The sequence of events in NER involves DNA damage recognition followed by an incision, damage removal and repair synthesis. DNA damage recognition in TCR involves initiation by CSB after the RNA Polymerase II complex encounters damage. In GGR damage is recognized by XPE or XPC. Pre-incision events involve XPA/RPA. The TFIIH complex, including the XPB and XPD helicases, unwinds the DNA helix resulting in an open conformation. XPG is then recruited to the complex and binds to TFIIH and RPA. The Cockayne syndrome proteins, CSA and CSB, are also involved in TCR and may function in shifting TFIIH from a transcription to repair complex. Dual incision involves XPG performing the initial cleavage 3′ to the site of DNA damage, followed by 5′ cleavage through the action or the XPF nuclease. Repair synthesis is performed by DNA Polymerase δ or ε with PCNA, and ligation to seal the gap involves DNA ligase I. Three main classes of diseases resulting from disruption of NER are listed, and pathway components that are disrupted in the respective diseases are represented in corresponding colors.
Xeroderma pigmentosum and related disorders
Mutations in various components of the NER pathway can lead to the human syndromes xeroderma pigmentosum (XP), Cockayne syndrome (CS), and trichothyrodystrophy (TTD)132, 133, 135. Although these diseases are clinically distinct, they do show some phenotypic similarities; in fact, in some cases different mutations in the same NER protein can lead to different syndromes (Fig. 5).
Eight different XP complementation groups (arising from mutations in the XPA-XPG NER components, or a variant polymerase XPV) resulting from mutations in different repair factors are known. Around 30% of individuals with XP develop neurodegeneration that results in global brain atrophy135, 136. Neurological symptoms appear from around 4 years of age and involve cognitive impairment followed by cerebellar-related problems such as dysarthria, balance disturbances, and sensorineural hearing loss; progressive neuropathy is evident from the second decade135, 137, 138. Increased cognitive impairment and corticospinal degeneration occurs progressively with age and at death almost all XPA individuals have severe neurological problems137, 139. This neuropathology is observed in individuals from XP subgroups in which the mutations affect TCR132, 134. Notably, XPC individuals which are defective in GGR but not TCR have little overt neurological impairment although mild brain atrophy is present137, and a brain tumour has been reported140.
DNA repair defects in CS involve the TCR component of NER and individuals with CS show progressive and severe neuropathology. It should be noted that the neuropathology in CS is quite different and distinct to that seen in XP. The neurological problems present in CS include profound microcephaly, mental retardation and progressive brain atrophy. Importantly, dysmyelination and calcification of the basal ganglia are also characteristic of CS, but are absent from XP141, 142. Two CS complementation groups result from mutations in two different genes, CSA or CSB, which function to promote the repair of DNA lesions encountered by the Pol II complex. Both CSA, which encodes a WD-repeat-containing protein143, and CSB, which encodes a helicase144, interact with TFIIH121, 143. CS can also result from certain mutations that occur in XPB, XPD or XPG28, 135, 145. Because neuropathology is different between XP and CS then something additional to simply defective TCR accounts for this difference. The specific mutations leading to CS probably result in more general and substantial transcriptional defects than those involved in XP, thereby accounting for CS-specific neuropathology. Another possibility is a defect in a sub-branch of GGR termed domain-associated repair that is important for repair of the non-transcribed strand of genes, which has been suggested to be particularly important in neurons133, 146. Clearly, ongoing studies will elaborate the interplay between NER factors and the basis for the resultant phenotypes specific components are disrupted.
TTD is also a disease associated with defective TCR that arises from certain mutations of the XPD helicase (or more rarely XPB or TTDA) and is characterized by sulphur-deficient brittle hair, and phenotypic similarities to XP and CS135. Sensitivity to UV light is observed in ∼50% of TTD individuals, but similar to CS, cancer predisposition is absent. The neurological abnormalities observed in TTD include microcephaly, mental retardation, deafness and ataxia147. Like CS, a striking aspect of the neuropathology found in TTD is dysmyelination135, 141, 148. Recently, the neurological defects associated with TTD were shown to involve deregulated expression of brain-specific thyroid hormone receptor-dependent genes134. These results suggested that the thyroid hormone pathway is a major target in TTD, which is consistent with deficits associated with hypomyelination, learning disorders and motor dysfunction. The pleiotropic nature of TTD-associated neurological defects probably results from impairment of other transcription factors in addition to thyroid hormone signaling.
Further insights into the impacts of NER and brain function have come from mouse models in which NER genes have been disrupted149. Inactivation of XPG revealed a loss of cerebellar purkinje cells150 while dual inactivation of CSB and XPA lead to cerebellar atrophy and loss of granule cells151, underscoring the importance of this pathway for brain function.
[8] Other disorders related to DNA repair deficits
A variety of other syndromes including Fanconi anemia (FA), Werner syndrome (WS), Bloom syndrome (BLM) and Rothmund Thomson syndrome (RTS) also result from DNA repair defects (Table 1). FA can result from disruption in any one of a large number of DNA repair factors required to remove cross-links in DNA strands, and currently there are 12 FA complementation groups152-154. FA presents with similar neuropathology to NBS, with microcephaly as a hallmark feature. Some subgroups of FA (FANC D1 and N) result from mutations in BRCA2, or its interacting partner PALB1, and are defective in DNA DSB repair152, 155, 156. Furthermore the D1 subgroup features brain tumors157. Given the link between DNA repair deficiency and tumorigenesis, it is somewhat surprising that brain tumors are not more frequent in DNA repair syndromes (Text Box 2).
Text Box 2: Neurodegeneration vs. Brain Tumors
Given the link between DNA damage and tumorigenesis, it might be expected that DNA repair syndromes affecting the nervous system would also present with brain tumors. While this does happen85, 86, 157, it is more frequent in these syndromes that tumors occur in tissues outside of the brain. This probably reflects the tissue-specific consequences of gene loss. For example, xeroderma pigmentosum (XP) results from defects in different genes in the nucleotide excision repair (NER) pathway, which can lead to cancer predisposition or neurodegeneration or both. Defects that lead to neuropathology generally affect transcription-coupled repair132, 134, while in tumor prone XP individuals, NER defects impact global genomic repair (GGR). During nervous system development DNA damage normally repaired by GGR, which could lead to tumors, may instead be eliminated by apoptosis. However, brain tumors can occur at low frequency in XP140.
Syndromes resulting from DNA strand break response defects such as ataxia telangiectasia (A-T), reflect the tissue specific signaling role for ATM (ataxia telangiectasia, mutated), in which a primary function is to activate apoptosis of non-replicating DNA-damaged neural cells58. In the immune system, where tumors mostly arise in A-T, ATM functions to maintain fidelity of immunoglobin DNA rearrangements that prevent oncogenic rearrangements53. However, other DNA repair syndromes such as Fanconi anemia (the FANCD1 subgroup with BRCA2-BReast Cancer Associated 2-mutations) develop medulloblastoma brain tumors157 (located in the cerebellum), reflecting the occurrence of DNA damage in rapidly proliferating cells of the developing cerebellum. This is also the case for Nijmegen breakage syndrome, in which loss of NBS1 leads to microcephaly and can also result in medulloblastoma85, 86. In these cases, medulloblastoma may occur because the DNA repair defect is quite substantial and affects the rapidly proliferating and abundant granule neuron precursors, which have a relatively high chance of acquiring mutations leading to transformation.
BLM, WS and RTS result from defective RECQ4-related helicase function158-161. The resultant neuropathology in these syndromes is not well characterized, although the syndromes produce quite dramatic and varying phenotypes outside of the nervous system, including cancer, progeria and proportional body size defects. In the case of WS, a neurological involvement has been reported but is controversial162-164. Why mutations in these helicases don't more substantially impact the nervous system is not clear. Their role in replication and their link to HR, suggests an important function in genomic integrity checks during neural precursor proliferation. Perhaps neurological disease is avoided in these syndromes by apoptotic elimination of damaged neural progenitors during development, or their role is not central to neural stem/progenitor replication and maintenance.
However, mutations in another helicase, the ortholog of the yeast Sen1p helicase termed Senataxin, results in ataxia with oculomotor apraxia 2 (AOA2), a syndrome characterized by pronounced cerebellar degeneration, oculomotor apraxia and sensory motor neuropathy165-170. Additionally, different mutations in this gene can lead to a form of amyotropic lateral sclerosis171. AOA2 is characterized by a specific defect in the response to certain types of DNA DSBs172, but is not sensitive to ionizing radiation173.
Furthermore, while distinct DNA repair pathways respond to specific DNA lesions, interplay between different DNA repair pathways is likely to provide fine-tuning of the DNA damage response. For example, pathways that deal with DNA inter-strand cross-links to prevent FA can also be involved in repair of DSBs154, and SSB repair factors also interact with DSB repair factors174, suggesting important interactions and cross-talk between DNA repair pathways. Unraveling the biology of DNA repair pathways and their role in disease prevention will benefit from refinements and ongoing work using mouse genetics.
Finally, the biochemical defect for many human diseases characterized by ataxia or microcephaly is unknown175, although in some cases sensitivity towards DNA damaging agents suggest the cause is a defect in DNA repair176. Thus, it is likely that the spectrum of DNA repair deficient diseases will continue to expand, as the respective disease-causing mutations in these syndromes are uncovered.
[9] Tissue specificity of repair pathways
Historically, information about DNA repair pathways that impact the nervous system has been obtained from individuals with DNA repair syndromes. Using cells derived from A-T, XP and FA individuals, defects in the response to ionizing radiation, UV light and cross-linking agents were identified, and these cells have continued to provide important biochemical insights into these DNA repair pathways. However, with an increased understanding of the molecular basis of the DNA damage response, and the development of mouse models of DNA repair deficiency, it has become apparent that many of the key signaling pathways that underpin the response to DNA damage exhibit a striking tissue or cell-type specificity177. For example, differential radiosensitivity was found within the brains of mice lacking ATM, as immature postmitotic neural cells were resistant to IR while proliferating progenitors were susceptible58, 60. Inactivation of NBS1 in the brain markedly affected the cerebellum178 while loss of Mdc1, despite its central role in the DNA damage response179, 180, resulted in a relatively modest phenotype181. These findings illustrate the challenges of translating in vitro biochemical data to a physiological setting.
[10] Why is the nervous system particularly sensitive to DNA damage?
The nervous system is very sensitive to DNA damage, particularly in comparison to other non-replicating cell types. For example, in DNA SSBR deficiencies the neurological symptoms are almost the exclusive presentation of the disease. There are some unique properties of the nervous system that may account for this relative sensitivity. The brain metabolizes around 20% of consumed oxygen, but contains a lower capacity than other body parts to neutralize reactive oxygen species, and neurons are particularly susceptible to oxidative stress182. This high oxidative load presumably generates free radicals that will increase DNA damage, particularly DNA SSBs, in the mature nervous system. Therefore, cells with defective SSBR will appear relatively more susceptible to this insult. As SSBs may subsequently interfere with the transcriptional machinery this could eventually result in cell death16, 132, 183.
The cerebellum is a prominent target of many human DNA repair syndromes raising the question of why this organ is so uniquely sensitive to DNA damage (Text Box 1). A notable feature of the cerebellum is its extended postnatal development7. This period of rapid cell proliferation will generate replication stress associated DNA damage that may impact granule neurons and perhaps indirectly other cerebellar cell types. Purkinje cells in the cerebellum are selectively sensitive to oxidative stress184, 185. Antioxidants can also promote survival of cultured ATM-deficient Purkinje cells186.
There are many unresolved issues regarding the apparent vulnerability of the nervous system. Because DNA repair syndromes are congenital it is important to know what periods during neural development are vulnerable and what are the on-going requirements for DNA repair pathways in mature neural compartments. Some DNA repair syndromes have a relatively later onset, questioning whether this reflects an active DNA repair requirement in postmitotic neurons, or if this is still a result of accrued developmental damage.
[11] Mitochondrial DNA repair
The mitochondria play a key role in nervous system function187, 188. Mitochondria contain DNA (mtDNA) that encodes 13 proteins required for the respiratory chain and 24 RNA genes, although proteins encoded by nuclear genes are also present in this organelle. Damage to the mtDNA can readily occur, as the mitochondrial genome is located on the inner membrane, a major site of ROS generation189. The primary DNA repair mechanism operative in the mitochondria is the BER/SSBR pathway, and there are many specific mitochondrial versions of components of this pathway (which are encoded by nuclear DNA)190, 191. A variety of DNA lesions including point mutations and large-scale deletion of mtDNA can lead to mitochondrial dysfunction and cellular apoptosis191, 192. In the nervous system, compromised mitochondrial function has been linked to neurodegeneration including Alzheimer's and Parkinson's disease 1, 187, 189, 191. Therefore, DNA repair defects that impact mtDNA would conceivably be a significant harbinger of neurological disease. It will therefore be important to determine the contribution of mitochondrial dysfunction towards neuropathology of human syndromes resulting from DNA repair defects.
[12] Insights from Neuropathology
Generally speaking, the consequences of DNA repair defects are microcephaly or neurodegeneration. While both pathologies ultimately result from cell loss, the fundamental underpinnings are different. Microcephaly results from cell loss or proliferation defects during neurogenesis. This may reflect increased apoptosis as a result of a failure to repair replication errors, or from unrepaired DNA damage during differentiation. Data from mouse models indicate that defects in DSBR such as LIG4 or BRCA2 deficiency markedly affects neurogenesis by causing widespread apoptosis during development that can result in microcephaly12, 13, 193. This is probably the case in human microcephaly such as the LIG4 syndrome48. DNA repair is important for maintenance of stem cells, and loss of these could also contribute to microcephaly194, 195. Stem cells may be a particularly susceptible population in other syndromes that feature microcephaly such as ATR-Seckel syndrome, as defects in ATR are known to impact replication and stem leading to cell loss or arrest62, 194-196. This could also be the case for FA-associated microcephaly as the FA pathway is required for maintenance of neural stem and progenitor cells during development and in the adult197.
Contrasting microcephaly is the neurodegenerative phenotype typified by A-T and other diseases with similar clinical presentation such as AOA1 and A-TLD. Early neurological problems in A-T point to a developmental requirement for ATM during nervous system development. This may relate to the need for ATM to remove DNA damaged cells early after cell cycle exit to prevent their incorporation into the nervous system58, 60. Diseases such as AOA2 or SCAN1 have a generally later onset, possibly because of backup DNA repair during development, while postmitotic neurons succumb to unrepaired damage, probably as a result of transcriptional interference16, 22. Additionally, some subgroups of XP (notably XPA mutations) also show severe neurodegeneration, but in many cases are also associated with microcephaly137. This neuropathology may reflect both developmental neurogenesis defects coupled with ongoing cell loss in the mature brain. Thus, cell loss arising from DNA damage during development leads to microcephaly, while the progressive degenerative syndromes may reflect cell loss after birth as a consequence of accumulated damage during development, or progressive loss from ongoing DNA damage that impacts transcription.
[13] Conclusions and perspectives
Many different cell types integrate function in the nervous system, and this diversity presents a challenge for understanding how the dynamics of the DNA damage response ensures homeostasis in the brain. DNA repair syndromes are congenital and as such raise important questions from a developmental perspective, such as how much of the eventual neuropathology is already set in place at birth? Understanding the consequences of DNA damage and associated neuropathology may involve other aspects of cellular homeostasis. For example, mitochondrial dysfunction is increasingly being linked to neurological disease, and as mitochondrial DNA is susceptible to damage, DNA repair defects could substantially impact this organelle in the brain192, 198. Additionally, the mechanism of cell death in neurodegenerative syndromes is not fully understood and could involve abortive cell cycle reentry as has been suggested for cell loss in Alzheimer's disease and has been reported for A-T199-201. Therefore, these additional aspects of DNA damage may be important contributors to neuropathology in DNA repair syndromes.
Future challenges include the following considerations; are both neurons and glia equally susceptible to loss of DNA damage responses? What is the basis for the relative impact of defective repair in specific cell types and different neural tissues? What is the actual on-going requirement for DNA repair and what are the relevant DNA repair pathways in mature neural compartments? If oxidative stress is important as an etiological agent, will antioxidants be effective in preventing cell loss? Resolving these issues will have important implications for our understanding of DNA repair syndromes and subsequently developing therapies for treatment of the neuropathology associated with these diseases.
Acknowledgments
I thank the NIH and ASLAC for financial support, and laboratory members for discussions and comments. I also thank a referee for providing helpful suggestions regarding NER diseases. Space constraints limited the number of primary research papers cited.
Glossary terms
- Neuroepithelia
A layer of proliferating neuroepithelial cells that makes up the neural plate and neural tube
- Oxidative load
The amount of oxidative stress in the form of free radicals or reactive oxygen species encountered by a tissue
- Free radicals
Molecules containing unpaired electrons that are unstable and reactive that can damage cellular components such as DNA and can be produced during metabolism
- Helical distortions
Topological perturbation of the DNA double helix that can register as damage to DNA repair machinery
- Non-homologous end joining
One of two distinct biochemical pathways for the repair of DNA double strand breaks which functions to modify non-compatible termini and directly ligate the broken DNA ends
- Homologous recombination repair
One of two distinct biochemical pathways for the repair of DNA double strand breaks which functions in S or G2 of the cell cycle and uses sister chromatid DNA as an undamaged template
- Cell-cycle checkpoint
A biochemical signaling event that is activated following stimuli such as DNA damage which pauses the cell cycle to allow time for recovery from the insult and maintain cellular homeostasis
- Hypomorphic mutation
A mutation that does not fully eliminate the function of a gene product and typically gives a less severe phenotype than a loss-of-function mutation
- Complementation group
The functional correction of a defective multiprotein complex by reintroduction of a missing or defective component from cellular extracts of another individual with the same disease
Biography
About the Author: Peter McKinnon worked on the biochemical basis of the neurodegenerative syndrome ataxia telangiectasia while a graduate student in Australia. He then took a postdoctoral position in molecular neurobiology at the Roche Institute of Molecular Biology where he gained experience using mouse genetics to study brain function. As a faculty member at St. Jude Children's Research Hospital his lab studies the DNA damage response in the mammalian brain and how this pathway functions to prevent brain tumors and neurodegeneration.
Footnotes
Resources:
http://neuromuscular.wustl.edu/ataxia/dnarep.html
http://www.pathology.washington.edu/research/werner/database/index.html
OMIM disease links:
References
- 1.Bender A, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–7. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 2.Katyal S, McKinnon PJ. DNA strand breaks, neurodegeneration and aging in the brain. Mech Ageing Dev. 2008;129:483–91. doi: 10.1016/j.mad.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kraytsberg Y, et al. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet. 2006;38:518–20. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 4.Nouspikel T, Hanawalt PC. When parsimony backfires: neglecting DNA repair may doom neurons in Alzheimer's disease. Bioessays. 2003;25:168–73. doi: 10.1002/bies.10227. [DOI] [PubMed] [Google Scholar]
- 5.Chan WY, Lorke DE, Tiu SC, Yew DT. Proliferation and apoptosis in the developing human neocortex. Anat Rec. 2002;267:261–76. doi: 10.1002/ar.10100. [DOI] [PubMed] [Google Scholar]
- 6.Dehay C, Kennedy H. Cell-cycle control and cortical development. Nat Rev Neurosci. 2007;8:438–50. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- 7.Goldowitz D, Hamre K. The cells and molecules that make a cerebellum. Trends Neurosci. 1998;21:375–82. doi: 10.1016/s0166-2236(98)01313-7. [DOI] [PubMed] [Google Scholar]
- 8.Wang VY, Zoghbi HY. Genetic regulation of cerebellar development. Nat Rev Neurosci. 2001;2:484–91. doi: 10.1038/35081558. [DOI] [PubMed] [Google Scholar]
- 9.Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–60. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 10.Zhang CL, Zou Y, He W, Gage FH, Evans RM. A role for adult TLX-positive neural stem cells in learning and behaviour. Nature. 2008;451:1004–7. doi: 10.1038/nature06562. [DOI] [PubMed] [Google Scholar]
- 11.Toni N, et al. Neurons born in the adult dentate gyrus form functional synapses with target cells. Nat Neurosci. 2008;11:901–7. doi: 10.1038/nn.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barnes DE, Stamp G, Rosewell I, Denzel A, Lindahl T. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr Biol. 1998;8:1395–8. doi: 10.1016/s0960-9822(98)00021-9. [DOI] [PubMed] [Google Scholar]
- 13.Gao Y, et al. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]; Refs 12 and 13 showed for the first time that repair of DNA DSBs was critical for homeostasis of the developing nervous system, thereby linking DNA damage to neurogenesis.
- 14.Lee Y, McKinnon PJ. Responding to DNA double strand breaks in the nervous system. Neuroscience. 2007;145:1365–74. doi: 10.1016/j.neuroscience.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 15.Karanjawala ZE, Murphy N, Hinton DR, Hsieh CL, Lieber MR. Oxygen metabolism causes chromosome breaks and is associated with the neuronal apoptosis observed in DNA double-strand break repair mutants. Curr Biol. 2002;12:397–402. doi: 10.1016/s0960-9822(02)00684-x. [DOI] [PubMed] [Google Scholar]; This paper provided evidence that oxidative stress can be an agent that leads to increased DNA breaks in the developing nervous system.
- 16.Saxowsky TT, Doetsch PW. RNA polymerase encounters with DNA damage: transcription-coupled repair or transcriptional mutagenesis? Chem Rev. 2006;106:474–88. doi: 10.1021/cr040466q. [DOI] [PubMed] [Google Scholar]
- 17.Zhou W, Doetsch PW. Effects of abasic sites and DNA single-strand breaks on prokaryotic RNA polymerases. Proc Natl Acad Sci U S A. 1993;90:6601–5. doi: 10.1073/pnas.90.14.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brooks PJ. The 8,5′-cyclopurine-2′-deoxynucleosides: candidate neurodegenerative DNA lesions in xeroderma pigmentosum, and unique probes of transcription and nucleotide excision repair. DNA Repair (Amst) 2008;7:1168–79. doi: 10.1016/j.dnarep.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crowe SL, Movsesyan VA, Jorgensen TJ, Kondratyev A. Rapid phosphorylation of histone H2A.X following ionotropic glutamate receptor activation. Eur J Neurosci. 2006;23:2351–61. doi: 10.1111/j.1460-9568.2006.04768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovtun IV, et al. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–52. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMurray CT. Hijacking of the mismatch repair system to cause CAG expansion and cell death in neurodegenerative disease. DNA Repair (Amst) 2008;7:1121–34. doi: 10.1016/j.dnarep.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–31. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 23.Fousteri M, Mullenders LH. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 2008;18:73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- 24.Kulkarni A, Wilson DM., 3rd The involvement of DNA-damage and -repair defects in neurological dysfunction. Am J Hum Genet. 2008;82:539–66. doi: 10.1016/j.ajhg.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKinnon PJ, Caldecott KW. DNA strand break repair and human genetic disease. Annu Rev Genomics Hum Genet. 2007;8:37–55. doi: 10.1146/annurev.genom.7.080505.115648. [DOI] [PubMed] [Google Scholar]
- 26.Rass U, Ahel I, West SC. Defective DNA repair and neurodegenerative disease. Cell. 2007;130:991–1004. doi: 10.1016/j.cell.2007.08.043. [DOI] [PubMed] [Google Scholar]
- 27.Subba Rao K. Mechanisms of disease: DNA repair defects and neurological disease. Nat Clin Pract Neurol. 2007;3:162–72. doi: 10.1038/ncpneuro0448. [DOI] [PubMed] [Google Scholar]
- 28.Sugasawa K. Xeroderma pigmentosum genes: functions inside and outside DNA repair. Carcinogenesis. 2008;29:455–65. doi: 10.1093/carcin/bgm282. [DOI] [PubMed] [Google Scholar]
- 29.Frappart PO, McKinnon PJ. Ataxia-telangiectasia and related diseases. Neuromolecular Med. 2006;8:495–511. doi: 10.1385/NMM:8:4:495. [DOI] [PubMed] [Google Scholar]
- 30.McKinnon PJ. ATM and ataxia telangiectasia. EMBO Rep. 2004;5:772–6. doi: 10.1038/sj.embor.7400210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perlman S, Becker-Catania S, Gatti RA. Ataxia-telangiectasia: diagnosis and treatment. Semin Pediatr Neurol. 2003;10:173–82. doi: 10.1016/s1071-9091(03)00026-3. [DOI] [PubMed] [Google Scholar]
- 32.Taylor AM, et al. Ataxia telangiectasia: a human mutation with abnormal radiation sensitivity. Nature. 1975;258:427–9. doi: 10.1038/258427a0. [DOI] [PubMed] [Google Scholar]
- 33.Savitsky K, et al. A Single Ataxia Telangiectasia Gene with a Product Similar to PI-3 Kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 34.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–68. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 35.Shiloh Y. The ATM-mediated DNA-damage response: taking shape. Trends Biochem Sci. 2006;31:402–10. doi: 10.1016/j.tibs.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 36.Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amst) 2007;6:923–35. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 37.West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4:435–45. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 38.Limbo O, et al. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell. 2007;28:134–46. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–14. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeda S, Nakamura K, Taniguchi Y, Paull TT. Ctp1/CtIP and the MRN complex collaborate in the initial steps of homologous recombination. Mol Cell. 2007;28:351–2. doi: 10.1016/j.molcel.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 41.Wyman C, Ristic D, Kanaar R. Homologous recombination-mediated double-strand break repair. DNA Repair (Amst) 2004;3:827–33. doi: 10.1016/j.dnarep.2004.03.037. [DOI] [PubMed] [Google Scholar]
- 42.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712–20. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 43.Bassing CH, Alt FW. The cellular response to general and programmed DNA double strand breaks. DNA Repair (Amst) 2004;3:781–96. doi: 10.1016/j.dnarep.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 44.Lees-Miller SP, Meek K. Repair of DNA double strand breaks by non-homologous end joining. Biochimie. 2003;85:1161–73. doi: 10.1016/j.biochi.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 45.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–13. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 46.Buck D, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–99. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 47.O'Driscoll M, et al. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol Cell. 2001;8:1175–85. doi: 10.1016/s1097-2765(01)00408-7. [DOI] [PubMed] [Google Scholar]; This paper and reference 46 reports the link between loss of NHEJ function and neuropathology.
- 48.O'Driscoll M, Gennery AR, Seidel J, Concannon P, Jeggo PA. An overview of three new disorders associated with genetic instability: LIG4 syndrome, RS-SCID and ATR-Seckel syndrome. DNA Repair (Amst) 2004;3:1227–35. doi: 10.1016/j.dnarep.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 49.Girard PM, Kysela B, Harer CJ, Doherty AJ, Jeggo PA. Analysis of DNA ligase IV mutations found in LIG4 syndrome patients: the impact of two linked polymorphisms. Hum Mol Genet. 2004;13:2369–76. doi: 10.1093/hmg/ddh274. [DOI] [PubMed] [Google Scholar]
- 50.Celeste A, et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675–9. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 51.Sedelnikova OA, Pilch DR, Redon C, Bonner WM. Histone H2AX in DNA damage and repair. Cancer Biol Ther. 2003;2:233–5. doi: 10.4161/cbt.2.3.373. [DOI] [PubMed] [Google Scholar]
- 52.Ward I, Chen J. Early events in the DNA damage response. Curr Top Dev Biol. 2004;63:1–35. doi: 10.1016/S0070-2153(04)63001-8. [DOI] [PubMed] [Google Scholar]
- 53.Bredemeyer AL, et al. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–70. doi: 10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
- 54.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–45. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 55.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–23. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 56.Lavin MF, Kozlov S. ATM activation and DNA damage response. Cell Cycle. 2007;6:931–942. doi: 10.4161/cc.6.8.4180. [DOI] [PubMed] [Google Scholar]
- 57.Wood JL, Chen J. DNA-damage checkpoints: location, location, location. Trends Cell Biol. 2008;18:451–5. doi: 10.1016/j.tcb.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 58.Herzog KH, Chong MJ, Kapsetaki M, Morgan JI, McKinnon PJ. Requirement for Atm in ionizing radiation-induced cell death in the developing central nervous system. Science. 1998;280:1089–91. doi: 10.1126/science.280.5366.1089. [DOI] [PubMed] [Google Scholar]; Together with ref 60 showed that ATM function was critical for DNA damage induced apoptosis in immature differentiating neural cells, but not in proliferating cells.
- 59.Jeffers JR, et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–8. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 60.Lee Y, Chong MJ, McKinnon PJ. Ataxia telangiectasia mutated-dependent apoptosis after genotoxic stress in the developing nervous system is determined by cellular differentiation status. J Neurosci. 2001;21:6687–93. doi: 10.1523/JNEUROSCI.21-17-06687.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takai H, et al. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. Embo J. 2002;21:5195–205. doi: 10.1093/emboj/cdf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–27. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–6. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 64.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 65.de Klein A, et al. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr Biol. 2000;10:479–82. doi: 10.1016/s0960-9822(00)00447-4. [DOI] [PubMed] [Google Scholar]
- 66.Jazayeri A, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 67.Burma S, Chen DJ. Role of DNA-PK in the cellular response to DNA double-strand breaks. DNA Repair (Amst) 2004;3:909–18. doi: 10.1016/j.dnarep.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 68.Gurley KE, Kemp CJ. Synthetic lethality between mutation in Atm and DNA-PK(cs) during murine embryogenesis. Curr Biol. 2001;11:191–194. doi: 10.1016/s0960-9822(01)00048-3. [DOI] [PubMed] [Google Scholar]
- 69.Sedgwick RP, Boder E. In: Handbook of Clinical Neurology. Vinken P, Bruyn G, Klawans H, editors. Elsevier; New York: 1991. pp. 347–423. [Google Scholar]
- 70.Gatti RA, et al. The pathogenesis of ataxia-telangiectasia. Learning from a Rosetta Stone. Clin Rev Allergy Immunol. 2001;20:87–108. doi: 10.1385/CRIAI:20:1:87. [DOI] [PubMed] [Google Scholar]
- 71.Paula-Barbosa MM, et al. Cerebellar Cortex Ultrastructure in Ataxia-telangiectasia. Annals of Neurology. 1983;13:297–302. doi: 10.1002/ana.410130312. [DOI] [PubMed] [Google Scholar]
- 72.Vinters HV, Gatti RA, Rakic P. In: Ataxia-telangiectasia: Genetics, Neuropathology, and Immunology of a degenerative Disease of Childhood. Gatti RA, Swift M, editors. Alan R. Liss, Inc; New York: 1985. pp. 233–235. [Google Scholar]
- 73.Farina L, et al. Ataxia-telangiectasia: MR and CT findings. J Comput Assist Tomogr. 1994;18:724–7. [PubMed] [Google Scholar]
- 74.Sardanelli F, et al. Cranial MRI in ataxia-telangiectasia. Neuroradiology. 1995;37:77–82. doi: 10.1007/BF00588526. [DOI] [PubMed] [Google Scholar]
- 75.Tavani F, et al. Ataxia-telangiectasia: the pattern of cerebellar atrophy on MRI. Neuroradiology. 2003;45:315–9. doi: 10.1007/s00234-003-0945-9. [DOI] [PubMed] [Google Scholar]
- 76.Barbieri F, et al. Is the sensory neuropathy in ataxia-telangiectasia distinguishable from that in Friedreich's ataxia? Morphometric and ultrastructural study of the sural nerve in a case of Louis Bar syndrome. Acta Neuropathol (Berl) 1986;69:213–9. doi: 10.1007/BF00688296. [DOI] [PubMed] [Google Scholar]
- 77.Lelli S, Trevisan C, Negrin P. Ataxia-teleangectasia: neurophysiological studies in 8 patients. Electromyogr Clin Neurophysiol. 1995;35:311–5. [PubMed] [Google Scholar]
- 78.Carney JP, et al. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–86. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 79.Stewart GS, et al. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–87. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]; This paper and ref 78 identify mutations of Mre11 in an A-T like syndrome thereby clarifying the link between DNA damage and neurodegeneration.
- 80.Taylor AM, Groom A, Byrd PJ. Ataxia-telangiectasia-like disorder (ATLD)-its clinical presentation and molecular basis. DNA Repair (Amst) 2004;3:1219–25. doi: 10.1016/j.dnarep.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 81.Saar K, et al. The gene for the ataxia-telangiectasia variant, Nijmegen breakage syndrome, maps to a 1-cM interval on chromosome 8q21. Am J Hum Genet. 1997;60:605–10. [PMC free article] [PubMed] [Google Scholar]
- 82.Weemaes CM, et al. A new chromosomal instability disorder: the Nijmegen breakage syndrome. Acta Paediatr Scand. 1981;70:557–64. doi: 10.1111/j.1651-2227.1981.tb05740.x. [DOI] [PubMed] [Google Scholar]
- 83.Bekiesinska-Figatowska M, et al. Cranial MRI in the Nijmegen breakage syndrome. Neuroradiology. 2000;42:43–7. doi: 10.1007/s002340050011. [DOI] [PubMed] [Google Scholar]
- 84.Digweed M, Sperling K. Nijmegen breakage syndrome: clinical manifestation of defective response to DNA double-strand breaks. DNA Repair (Amst) 2004;3:1207–17. doi: 10.1016/j.dnarep.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 85.Bakhshi S, et al. Medulloblastoma with adverse reaction to radiation therapy in nijmegen breakage syndrome. J Pediatr Hematol Oncol. 2003;25:248–51. doi: 10.1097/00043426-200303000-00013. [DOI] [PubMed] [Google Scholar]
- 86.Distel L, Neubauer S, Varon R, Holter W, Grabenbauer G. Fatal toxicity following radio- and chemotherapy of medulloblastoma in a child with unrecognized Nijmegen breakage syndrome. Med Pediatr Oncol. 2003;41:44–8. doi: 10.1002/mpo.10275. [DOI] [PubMed] [Google Scholar]
- 87.Maser RS, Zinkel R, Petrini JH. An alternative mode of translation permits production of a variant NBS1 protein from the common Nijmegen breakage syndrome allele. Nat Genet. 2001;27:417–21. doi: 10.1038/86920. [DOI] [PubMed] [Google Scholar]
- 88.Carson CT, et al. The Mre11 complex is required for ATM activation and the G(2)/M checkpoint. Embo J. 2003;22:6610–6620. doi: 10.1093/emboj/cdg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Uziel T, et al. Requirement of the MRN complex for ATM activation by DNA damage. Embo J. 2003;22:5612–21. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hopfner KP, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–6. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 91.Moreno-Herrero F, et al. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–3. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- 92.Wiltzius JJ, Hohl M, Fleming JC, Petrini JH. The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat Struct Mol Biol. 2005;12:403–7. doi: 10.1038/nsmb928. [DOI] [PubMed] [Google Scholar]; Together with refs 90 and 91 this work provides important details of how MRN functions at a DNA double strand break.
- 93.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–11. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 94.Fernandes N, et al. DNA damage-induced association of ATM with its target proteins requires a protein interaction domain in the N terminus of ATM. J Biol Chem. 2005;280:15158–64. doi: 10.1074/jbc.M412065200. [DOI] [PubMed] [Google Scholar]
- 95.You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol. 2005;25:5363–79. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes Dev. 2004;18:1423–1438. doi: 10.1101/gad.1200304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 98.Almeida KH, Sobol RW. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair (Amst) 2007;6:695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–50. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wilson DM, 3rd, Bohr VA. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair (Amst) 2007;6:544–59. doi: 10.1016/j.dnarep.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 101.Date H, et al. Early-onset ataxia with ocular motor apraxia and hypoalbuminemia is caused by mutations in a new HIT superfamily gene. Nat Genet. 2001;29:184–188. doi: 10.1038/ng1001-184. [DOI] [PubMed] [Google Scholar]
- 102.el-Khamisy SF, Caldecott KW. DNA single-strand break repair and spinocerebellar ataxia with axonal neuropathy-1. Neuroscience. 2007;145:1260–6. doi: 10.1016/j.neuroscience.2006.08.048. [DOI] [PubMed] [Google Scholar]
- 103.Moreira MC, et al. The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nat Genet. 2001;29:189–193. doi: 10.1038/ng1001-189. [DOI] [PubMed] [Google Scholar]; Together with ref 101, identified the gene mutated in AOA1 as Aprataxin and implicates DNA repair deficiency as the potential cause of AOA1.
- 104.Onodera O. Spinocerebellar ataxia with ocular motor apraxia and DNA repair. Neuropathology. 2006;26:361–7. doi: 10.1111/j.1440-1789.2006.00741.x. [DOI] [PubMed] [Google Scholar]
- 105.Takashima H, et al. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat Genet. 2002;32:267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 106.Aicardi J, et al. Ataxia-ocular motor apraxia: a syndrome mimicking ataxia-telangiectasia. Ann Neurol. 1988;24:497–502. doi: 10.1002/ana.410240404. [DOI] [PubMed] [Google Scholar]
- 107.El-Khamisy SF, et al. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434:108–13. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]; Provides the important connection between defective DNA SSBR and neurodegeneration.
- 108.Connelly JC, Leach DR. Repair of DNA covalently linked to protein. Mol Cell. 2004;13:307–16. doi: 10.1016/s1097-2765(04)00056-5. [DOI] [PubMed] [Google Scholar]
- 109.Interthal H, et al. SCAN1 mutant Tdp1 accumulates the enzyme--DNA intermediate and causes camptothecin hypersensitivity. Embo J. 2005;24:2224–33. doi: 10.1038/sj.emboj.7600694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Miao ZH, et al. Hereditary ataxia SCAN1 cells are defective for the repair of transcription-dependent topoisomerase I cleavage complexes. DNA Repair (Amst) 2006;5:1489–1494. doi: 10.1016/j.dnarep.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 111.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–40. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 112.Zhou T, et al. Deficiency in 3′-phosphoglycolate processing in human cells with a hereditary mutation in tyrosyl-DNA phosphodiesterase (TDP1) Nucleic Acids Res. 2005;33:289–297. doi: 10.1093/nar/gki170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hirano R, et al. Spinocerebellar ataxia with axonal neuropathy: consequence of a Tdp1 recessive neomorphic mutation? Embo J. 2007;26:4732–43. doi: 10.1038/sj.emboj.7601885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Katyal S, et al. TDP1 facilitates chromosomal single-strand break repair in neurons and is neuroprotective in vivo. EMBO J. 2007;26:4720–31. doi: 10.1038/sj.emboj.7601869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ahel I, et al. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–6. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]; Identifies the specific endogenous DNA lesion that requires Aprataxin for removal thereby preventing neurodegeneration.
- 116.Seidle HF, Bieganowski P, Brenner C. Disease-associated mutations inactivate AMP-lysine hydrolase activity of Aprataxin. J Biol Chem. 2005;280:20927–20931. doi: 10.1074/jbc.M502889200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rass U, Ahel I, West SC. Actions of aprataxin in multiple DNA repair pathways. J Biol Chem. 2007;282:9469–74. doi: 10.1074/jbc.M611489200. [DOI] [PubMed] [Google Scholar]
- 118.Le Ber I, et al. Frequency and phenotypic spectrum of ataxia with oculomotor apraxia 2: a clinical and genetic study in 18 patients. Brain. 2004;127:759–67. doi: 10.1093/brain/awh080. [DOI] [PubMed] [Google Scholar]
- 119.Sugawara M, et al. Purkinje cell loss in the cerebellar flocculus in patients with ataxia with ocular motor apraxia type 1/early-onset ataxia with ocular motor apraxia and hypoalbuminemia. Eur Neurol. 2008;59:18–23. doi: 10.1159/000109256. [DOI] [PubMed] [Google Scholar]
- 120.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–69. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 121.Laine JP, Egly JM. When transcription and repair meet: a complex system. Trends Genet. 2006;22:430–6. doi: 10.1016/j.tig.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 122.Schumacher B, Garinis GA, Hoeijmakers JH. Age to survive: DNA damage and aging. Trends Genet. 2008;24:77–85. doi: 10.1016/j.tig.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 123.Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–5. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 124.Sugasawa K, et al. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 2001;15:507–21. doi: 10.1101/gad.866301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Coin F, Oksenych V, Egly JM. Distinct roles for the XPB/p52 and XPD/p44 subcomplexes of TFIIH in damaged DNA opening during nucleotide excision repair. Mol Cell. 2007;26:245–56. doi: 10.1016/j.molcel.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 126.Fan L, et al. Conserved XPB core structure and motifs for DNA unwinding: implications for pathway selection of transcription or excision repair. Mol Cell. 2006;22:27–37. doi: 10.1016/j.molcel.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 127.Fan L, et al. XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell. 2008;133:789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper, together with ref 123-126 provides critical insights for understanding the early events occurring during NER.
- 128.Liu H, et al. Structure of the DNA repair helicase XPD. Cell. 2008;133:801–12. doi: 10.1016/j.cell.2008.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wolski SC, et al. Crystal structure of the FeS cluster-containing nucleotide excision repair helicase XPD. PLoS Biol. 2008;6:e149. doi: 10.1371/journal.pbio.0060149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mu D, Hsu DS, Sancar A. Reaction mechanism of human DNA repair excision nuclease. J Biol Chem. 1996;271:8285–94. doi: 10.1074/jbc.271.14.8285. [DOI] [PubMed] [Google Scholar]
- 131.Sijbers AM, et al. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86:811–22. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- 132.Cleaver JE. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat Rev Cancer. 2005;5:564–73. doi: 10.1038/nrc1652. [DOI] [PubMed] [Google Scholar]; Important review of the relationship between cancer and neurodegeneration in XP.
- 133.Nouspikel T. Nucleotide excision repair and neurological diseases. DNA Repair (Amst) 2008;7:1155–67. doi: 10.1016/j.dnarep.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 134.Compe E, et al. Neurological defects in trichothiodystrophy reveal a coactivator function of TFIIH. Nat Neurosci. 2007;10:1414–22. doi: 10.1038/nn1990. [DOI] [PubMed] [Google Scholar]; Shows how deregulated TF11H can lead to abnormalities in stabilization of thyroid hormone receptor with DNA-responsive elements and the implications of this for neuropathology found in TTD.
- 135.Kraemer KH, et al. Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: a complex genotype-phenotype relationship. Neuroscience. 2007;145:1388–96. doi: 10.1016/j.neuroscience.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mimaki T, et al. Neurological manifestations in xeroderma pigmentosum. Ann Neurol. 1986;20:70–5. doi: 10.1002/ana.410200112. [DOI] [PubMed] [Google Scholar]
- 137.Anttinen A, et al. Neurological symptoms and natural course of xeroderma pigmentosum. Brain. 2008;131:1979–89. doi: 10.1093/brain/awn126. [DOI] [PubMed] [Google Scholar]
- 138.Robbins JH, Kraemer KH, Merchant SN, Brumback RA. Adult-onset xeroderma pigmentosum neurological disease--observations in an autopsy case. Clin Neuropathol. 2002;21:18–23. [PubMed] [Google Scholar]
- 139.Sidwell RU, et al. A novel mutation in the XPA gene associated with unusually mild clinical features in a patient who developed a spindle cell melanoma. Br J Dermatol. 2006;155:81–8. doi: 10.1111/j.1365-2133.2006.07272.x. [DOI] [PubMed] [Google Scholar]
- 140.Giannelli F, Avery J, Polani PE, Terrell C, Giammusso V. Xeroderma Pigmentosum and medulloblastoma: chromosomal damage to lymphocytes during radiotherapy. Radiat Res. 1981;88:194–208. [PubMed] [Google Scholar]
- 141.Brooks PJ, Cheng TF, Cooper L. Do all of the neurologic diseases in patients with DNA repair gene mutations result from the accumulation of DNA damage? DNA Repair (Amst) 2008;7:834–48. doi: 10.1016/j.dnarep.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Itoh M, et al. Neurodegeneration in hereditary nucleotide repair disorders. Brain Dev. 1999;21:326–33. doi: 10.1016/s0387-7604(99)00033-9. [DOI] [PubMed] [Google Scholar]
- 143.Henning KA, et al. The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell. 1995;82:555–64. doi: 10.1016/0092-8674(95)90028-4. [DOI] [PubMed] [Google Scholar]
- 144.van der Horst GT, et al. Defective transcription-coupled repair in Cockayne syndrome B mice is associated with skin cancer predisposition. Cell. 1997;89:425–35. doi: 10.1016/s0092-8674(00)80223-8. [DOI] [PubMed] [Google Scholar]
- 145.Ito S, et al. XPG stabilizes TFIIH, allowing transactivation of nuclear receptors: implications for Cockayne syndrome in XP-G/CS patients. Mol Cell. 2007;26:231–43. doi: 10.1016/j.molcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 146.Nouspikel T, Hanawalt PC. Terminally differentiated human neurons repair transcribed genes but display attenuated global DNA repair and modulation of repair gene expression. Mol Cell Biol. 2000;20:1562–70. doi: 10.1128/mcb.20.5.1562-1570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kraemer KH, Faghri S, Tamura D, Digiovanna JJ. Trichothiodystrophy: A systematic review of 112 published cases characterizes a wide spectrum of clinical manifestations. J Med Genet. 2008 doi: 10.1136/jmg.2008.058743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nance MA, Berry SA. Cockayne syndrome: review of 140 cases. Am J Med Genet. 1992;42:68–84. doi: 10.1002/ajmg.1320420115. [DOI] [PubMed] [Google Scholar]
- 149.Friedberg EC, Meira LB. Database of mouse strains carrying targeted mutations in genes affecting biological responses to DNA damage Version 7. DNA Repair (Amst) 2006;5:189–209. doi: 10.1016/j.dnarep.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 150.Sun XZ, Harada YN, Takahashi S, Shiomi N, Shiomi T. Purkinje cell degeneration in mice lacking the xeroderma pigmentosum group G gene. J Neurosci Res. 2001;64:348–54. doi: 10.1002/jnr.1085. [DOI] [PubMed] [Google Scholar]
- 151.Murai M, et al. Early postnatal ataxia and abnormal cerebellar development in mice lacking Xeroderma pigmentosum Group A and Cockayne syndrome Group B DNA repair genes. Proc Natl Acad Sci U S A. 2001;98:13379–84. doi: 10.1073/pnas.231329598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Reid S, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–4. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 153.Mirchandani KD, D'Andrea AD. The Fanconi anemia/BRCA pathway: a coordinator of cross-link repair. Exp Cell Res. 2006;312:2647–53. doi: 10.1016/j.yexcr.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 154.D'Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 155.Rahman N, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–7. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Erkko H, et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007 doi: 10.1038/nature05609. [DOI] [PubMed] [Google Scholar]
- 157.Offit K, et al. Shared genetic susceptibility to breast cancer, brain tumors, and Fanconi anemia. J Natl Cancer Inst. 2003;95:1548–51. doi: 10.1093/jnci/djg072. [DOI] [PubMed] [Google Scholar]; Identified medulloblastoma brain tumors as a feature of FA resulting from biallelic BRCA2 mutations.
- 158.Hunter N. The RecQ DNA helicases: Jacks-of-all-trades or master-tradesmen? Cell Res. 2008;18:328–30. doi: 10.1038/cr.2008.33. [DOI] [PubMed] [Google Scholar]
- 159.Hickson ID. RecQ helicases: caretakers of the genome. Nat Rev Cancer. 2003;3:169–78. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 160.Harrigan JA, Bohr VA. Human diseases deficient in RecQ helicases. Biochimie. 2003;85:1185–93. doi: 10.1016/j.biochi.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 161.van Brabant AJ, Stan R, Ellis NA. DNA helicases, genomic instability, and human genetic disease. Annu Rev Genomics Hum Genet. 2000;1:409–59. doi: 10.1146/annurev.genom.1.1.409. [DOI] [PubMed] [Google Scholar]
- 162.De Stefano N, et al. MR evidence of structural and metabolic changes in brains of patients with Werner's syndrome. J Neurol. 2003;250:1169–73. doi: 10.1007/s00415-003-0167-4. [DOI] [PubMed] [Google Scholar]
- 163.Kakigi R, Endo C, Neshige R, Kohno H, Kuroda Y. Accelerated aging of the brain in Werner's syndrome. Neurology. 1992;42:922–4. doi: 10.1212/wnl.42.4.922. [DOI] [PubMed] [Google Scholar]
- 164.Postiglione A, et al. Premature aging in Werner's syndrome spares the central nervous system. Neurobiol Aging. 1996;17:325–30. doi: 10.1016/0197-4580(96)00033-4. [DOI] [PubMed] [Google Scholar]
- 165.Arning L, et al. Identification and characterisation of a large Senataxin (SETX) gene duplication in ataxia with ocular apraxia type 2 (AOA2) Neurogenetics. 2008 doi: 10.1007/s10048-008-0139-z. [DOI] [PubMed] [Google Scholar]
- 166.Bassuk AG, et al. In cis autosomal dominant mutation of Senataxin associated with tremor/ataxia syndrome. Neurogenetics. 2007;8:45–9. doi: 10.1007/s10048-006-0067-8. [DOI] [PubMed] [Google Scholar]
- 167.Duquette A, et al. Mutations in senataxin responsible for Quebec cluster of ataxia with neuropathy. Ann Neurol. 2005;57:408–14. doi: 10.1002/ana.20408. [DOI] [PubMed] [Google Scholar]
- 168.Fogel BL, Perlman S. Novel mutations in the senataxin DNA/RNA helicase domain in ataxia with oculomotor apraxia 2. Neurology. 2006;67:2083–4. doi: 10.1212/01.wnl.0000247661.19601.28. [DOI] [PubMed] [Google Scholar]
- 169.Moreira MC, et al. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat Genet. 2004;36:225–7. doi: 10.1038/ng1303. [DOI] [PubMed] [Google Scholar]; Identified disruption of a helicase-like factor, Senataxin, as the basis for AOA2.
- 170.Nicolaou P, et al. A novel c.5308_5311delGAGA mutation in Senataxin in a Cypriot family with an autosomal recessive cerebellar ataxia. BMC Med Genet. 2008;9:28. doi: 10.1186/1471-2350-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chen YZ, et al. Senataxin, the yeast Sen1p orthologue: characterization of a unique protein in which recessive mutations cause ataxia and dominant mutations cause motor neuron disease. Neurobiol Dis. 2006;23:97–108. doi: 10.1016/j.nbd.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 172.Suraweera A, et al. Senataxin, defective in ataxia oculomotor apraxia type 2, is involved in the defense against oxidative DNA damage. J Cell Biol. 2007;177:969–79. doi: 10.1083/jcb.200701042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nahas SA, Duquette A, Roddier K, Gatti RA, Brais B. Ataxia-oculomotor apraxia 2 patients show no increased sensitivity to ionizing radiation. Neuromuscul Disord. 2007;17:968–9. doi: 10.1016/j.nmd.2007.06.464. [DOI] [PubMed] [Google Scholar]
- 174.Clements PM, et al. The ataxia-oculomotor apraxia 1 gene product has a role distinct from ATM and interacts with the DNA strand break repair proteins XRCC1 and XRCC4. DNA Repair (Amst) 2004;3:1493–502. doi: 10.1016/j.dnarep.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 175.Hassin-Baer S, et al. Absence of mutations in ATM, the gene responsible for ataxia telangiectasia in patients with cerebellar ataxia. J Neurol. 1999;246:716–719. doi: 10.1007/s004150050438. [DOI] [PubMed] [Google Scholar]
- 176.Gueven N, et al. A subgroup of spinocerebellar ataxias defective in DNA damage responses. Neuroscience. 2007;145:1418–25. doi: 10.1016/j.neuroscience.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 177.Frappart PO, McKinnon PJ. Mouse models of DNA double-strand break repair and neurological disease. DNA Repair (Amst) 2008;7:1051–60. doi: 10.1016/j.dnarep.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Frappart PO, et al. An essential function for NBS1 in the prevention of ataxia and cerebellar defects. Nat Med. 2005;11:538–44. doi: 10.1038/nm1228. [DOI] [PubMed] [Google Scholar]
- 179.Stucki M, et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–26. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 180.Stucki M, Jackson SP. gammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst) 2006;5:534–43. doi: 10.1016/j.dnarep.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 181.Lou Z, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 182.Barzilai A. The contribution of the DNA damage response to neuronal viability. Antioxid Redox Signal. 2007;9:211–8. doi: 10.1089/ars.2007.9.211. [DOI] [PubMed] [Google Scholar]
- 183.Ljungman M, Lane DP. Transcription - guarding the genome by sensing DNA damage. Nat Rev Cancer. 2004;4:727–37. doi: 10.1038/nrc1435. [DOI] [PubMed] [Google Scholar]
- 184.Cervos-Navarro J, Diemer NH. Selective vulnerability in brain hypoxia. Crit Rev Neurobiol. 1991;6:149–82. [PubMed] [Google Scholar]
- 185.Pulsinelli WA. Selective neuronal vulnerability: morphological and molecular characteristics. Prog Brain Res. 1985;63:29–37. doi: 10.1016/S0079-6123(08)61973-1. [DOI] [PubMed] [Google Scholar]
- 186.Chen P, et al. Oxidative stress is responsible for deficient survival and dendritogenesis in purkinje neurons from ataxia-telangiectasia mutated mutant mice. J Neurosci. 2003;23:11453–60. doi: 10.1523/JNEUROSCI.23-36-11453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.DiMauro S, Schon EA. Mitochondrial disorders in the nervous system. Annu Rev Neurosci. 2008;31:91–123. doi: 10.1146/annurev.neuro.30.051606.094302. [DOI] [PubMed] [Google Scholar]
- 188.Finsterer J. Central nervous system manifestations of mitochondrial disorders. Acta Neurol Scand. 2006;114:217–38. doi: 10.1111/j.1600-0404.2006.00671.x. [DOI] [PubMed] [Google Scholar]
- 189.de Souza-Pinto NC, Wilson DM, 3rd, Stevnsner TV, Bohr VA. Mitochondrial DNA base excision repair neurodegeneration. DNA Repair (Amst) 2008;7:1098–109. doi: 10.1016/j.dnarep.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lakshmipathy U, Campbell C. Mitochondrial DNA ligase III function is independent of Xrcc1. Nucleic Acids Res. 2000;28:3880–6. doi: 10.1093/nar/28.20.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Weissman L, de Souza-Pinto NC, Stevnsner T, Bohr VA. DNA repair, mitochondria, and neurodegeneration. Neuroscience. 2007;145:1318–29. doi: 10.1016/j.neuroscience.2006.08.061. [DOI] [PubMed] [Google Scholar]
- 192.Druzhyna NM, Wilson GL, LeDoux SP. Mitochondrial DNA repair in aging and disease. Mech Ageing Dev. 2008;129:383–90. doi: 10.1016/j.mad.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Frappart PO, Lee Y, Lamont J, McKinnon PJ. BRCA2 is required for neurogenesis and suppression of medulloblastoma. EMBO J. 2007;26:2732–42. doi: 10.1038/sj.emboj.7601703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Nijnik A, et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447:686–90. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- 195.Rossi DJ, et al. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447:725–9. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- 196.Ruzankina Y, et al. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1:113–26. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Together with refs 194 and 195, this work showed the importance of DNA repairs in the maintenance of stem cell populations.
- 197.Sii-Felice K, et al. Fanconi DNA repair pathway is required for survival and long-term maintenance of neural progenitors. EMBO J. 2008;27:770–81. doi: 10.1038/emboj.2008.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Yang JL, Weissman L, Bohr VA, Mattson MP. Mitochondrial DNA damage and repair in neurodegenerative disorders. DNA Repair (Amst) 2008;7:1110–20. doi: 10.1016/j.dnarep.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kruman II, et al. Cell cycle activation linked to neuronal cell death initiated by DNA damage. Neuron. 2004;41:549–61. doi: 10.1016/s0896-6273(04)00017-0. [DOI] [PubMed] [Google Scholar]
- 200.Yang Y, Herrup K. Loss of neuronal cell cycle control in ataxia-telangiectasia: a unified disease mechanism. J Neurosci. 2005;25:2522–9. doi: 10.1523/JNEUROSCI.4946-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Yang Y, Herrup K. Cell division in the CNS: protective response or lethal event in post-mitotic neurons? Biochim Biophys Acta. 2007;1772:457–66. doi: 10.1016/j.bbadis.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]