Abstract
Influenza A virus is a serious public health threat. Most recently the 2009/H1N1 pandemic virus had an inherent ability to evade the host's immune surveillance through genetic drift, shift, and genomic reassortment. Immune characterization of 2009/H1N1 utilized monoclonal antibodies, neutralizing sera, and proteomics. Increased age may have provided some degree of immunity, but vaccines against seasonal influenza viruses seldom yield cross-reactive immunity, exemplified by 2009/H1N1. Nonetheless, about 33% of individuals, over the age of 60, had cross-reactive neutralizing antibodies against 2009/H1N1, whereas only 6-9% young adults had these antibodies. Children characteristically had no detectable immunity against 2009/H1N1. Taken together, these observations suggest some degree of immune transference with at least certain strains of virus that have afflicted the human population in past decades. Because internal influenza proteins may exhibit less antigenic variation, it is possible that prior exposure to diverse strains of influenza virus provide some immunity to novel strains, including the recent pandemic strain (swine-avian A/H1N1). Current trends in immunological studies – specifically the modulation of cellular immune surveillance provided by TH17 and Tregs – also support the need for additional proteomic research for characterizing novel translational evidence-based treatment interventions based on cytokine function to help defeat the virus. Timely and critical research must characterize the impact of genetics and epigenetics of oral and systemic host immune surveillance responses to influenza A virus. The continued development and application of proteomics and gene expression across viral strains and human tissues increases our ability to combat the spread of influenza epidemics and pandemics.
Keywords: A/H1N1 2009, cytokines, TH17, Tregs, cellular immunity, epigenetics, comparative effectiveness, efficacy research
Background & Description
The inherent ability of the 2009/H1N1 pandemic influenza virus to evade immune surveillance through genetic drift, shift, and genomic reassortment is a serious public health threat [1]. Reactivity studies demonstrate species-specific antigenic differences in virulence as well as immune surveillance. The ‘avianlike’ swine viruses, including the recent pandemic strain (swine-avian A/H1N1) are characteristically most similar to avian viruses, and antigenic drift is more marked in ‘avian-like’ swine viruses than in classical swine strains [1].
Humoral Immunity
Phage expression library molecular methods serve to characterize influenza antigens and antibodies. Immune characterization methods used wholegenome- fragment phage display libraries expressing fragments of 15-350 amino acids traversing all the proteins of influenza A/Vietnam/1203/2004 (H5N1). These oligopeptides induced B cell responses using neutralizing sera and monoclonal antibodies following full recovery from H5N1 infection. This approach yielded two broadly neutralizing human monoclonal antibodies with conformation-dependent epitopes. Sera of patients convalescing from H5N1 H5 HA, without any cross-reactivity with sera from H5N1-naïve or H1N1- /H3N2-seropositive controls. Additional promising epitopes include the neuraminidase catalytic site M2 ectodomain, and the PB1-F2 virulence factor. The germline gene VH1-69 yields a common neutralizing mechanism specific for the HA fusion domain. This region exhibits less neutralization escape, and could be a promising strategy for broad-spectrum therapeutic protection against pandemic influenza threats [2].
Cellular Immune Surveillance
A wider use of influenza vaccines could control the severity of influenza pandemics. Moreover, comparative effectiveness and efficacy research increasingly indicates that humoral immunity alone may be insufficient to defeat influenza virus. Cellular immune events, mediated by sub-populations of both CD4+ and CD8+ T lymphocytes, contribute important surveillance against influenza virus, via cytokine modulation and the cross-reactivity that exists with virus-specific cytotoxic T lymphocytes (CTL, CD8+CD38+). For epitopes on NP and M1, CTL-based assays cross-reacted with viral strains from patient infections that were up to 9 years apart [3]. The 1977/8 influenza A/H1N1 infections occurred in otherwise healthy children and young adults in up to 50% of the cases. In fact, actuarial inferences from epidemiological data suggest that people infected with the 1945-55 influenza A/H1N1 developed immune surveillance against that later strain, which persisted as immune memory [4].
Incidences for H5N1 infection and fatality rates are lower for the older than 40- year age group (10.9% and 32%, respectively) than less than 40-year age group (89.1% and 59%, respectively). While these differences may be due to exposure to seasonal infection or vaccination, and the higher fatality rate may be due to differences in virus replication rates, the persistence of antigenspecific CD8+ T cells confers the ability of CTL's to clear infection and assists in reducing viral pathogenesis [5]. Greater than 97% of US H1N1 recovered isolates are from patients who are younger than 26 years old. For people over the age of 35, the recovery rate of H1N1 viruses drops to 4-5%. In the elderly, T-cell responses correlate better with vaccine protection than humoral immunity [6].
Viral infection leads to increased production of several cytokines, which are finely controlled by T cell sub-populations of TH17 and regulatory T cells (Tregs). The more virulent H5N1 influenza strain results in cytokine ‘storms’, characterized by sharp “great”-elevations of cytokine cascades [7]. IL-21, which is produced by TH17, may contribute to the mechanism by which CD4+ cells orchestrate the immune system response to viral infections, and specifically counteracts influenza infection in a mouse model. Without IL-21, CD4+ and CD8+ T cell responses are rapidly abrogated during acute and chronic viral infection, and viral clearance is severely impaired. IL-21 stimulates activation and proliferation of natural killer cells and CTL's via IL- 21 receptor-mediated activation of the STAT3 pathway [7, 8]. These modulatory effects by TH17 on one hand seem to be modulated by Tregs on the other, such that, for instance, IL-12 enhances anti-viral immune response, and the related cytokine, IL-28B (Type III anti-viral λFN family: with IFN-λ1, IFN-λ2, and IFN-λ3, e IL-29, IL-28A, and IL-28B, respectively) has substantial influence in adaptive immune responses as a vaccine adjuvant [9]. Presently, well-controlled experiments are used to characterize the cytokine storms in influenza pandemic, particularly in terms of TH17 and Tregs modulation. The current trends of knowledge depicted in Figure 1, opens novel avenues for clinical verification and Comparative Effectiveness and Efficacy Research and Analysis for (evidence-based) Practice (CEERAP), in order to utilize the best available evidence for ensuring most successful influenza management.
Figure 1.
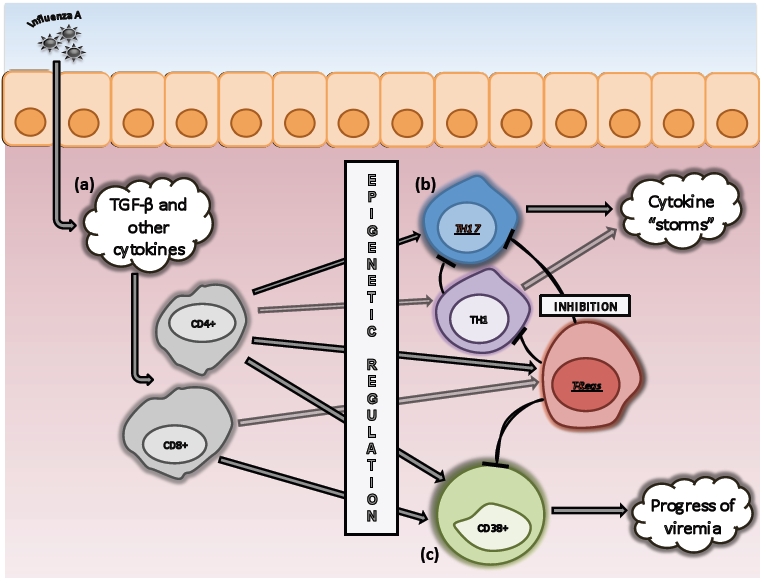
Tregs/TH17 Modulatory Balance in H1N1 Infection. (a) Cellular immune surveillance to H1N1 infection engages the release of a spectrum of cytokines (IFN-γ, TNF-α, IL015, IL-12p70, IL-8, IL-9, IL-6, IL-21, and TGF- β) within the microenvironment [7, 11], which act in concert to regulate T cell subset activation, maturation and differentiation. TGF-β plays a critical role in the regulation of both TH17 and Tregs (CD4+CD25+FoxP3+), which blunt T cell activation and TH17 generation and activity. This dual signaling function of TGF-β suggests that the ultimate balance between these subsets in cellular immune surveillance to influenza is likely due to epigenetic factors in the microenvironment [11] such as consequential to, but not limited to pregnancy [12] and immune senescence [10]. (b) While the proportion of TH17 cells may be relatively low in influenza patients, compared to control subjects, the secretion of TH17 and TH1 cytokines is vigorous, and characteristically the reported cytokine “storms” [7]. (c) The cytokine “storms” cascade leads to an increase in CD8+CD38+ CTL's, whose number and proportion correlate with the stage of infection, as they are responsible for the elimination of virally infected host cells.
Impact of Immunology on Clinical Outcome
In most cases, patients with H1N1 influenza have self-limited illness. However, some patients, especially those with co-morbidities, are more susceptible to influenza complications. In addition, the fine regulation of cellular immunity requires both vigorous effector responses and the maintenance of regulatory T subsets, such as TH17 and Tregs. Any disturbances in the balance between these two opposite activities of immune system, including tumor microenvironment, pregnancy or immune senescence, can seriously jeopardize this balance, and profoundly affects epigenetic regulatory controls of cellular immune surveillance [10– 12]. The latter actually provides a suggestive rationale to account for the anomaly of the 2009 influenza pandemic that exhibited higher lethality in younger patients than in the elderly, as noted above, because it is conceivable that the Tregs/TH17 ratio may be biased toward Tregs with age, thus yielding to a suppression and inhibition of TH17- mediated responses during senescence [10].
Pregnancy is a known risk of severe influenza. In the 2009 H1N1 pandemic, pregnancy led to increased risk of morbidity and mortality. In the US, most deaths in this population were due to pneumonia and respiratory failure. Because of the particular cellular immune conditions of pregnancy, vaccine failure is not uncommon in pregnant patients. To date, the corresponding efficacy of immune response to H1N1 monovalent vaccine remains unclear in this population. One mechanism may involve shifting immunity away from cell-medicated immunity towards humeral immunity, which could impair vaccine response. This may occur in association with occasional immunoglobulin G2 subclass deficiency in pregnant women with severe influenza. H1N1 may behave the same way in HIV infected population as in general population. Severe influenza seems to occur in late or advance diseases. Cellular immune response to the influenza virus appears to be lower in HIV-seropositive patients than general population.
In conclusion, current trends suggest that immunological studies support the need for additional proteomic research for characterizing novel treatment interventions based on chemokine and cytokine functions to help defeat the influenza virus. Timely and critical research must characterize the impact of genetics and epigenetics of oral and systemic host immune surveillance responses to H1N1. The continued development and application of proteomics and gene expression to diagnosis and characterization of influenza viruses and host responses across viral strains and human tissues ensure our ability to combat the spread of influenza epidemics and pandemics by means of carefully articulated research synthesis designs for developing and testing Translational Comparative Effectiveness and Efficacy Research and Analysis for Practice (TCEERAP) interventions.
Footnotes
Citation:Barkhordarian et al, Bioinformation 6(1): 39-40 (2011)
References
- 1.P Shapshak, et al. Mol Diag and Ther. 2011 in press. [Google Scholar]
- 2.S Khurana, et al. PLoS Med. 2009;6:e1000049. doi: 10.1371/journal.pmed.1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.PG Thomas, et al. Emerg Infect Dis. 2006;12:48. doi: 10.3201/eid1201.051237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WP Glezen, et al. Am J Epidemiol. 1991;133:296. doi: 10.1093/oxfordjournals.aje.a115874. [DOI] [PubMed] [Google Scholar]
- 5.Z Xing, CJ Cardona. Emerg Infect Dis. 2009;15:1847. doi: 10.3201/eid1511.090685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.JE McElhaney, et al. J Immunol. 2006;176:6333. doi: 10.4049/jimmunol.176.10.6333. [DOI] [PubMed] [Google Scholar]
- 7.JF Bermejo-Martin, et al. Crit Care. 2009;13:R201. doi: 10.1186/cc8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.H Elsaesser, et al. Science. 2009;324:1569. [Google Scholar]
- 9.MP Morrow, et al. Blood. 2009;113:5868. [Google Scholar]
- 10.P Trzonkowski, T In Fulop. Handbook on Immunosenescence,Springer. 2009:343. [Google Scholar]
- 11.OO Oluwadara, et al. Bioinformation. 2011;5:285. [Google Scholar]
- 12.MH Ramchandani, et al. Bioinformation. 2011;5:300. [Google Scholar]


