Abstract
The symbiosis of the pea aphid Acyrthosphion pisum with the bacterium Buchnera aphidicola APS represents the best-studied insect obligate symbiosis. Here we present a refined picture of this symbiosis by linking pre-genomic observations to new genomic data that includes the complete genomes of the eukaryotic and prokaryotic symbiotic partners. In doing so, we address four issues central to understanding the patterns and processes operating at the A. pisum/Buchnera APS interface. These four issues include: (1) lateral gene transfer, (2) host immunity, (3) symbiotic metabolism, and (4) regulation.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-011-0645-2) contains supplementary material, which is available to authorized users.
Keywords: Buchnera, Symbiosis, Amino acid metabolism, Bacteriocyte, Regulation, Lateral gene transfer
Introduction
Endosymbiosis, which is common in insects, has contributed to the ecological and evolutionary success of many taxa by provisioning novel traits, thereby facilitating exploitation of otherwise inaccessible niches [1–3]. The best-studied insect obligate symbiosis is the symbiosis of the pea aphid Acyrthosphion pisum with the bacterium Buchnera aphidicola APS [4].
The aphid/Buchnera symbiosis is primarily a nutritional symbiosis. Aphids feed on plant phloem sap, an unbalanced diet that is deficient in essential amino acids (protein amino acids that cannot be synthesized de novo by animals). To utilize phloem sap as their sole dietary component, most aphids are critically dependent on an ancient symbiosis with the bacterium B. aphidicola. Buchnera colonies are housed inside large specialized aphid cells called bacteriocytes [5]. Studies involving symbiont removal and/or manipulative artificial diet experiments (e.g., [6–8]) coupled more recently with whole genome sequencing of both Buchnera and A. pisum [9–11] demonstrate that the nutritional needs of aphids are fully compensated by the biosynthetic capabilities of the holosymbiont (aphid + Buchnera). In sum, the aphid/Buchnera association is obligate and mutualistic with neither partner being able to reproduce in the absence of the other [12–16].
Unfortunately, the intimacy and obligacy of the aphid/Buchnera symbiosis hampers experimental studies as sophisticated as those in free-living model organisms or even those in aphid secondary symbionts (reviewed in [17, 43], Pontes [18]). However, recent advances in genomics have enabled biological studies of many uncultivable organisms, with exploration of the aphid/Buchnera symbiosis being no exception. Given the February 2010 publication of the draft genome sequence of the pea aphid, Acyrthosiphon pisum, and a decade-earlier publication of the complete genome sequence of B. aphidicola APS [9, 10], we are now able to study important and previously intractable questions in symbiosis by utilizing the complete genomic information of both symbiotic partners. The goal of this review is to present a refined picture of the aphid/Buchnera symbiosis by linking pre-genomic observations to new genomic data that includes the complete genomes of the eukaryotic and prokaryotic symbiotic partners. Following a biological and historical overview of the A. pisum/Buchnera APS symbiosis, we address four issues central to understanding the patterns and processes operating at the symbiotic interface. These four issues include: (1) lateral gene transfer, (2) host immunity, (3) symbiotic metabolism, and (4) regulation.
Overview of aphid/Buchnera symbiosis with historical background
Almost all of 4,000 extant species of aphid (Homoptera: Aphididae), with a few exceptions, harbor an obligate bacterial symbiont called B. aphidicola [19–22]. Buchnera constitute a monophyletic group in the γ-subdivision of the Proteobacteria with the group of bacteria that includes Escherichia coli as their closest relatives [23, 24]. The molecular phylogenies of Buchnera and aphids are strictly congruent, suggesting a single original infection in the common ancestor of extant aphid species, followed by co-speciation of aphids and Buchnera [5]. The origin of the symbiosis is dated to 160–280 MYA [22].
For most of the aphid lifecycle, Buchnera are restricted to huge polyploid aphid cells called bacteriocytes [25]. These cells are grouped into an organ-like structure called the mycetome, or bacteriome, which consists of two halves extending along the body and joining above the hindgut (Fig. 1b). Small, flattened cells called sheath cells form a thin casing around bacteriocytes [2]. The symbionts are passed directly from mother to offspring by transovarial transfer [2, 25] (Fig. 1a).
Fig. 1.
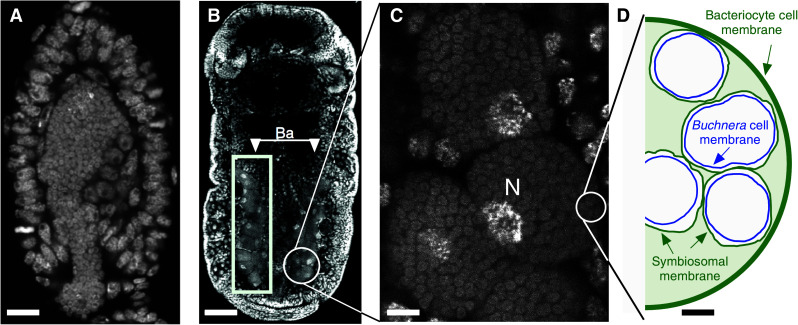
DNA-stained confocal microscope images of aphid embryos. a Incorporation of the maternal endosymbiotic bacteria to a stage 7 embryo. The bacteria are visible as small round cells. Anterior is to top. Scale bar 14 μm. b Aggregates of large aphid bacteriocyte cells that run perpendicular to the main axis of a stage 16 embryo are clearly visible (Ba). Scale bar 50 μm. c Magnified view of aphid bacteriocyte cells showing the large polyploid bacteriocyte cell nucleus (N) and a high density of Buchnera within the cell. Scale bar 6 μm. d Cartoon of the aphid bacteriocyte. The two aphid-derived membranes are shown in green, and the Buchnera cell membrane is shown in blue (drawn to scale from Fig. 1 of Baumann et al. [5]). Scale bar 2 μm. Parthenogenetic viviparous embryos were stained with TO-PRO3 (a) or DAPI (b, c). Composite figure and cartoon courtesy of Daniel R. G. Price. Confocal microscopy images: Shuji Shigenobu (a), James Baker (b, c)
The structure of bacteriome was identified in the middle of 19th century, and yet the function of this organ was misunderstood for a long time. Huxley named it the “pseudovitellus”, thinking that the granules (actually Buchnera) were yolk-like materials [26]. It was another 52 years before Pierantoni and Sule in 1910 demonstrated that the “pseudovitellus” is, in fact, a symbiotic organ populated with microorganisms [27, 28]. Mycetome biology was pioneered by Buchner and his students [29], whose extensive light-microscopy work was summarized in his monograph that describes many insect/microbe symbioses including those of aphids [2, 30]. Subsequent to the development of electromicroscopy, ultrastructural studies of bacteriocytes advanced our understanding of the aphid/Buchnera symbiosis (reviewed in [31]) by demonstrating that while bacteriocytes are highly specialized cells for housing symbionts and unique in that their cytoplasm is filled with tens of thousands of Buchnera cells, they contain a normal complement of eukaryotic organelles that include mitochondria, an endoplasmic reticulum (ER), and Golgi apparatus. Further electron microscopy revealed that three discrete membranes separate the cytoplasm of Buchnera from the cytoplasm of the bacteriocyte [32]. Two of the membranes are the inner and outer Buchnera membranes that exhibit a typical Gram-negative bacterial structure [33]. The third membrane that generally individually envelopes each Buchnera cell is known as the symbiosomal membrane and is of host origin (Fig. 1c).
The nutritional basis of the aphid/Buchnera symbiosis was postulated almost 60 years ago by Buchner [30], tested using artificial diet assays and generation of aposymbiotic aphids in the 1960s–1990s (e.g., see Fig. 2 generated from [34–36]) and finally irrevocably confirmed in the past decade with whole genome sequencing of multiple lineages of Buchnera and most recently the pea aphid, A. pisum [9, 10, 37–39]. While the nutritional basis of the aphid/Buchnera symbioses is now well appreciated, the regulatory processes and structures that mediate the intimate interactions between aphids and Buchnera have largely been neglected. In most part this neglect stems from the fact that until now the aphid model has been insufficiently equipped with the genetic and molecular tools necessary for characterization of regulatory processes and structures operating at the symbiotic interface. That said, the last 40+ years of research have left us with a comprehensive blueprint of the amino acid metabolism of this symbiosis, a blueprint that provides the foundation for hypothesis-testing research focused on characterization of the regulatory processes and structures operating at the symbiotic interface.
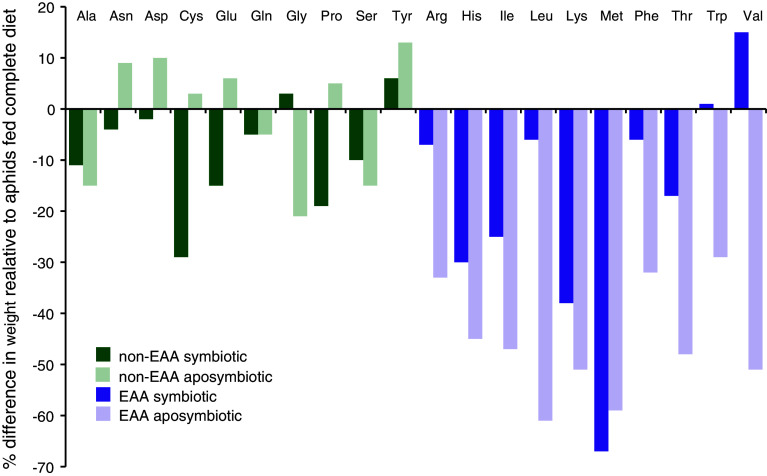
Fig. 2.
Results of Mittler [34] single amino acid elimination experiment demonstrating Buchnera provisioning of essential amino acids. Nonessential amino acids (listed alphabetically) are represented by blue bars, while essential amino acids (also alphabetically listed) are shown in pink. Dark colors show data from symbiotic Myzus persicae while light colors show data from aposymbiotic M. persicae. While the data are largely qualitative, they are representative of the types of early data that established the nutritional provisioning of essential amino acids by Buchnera to their aphid hosts. Data are taken from 3day/apterous columns in Table 2 of [34]
Whole genome sequences of both symbiotic partners
The aphid/Buchnera symbiosis is the first obligately symbiotic partnership for which both the host and symbiont have fully sequenced genomes [9, 10] [the second, that of the human body louse, Pediculus humanus humanus and its endocellular bacterial symbiont Candidatus Riesia pediculicola [40], was published in July 2010 [41] but lacks the extensive body of pre-genomic work that benefits study of the aphid/Buchnera symbiosis].
Whole genome sequence of the symbiont, Buchnera APS
The genome of Buchnera from A. pisum (Buchnera APS) comprises an AT-rich circular chromosome and two small plasmids. At only 641 kb, the chromosome of Buchnera APS is less than one-seventh the size of those of free-living relatives such as E. coli (K-12 strain, 4.6 Mb) and close to the theoretical minimum genome size for any living organism [42]. The small genome encodes 583 protein-coding genes, a reduced gene repertoire that is essentially a subset of that of E. coli, with almost no genes unique to Buchnera and a pattern of genome reduction that is now known to be typical of obligate, intracellular, mutualistic bacteria [43–45]. As also tends to be typical of the genomes of obligate intracellular symbionts, the Buchnera genome does not contain any genes acquired by lateral gene transfer (LGT) [10]. Remarkably, Buchnera lacks “apparently essential” genes for cellular life; missing genes include those necessary for biosynthesis of cell-surface components, including lipopolysaccharides and phospholipids, regulatory genes, and genes involved in cellular defense [10]. In fact, the gene repertoire of Buchnera is so specialized to intracellular life that Buchnera cannot survive outside the eukaryotic cell. However, despite its massive gene losses, Buchnera retain genes for the biosynthesis of nutrients and other metabolites required by the host; the details of which are discussed in “Discussion of four issues central to understanding the patterns and processes operating at interface of the aphid/Buchnera symbiosis” below.
In addition, to the complete genome sequence of Buchnera APS from A. pisum [10], the complete Buchnera genomes of three other aphid species [37–39] as well as eight more strains of Buchnera from A. pisum have been sequenced [9, 46]. What emerges from these genomes is a fine-scale understanding of the ongoing process and rates of symbiont genome reduction.
Draft genome assembly of the host, A. pisum
Acyrthosiphon pisum with 34,604 predicted genes in its 464-Mb draft genome assembly (Acyr_1.0) has the highest gene count of any insect sequenced to date [9, 46]. This high gene count reflects both extensive gene duplications and the presence of aphid-specific orphan genes. Gene duplications were detected for >2,000 gene families and orphan genes comprise 20% of the total number of genes in the genome. The role, if any, played by gene duplication and the occurrence of so many orphan genes in the evolution of the aphid/Buchnera symbiosis is not known, but there are indications that at least some of these genes are associated with metabolic, structural, and developmental innovations related to the symbiosis [47]. On the other hand, the pea aphid genome lacks some genes highly conserved in other animals including unusual losses of immunity genes [48] and the entire urea cycle [11].
Discussion of four issues central to understanding the patterns and processes operating at interface of the aphid/Buchnera symbiosis
Lateral gene transfer
The massive gene losses associated with genome evolution in obligate intracellular bacterial symbionts including Buchnera coupled with patterns of eukaryotic genome evolution associated with acquisition of mitochondria and chloroplasts [49] and knowledge that aphid genomes have experienced numerous bouts of transposition of mitochondrial genes to the nuclear genome [51], resulted in the formulation of the expectation that genes of Buchnera origin would be found in the A. pisum genome. Thus, one of the more surprising results of the pea aphid genome project was that the A. pisum genome contains no genes of Buchnera origin [9, 52]; a fact that rules out lateral gene transfer to the host genome as a driver of genome size reduction in Buchnera and one indicative of differences in the patterns and processes of genome reduction in Buchnera as opposed to organellar genomes.
While the A. pisum genome does not contain genes of Buchnera origin, unlike Buchnera, it has acquired genes by lateral gene transfer from three sources. First, 56 mitochondrial genes were detected in the A. pisum genome, although all of them are pseudogenized [9]. Second, the A. pisum genome contains functional genes of fungal origin encoding the entire pathway for carotenoid biosynthesis [53]. Third, 12 genes acquired from bacteria were identified [9, 52]. Of the nine bacterial genes that appear to be intact, all of them were closely related to genes of the α-proteobacteria (Buchnera is a γ-proteobacterium) and were demonstrated by phylogenetic analysis not to be of Buchnera origin.
Of the three groups of laterally acquired genes in the A. pisum genome gene expression, work indicates that members of the third group, those of α-proteobacterial origin, are functionally important at the symbiotic interface [54]. In particular, LdcA (LD-carboxypeptidase), AmiD (N-acetylmuramoyl-l-alanine amidase) and the RlpA (rare lipoprotein A) family that is represented by five A. pisum paralogs (RlpA1-5), are up-regulated in the bacteriocytes relative to whole insects. Homology-based annotation indicates that LdcA and AmiD encode enzymes involved in metabolism of peptidoglycan, a major component of bacterial membranes including those of Buchnera [33], while the function of RlpA is unknown. Recently, Nikoh and Nakabachi [55] proposed that LdcA and AmiD may have compensatory functions to support the survival of Buchnera and eventually control the proliferation of the symbionts, while A. pisum whole-genome sequencing indicates that the RlpA paralogs contain N-terminal eukaryotic-type signal peptides, suggesting that the bacterial genes have fused with an eukaryotic signal peptide and can thereby function in the context of the host eukaryotic cell [55]. The N-terminal eukaryotic-type signal peptides are predicted to target secretory pathway proteins, a prediction that future protein localization studies should address. In the context of our goal to characterize the processes and structures at the aphid/Buchnera symbiotic interface, we hypothesize that the RlpA proteins are targeted to Buchnera via the symbiosomal membrane, a probable scenario given that the space Buchnera inhabit is topologically outside of the host cell (Fig. 1d) and that symbiosomal membranes sometimes show close association with bacteriocyte endoplasmic reticula (a key secretion pathway structure in eukaryotic cells) [31, 56].
In sum, A. pisum laterally acquired genes of mitochondrial, fungal, and α-proteobacterial origin. While the fungal acquisition of carotenoid biosynthesis underlies the color polymorphism in pea aphids [53], the functional role of laterally acquired α-proteobacterial genes remains to be elucidated. That said, studies of gene expression at the aphid/Buchnera interface coupled with examination of gene structure suggest that the genes of α-proteobacterial origin play critical roles in symbiotic regulation.
Host immunity
Most insects have an innate immune system to defend themselves against a wide range of pathogenic and parasitic organisms. Sustainable accommodation of Buchnera within the host body raises the possibility of a modification of the immune response in aphids. Consistent with this reasoning, A. pisum appears to have only limited immune capacity as displayed by reduced lysozyme-like activity in aphids stabbed with a bacteria-contaminated needle, no detectable activity against live bacteria in hemolymph assays and no detectable induction of gene expression in response to bacterial challenge [57]. The apparently reduced immune capacity of aphids is consistent with the discovery that the A. pisum genome is missing many genes central to immune function in other insects [9, 48]. While orthologs of key components of the immune-related Toll, Jak/Stat, and JNK signaling pathways are present in the pea aphid genome, genes for other immune response pathways are absent. Specifically, the immunodeficiency (IMD) pathway is missing the genes IMD, dFADD, Dredd, and Relish, while all the IMD pathway genes are intact in genomes of other sequenced insects. In addition to signaling pathways, some genes for microbial recognition are missing. Peptidoglycan receptor proteins (PGRPs) are critical components for detecting bacterial invasion following activation of the IMD and Toll pathways and are highly conserved from insects to mammals, yet no PGRPs were identified in the A. pisum genome. Furthermore, the gene repertoire of innate immunity effector molecules, such as antimicrobial peptides, is also limited. Taken together, the strikingly reduced immunity-related gene repertoire of A. pisum facilitates speculation that the reduced immune capacity of aphids may account for the evolutionary success of aphids in acquiring beneficial symbionts such as Buchnera [96].
Symbiotic metabolism
As is the case for all insect/microbial nutritional symbioses, nutrient provisioning by Buchnera to the host aphid represents the heart of the symbiosis and is likely the sole reason that such symbioses have persisted through evolutionary time. The nine panels of Fig. 3 present a graphic review of the aphid/Buchnera amino acid metabolism literature that combined with the following text will, we hope, provide the reader with a clear understanding of both the intimacy and complementarity of amino acid synthesis and upgrading in the aphid/Buchnera symbiosis.
Fig. 3.
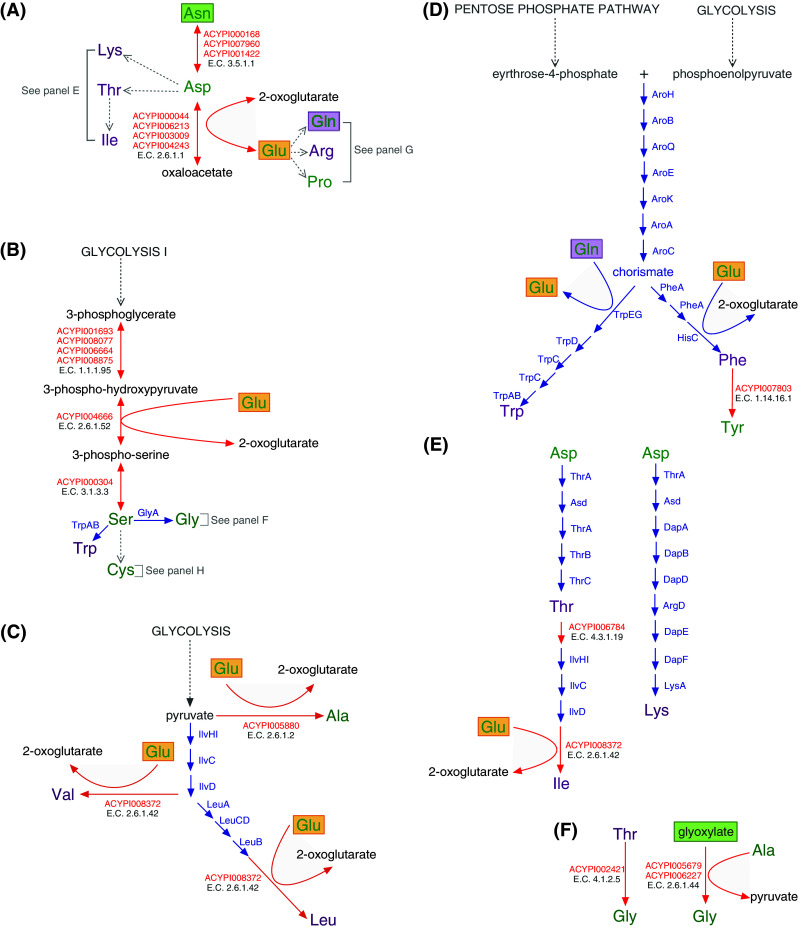
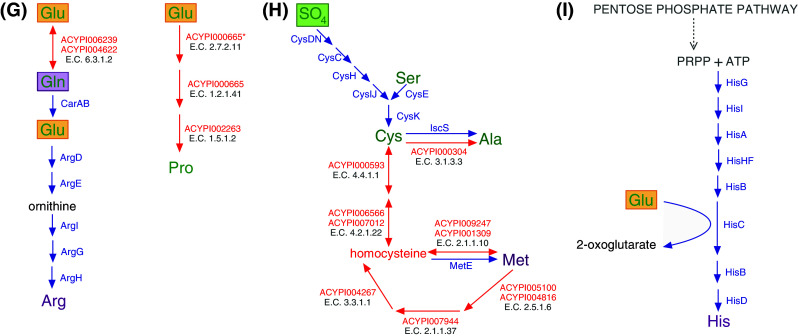
Blueprint of amino acid biosynthesis and nutritional upgrading in the A. pisum/Buchnera APS symbiosis. Essential amino acids are shown in purple, while non-essential amino acids are in green. A. pisum enzymes are shown with red arrows, while Buchnera enzymes are shown with blue arrows. Green boxes around metabolites indicate a phloem sap source and purple boxes highlight the importance of glutamine (Gln), which is a dominant amino acid in hemolymph [64] actively taken up by bacteriocytes and converted to glutamate (Glu, orange boxes). Glutamate then actively crosses the symbiosomal membrane [64, 65] where it serves as an amino donor and metabolic precursor for the synthesis of nonessential amino acids. a Asparagine (Asn), the dominant amino acid in the phloem sap diet of the pea aphid [62], aspartate (Asp), glutamate (Glu) and glutamine (Gln) are the four amino acids of central importance to the aphid/Buchnera symbiosis; at least one of these four is required as the primary source of all protein amino acids for the holosymbiont [35]. b–d Both A. pisum and Buchnera APS retain glycolysis and the pentose phosphate pathway and thus the ability to synthesize 3-phosphoglycerate, phosphoenolpyruvate, pyruvate, and eyrthrose-4-phosphate. e Aspartate (Asp) is transported across the Buchnera APS membrane where it serves as the metabolic precursor of the essential amino acids isoleucine (Ile), lysine (Lys), and threonine (Thr). Synthesis of the branched-chain essential amino acids valine (Val), leucine (Leu), and isoleucine (Ile) requires within-pathway metabolic collaboration between symbiotic partners (c, e) [11]. f In addition to the ability of Buchnera APS to synthesize glycine (Gly) from serine (b), two pathways for the biosynthesis of glycine are retained by A. pisum. g Both A. pisum and Buchnera APS retain the ability to synthesize glutamate from glutamine. Glutamate serves as the metabolic precursor of proline and arginine. Arginine (Arg), the tenth essential amino acid is synthesized by Buchnera APS where A. pisum has uniquely lost the metabolic capability to synthesize this amino acid [9, 11]. *The annotation of ACYPI000665 from KEGG (http://www.genome.jp/kegg/). h The non-essential amino acid cysteine (Cys) is synthesized by Buchnera APS from phloem sap provisioned sulfate and A. pisum synthesized serine (b). Buchnera APS has lost the capacity to synthesize the essential amino acid methionine (Met), yet strong physiological evidence exists that demonstrates that the holosymbiont possesses the capacity for methionine biosynthesis [49]. The pathway for methionine biosynthesis presented here is that of [9, 11]. The non-essential amino acid alanine (Ala) can be synthesized by both Buchnera APS and A. pisum (from Cys shown here and from pyruvate, c). i Both A. pisum and Buchnera APS retain the pentose phosphate pathway and the ability to synthesize phosphoribosyl pyrophosphate (PRPP) from ribose-5-phosphate using ribose-phosphate diphosphokinase (E.C. 2.7.6.1, A. pisum: ACYPI006288, Buchnera APS: PrsA, KEGG (http://www.genome.jp/kegg/). With the exception of the cases indicated, pathways presented were constructed using the AcypiCyc Acyrthosiphon pisum (genome paper version) and Buchnera aphidicola APS (Acyrthosiphon pisum) databases (http://acypicyc.cycadsys.org)
Plant phloem sap, the sole dietary component of aphids, is a nutritionally unbalanced diet deficient in essential amino acids. Essential amino acids (shown in purple in Fig. 3) are the protein amino acids that cannot be synthesized de novo by animals and include histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, valine, and for A. pisum, which is missing a complete urea cycle, arginine [9, 11]. Phloem sap differs both qualitatively and quantitatively in amino acid composition from plant species to plant species [58] and is even known to fluctuate diurnally [59] and developmentally [60]. While the phloem sap of most species contains at least trace amounts of all 20 protein amino acids, a subset of only four (asparagine, aspartate, glutamate, and glutamine), in varying combinations, represent the dominant amino acids [58]. Early artificial diet experiments in Myzus persicae demonstrated that omission of aspartate, glutamate, asparagine, and glutamine as a group essentially prevented growth while their individual omission from artificial diets had no effect [35]. On examination of Fig. 3, built from the full-genome sequences of A. pisum and Buchnera APS, the metabolic basis of this early observation is very clear; at least one of these four amino acids is essential as the primary source of all protein amino acids in the aphid/Buchnera symbiosis. In the case of the phloem sap diet of A. pisum, the dominant amino acid is asparagine, which has been demonstrated to comprise up to an amazing 70% of the total amino acid content of alfalfa phloem [61]. The dominance of asparagine in the phloem sap diet of A. pisum [62] renders it the main source of protein amino acids for the A. pisum/Buchnera APS symbiosis and is indicated by the green box of Fig. 3a.
Whole-genome analysis of A. pisum indicates that following ingestion, asparagine is converted by the enzyme asparaginase (E.C. 3.5.1.1) to aspartate, which is then transaminated to oxaloacetate by aspartate transaminase (E.C. 2.6.1.1), thus liberating glutamate [11]. Aspartate and glutamate are precursors for the synthesis of four essential (arginine, isoleucine, lysine, and threonine) and two nonessential amino acids (glutamine and proline) (Fig. 3a). In addition, glutamate functions as an amino donor in the synthesis of five essential amino acids, histidine, isoleucine, leucine, phenylalanine, and valine and three non-essential amino acids, alanine, serine, and tyrosine (Fig. 3b–e, i). The nonessential amino acid serine is then processed largely by Buchnera APS into three nonessential amino acids (alanine, cystine, and glycine) and two essential amino acids (methionine (but see Fig. 3h) and tryptophan) [10] (and note that the nonessential amino acids alanine (Fig. 3b), cysteine (Fig. 3h), and glycine (Fig. 3f) can also be synthesized from amino acid and phloem sap precursors by A. pisum). Finally, in the biosynthesis of the essential amino acid tryptophan, glutamine (which is synthesized by the aphid from glutamate, Fig. 3g) serves as the amino donor. Thus, in A. pisum, asparagine functions as a metabolic precursor for the synthesis of all protein amino acids.
One of the most striking and unprecedented features of amino acid metabolism in the A. pisum/Buchnera APS symbiosis is the nature of the metabolic collaboration between symbiotic partners. By the time Shigenobu et al. [10] published the genome sequence of Buchnera APS, numerous dietary and symbiont manipulation experiments, some of which included isotopic labeling of amino acid precursors, had established that Buchnera functions in the synthesis of arginine, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, and valine (e.g. [36, 63, 64]). Further, some had even implicated Buchnera in the synthesis of the nonessential amino acid tyrosine [36]. Thus, it was not surprising that completion of the first Buchnera genome in 2000 demonstrated that despite extensive gene loss and a significant reduction in genome size, Buchnera retains almost all of the genes necessary for the synthesis of all ten essential amino acids but has (with the exception of glycine, cysteine, and possibly tyrosine) lost the ability to synthesize nonessential amino acids [10]. While the Buchnera APS genome was found to retain almost all of the genes necessary for essential amino acid biosynthesis, some important genes were missing. These included IlvE, the branch-chain amino acid transaminase, IlvA, the first gene in the pathway of isoleucine biosynthesis from threonine as well as MetB and MetC, the next-to-last two genes in the methionine biosynthesis pathway from aspartate through homoserine. The absence of these genes led Shigenobu et al. to propose that the these apparently missing reactions must be mediated by other genes encoded by the Buchnera APS genome, e.g., that the transamination reaction that is typically mediated by IlvE, the branched-chain amino acid transaminase (E.C. 2.6.1.42), is mediated by another one of the five Buchnera encoded transaminases (see Table 2 of [11] for a complete list of the Buchnera APS encoded transaminases). Thus, on completion of the Buchnera APS genome, the blueprint of aphid/Buchnera symbiosis certainly included the notion of metabolic complementarity, but the complementarity was understood to partition whole metabolic pathways. Annotation of metabolism-related genes in the A. pisum genome in 2009 by members of the International Aphid Genomics Consortium resulted in a more complex blueprint of amino acid metabolism in the A. pisum/Buchnera APS symbiosis and the realization that the complementarity of nutritional metabolism in the symbiosis is more intimate than previously envisioned [9, 11]. The current blueprint includes five cases of metabolic complementarity that involve within-pathway collaboration between symbiotic partners. These cases include the synthesis of four essential amino acids, leucine and valine (Fig. 3c), isoleucine (Fig. 3e), and methionine (Fig. 3h) and one non-essential amino acid, tyrosine (Fig. 3d).
Perhaps the most rewarding aspect of assembling the genomic blueprint of amino acid metabolism in the aphid/Buchnera symbiosis (Fig. 3) is the fact that it facilitates reinterpretation of the extensive body of pre-genome research and thus leaves us with some important insights into the compartmentalization of amino acid biosynthesis not only between symbiotic partners (as described above) but also within cellular and subcellular compartments. Thus, in the case of the A. pisum/Buchnera APS symbiosis, the picture that emerges is that asparagine, the dominant amino acid in the phloem sap diet of A. pisum [62] is converted, by the aphid to glutamine through aspartate and glutamate such that the dominant amino acids in A. pisum hemolymph are asparagine and glutamine [64]. Glutamine is then actively taken up by bacteriocytes [64] where it is converted by glutamate-ammonia ligase (E.C. 6.3.1.2), an enzyme involved in nitrogen recycling and one of the five most highly expressed genes in A. pisum bacteriocytes [54], to glutamate. Then the symbiosomal membrane encased Buchnera cells, which do not readily uptake glutamine, actively and apparently limitlessly uptake glutamate [64, 65]. Significantly, the work of Sasaki and Ishikawa [64] coupled with the genomic blueprint of amino acid metabolism presented in Fig. 3 indicates that the symbiosomal space [the subcellular space that surrounds each Buchnera cell, bounded on the outside by the symbiosomal membrane and on the inside by the outer membrane of Buchnera (Fig. 1)] is a metabolically active space in which the aphid minimally synthesizes alanine, aspartate, and proline. Further, the coupling of these data sets confirms that Buchnera is able to uptake glutamate that is then utilized as an amino donor in the biosynthesis of essential amino acids such as phenylalanine. Finally, work with symbiotic and aposymbiotic A. pisum raised on artificial diet, coupled with the literature described here and the genomic blueprint of amino acid metabolism presented in Fig. 3 strongly indicates that the above proposed pathway from asparagine in phloem sap to glutamine in hemolymph/bacteriocytes and glutamate in the symbiosomal space is the main route of nitrogen upgrading in the A. pisum/Buchnera symbiosis.
Regulation
Possibly the most exciting questions in symbiosis research concern regulation, in particular, the mechanisms by which genes encoded by the genomes of the holosymbiont are cooperatively regulated. Demonstration that the relative biomass of symbiotic partners is developmentally and environmentally predictable resulted in the inference that the aphid/Buchnera symbiosis is tightly regulated (e.g., [66–71] and prompted a large body of physiological work aimed at demonstrating regulation of the symbiosis by quantifying the relationship between nutrient demand and supply (e.g., [6, 7, 72]). Most recently, advances in molecular and genomic technologies mean that we can study regulation and regulatory processes on a genome-wide scale at transcriptional, translational and even post-translational levels. What follows is largely a review of studies concerning regulation of the aphid/Buchnera symbiosis in the era of genomics that currently focuses on three aspects: (a) regulatory gene content, (b) transcription, and (c) other genetic control. We then summarize them in “Experimental evidence indicates that host aphids play the dominant role in symbiotic regulation”.
Regulatory gene content
Whole-genome analysis of Buchnera APS revealed that in contrast to free-living bacteria such as E. coli, whose genome encodes 233 regulatory proteins [73], regulatory proteins are almost completely missing from the Buchnera genome [10]. While some parasitic bacteria with small genomes, such as Mycoplasma [74] and Rickettsia [75], have also lost large parts of their regulatory systems, Buchnera is unique in the extent to which it has lost regulatory genes. For example, while Buchnera Sg., retains a single-functional amino acid biosynthesis regulatory gene (metR) [76], no functional transcriptional regulator of amino-acid biosynthesis is present in the Buchnera APS genome despite the conservation of many genes for amino-acid biosynthesis [76, 77]. In addition, Buchnera lacks two-component regulatory systems, which generally control gene expression in response to environmental changes [78] and transcriptional attenuation systems [10, 79].
In contrast to the scarcity of regulatory genes in the Buchnera genome, the pea aphid possesses a complement of metazoan regulatory genes with greater gene family diversity than that seen in other sequenced arthropods [9]. Transcription factors [80] and genes for signal transduction pathways [80], chromatin remodeling [81], miRNA pathway [82], and DNA methylation [83] are well conserved, with some of them exhibiting unusual aphid-specific gene duplications. Some transcription factors such as Distal-less were found to be expressed in the bacteriocytes [84]. Yet, to date, studies of regulation in aphids are in their infancy and it remains largely unknown how these genes are involved in symbiotic regulation.
Transcription
Does the scarcity of regulatory genes in the Buchnera gene repertoire incur reduced regulatory capacities? To address this question, several laboratories have used microarrays to obtain transcriptomic profiles of Buchnera [4, 76, 77, 85–87]. Collectively, these transcriptomic data demonstrate that Buchnera gene expression changes in response to environmental perturbation are subtle (reviewed in [21]). For example, only a small fraction of Buchnera genes responded to heat-stress treatments [87] and yet almost all of the 20 orthologs of heat shock genes in the Buchnera genome are constitutively overexpressed under non-stress conditions. A striking example is the GroEL homolog, which was initially named ‘symbionin’ due to its extremely high representation in protein analysis of symbiotic aphids in comparison to Buchnera-free (aposymbiotic) aphids [88]. Similarly, only a few transcriptional changes were observed in response to changes in host amino acid requirements [76, 77]. Thus, overwhelmingly, microarray-generated transcriptomic studies of Buchnera demonstrate that the transcriptional dynamics of Buchnera are static.
Expressed sequence tag-based transcriptional studies were the first DNA-based studies to truly focus on the aphid/Buchnera symbiotic interface. In 2005, Nakabachi et al. [54] published a study describing the eukaryotic mRNA population of the A. pisum bacteriocyte. In doing so they identified 337 unigenes. Subsequent comparison of ESTs among cDNA libraries prepared from various tissues and organs revealed that bacteriocytes are rich in unique (tissue-specific) transcripts [89]; among the 315 transcripts expressed in bacteriocytes, 58 exhibited statistically significant bacteriocyte-specific expression (see Supplemental Online Material, Table S1 for an updated annotation of these genes). The 58 bacteriocyte-specific transcripts were mostly associated with the utilization of essential amino acids, biosynthesis of nonessential amino acids, and transport, but also included laterally acquired genes of α-proteobacterial origin such as RlpA family members and LdcA (see above). Interestingly, the most abundant transcripts in the bacteriocyte were from an i-type lysozyme, Lys1, whose expression level was 25-fold that of ribosomal proteins. While c-type lysozymes have been demonstrated to possess lytic activities in many organisms (reviewed in [48]), the bacteriolytic activities of i-type lysozymes have not been characterized. Therefore, it is unknown whether A. pisum Lys1 has bacteriolytic activity. Lysosomal break-down of Buchnera cells has been observed in ultrastructural studies [90] and thus, given its high level of bacteriocyte expression, it seems reasonable to hypothesize that Lys1 has bacteriolytic activity and that the host uses it to regulate its Buchnera population. It is, of course, also possible that Lys1 serves a non-bacteriolytic function at the A. pisum/Buchnera symbiotic interface and resolution of these alternatives requires experimental work.
Other genetic control
It is not uncommon to find that key enzymes in the genomes of nutritional bacterial symbionts are located on plasmids (e.g., those of B. aphidicola [10] and Candidatus Riesia pediculicola, the bacterial symbiont of the human body louse [41]). In the Buchnera genomes of many aphid species, genes for the biosynthesis of the essential amino acids leucine and tryptophan are located on circular plasmids [79, 91, 92]. The leucine plasmid, pLeu, encodes four genes for leucine biosynthesis (LeuABCD), while the tryptophan plasmid encodes anthranilate synthase (EC:4.1.3.27; trpEG), the first as well as the rate-limiting enzyme in the tryptophan biosynthetic pathway. The plasmid localization of these amino acid biosynthesis genes was originally interpreted as a mechanism by which Buchnera could overproduce leucine and tryptophan, as not only are plasmids typically found at high copy number in bacterial cells, the plasmids themselves sometimes also contain multiple tandem operon duplications [4]. However, as reviewed by Moran et al. [4], this interpretation is complicated by more-recent observations including the discovery that Buchnera cells contain many chromosomal copies [93], that the ratio of plasmid copy number to chromosome copy number is often much less than one [4] that at least some of the tandem copies of TrpEG are usually psuedogenized [10] and finally, that, in the case of tryptophan, amino acid production may not be correlated with the degree of trpEG amplification [94]. Thus, Moran et al. [4] conclude that the possible engagement of plasmids in regulation of amino acid production is complicated and requires further investigation.
Experimental evidence indicates that host aphids play the dominant role in symbiotic regulation
In taking a step back from the past 40+ years of research on the aphid/Buchnera symbiosis, the image that is beginning to emerge is one in which the host and not the symbiont plays the dominant role in regulating the symbiosis. Indeed, recent in silico analyses paint a picture that is consistent with this image. Thomas et al. [95] reconstructed the Buchnera APS metabolic network of 196 genes, 140 compounds, and 263 reactions, and compared it to that of E. coli. In doing so, they found that changes in carbon and nitrogen input impact essential amino acid production, even in the absence of Buchnera APS transcriptional regulation, suggesting that host aphids by modification of their feeding behavior and regulatory manipulation of supply of carbon and nitrogen to Buchnera can regulate the symbiosis and ultimately, among other metabolites, the supply of essential amino acids. Thus, informed by recent genomic and transcriptomic studies, it is clear that elucidation of the mechanisms by which the symbiosis is regulated needs to focus on the aphid host’s ability to behaviorally, transcriptionally, post-transcriptionally, and translationally, regulate the supply of Buchnera metabolic precursors.
Summary
The genetic complementarity of amino acid biosynthesis genes clearly demonstrates that the A. pisum/Buchnera metabolic collaboration operates at a genomic level. In this way, the genomes of both partners have been shaped by the symbiosis.
The genome of Buchnera has experienced massive gene losses through its symbiosis with aphids and while Buchnera lacks some apparently essential genes, such as those for phospholipid biosynthesis, it retains genes for the biosynthesis of nutrients required by the host.
The A. pisum immune system is reduced. This unusual loss may account for the evolutionary success of aphids in acquiring Buchnera.
There is no lateral gene transfer from Buchnera to the aphid genome.
There are functional lateral gene transfers from non-Buchnera bacteria to the aphid genome. Some of these bacterial laterally acquired genes are highly expressed in bacteriocytes, suggesting important roles in the aphid/Buchnera symbiosis.
Likely because of the almost complete absence of regulatory genes in the Buchnera genome, the transcriptional dynamics of Buchnera is static.
The host appears to play the dominant role in regulating the symbiosis.
Concluding remarks
The most pressing unresolved questions in studies of symbiosis concern understanding how the symbiotic partners are integrated to generate the holosymbiont. The full genome sequences of Buchnera APS and A. pisum clearly demonstrate realization of this mutualism at the genomic level, yet our current knowledge of the genetic and mechanistic basis of holosymbiotic integration is truly limited. For example, parallel analysis of the full genome sequences of both partners resulted in the inference of metabolic integration in amino acid biosynthesis, yet it remains unclear how the metabolites and intermediates are exchanged across the aphid host’s symbiosomal membrane and the symbiont’s inner and outer bacterial membranes. In other words, the cellular and sub-cellular integration of the symbiosis remain to be characterized. Another class of integration that is somewhat outside the scope of this review, but one that is in need of focused research efforts, concerns the 150-MY-old vertical transmission of symbiont to host that we term developmental integration. In this context it is known that transovarial bacterial infection and host oogenesis or embryogenesis are well coordinated [2, 25, 84] but the underlying molecular mechanisms regulating developmental integration remain elusive. Future research efforts focused on understanding how the symbiotic partners are integrated to generate the holosymbiont will be facilitated by the International Aphid Genomics Consortium’s focused efforts to continue development of community resources in four key areas: (1) improvement of the current pea aphid genome annotation; (2) the sequencing of new aphid genomes (which will facilitate examination of the ways in which holosymbiotic integration has been achieved in a range of aphid taxa); (3) the development of the AphidAtlas, a gene atlas for the aphid that includes generation of a controlled anatomical vocabulary and deep characterization of transcripts and proteins; and, (4) the development of bioinformatic analyses, tools, and services [96].
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
Support for SS came from KAKENHI Grant-in-Aid for Young Scientists A (#22687018). ACCW thanks Angela Douglas, Nancy Moran, and members of the Wilson Lab, especially Dan Price and Philip Kushlan, for useful discussions. Figure 1 was prepared by DRG Price.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Note added in proof
Our literature survey ended in October 2010. During proof editing an important paper of relevance to this review was published. Hansen and Moran (2011) performed a deep-sequencing transcriptome analysis of the pea aphid bacteriocyte revealing a finer view of aphid/Buchnera symbiotic metabolism. Most significantly, they annotated a previously unidentified A. pisum gene, glutamate synthase (GltS). GltS, along with glutamine synthase (GS), is highly upregulated in bacteriocyte tissue. Importantly, GltS and GS are potentially working together in the bacteriocyte recycling waste nitrogen from ammonia via the GOGAT cycle.
Reference
Hansen AK, Moran NA (2011) Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc Natl Acad Sci USA 108(7):2849–2854
References
- 1.Ishikawa H (2003) Insect symbiosis: an introduction. In: Bourtzis K, Miller TA (ed) Insect symbiosis. CRC Press, Boca Raton
- 2.Buchner P. Endosymbiosis of animals with plant microorganisms. New York: Interscience; 1965. [Google Scholar]
- 3.Moran NA. Symbiosis as an adaptive process and source of phenotypic complexity. Proc Natl Acad Sci USA. 2007;104(Suppl 1):8627–8633. doi: 10.1073/pnas.0611659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moran NA, Plague GR, Sandström JP, Wilcox JL. A genomic perspective on nutrient provisioning by bacterial symbionts of insects. Proc Natl Acad Sci USA. 2003;100(2):14543–14548. doi: 10.1073/pnas.2135345100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumann P, Baumann L, Lai C-Y, Rouhbakhsh D, Moran NA, et al. Genetics, physiology, and evolutionary relationships of the genus Buchnera: intracellular symbionts of aphids. Annu Rev Microbiol. 1995;49:55–94. doi: 10.1146/annurev.mi.49.100195.000415. [DOI] [PubMed] [Google Scholar]
- 6.Douglas AE, Minto LB, Wilkinson TL. Quantifying nutrient production by the microbial symbionts in an aphid. J Exp Biol. 2001;204:349–358. doi: 10.1242/jeb.204.2.349. [DOI] [PubMed] [Google Scholar]
- 7.Douglas AE, Prosser WA. Synthesis of the essential amino acid tryptophan in the peas aphids (Acyrthosiphon pisum) symbiosis. J Insect Physiol. 1992;38:565–568. doi: 10.1016/0022-1910(92)90107-O. [DOI] [Google Scholar]
- 8.Gunduz EA, Douglas AE. Symbiotic bacteria enable insect to use a nutritionally inadequate diet. Proc R Soc B-Biol Sci. 2009;276:987–991. doi: 10.1098/rspb.2008.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.International Aphid Genomics Consortium Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010;8:e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 11.Wilson ACC, Ashton PD, Calevro F, Charles H, Colella S, et al. Genomic insight into the amino acid relations of the pea aphid Acyrthosiphon pisum with its symbiotic bacterium Buchnera aphidicola . Insect Mol Biol. 2010;19:249–258. doi: 10.1111/j.1365-2583.2009.00942.x. [DOI] [PubMed] [Google Scholar]
- 12.Douglas AE. Requirement of pea aphids (Acyrthosiphon pisum) for their symbiotic bacteria. Entomol Exp Appl. 1992;65:195–198. [Google Scholar]
- 13.Dunbar HE, Wilson ACC, Ferguson NR, Moran NA. Aphid thermal tolerance is governed by a point mutation in bacterial symbionts. PLoS Biol. 2007;5:1006–1015. doi: 10.1371/journal.pbio.0050096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mittler TE. Some effects on the aphid Myzus persicae of ingesting antibiotic incorporated into artificial diets. J Insect Physiol. 1971;17:1333–1347. doi: 10.1016/0022-1910(71)90198-3. [DOI] [Google Scholar]
- 15.Montllor CB, Maxmen A, Purcell AH. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol Entomol. 2002;27:189–195. doi: 10.1046/j.1365-2311.2002.00393.x. [DOI] [Google Scholar]
- 16.Srivastava PN, Auclair JL. Effects of antibiotics on feeding and development of pea aphid, Acyrthosiphon pisum (Harris) (Homoptera-Aphididae) Can J Zool-Revue Canadienne De Zoologie. 1976;54:1025–1029. doi: 10.1139/z76-115. [DOI] [Google Scholar]
- 17.Degnan PH, Leonardo TE, Cass BN, Hurwitz B, Stern D, et al. Dynamics of genome evolution in facultative symbionts of aphids. Environ Microbiol. 2010;12:2060–2069. doi: 10.1111/j.1462-2920.2009.02085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pontes M, Smith K et al (2009) Insect facultative symbionts: biology, culture, and genetic modification. Insect symbiosis vol. 3. CRC Press, Boca Raton
- 19.Brinza L, Vinuelas J, Cottret L, Calevro F, Rahbe Y, et al. Systemic analysis of the symbiotic function of Buchnera aphidicola, the primary endosymbiont of the pea aphid Acyrthosiphon pisum . C R Biol. 2009;332:1034–1049. doi: 10.1016/j.crvi.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Douglas AE. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera . Annu Rev Entomol. 1998;43:17–37. doi: 10.1146/annurev.ento.43.1.17. [DOI] [PubMed] [Google Scholar]
- 21.Moran NA, Degnan PH. Functional genomics of Buchnera and the ecology of aphid hosts. Mol Ecol. 2006;15:1251–1261. doi: 10.1111/j.1365-294X.2005.02744.x. [DOI] [PubMed] [Google Scholar]
- 22.Moran NA, Munson MA, Baumann P, Ishikawa H. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc R Soc Lond Ser B. 1993;253:167–171. doi: 10.1098/rspb.1993.0098. [DOI] [Google Scholar]
- 23.Munson MA, Baumann P, Clark MA, Baumann L, Moran NA, et al. Evidence for the establishment of aphid-eubacterium endosymbiosis in an ancestor of four aphid families. J Bacteriol. 1991;173:6321–6324. doi: 10.1128/jb.173.20.6321-6324.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munson MA, Baumann P, Kinsey MG. Buchnera gen. nov. and Buchenera aphidicola sp. nov., a taxon consisting of the mycetocyte-associated, primary endosymbionts of aphids. Int J Syst Bacteriol. 1991;41:566–568. doi: 10.1099/00207713-41-4-566. [DOI] [Google Scholar]
- 25.Miura T, Braendle C, Shingleton AW, Sisk G, Kambhampati S, et al. A comparison of parthenogenetic and sexual embryogenesis of the pea aphid Acyrthosiphon pisum (Hemiptera: Aphidoidea) J Exp Zool B Mol Dev Evol. 2003;295:59–81. doi: 10.1002/jez.b.3. [DOI] [PubMed] [Google Scholar]
- 26.Huxley TH. On the agamic reproduction and morphology of aphids. Trans Linn Soc Lond. 1858;22:193–236. doi: 10.1111/j.1096-3642.1856.tb00092.x. [DOI] [Google Scholar]
- 27.Pierantoni U. Origine e struttura del corpo ovale del Dactylopius citri e del corpo verde dell’Aphis brassicae. Boll Soc Nat Napoli. 1910;24:1–43. [Google Scholar]
- 28.Sulc K. “Pseudovitellus” und ähnliche Gewebe der Homopteren sind Wohnstatten symbiontischer Saccharomyceten. Sitzber Bohm Ges Wiss. 1910;25:108–134. [Google Scholar]
- 29.Sapp J. Paul Buchner (1886–1978) and hereditary symbiosis in insects. Int Microbiol. 2002;5:145–150. doi: 10.1007/s10123-002-0079-7. [DOI] [PubMed] [Google Scholar]
- 30.Buchner P. Endosymbiose der Tiere mit Pflanzlichen Mikroorganismen. Basel: Birkhäuser; 1953. [Google Scholar]
- 31.Houk EJ, Griffiths GW. Intracellular symbiotes of the Homoptera. Annu Rev Entomol. 1980;25:161–187. doi: 10.1146/annurev.en.25.010180.001113. [DOI] [Google Scholar]
- 32.McLean DL, Houk EJ. Phase contrast and electron microscopy of the mycetocytes and symbiotes of the pea aphid, Acyrthosiphon pisum . J Insect Physiol. 1973;19(625–629):631–633. [Google Scholar]
- 33.Houk EJ, Griffiths GW, Hadjokas NE, Beck SD. Peptidoglycan in the cell wall of the primary intracellular symbiote of the pea aphid. Science. 1977;198:401–403. doi: 10.1126/science.198.4315.401. [DOI] [PubMed] [Google Scholar]
- 34.Mittler TE. Dietary amino acid requirements of the aphid Myzus persicae affected by antibiotic uptake. J Nutr. 1971;101:1023–1028. doi: 10.1093/jn/101.8.1023. [DOI] [PubMed] [Google Scholar]
- 35.Dadd RH, Krieger DL. Dietary amino acid requirements of aphid Myzus persicae . J Insect Physiol. 1968;14:741–764. doi: 10.1016/0022-1910(68)90186-8. [DOI] [Google Scholar]
- 36.Strong FE, Sakamoto SS. Some amino acid requirements of the green peach aphid, Myzus persicae (Sulzer), determined with glucose-U-C-14. J Insect Physiol. 1963;9:875–879. doi: 10.1016/0022-1910(63)90046-5. [DOI] [Google Scholar]
- 37.Pérez-Brocal V, Gil R, Ramos S, Lamelas A, Postigo M, et al. A small microbial genome: the end of a long symbiotic relationship? Science. 2006;314:312–313. doi: 10.1126/science.1130441. [DOI] [PubMed] [Google Scholar]
- 38.Tamas I, Klasson L, Canbäck B, Näslund AK, Eriksson A-S, et al. 50 million years of genomic stasis in endosymbiotic bacteria. Science. 2002;296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- 39.van Ham RCHJ, Kamerbeek J, Palacios C, Rausell C, Abascal F, et al. Reductive genome evolution in Buchnera aphidicola . Proc Natl Acad Sci USA. 2003;100:581–586. doi: 10.1073/pnas.0235981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sasaki-Fukatsu K, Koga R, Nikoh N, Yoshizawa K, Kasai S, et al. Symbiotic bacteria associated with stomach discs of human lice. Appl Environ Microbiol. 2006;72:7349–7352. doi: 10.1128/AEM.01429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirkness EF, Haas BJ, Sun W, Braig HR, Perotti MA, et al. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc Natl Acad Sci. 2010;107:12168–12173. doi: 10.1073/pnas.1003379107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maniloff J. The minimal cell genome: “on being the right size”. Proc Natl Acad Sci USA. 1996;93:10004–10006. doi: 10.1073/pnas.93.19.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 44.Moya A, Pereto J, Gil R, Latorre A. Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat Rev Genet. 2008;9:218–229. doi: 10.1038/nrg2319. [DOI] [PubMed] [Google Scholar]
- 45.Toft C, Andersson SGE. Evolutionary microbial genomics: insights into bacterial host adaptation. Nature Reviews Genetics. 2010;11:465–475. doi: 10.1038/nrg2798. [DOI] [PubMed] [Google Scholar]
- 46.Moran NA, McLaughlin HJ, Sorek R. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science. 2009;323:379–382. doi: 10.1126/science.1167140. [DOI] [PubMed] [Google Scholar]
- 47.Huerta-Cepas J, Dopazo H, Dopazo J, Gabaldón T. The human phylome. Genome Biol. 2007;8:R109. doi: 10.1186/gb-2007-8-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gerardo NM, Altincicek B, Anselme C, Atamian H, Barribeau SM, et al. Immunity and other defenses in pea aphids, Acyrthosiphon pisum . Genome Biol. 2010;11:R21. doi: 10.1186/gb-2010-11-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dyall SD, Brown MT, Johnson PJ. Ancient invasions: from endosymbionts to organelles. Science. 2004;304:253–257. doi: 10.1126/science.1094884. [DOI] [PubMed] [Google Scholar]
- 50.Douglas AE. Sulphate utilization in an aphid symbiosis. Insect Biochem. 1988;18(6):599–605. [Google Scholar]
- 51.Sunnucks P, Hales DF. Numerous transposed sequences of mitochondrial cytochrome oxidase I-II in aphids of the genus Sitobion (Hemiptera: Aphididae) Mol Biol Evol. 1996;13:510–523. doi: 10.1093/oxfordjournals.molbev.a025612. [DOI] [PubMed] [Google Scholar]
- 52.Nikoh N, McCutcheon JP, Kudo T, Miyagishima S-y, Moran NA, et al. Bacterial genes in the aphid genome: absence of functional gene transfer from Buchnera to its host. PLoS Genet. 2010;6:e1000827. doi: 10.1371/journal.pgen.1000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moran NA, Jarvik T. Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science. 2010;328:624–627. doi: 10.1126/science.1187113. [DOI] [PubMed] [Google Scholar]
- 54.Nakabachi A, Shigenobu S, Sakazume N, Shiraki T, Hayashizaki Y, et al. Transcriptome analysis of the aphids bacteriocyte the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera . Proc Natl Acad Sci USA. 2005;102:5477–5482. doi: 10.1073/pnas.0409034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nikoh N, Nakabachi A. Aphids acquired symbiotic genes via lateral gene transfer. BMC Biol. 2009;7:12. doi: 10.1186/1741-7007-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Griffiths GW, Beck SD. Ultrastructure of pea aphid mycetocytes: evidence for symbiote secretion. Cell Tissue Res. 1975;159:351–367. doi: 10.1007/BF00221782. [DOI] [PubMed] [Google Scholar]
- 57.Altincicek B, Gross J, Vilcinskas A. Wounding-mediated gene expression and accelerated viviparous reproduction of the pea aphid Acyrthosiphon pisum . Insect Mol Biol. 2008;17:711–716. doi: 10.1111/j.1365-2583.2008.00835.x. [DOI] [PubMed] [Google Scholar]
- 58.Dinant S, Bonnemain JL, Girousse C, Kehr J. Phloem sap intricacy and interplay with aphid feeding. C R Biol. 2010;333:504–515. doi: 10.1016/j.crvi.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 59.Winter H, Lohaus G, Heldt HW. Phloem transport of amino-acids in relation to their cytosolic levels in barley leaves. Plant Physiol. 1992;99:996–1004. doi: 10.1104/pp.99.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Corbesier L, Havelange A, Lejeune P, Bernier G, Perilleux C. N content of phloem and xylem exudates during the transition to flowering in Sinapis alba and Arabidopsis thaliana . Plant Cell Environ. 2001;24:367–375. doi: 10.1046/j.1365-3040.2001.00683.x. [DOI] [Google Scholar]
- 61.Girousse C, Bonnemain JL, Delrot S, Bournoville R. Sugar and amino-acid-composition of phloem sap of Medicago sativa—a comparative-study of 2 collecting methods. Plant Physiol Biochem. 1991;29:41–48. [Google Scholar]
- 62.Sandström J, Pettersson J. Amino acid composition of phloem sap and the relation to intraspecific variation in pea aphid (Acyrthosiphon pisum) performance. J Insect Physiol. 1994;40:947–955. doi: 10.1016/0022-1910(94)90133-3. [DOI] [Google Scholar]
- 63.Sasaki T, Hayashi H, Ishikawa H. Growth and reproduction of the symbiotic and aposymbiotic pea aphids, Acyrthosiphon pisum maintained on artificial diets. J Insect Physiol. 1991;37:749–756. doi: 10.1016/0022-1910(91)90109-D. [DOI] [Google Scholar]
- 64.Sasaki T, Ishikawa H. Production of essential amino-acids from glutamate by mycetocyte symbionts of the pea aphid, Acyrthosiphon pisum . J Insect Physiol. 1995;41:41–46. doi: 10.1016/0022-1910(94)00080-Z. [DOI] [Google Scholar]
- 65.Whitehead LF, Douglas AE. A metabolic study of Buchnera, the intracellular bacterial symbionts of the pea aphid Acyrthosiphon pisum . J Gen Microbiol. 1993;139:821–826. [Google Scholar]
- 66.Baumann L, Baumann P. Growth Kinetics of the Endosymbiont Buchnera aphidicola in the Aphid Schizaphis graminum . Appl Environ Microbiol. 1994;60:3440–3443. doi: 10.1128/aem.60.9.3440-3443.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Whitehead L, Douglas A. Populations of symbiotic bacteria in the parthenogenetic pea aphid (Acyrthosiphon pisum) symbiosis. Proc R Soc Lond B Biol Sci. 1993;254:29–32. doi: 10.1098/rspb.1993.0122. [DOI] [Google Scholar]
- 68.Wilkinson TL, Douglas AE. Host cell allometry and regulation of the symbiosis between pea aphids, Acyrthosiphon pisum, and bacteria, Buchnera . J Insect Physiol. 1998;44:629–635. doi: 10.1016/S0022-1910(98)00030-4. [DOI] [PubMed] [Google Scholar]
- 69.Hongoh Y, Ishikawa H. Changes of mycetocyte symbiosis in response to flying behavior of alatiform aphid (Acyrthosiphon pisum) Zool Sci. 1994;11:731–735. [Google Scholar]
- 70.Nishikori K, Kubo T, Morioka M. Morph-dependent expression and subcellular localization of host serine carboxypeptidase in bacteriocytes of the pea aphid associated with degradation of the endosymbiotic bacterium Buchnera . Zool Sci. 2009;26:415–420. doi: 10.2108/zsj.26.415. [DOI] [PubMed] [Google Scholar]
- 71.Nishikori K, Morioka K, Kubo T, Morioka M. Age- and morph-dependent activation of the lysosomal system and Buchnera degradation in aphid endosymbiosis. J Insect Physiol. 2009;55:351–357. doi: 10.1016/j.jinsphys.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 72.Febvay G, Rahbe Y, Rynkiewicz M, Guillaud J, Bonnot G. Fate of dietary sucrose and neosynthesis of amino acids in the pea aphid, Acyrthosiphon pisum, reared on different diets. J Exp Biol. 1999;202:2639–2652. doi: 10.1242/jeb.202.19.2639. [DOI] [PubMed] [Google Scholar]
- 73.Serres MH, Goswami S, Riley M. GenProtEC: an updated and improved analysis of functions of Escherichia coli K-12 proteins. Nucleic Acids Res. 2004;32:D300–D302. doi: 10.1093/nar/gkh087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, et al. The minimal gene complement of Mycoplasma genitalium . Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 75.Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UC, et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 76.Moran NA, Dunbar HE, Wilcox JL. Regulation of transcription in a reduced bacterial genome: nutrient-provisioning genes of the obligate symbiont Buchnera aphidicola . J Bacteriol. 2005;187:4229–4237. doi: 10.1128/JB.187.12.4229-4237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reymond N, Calevro F, Viñuelas J, Morin N, Rahbé Y, et al. Different levels of transcriptional regulation due to trophic constraints in the reduced genome of Buchnera aphidicola APS. Appl Environ Microbiol. 2006;72:7760–7766. doi: 10.1128/AEM.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hoch JA. Two-component and phosphorelay signal transduction. Curr Opin Microbiol. 2000;3:165–170. doi: 10.1016/S1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 79.Lai CY, Baumann L, Baumann P. Amplification of trpEG: adaptation of Buchnera aphidicola to an endosymbiotic association with aphids. Proc Natl Acad Sci USA. 1994;91:3819–3823. doi: 10.1073/pnas.91.9.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shigenobu S, Bickel RD, Brisson JA, Butts T, Chang C-C, et al. Comprehensive survey of developmental genes in the pea aphid, Acyrthosiphon pisum: frequent lineage-specific duplications and losses of developmental genes. Insect Mol Biol. 2010;19:47–62. doi: 10.1111/j.1365-2583.2009.00944.x. [DOI] [PubMed] [Google Scholar]
- 81.Rider SD, Srinivasan DG, Hilgarth RS. Chromatin-remodelling proteins of the pea aphid, Acyrthosiphon pisum (Harris) Insect Mol Biol. 2010;19(Suppl 2):201–214. doi: 10.1111/j.1365-2583.2009.00972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jaubert-Possamai S, Rispe C, Tanguy S, Gordon K, Walsh T, et al. Expansion of the miRNA Pathway in the hemipteran insect Acyrthosiphon pisum . Mol Biol Evol. 2010;27:979–987. doi: 10.1093/molbev/msp256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Walsh TK, Brisson JA, Robertson HM, Gordon K, Jaubert-Possamai S, et al. A functional DNA methylation system in the pea aphid, Acyrthosiphon pisum . Insect Mol Biol. 2010;19(Suppl 2):215–228. doi: 10.1111/j.1365-2583.2009.00974.x. [DOI] [PubMed] [Google Scholar]
- 84.Braendle C, Miura T, Bickel R, Shingleton AW, Kambhampati S, et al. Developmental origin and evolution of bacteriocytes in the aphid-Buchnera symbiosis. PLoS Biol. 2003;1:E21. doi: 10.1371/journal.pbio.0000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bermingham J, Rabatel A, Calevro F, Viñuelas J, Febvay G, et al. Impact of host developmental age on the transcriptome of the symbiotic bacterium Buchnera aphidicola in the pea aphid (Acyrthosiphon pisum) Appl Environ Microbiol. 2009;75:7294–7297. doi: 10.1128/AEM.01472-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Viñuelas J, Calevro F, Remond D, Bernillon J, Rahbé Y, et al. Conservation of the links between gene transcription and chromosomal organization in the highly reduced genome of Buchnera aphidicola . BMC Genomics. 2007;8:143. doi: 10.1186/1471-2164-8-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wilcox JL, Dunbar HE, Wolfinger RD, Moran NA. Consequences of reductive evolution for gene expression in an obligate endosymbiont. Mol Microbiol. 2003;48:1491–1500. doi: 10.1046/j.1365-2958.2003.03522.x. [DOI] [PubMed] [Google Scholar]
- 88.Ishikawa H. Host symbiont interactions in the protein-synthesis in the pea aphid, Acyrthosiphon pisum . Insect Biochem. 1982;12:613–622. doi: 10.1016/0020-1790(82)90048-8. [DOI] [Google Scholar]
- 89.Sabater-Muñoz B, Legeai F, Rispe C, Bonhomme J, Dearden PK, et al. Large-scale gene discovery in the pea aphid Acyrthosiphon pisum (Hemiptera) Genome Biol. 2006;7:R21. doi: 10.1186/gb-2006-7-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Griffiths GW, Beck SD. Intracellular symbiotes of the pea aphid, Acyrthosiphon pisum . J Insect Physiol. 1973;19:75–84. doi: 10.1016/0022-1910(73)90223-0. [DOI] [Google Scholar]
- 91.Bracho AM, Martínez-Torres D, Moya A, Latorre A. Discovery and molecular characterization of a plasmid localized in Buchnera sp. bacterial endosymbiont of the aphid Rhopalosiphum padi . J Mol Evol. 1995;41:67–73. doi: 10.1007/BF00174042. [DOI] [PubMed] [Google Scholar]
- 92.Rouhbakhsh D, Lai CY, von Dohlen CD, Clark MA, Baumann L, et al. The tryptophan biosynthetic pathway of aphid endosymbionts (Buchnera): genetics and evolution of plasmid-associated anthranilate synthase (trpEG) within the aphididae. J Mol Evol. 1996;42:414–421. doi: 10.1007/BF02498635. [DOI] [PubMed] [Google Scholar]
- 93.Komaki K, Ishikawa H. Intracellular bacterial symbionts of aphids possess many genomic copies per bacterium. J Mol Evol. 1999;48:717–722. doi: 10.1007/PL00006516. [DOI] [PubMed] [Google Scholar]
- 94.Birkle LM, Minto LB, Douglas AE. Relating genotype and phenotype for tryptophan synthesis in an aphid-bacterial symbiosis. Physiol Entomol. 2002;27:302–306. doi: 10.1046/j.1365-3032.2002.00301.x. [DOI] [Google Scholar]
- 95.Thomas GH, Zucker J, Macdonald SJ, Sorokin A, Goryanin I, et al. A fragile metabolic network adapted for cooperation in the symbiotic bacterium Buchnera aphidicola . BMC Syst Biol. 2009;3:24. doi: 10.1186/1752-0509-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.International Aphid Genomics Consortium (2010) Aphid White Paper II: proposal to complete development of the aphid model. http://www.aphidbase.com/aphidbase/news/aphid_white_paper_ii
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.