Abstract
The protruding (P) domain of norovirus VP1 is responsible for immune recognition and host receptor interaction. Our previous studies have demonstrated that a modification of the ends of the P domain affects the conformation and/or function of the P protein. An expression of the P domain with or without the hinge, or with an additional cysteine at either ends of the P protein resulted in P dimmers and/or P particles. Here we report a new type of subviral particle, the small P particles, through a further modification, either an addition of the Flag-tag or a change of the arginine-cluster, at the C-terminus of the cysteine-containing P domain. Gel-filtration and cryo-EM studies showed that the small P particles are tetrahedrons formed by 6 P dimers or 12 P monomers that is half-size of the P particles. Fitting of the crystal structure of the P domain into the cryo-EM density map of the particle indicated similar conformations of the P dimers as those in P particles. The small P particles bind human HBGAs and are antigenically reactive similar to their parental VLPs and P particles. These data suggest that the C-terminus of the P domain is an important factor in the formation of the P particles. Further elucidation of the mechanism of these modifications in the P particle formation would be important in structure biology and morphogenesis of noroviruses. The small P particles may also be a useful alternative in study of norovirus-host interaction and vaccine development for noroviruses.
Introduction
Noroviruses are the most important viral pathogens causing epidemics acute gastroenteritis in all ages in both developing and developed countries (Green, Chanock, and Kapikian, 2001; Tan, Farkas, and Jiang, 2008; Tan and Jiang, 2007). They are small, round, and nonenveloped RNA viruses within the family Caliciviridae. The norovirus capsid is constituted by a single major structural protein, the capsid protein (VP1), although another minor structural protein (VP2) has also been proposed (Glass et al., 2000) that may affect the stability of the viral structures.
The crystal structure of a recombinant norovirus capsid displays an icosahedral symmetry containing 180 copies of VP1 that further organized into dimers and pentamers of dimer (Prasad et al., 1999). Each VP1 can be divided into two major domains, forming the interiors shell (S) and the exterior protrusion (P) of the capsid, linked by a short, flexible hinge. In vitro expression of full-length VP1 through recombinant baculoviruses in insect cells results in spontaneous assembly of empty virus-like particles (VLPs) that are structurally and antigenically indistinguishable from the authentic viruses. Since human noroviruses are uncultivable in the laboratory, VLPs have been used as an important reagent in diagnosis, study of virus-host interaction, immunology and vaccine development for noroviruses.
The P domain can be further divided into P1 and P2 subdomains, constituting the leg and the head of the protruding P dimer, respectively (Prasad et al., 1999). The P2 subdomain is involved in host interactions related to the human histo-blood group antigen (HBGA) receptors (Bu et al., 2008; Cao et al., 2007; Choi et al., 2008; Tan et al., 2003; Tan and Jiang, 2010; Tan et al., 2008d). Crystallography studies also revealed extensive intermolecular interactions between two monomers in a P dimer (Bu et al., 2008; Cao et al., 2007; Choi et al., 2008; Prasad et al., 1999), suggesting a high tendency of dimerization and further complex formation. The recently reported P particles of noroviruses may be a result of these intermolecular interactions (Tan et al., 2008a; Tan, Hegde, and Jiang, 2004; Tan et al., 2010; Tan and Jiang, 2005a; Tan and Jiang, 2005b; Tan and Jiang, 2007; Tan, Meller, and Jiang, 2006).
The cryo-EM study of norovirus P particle revealed an octahedral symmetry containing 24 P monomers or 12 P dimers (Tan et al., 2008a; Tan and Jiang, 2005b). Fitting of the crystal structures of the P domain into the cryo-EM density map of the P particle revealed similar orientation of the P dimer in the P particle as that in VLP, in which the P1 domains are inward forming the core of the P particle, while the P2 domains are outward forming the surface of the P particle (Tan et al., 2008a). The carbohydrate binding sites are exposed on the surface, consistent with the fact that P particle binds to HBGA receptors (Tan et al., 2008a; Tan and Jiang, 2005b). Similarly, free P dimer is also able to bind HBGAs, although the binding affinity is lower than that of the P particles and VLPs (Tan, Hegde, and Jiang, 2004; Tan and Jiang, 2005b).
We reported here the identification of another subviral P particle of noroviruses, the small P particle. It is formed after further modifications at the C-terminus of the P domain. This small P particle is only half size of the P particle, containing 6 P dimers, in a tetrahedral symmetry, but revealed similar antigenic and HBGA binding properties as that of the parental VLP and P particle and, thus, could be another alternative in norovirus research. Elucidation of this particle will add useful information in our understanding of the structure and morphogenesis of noroviruses.
Materials and methods
Production of P domain recombinant proteins
The previously made P particle expression construct of VA387 (GII.4) (mutant CNGRC-P) (Tan and Jiang, 2005b) containing the plasmid pGEX-4T-1 (GST-gene fusion system; GE Healthcare Life Sciences) was modified to generate four new constructs: 1) A flag tag (DYKDDDDK) was added to the C-terminus of the P domain (P-flag), 2) the three consecutive arginines [R-cluster, (Tan, Meller, and Jiang, 2006)] at the C-terminus of the P domain were modified into RPRPRP to avoid the multiple trypsin recognition sites (P-RP), 3) the R-cluster was replaced by three consecutive, positive charged histidines (P-HHH), and 4) the R-cluster and the last two residue (RRRAL) of the P domain were deleted (P-RRRΔ, Fig 1A). In all four constructs the short peptide (CNGRC) at the N-terminus was modified to CNGHC to avoid the trypsin recognition site (R•C). To further confirm role of the R-cluster in the P and small P particle formation, a 5th construct was made by shifting the position of the cystein-containing peptide (CDCRGDCFC) to the C-terminus of the P-RRRΔ mutant (P-RRRΔ-2, Fig 1A). The modified constructs were expressed in E. coli strain BL21 as described previously (Tan, Hegde, and Jiang, 2004; Tan and Jiang, 2005b; Tan, Meller, and Jiang, 2006). Briefly, expression was performed at room temperature (~25 °C) overnight following an induction with 0.25 mM isopropyl-β-D-thiogalactopyranoside (IPTG). Purification of the recombinant GST-P domain fusion protein was carried out using Glutathione Sepharose 4 Fast Flow (GE Healthcare Life Sciences). The P protein was released from GST by thrombin (GE Healthcare Life Sciences) cleavage at room temperature for 16 hours. Further purification of the P protein was performed through a gel filtration chromatography.
Fig 1.
Production and identification of the small P particles. (A) The expression constructs of the five recombinant P proteins were schematically illustrated with indications of the GST (Glutathione S-transferase), the P domain, the cystein-containing peptide (circled C), the flag tag (circled F), the C-terminal arginine cluster (RRR) and its modification forms, RPRPRP, HHH, and the deletion of the R-cluster (RRRΔ). The thrombin recognition site was also indicated. (B to H) Analyses of the C-terminal modified P protein mutants and their parental P particles by SDS PAGE (B and D to H), Western blot (B, right), gel filtrations (C to H). (B) The P-flag chimeric protein was separated on an SDS PAGE (left) and then analyzed by Western blot (right). Lanes 1, 3, 5, and 7 were the unmodified P particles as controls, while lanes 2, 4, 6, and 8 were the P-flag chimeric protein. Lanes 5 and 6 were detected by hyperimmune serum against norovirus VLP (VA387), whereas lanes 7 and 8 by M1 monoclonal antibody against flag tag. M was prestained protein marker (low range, Bio-Rad). (C) A gel filtration curve of the parental P particle shows a single major peak of the P particle and a minor peak of the P dimer. (D and H) Gel filtration curves of the P-flag (D), P-RP (E), P-HHH (F), P-RRRΔ (G) and PRRRΔ-2 (H) proteins show either two major peaks with similar retention (D and E, upper panels) or a single major peak of P dimer (F to H). SDS-PAGE analyses of the peak fractions were also showed either in the lower panels (D and E) or on the right side of the gel filtration curves (F to H). The gel filtration columns [Superdex 200, either HiLoad 16/60 (C to F) or 10/30 GL (G and H)] were calibrated by the Gel Filtration Calibration Kit and the defined wild type P particle (830 kDa) and P dimer (70kDa) of norovirus (VA387).
Gel filtration chromatography
was performed for further purification and analysis of the P proteins through a fast performance liquid chromatography (FPLC) system (GE Healthcare Life Sciences) as described previously (Tan, Hegde, and Jiang, 2004; Tan and Jiang, 2005b; Tan, Meller, and Jiang, 2006). Briefly, the affinity column purified P protein was loaded on a size exclusion column [Superdex 200 with either 120 ml (HiLoad 16/60, Prep Grade) or 24 ml (10/30 GL) bed volume, GE Healthcare Life Sciences] powered by an AKTA FPLC system (model 920, GE Healthcare Life Sciences). The column Superdex 200 has a separation range of 3 to 600-kDa globular proteins. The molecular weight (MW) of the eluted fractions was calibrated by Gel Filtration Calibration Kit containing proteins with MWs of 2,000 kDa (void), 669 kDa, 440 kDa, 232 kDa, 147 kDa, 67 kDa, 47 kDa. Alternatively, gel filtration column can be calibrated by the defined wild type P particle (830 kDa) and the P dimer (69 kDa) of VA387 (Tan, Hegde, and Jiang, 2004; Tan and Jiang, 2005b). The proteins of interested fractions were further analyzed by a SDS PAGE and a Western blot.
SDS PAGE and Western blot
Quality of the P protein was analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS PAGE) using a 10% gel and a standard electrophoresis procedure. The specific compositions of the P protein with flag tag were detected by a Western blot as described elsewhere (Tan, Hegde, and Jiang, 2004; Tan et al., 2004) using our home made hyperimmune serum (1:3333) against norovirus VLP (VA387) and the monoclonal antibody M1 (1:400) (Sigma Aldrich, St. Louis, MO) against the flag tag, respectively. Blotted membrane was blocked by 5% nonfat milk. Secondary antibody-HRP (horseradish peroxidase) conjugate (1:5,000) was used and the HRP was detected by ECL Eastern Blotting Detection Reagents (GE Healthcare, Buckinghamshire, England). The ECL signals were captured by Hyperfilm ECL (GE Healthcare, Buckinghamshire, England).
Electron microscopy (EM)
Gel filtration-purified small P particles were prepared for EM examination using 1% ammonium molybdate for a negative stain. Specimens were examined using an EM10 C2 microscope (Zeiss, Germany) at 80 kV at a magnification of × 80,000.
Electron cryo-microscopy (cryo-EM) imaging
Same procedure that previously used for P particle analysis (Tan et al., 2008b; Tan et al., 2010) was adapted here. Briefly, a liquots (3–4 µl) of the gel-filtration-purified small P particle solution were flash frozen onto Quantifoil grids in liquid ethane cooled by liquid N2. The sample grids were loaded into the microscope and low dose images (~20e/A2) were recorded on films using CM200 cryo-microscope with a field emission gun operating at 200 KV. The images were taken at nominal magnification of × 50,000 and in the defocus range of 2.0 to 4.0 µm. The micrographs were selected and digitized using a Nikon Super CoolScan 9000ED scanner at step size of 6.35 µm /pixel. The scanned images were binned resulting in the final sampling of the images at 2.49Å/pixel for further image processing and 3-D reconstruction.
Cryo-EM image processing and 3-D reconstruction
The images of the small P Particles were selected using EMAN’s boxer program (Ludtke, Baldwin, and Chiu, 1999). The selected images were manually filtered to exclude false positive. The EMAN’s ctfit program (Ludtke, Baldwin, and Chiu, 1999) was used to manually determine the contrast-transfer-function (CTF) parameters associated with the set of particle images originating from the same micrograph. 30788 images were chosen for processing after CTF phase-corrected. Initial model of the small P particles were created using EMAN’s startoct program (Ludtke, Baldwin, and Chiu, 1999). Then the EMAN’s refine program (Ludtke, Baldwin, and Chiu, 1999; Ludtke et al., 2001) was used to iteratively determine the center and orientation of the raw particles and reconstruct the 3-D maps from the 2-D images by the EMAN make3d program until convergence. Tetrahedron symmetry was imposed during reconstruction of the small P particles. The resolution of the reconstructions was measured by splitting the data set into two halves, even and odd, whose 3D maps were then generated and compared, indicating that the resolution resulted in 0.5 Fourier shell correlation (Heel, 1987).
Electron cryomicroscopic model evaluation and analysis
The crystal structures of noroviral P domains of VA387 (Cao et al., 2007) were fitted into the 3D structure of the small P particle using Chimera software (Pettersen E. F, 2004). Simple rigid body motion was considered to find the best matching of the x-ray structure to the 3D structure of small P particles. No steric crash was seen when the fitted dimer was duplicated symmetrically to produce tetrahedral structure.
Enzyme-linked Immunosorbent Assay (EIA)
was used to detect the flag tag on the P dimer and the small P particle without a denature treatment. P proteins at indicated concentrations were coated on a 96-well microtiter plate (Dynex Immulon; Dynatech, Franklin, MA) at 4°C overnight. After blocked by 5% nonfat milk, monoclonal antibody M1 (1:400) (Sigma Aldrich) against the flag tag was incubated with the coated proteins. The bound M1 antibody was detected by a secondary antibodies-HRP conjugate as described elsewhere (Huang et al., 2003; Tan et al., 2004).
HBGA binding assay
Saliva-based HBGA binding assays were performed basically as described elsewhere (Huang et al., 2003; Huang et al., 2005). Briefly, boiled saliva samples with defined HBGA phenotypes were diluted 1000x and coated on 96-well microtiter plates (Dynex Immulon; Dynatech, Franklin, MA). After blocked by 5% nonfat milk, VLPs, P particles, small P particles, and P dimers of VA387 (GII.4) were added to incubate with the saliva. The bound VLP/P particle/small P article/P dimer were detected using a rabbit anti-VA387 VLP antiserum (1:3,300), followed by an addition of the secondary HRP-conjugated goat anti-rabbit IgG (ICN, Aurora/OH).
Results
Production of P domains with C-terminal modifications
In an attempt to present a small foreign epitope by the norovirus P particle (Tan et al., 2010), a fusion protein of the P domain of VA387 (GII.4) with a flag tag (DYKDDDDK) at the C-terminus of the protein was made (Fig 1A). A cysteine-containing peptide (CNGHC) was also included at the N-terminus of the P protein to facilitate the P particle formation (Tan and Jiang, 2005b). The resulting chimeric protein (P-flag) had an expected size of ~35 kDa and migrated slightly slower than the unmodified P protein (Fig 1B, comparing lane 1 with lane 2, lane 3 with 4). The P-flag protein was produced in E. coli with a yield of 8–10 mg/liter bacterial culture at purity of 85–90% after affinity column purification. Western blot analysis using a hyperimmune serum against norovirus VLP (VA387, GII.4) and a monoclonal antibody M1 (Sigma Aldrich) against the flag peptide confirmed the presence of the flag-tag in the P protein (Fig 1B, right penal). In addition, in an attempt to delete the trypsin recognition sites in the arginine-cluster (Tan, Meller, and Jiang, 2006), the motif RRR at the C-terminus of the P domain was modified to RPRPRP (Fig 1A). The resulting P protein (P-RP) showed a similar size, yield and mobility on a SDS-PAGE as that of the unmodified P protein (Fig 1E, data not shown).
The modified P proteins form a new type of subviral particle
Gel filtration analyses of the P-flag and P-RP proteins revealed two major peaks at ~400 and ~70 kDa, respectively. SDS PAGE and Western blot analyses using antibodies specific to norovirus VLP and flag-tag confirmed the presence of the P-flag or P-RP proteins in the two peaks (Fig 1D and E, data not shown). The second peaks were clearly dimers because of their similar size (~70 kDa) to the unmodified P dimer [Fig 1C, D and E, (Tan, Hegde, and Jiang, 2004; Tan and Jiang, 2005b)]. The first peak at ~400 kDa apparently contains a protein complex in about a half size of the P particles (~830 kDa) (Fig 1, compare C with D and E), and therefore must represent a new type of subviral particle of norovirus.
Above results prompted us to further study the role of the C-terminus in the P domain complex formation. Three more P domain mutants with modifications at the R-cluster were made, one had a replacement of the R-cluster (RRR) by three histidines (P-HHH), another had a deletion of the whole R-cluster (P-RRRΔ), and the third one had a deletion of the R-cluster and the cystein-containing peptide (CDCRGDCFC) was fused to the C-terminus of the P protein (P-RRRΔ-2, Fig 1A). While high yield of P proteins (>15 mg/liter bacteria culture) were produced, these three P proteins formed neither P particles nor the 400 kDa complexes as shown in a gel filtration. Interestingly, a common minor peak at ~200 kDa was seen for all three P proteins in addition to the dimer peaks (Fig 1F to H). These results extended our previous conclusion on the requirement of the R-cluster in the P particle formation (Tan, Meller, and Jiang, 2006) to the 400 kDa P complex .
The small P particle is tetrahedral Symmetry
EM examination of the first peak of the P-flag and PRP proteins revealed particles with square and triangle shapes (Fig 2A and B). These particles with a diameter of 10–15 nm were significantly smaller than the P particles (Tan et al., 2008a; Tan and Jiang, 2005b) and were named small P particles. Similar structures were also observed by cryo-EM on frozen hydrated grids (Fig 2C). After processing of 30,788 two-D images (Fig 2D), the three-D structure of the small P particle was reconstructed to a resolution of ~7.6Å (Fig 3). The small P particle exhibits a tetrahedral symmetry with a diameter of ~14 nm and a center cavity. Each small particle is composed of 12 P monomers or 6 dimers that is a half size of the P particle (Tan and Jiang, 2005b). This observation is consistent with the gel filtration data (Fig 1D). The typical triangle and square orientations of the small P particles were showed in Fig 3A and E, respectively.
Fig 2.
Electron microscopy and cryo-electron microscopy of small P particles. (A and B) Micrographs of the small P particles by electron microscopy showed shapes of the small P particles of P-flag (A) and P-RP (B) mutants. The white square regions show the typical square and triangle shapes of the small P particle. (C and D) Cryo-EM image processing of the small P particle. (C) A micrograph of the small P particles of VA387 (P-flag) that was taken at 4.5 µm underfocus. The squares indicate the typical small P particle images for image processing and 3-D structure reconstruction. (D) 3-D model projections and the corresponding class averages of the small P particle images are shown in multiple views.
Fig 3.
The structures of the small P particles. (A to C) The 3-D structure of the small P particle (P-flag) viewed at the top (A), side (B), and 60° turn clockwise from the side view (C), showing a tetrahedral symmetry with a center cavity. (D) Fitting of the crystal structure (ribbon model) of VA387 P domain in the cryo-EM density map of the small P particle. (E and F) The top of a P dimer subunit (rectangle) of the small P particle was shown by two P2 subdomains (ribbon model, purple and blue) (E) and the rectangular region is enlarged in (F) with indications of the residues T344 and R345 (green) as location of the HBGA-binding site.
Fitting of the crystal structure of norovirus P domain of VA387 (GII.4) (Cao et al., 2007) into the cryo-EM density map of a small P particle (Fig 3 D and E) showed a similar orientation of the P dimers to that of VLPs and P particles. The P1 subdomain or the legs of the P dimer is located near the center cavity of the small P particle, whereas the P2 subdomain or the head of the P dimer is outward from the center constituting the outer surface of the small P particle. The HBGA-binding interfaces (Cao et al., 2007; Tan et al., 2008d) are located at the top of each P dimer subunit, corresponding to the outermost surface of the small P particle (Fig 3E and F), as shown by residue T344 and R345, two important amino acids at the central region of the carbohydrate binding sites of VA387 (Cao et al., 2007; Tan et al. 2008c) (Fig 3F).
The flag-tag of the small P particles is accessible to antibody
The small P particles were strongly reactive with the M1 antibody specific to the flag-tag in an EIA, similar to that observed for the P-flag dimers (Fig 4A). This result indicated that the C-terminally attached flag tag is well presented on the small P particles. The readily accessibility of the flag tag to antibody may provide a purification strategy of the small P particles through an affinity chromatography using flag-tag specific antibodies (M1 and M2), making the small P particle a potentially useful reagent for norovirus research.
Fig 4.
The immune reactivity of the small P particles to the flag tag antibody (A) and bindings of the P domain complexes and VLP to HBGA receptors (B to G). (A) The small P particle of the P-flag (small P P) reacted to M1 monoclonal antibody against flag tag in EIAs. The P dimer of P-flag (dimer) and the parental P particle without flag tag (CNGRC-P) served as positive and negative controls, respectively. (B to G) The small P particles of P-flag (B) and P-RP (C) bound to HBGAs, while the P-HHH mutant did not (D). The HBGA-bindings of the parental VLP (E), P particle (F), and P dimer (G) were shown for comparison. Y-axis indicates the signal intensity (OD450), whereas X-axis shows protein concentrations (ng/µl) used for the assays. The protein concentrations were selected to produce maximum and minimum binding activities. O, A, and B represent the saliva samples of types O, A and B secretor, while N represents the saliva of non-secretor. All data shown in this figure were averages of triplicate experiments.
The small P particles maintain similar antigenicity as their parental VLPs
Similar to the P particles (Tan et al., 2008a; Tan and Jiang, 2005b), the small P particles revealed strong reactivities with hyperimmune antibodies against their parental VLPs in a Western blot analysis (Fig 1B) and an antigen detecting Elisa (Fig 4 B and C). Thus, the small P particles maintain similar antigenicity as their parental P particles and VLPs.
The Small P particle binds to HBGA receptors
Saliva-based HBGA binding assays showed that the small P particles of both P-flag and P-RP were able to bind to HBGAs (Fig 4 B and C). Although the binding affinities were lower than that of VLPs and P particles (Fig 4E and F), they were significantly higher than that of the P dimers (Fig 4G). We also noted that bindings of all three P domain complexes (P particles, small P particles and P dimers) to saliva of type O secretor were clearly weaker compared with that of VLPs, suggesting minor structural or conformational differences at the carbohydrate binding sites between VLP and these P domain complexes. None of the three R-cluster deletion mutants (P-HHH, P-RRRΔ and P-RRRΔ-2) at the indicated concentration range revealed specific binding to HBGAs in the saliva binding assays (Fig 4D and data not shown), supporting the previous observation that the R-cluster is important for binding to HBGA receptors (Tan, Meller, and Jiang, 2006).
Discussion
In this study we reported the identification and characterization of a novel subviral particle, the small P particle of norovirus. It is formed by the P domains with a cysteine containing peptide at the N-terminus and a further modification at the C-terminus, either an addition of the flag tag or a modification of the arginine-cluster (RRR) into RPRPRP. The small P particle contains 12 P monomers that organize into 6 P dimers, which is a half size of the P particles. Each P dimer subunit has a similar orientation in the P and the small P particles, with the carbohydrate binding sites on the outermost surface of each particle. The small P particles bind the HBGAs similar to their parental P particles and VLPs. Since the small P particles are efficiently produced in E. coli, easily purified, they may be a valuable alternative for study of host-pathogen interaction, immunology, diagnosis, and vaccine development for noroviruses.
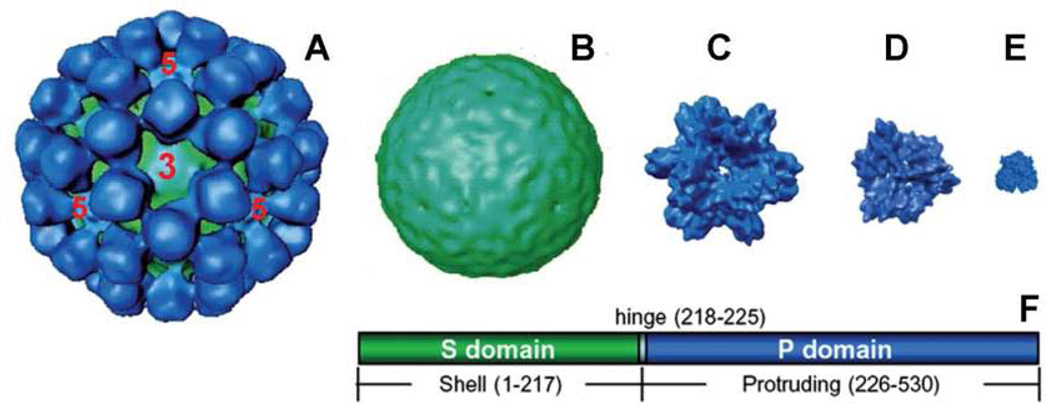
Different complexes of full-length and truncated norovirus VP1 have been reported, including VLPs formed by 180 full-length VP1s (Prasad et al., 1999), the small VLPs by 60 full-length VP1s (White, Hardy, and Estes, 1997), the thin-layer S particles by 180 S domains (Bertolotti-Ciarlet et al., 2002), the P particles by 24 P domains (Tan et al., 2008b; Tan and Jiang, 2005b), and free P dimers (Bu et al., 2008; Cao et al., 2007; Choi et al., 2008; Tan, Hegde, and Jiang, 2004). This report added the small P particle formed by 12 P monomers, making total of 6 different complexes in the list (Fig 5). All these complexes assemble spontaneously, indicating a high tendency of intermolecular interaction among individual VP1s and P domains. Some of these complexes, including the VLPs (equivalent to the authentic virion) and the soluble P dimers (Greenberg et al., 1981; Hardy et al., 1995; Tan, Meller, and Jiang, 2006), are apparently presented in vivo, while others, including the small VLP, the S, P and the small P particles, are possibly artificial, occurring only in the laboratories. Further studies of the structure-function relationships of these particles would be significant in structural biology and morphogenesis and pathogenesis of noroviruses.
Fig 5.
Structures (surface models) of the five well-defined complexes that formed by full-length or truncated norovirus VP1. (A) VLP (180mer, Φ=~37 nm); (B) S particle (180mer, Φ=~20 nm); (C) P particle (24mer, Φ=~20 nm); (D) small P particle (12mer, Φ=~14 nm); (E) P dimer (Φ=~6 nm); (F) the linear structure of norovirus VP1 with indications of shell (S, green) and protruding (P, blue) domains that are linked by a short hinge. Numbers are based on Norwalk virus VP1. The number 3 and 5 in (A) indicates the icosahedral three- and fivefold axes, respectively.
The full-size VLPs (Fig 5A) represent the native structure of norovirus capsid. The crystal structures of recombinant VLPs of the prototype Norwalk virus (Prasad et al., 1999) showed multiple intermolecular interactions in both the S and P domains of the VP1 that must be the major driving force of the dimerization and capsid formation. This also explains the fact that the P proteins assemble spontaneously into dimers and P particles (Bu et al., 2008; Cao et al., 2007; Choi et al., 2008; Tan, Hegde, and Jiang, 2004; Tan and Jiang, 2005b). Intermolecular contacts in the P domain have been observed among P1 subdomains of the five P dimers around the icosahedral fivefold axis and among the P2 subdomains of the six P dimers around the threefold axis (Fig 5A) (Hutson, Atmar, and Estes, 2004; Prasad et al., 1999). These interactions may be the major forces of the formation of the P particle and small P particles of noroviruses (Fig 5C and D).
While the P domain alone can form P particles, an addition of cystein residues at either end of the P domain stabilizes the P particle formation (Tan and Jiang, 2005b). In this study we demonstrated that a further modification at the C-terminus of the P domain, either through an addition of a flag tag (P-flag) or a change of the arginine-cluster (P-RP), lead to the formation of the small P particles. These modifications apparently did not affect the dimerization of the P domain because P dimers remained the basic units of these particles. The mechanism of different modifications resulting in variable P particles remains unknown, but these modifications are likely to affect the ways of the intermolecular interactions among the P dimers (Prasad et al., 1999). Since the octahedral P particle can assemble spontaneously by the P protein alone without extra modification (Tan and Jiang, 2005b), the inter-dimer interactions among the 12 P dimer subunits of the P particle may represent more natural interactions in native viral particles, which occur in the legs of the arch-like P dimers (Tan et al., 2008a). The addition of cysteine residues to the ends of P domain apparently stabilizes or strengthens these interactions (Tan et al., 2008a; Tan and Jiang, 2005b). In the case of the small P particle, the modifications at the C-terminus of the P domain may weaken some original, but create some new interactions, resulting in a different P particle, as a balance of the new interaction.
The arginine cluster has been shown to be important for P particle formation and HBGA binding of the P proteins (Tan, Meller, and Jiang, 2006). The recently solved crystal structure of the P dimer of murine norovirus 1 (MNV-1) has revealed a direct role of the C-terminus of the P domain in the inter-P dimer interaction (Taube et al., 2010), supporting the previous observations. In this study we further demonstrated that modifications at (P-RP) or near (P-flag) the R-cluster lead to the shift from the P particle to the small P particle. However, additional modifications of the R-clusters, for example, a replacement of the R-cluster (RRR) by a histidine cluster (P-HHH) did not result in P- or small P particle formation and the proteins did not bind HBGAs. This result suggests that the positive charge of the R-cluster may not be the major factor for P particle formation and HBGA binding. In addition, we found that the role of the R-cluster in the P particle formation cannot be replaced by a simple addition of cysteine residues to either end of the P protein (P-RRRΔ and P-RRRΔ-2). These data indicate a crucial role of the R-cluster in the P- and small P particle formation and HBGA-binding, which need to be defined in future.
The presence of the minor peak at ~200 kDa of the R-cluster mutants (P-HHH, P-RRRΔ and P-RRRΔ-2) raised a question whether these are new type of P complexes. The estimated molecular weight by gel filtration suggests that this putative complex contains 6 P monomers or 3 P dimers. According to the crystal structures of the Norwalk virus capsid, VP1 dimers, as well as pentamers and decamer of VP1 dimers were proposed as intermediates in the assembly pathway of norovirus capsids (Prasad et al., 1999). The observed ~200 kDa complexes (trimer of the P dimer) in this study could represent such intermediates or building blocks in pathway of the P- and small P particle assembly. In the capsid formation multiple interactions among S domains are the major contribution for the icosahedral capsid assembly. In the P- and small P particle formation the interactions among the P domains are less and simpler, resulting in much smaller particles of 6mer and 12mer of the P dimers. Further study on this new complex would be helpful in understanding the morphogenesis of the subviral particle.
The continual inability to cultivate noroviruses highlights the need of in vitro production of viral capsid protein as a surrogate for study of virus-host interaction and vaccine development. In addition to the full-size VLPs, we now have variable alternatives in production of subviral particles in different sizes made from the S and P domains, respectively. Among these subviral particles the P particle described in our previous studies and the small P particle reported here are particularly valuable because they can be easily produced in the E. coli system, which is cost effective compared with the VLPs that require a eukaryotic expression system. While the P particles may be more immunogenic because of the larger size, the small P particles may find unique applications such as penetrating to unique tissues or host cells because of the smaller size. Future characterization to explore the usefulness of these particles in variable fields such as virus/host interaction, immunology, morphogenesis, pathogenesis and vaccine development against noroviruses are necessary.
Acknowledgement
The research described in this article was supported by the National Institute of Health, the National Institute of Allergy and Infectious diseases (R01 AI37093 and R01 AI055649), and the Department of Defense (PR033018) to X.J. This study was also supported by an Institutional Clinical and Translational Science Award, NIH/NCRR Grant Number 1UL1RR026314-01 and the Infectious Disease Grant (Nipert Foundation) to M. T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bertolotti-Ciarlet A, White LJ, Chen R, Prasad BV, Estes MK. Structural requirements for the assembly of Norwalk virus-like particles. J Virol. 2002;76(8):4044–4055. doi: 10.1128/JVI.76.8.4044-4055.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu W, Mamedova A, Tan M, Xia M, Jiang X, Hegde RS. Structural basis for the receptor binding specificity of Norwalk virus. J Virol. 2008;82(11):5340–5347. doi: 10.1128/JVI.00135-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Lou Z, Tan M, Chen Y, Liu Y, Zhang Z, Zhang XC, Jiang X, Li X, Rao Z. Structural basis for the recognition of blood group trisaccharides by norovirus. J Virol. 2007;81(11):5949–5957. doi: 10.1128/JVI.00219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JM, Hutson AM, Estes MK, Prasad BV. Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus. Proc Natl Acad Sci U S A. 2008;105(27):9175–9180. doi: 10.1073/pnas.0803275105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass PJ, White LJ, Ball JM, Leparc-Goffart I, Hardy ME, Estes MK. Norwalk virus open reading frame 3 encodes a minor structural protein. J Virol. 2000;74(14):6581–6591. doi: 10.1128/jvi.74.14.6581-6591.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green K, Chanock R, Kapikian A. Human Calicivirus. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields Virology. 4th ed. Vol. 2. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 841–874. 2 vols. [Google Scholar]
- Greenberg HB, Valdesuso JR, Kalica AR, Wyatt RG, McAuliffe VJ, Kapikian AZ, Chanock RM. Proteins of Norwalk virus. J Virol. 1981;37(3):994–999. doi: 10.1128/jvi.37.3.994-999.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy ME, White LJ, Ball JM, Estes MK. Specific proteolytic cleavage of recombinant Norwalk virus capsid protein. J Virol. 1995;69(3):1693–1698. doi: 10.1128/jvi.69.3.1693-1698.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heel MV. Similarity measures between images. Ultramicroscopy. 1987;21:95–100. [Google Scholar]
- Huang P, Farkas T, Marionneau S, Zhong W, Ruvoen-Clouet N, Morrow AL, Altaye M, Pickering LK, Newburg DS, LePendu J, Jiang X. Noroviruses bind to human ABO, Lewis, and secretor histo-blood group antigens: identification of 4 distinct strain-specific patterns. J Infect Dis. 2003;188(1):19–31. doi: 10.1086/375742. [DOI] [PubMed] [Google Scholar]
- Huang P, Farkas T, Zhong W, Tan M, Thornton S, Morrow AL, Jiang X. Norovirus and histo-blood group antigens: demonstration of a wide spectrum of strain specificities and classification of two major binding groups among multiple binding patterns. J Virol. 2005;79(11):6714–6722. doi: 10.1128/JVI.79.11.6714-6722.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson AM, Atmar RL, Estes MK. Norovirus disease: changing epidemiology and host susceptibility factors. Trends Microbiol. 2004;12(6):279–287. doi: 10.1016/j.tim.2004.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128(1):82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Jakana J, Song JL, Chuang DT, Chiu W. A 11.5 A single particle reconstruction of GroEL using EMAN. J Mol Biol. 2001;314(2):253–262. doi: 10.1006/jmbi.2001.5133. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, G. T. D, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Prasad BV, Hardy ME, Dokland T, Bella J, Rossmann MG, Estes MK. X-ray crystallographic structure of the Norwalk virus capsid. Science. 1999;286(5438):287–290. doi: 10.1126/science.286.5438.287. [DOI] [PubMed] [Google Scholar]
- Tan M, Fang P, Chachiyo T, Xia M, Huang P, Fang Z, Jiang W, Jiang X. Noroviral P particle: Structure, function and applications in virus-host interaction. Virology. 2008a;382:115–123. doi: 10.1016/j.virol.2008.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Fang P, Chachiyo T, Xia M, Huang P, Fang Z, Jiang W, Jiang X. Noroviral P particle: structure, function and applications in virus-host interaction. Virology. 2008b;382(1):115–123. doi: 10.1016/j.virol.2008.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Farkas T, Jiang X. Molecular Pathogenesis of Human Norovirus. In: Yang DC, editor. Host Gene Responses to RNA Virus Infection. 1 ed. Singapore: World Scientific; 2008. in press. [Google Scholar]
- Tan M, Hegde RS, Jiang X. The P domain of norovirus capsid protein forms dimer and binds to histo-blood group antigen receptors. J Virol. 2004;78(12):6233–6242. doi: 10.1128/JVI.78.12.6233-6242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Huang P, Meller J, Zhong W, Farkas T, Jiang X. Mutations within the P2 domain of norovirus capsid affect binding to human histo-blood group antigens: evidence for a binding pocket. J Virol. 2003;77(23):12562–12571. doi: 10.1128/JVI.77.23.12562-12571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Huang P, Xia M, Fang P, Zhong W, Wei C, Jiang W, Jiang X. Norovirus P particle, a multifunctional vaccine platform. J Virol. 2010;84 doi: 10.1128/JVI.01835-10. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Jiang X. Norovirus and its histo-blood group antigen receptors: an answer to a historical puzzle. Trends Microbiol. 2005a;13(6):285–293. doi: 10.1016/j.tim.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Tan M, Jiang X. The p domain of norovirus capsid protein forms a subviral particle that binds to histo-blood group antigen receptors. J Virol. 2005b;79(22):14017–14030. doi: 10.1128/JVI.79.22.14017-14030.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Jiang X. Norovirus-host interaction: implications for disease control and prevention. Expert Rev Mol Med. 2007;9(19):1–22. doi: 10.1017/S1462399407000348. [DOI] [PubMed] [Google Scholar]
- Tan M, Jiang X. Norovirus gastroenteritis, carbohydrate receptors, and animal models. PLoS pathogens. 2010;6(8):e1000983. doi: 10.1371/journal.ppat.1000983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Meller J, Jiang X. C-terminal arginine cluster is essential for receptor binding of norovirus capsid protein. J Virol. 2006;80(15):7322–7331. doi: 10.1128/JVI.00233-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Xia M, Cao S, Huang P, Farkas T, Meller J, Hegde RS, Li X, Rao Z, Jiang X. Elucidation of strain-specific interaction of a GII-4 norovirus with HBGA receptors by site-directed mutagenesis study. Virology. 2008c;379:324–334. doi: 10.1016/j.virol.2008.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Xia M, Cao S, Huang P, Meller J, Hegde R, Li X, Rao Z, Jiang X. Elucidation of strain-specific interaction of a GII-4 norovirus with HBGA receptors by site-directed mutagenesis study. Virology. 2008d doi: 10.1016/j.virol.2008.06.041. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Zhong W, Song D, Thornton S, Jiang X. E. coli-expressed recombinant norovirus capsid proteins maintain authentic antigenicity and receptor binding capability. J Med Virol. 2004;74(4):641–649. doi: 10.1002/jmv.20228. [DOI] [PubMed] [Google Scholar]
- Taube S, Rubin JR, Katpally U, Smith TJ, Kendall A, Stuckey JA, Wobus CE. High-resolution x-ray structure and functional analysis of the murine norovirus 1 capsid protein protruding domain. J Virol. 2010;84(11):5695–5705. doi: 10.1128/JVI.00316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White LJ, Hardy ME, Estes MK. Biochemical characterization of a smaller form of recombinant Norwalk virus capsids assembled in insect cells. J Virol. 1997;71(10):8066–8072. doi: 10.1128/jvi.71.10.8066-8072.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]