Abstract
Purpose
Urine proteomics is emerging as a powerful tool for biomarker discovery. The purpose of this study is the development of a well characterized “real life” sample that can be used as reference standard in urine clinical proteomics studies.
Experimental design
We report on the generation of male and female urine samples that are extensively characterized by different platforms and methods (CE-MS, LC-MS, LC-MS/MS, GeLC-MS, and 2DE-MS) for their proteome and peptidome. In several cases analysis involved a definition of the actual biochemical entities, i.e. proteins/peptides associated with molecular mass and detected posttranslational modifications and the relative abundance of these compounds.
Results
The combination of different technologies allowed coverage of a wide mass range revealing the advantages and complementarities of the different technologies. Application of these samples in “inter-laboratory” and “inter-platform” data comparison is also demonstrated.
Conclusions and Clinical Relevance
These well characterized urine samples are freely available upon request to enable data comparison especially in the context of biomarker discovery and validation studies. It is also expected that they will provide the basis for the comprehensive characterization of the urinary proteome.
Keywords: clinical proteomics, proteome, standard, urine
Introduction
The usefulness of urine analysis in clinical proteomics has been advocated in several initial studies [1–3] and was demonstrated in several recent studies (reviewed in [4,5]). Urine has become a main target in clinical proteome analysis and is already applied in clinical settings [6].
The multi-parametric analysis enabling identification of valid biomarkers mandates the availability of datasets from several hundred to thousands of patients and controls, as observed in genomics [7]. Generally, the large datasets cannot be produced by a single laboratory. Hence, inter-laboratory comparison becomes essential, and the comparability of datasets will be the key success factor in clinical proteomics.
However, the current situation is far away from this need: comparability between laboratories and analytical platforms generally does not exist. The validation of platforms, if done at all, usually relies on the use of only a few highly purified proteins and synthetic peptides, providing a limited assessment of the technical variability of a platform. Importantly, such standards do not reflect the variability introduced during pre-analytical sample handling, associated with suppression effects, endogenous enzyme activity, adsorption etc. These factors are regularly encountered during analysis of complex samples.
Furthermore, the exact composition of the urinary proteome remains largely unknown. While extensive lists of proteins based on the detection of tryptic peptides have been presented [8,9], these usually lack measurements of abundance and precise information on the native peptides or proteins identified. For example, among several different fragments of collagen alpha-1(I) present in urine, some are disease-specific biomarkers, while others are not. However, most reports list the database hit, i.e. collagen alpha-1(I) chain precursor (a ~110 kDa pre-protein) that is in fact unlikely to be present as a full length protein in urine.
Thus, an essential step forward is the introduction of a well characterized urine sample that could be used as “standard”. This sample should (i) be representative of the currently identifiable normal urine proteome; (ii) be characterized in depth by different methods and platforms; (iii) enable to assess platform capability including pre-analytical steps, platform performance, and normalisation; (iv) enable comparison of datasets; and (v) be freely available to all laboratories working in the field of urinary proteomics.
Here, we report on the generation and extensive characterization of two reference “standard” samples, representing “normal” pooled urine samples from healthy females and males, respectively.
Materials and Methods
Urine samples
The two “standard” samples consisted of pooled midstream morning urine (collected between 8 and 10 am) from multiple collections from 7 male and 8 female healthy volunteers, respectively. There were no specific dietary or, in the case of females, menstrual cycle requirements (with the exception of absence of menstruation) for inclusion in the study. The use of pooled versus individual urine was favored due to the need for large volumes which renders the collection of the latter less practical and technically more demanding. All subjects in the study were healthy volunteers, only urine was collected and all samples were provided anonymous. All samples were collected in Germany, and under German law this study does not require IRB approval. Informed consent was obtained from participating individuals. The study complied with the guidelines of the Declaration of Helsinki (www.wma.net/en/30publications/10policies/b3/index.html). The collection in all cases followed the procedure that was used in several recent studies (e.g.; [10–12], full details on the collection protocol are also provided at www.eurokup.org: collection protocol for peptidomics analysis). This urine collection protocol is in agreement to a “standard protocol for urine collection” currently under development by the HUKPP and EuroKUP networks (for more information please visit: www.eurokup.org; www.hukpp.org). To ease later comparison with samples collected at other centres and in agreement with the recently reported suggestions for gel based urine proteomics [13,14], no protease inhibitors were added and the pH was not adjusted. In addition no phosphatase inhibitors were included. Demographical/clinical data on the volunteers are given in Table 1A. Collected samples (between 40 and 100 mL per collection) were frozen immediately at −20°C. Upon completion of collection, all frozen samples were thawed on ice, sonicated, combined (total volume > 2,500 mL per gender group), divided into several 1, 10, and 50 mL aliquots, and frozen again at −80°C. The urinary proteome is not affected significantly by up to 3 freeze/thaw cycles following initial freezing ([15] and unpublished data).
Table 1A.
Demographical data of subjects involved and concentration of clinically relevant analytes in the male and female urine standard. Upper part of the table in bold: Demographic characteristics include average age, body mass index (BMI), glomerular filtration rate (in ml/min, estimated based on the Cockroft-Gould), and diastolic and systolic blood pressure (in mmHg). Lower part of the table: protein/peptide identity (name or sequence), and average concentration (in ng/ml) and standard deviation are given. n.d.: not detectable. The urinary peptides (see sequences in italic type) could be quantified with use of external stable isotope-labelled peptide standards [11] Hydroxylated proline sites are annotated by a “p”. For further sequence information see Supplementary 'Detailed Tables').
| Analyte or Parameter | male | female |
|---|---|---|
| Age | 36 ± 6 | 28 ± 6 |
| BMI | 22.5 ± 1.1 | 22.4 ± 2.8 |
| GFR (CG) | 108 ± 6 | 104 ± 9 |
| Diastolic BP | 77 ± 4 | 71 ± 6 |
| Systolic BP | 126 ± 4 | 112 ± 7 |
| Creatinine | 879000 ± 26370 | 787000 ± 23610 |
| total protein | 14600 ± 365 | 18400 ± 460 |
| IgA | 557.7 ± 23.3 | 258.2 ± 14.1 |
| HAA-IgA | 0.224 ± 0.008 | 0.186 ± 0.012 |
| CD14 | 10.15 ± 0.36 | 16.37 ± 0.41 |
| Ngal | 2.20 ± 0.08 | 8.89 ± 0.36 |
| MGPRGPpGPpG | 1.82 ± 1.00 | 2.84 ± 1.42 |
| ApGDRGEpGPp | 23.86 ± 13.09 | 29.19 ± 12.34 |
| GDPGPPGpPGpPGpPAI | 18.01 ± 23.19 | 22.17 ± 13.11 |
| SpGSpGPDGKTGPPGpAG | 3.75 ± 1.4 | 6.29 ± 2.16 |
| PpGEAGKpGEQGVPGDLG | 0.79 ± 0.57 | 1.39 ± 1.02 |
| EAIPMSIPPEVKFNKPF | n.d. | n.d. |
| NGDDGEAGKpGRpGERGPpGP | 8.24 ± 6.09 | 21.47 ± 20.49 |
| DAGApGApGGKGDAGApGERGPpG | 23.01 ± 21.51 | 31.65 ± 16.65 |
| AGPpGEAGKpGEQGVpGDLGAPGP | 1.59 ± 0.93 | 3.28 ± 1.57 |
| AGPpGEAGKpGEQGVpGDLGApGP | 5.18 ± 2.89 | 12.45 ± 2.75 |
| ADGQpGAKGEpGDAGAKGDAGPpGPA | 8.70 ± 4.14 | 16.44 ± 7.32 |
| GKNGDDGEAGKpGRpGERGPpGPQ | 3.45 ± 2.99 | 11.09 ± 6.67 |
| TGPIGPpGPAGApGDKGESGPSGPAGPTG | 1.19 ± 1.12 | 3.01 ± 1.13 |
| PpGESGREGApGAEGSpGRDGSpGAKGDRGETGP | 56.04 ± 37.67 | 193.30 ± 87.88 |
| MIEQNTKSPLFMGKVVNPTQK | n.d. | n.d. |
2DE analysis: Processing, separation, and identification of urinary proteins
Fifty ml of urine sample was concentrated approximately 10-fold using the Millipore (Bedford, MA, USA) stir-cell apparatus with a PLBC Regenerated Cellulose membrane (3,000 Da cut-off) under nitrogen pressure (4.5 Bar). The protein content in the concentrated sample was determined to be 0.972 mg for the male sample and 1.242 mg for the female sample by the Bradford assay (Biorad, Hercules, CA, USA).
The concentrated urine samples were further subjected to protein precipitation using 7.5% TCA (trichloroacetic acid) and 0.1% NLS (N-lauroylsarcosine sodium salt [16]. Typical yields for this precipitation approach are 85–95% (Vlahou, unpublished data). Following an overnight incubation at −20°C, the samples were centrifuged at 10,000 g for 30 min. The supernatant was removed and the pellet washed twice with cold tetrahydrofuran (THF). The pellet was resuspended in 800 μL sample buffer (7 M urea, 2 M thiourea, CHAPS 4% w/v, DTE 1% w/v, 50 mM Tris pH 6.8), aliquoted in 150 μg aliquots and stored at −20°C.
Two-dimensional separation of urinary proteins was performed as described [17] with the following modifications: the samples were loaded on IEF strips pH 3–10 NL (18 cm; Biorad, Hercules, CA, USA). Four-hundred-fifty μg protein (for Coomassie staining) or 150 μg protein (for silver staining) was loaded onto the strips. Silver staining was performed according to Chevallet et al., Protocol B [18]. To confirm reproducibility of profiles, urinary proteins from male and female samples were analyzed in at least 3 replicates each. The gel with the female urine sample was used as master for the demonstration of spot identifications. Gels were scanned using GS-800 calibrated densitometer. Protein spots were excised manually or automatically using Proteineer Sp Protein picker (Bruker Daltonics, Bremen, Germany). Tryptic digest and Peptide Mass Fingerprinting (PMF) were performed as previously described [17]. In brief, peptide masses were analyzed by MALDI-TOF-TOF MS (Ultraflex, Bruker Daltonics, Bremen, Germany), peak lists were created with FlexAnalysis v2.2 software (Bruker), smoothing was applied with Savitzky-Golay algorithm (width 0.2 m/z, cycle number 1), and a signal/noise threshold ratio of 2.5 was allowed. Resolution for the mass range of collected data (900–3000 m/z) was at least 6000 and for the calibrants at least 8000. For calibration, internal standard peptides des-Arg-bradykinin (Sigma, 904.4681 Da) and adrenocorticotropic hormone fragment 18–39 (Sigma, 2465.1989 Da) included in the peptide mixture were utilized. The probability score with p < 0.05 defined by the software was used as the criterion for the affirmative protein identification. Peaks corresponding to trypsin auto-proteolysis, matrix, and keratin fragments were not considered for protein search (MALDI-TOF MS spectra as well as peak list excluded from the analysis are provided in the Supplementary “MALDI-TOF-MS PMF spectra” file).
For peptide matching (Mascot Server 2.2.01; Matrix Science, London, UK), the following two settings were used for the MALDI-TOF MS data analysis:
Standard parameters- Mascot search: Database: SwissProt; Taxonomy: Human; Enzyme: Trypsin; Mis-cleavages: 2; Mass tolerance: 25 ppm; Monoisotopic mass; Fixed modifications: carbamidomethylation (Cys); Variable modifications: Oxidation (Met).
Many variable modifications -Mascot search: Database: SwissProt; Taxonomy: Human; Enzyme: Trypsin; Mis-cleavages: 2; Mass tolerance: 25 ppm; Monoisotopic mass; Fixed modifications: none; Variable modifications: N-acetyl (protein), Cysteic acid (Cys), Oxidation (Met), Phospho (STY), Sulfonate (Met), Pyro-glu (N-term E), Pyro-glu (N-term Q).
The resulting protein and peptide identifications were manually filtered by detailed examination of the MALDI-TOF MS spectra for the occurrence of the peaks and comparison of the search results (1 and 2, above). A delta mass of 0.03 was used as threshold for peptide identification. Data were searched against human entries in the SwissProt (release 56.0; 20402 entries database). Analysis of the data using a sequence-scrambled version of Swiss-Prot generated by the decoy-generating script available at Matrix Science (http://www.matrixscience.com/downloads/decoy.pl.gz) and using the settings described above provided no identifications.
Additionally, tryptic digests of selected spots identified by MALDI-TOF-MS as albumin and serotransferin isoforms were also analyzed by CE-MS and similarly processed for their direct comparison to the former. The CE-MS identified peak lists are included in Supplementary `Detailed Tables', Spreadsheet `2DEspots-CEMS Analysis' and Supplementary Figure 1.
ELISA
CD14 levels were determined using Human sCD14 Quantikine ELISA Kit (R&D Systems, Minneapolis, MN) and NGAL levels were measured with NGAL ELISA kit (BioPorto, Gentofte, Denmark). Urinary concentration of IgA and galactose-deficient IgA1 were measured by capture ELISA as described previously [19].
Urine sample processing, nano-HPLC FTMS/MS, and database search for GeLC-MS
One mL of sample was deposited on a vivaspin 5kDa cut-off microconcentrator (Sartorius group) and centrifugated (5,000 × g) to a volume of ~100 μL. Two mL of deionized water (ELGA 18.2 MΩ) were added to the tube for desalting and the sample was centrifuged again to a volume of ~30 μL. The concentrated sample was completed to 75 μL with Laemmli buffer 5× to redissolve proteins which may have adsorbed on the ultrafiltration membrane. Thirty μL of the sample was deposited on a 4–12% Bis-Tris precast 1D gel (Invitrogen) and separated. Following gel staining with Coomassie blue, twenty-two bands were cut and proteins were oxidized using H2O2 before in-gel trypsin digestion. The resulting peptide mixtures were vacuum-dried.
Before analysis, the peptide mixtures corresponding to each gel band were resuspended in 15 μL H2O-Acetonitrile-formic acid (97.8:2:0.2) spiked with 0.1% TFA.
All Nano-HPLC FTMS/MS experiments were performed on a 7-tesla hybrid linear ion trap Fourier transform mass spectrometer LTQ-FT Ultra (Thermo Electron, Bremen, Germany) coupled to an Ultimate 3000 (LC-Packings, Amsterdam, Netherlands) high performance liquid chromatographer. Samples were loaded on an LC-Packing Acclaim Pepmap 100 C18 precolumn (300 μm ID, 5 mm long, 5 μm particles, 100 Ǻ pores) and separated on an LCPackings Pepmap nano-column (75 μm ID, 15 cm long, 3 μm particles, 100 Ǻ pores) coupled with a New Objective (Woburn, MA,USA) PicoTip electrospray emitter (30 μm). The nanoflow rate was set to 300 nL/min. Mobile phases consisted of (A) 0.1 % formic acid, 97.9 % water and 2 % acetonitrile (v/v/v) and (B) 0.08 % formic acid, 20 % water in 79.92 % acetonitrile (v/v/v). The loading buffer (C) was composed of 0.2 % formic acid 97.8 % water and 2 % acetonitrile (v/v/v). Ten μL of sample were loaded onto the precolumn during a 1 min injection event at a flow rate of 20 μL/min before the gradient was applied. The gradient profile was the following: from 4 % to 50 % B in 60 min; from 50 % to 90 % B in 5 min; constant 90 % B for 5 min and return to 4 % B in 1 min. The column was re-equilibrated for 28 min at 4% B between runs. MS data were acquired in the FTMS detection mode of operation (reduced profile) on a 450–1800 m/z range with resolution settings 50,000 at target 5e5. MS/MS spectra were obtained concomitantly with the FTMS detection for the 3 most abundant ions on the LTQ ion trap at 35% collision energy. Dynamic exclusion was set to avoid reselecting the same ion multiple times, using a 5 ppm tolerance window and 3 minutes exclusion with one repeat during the first minute.
The Mascot (version 2.2.03 Matrix Science, London, UK) engine was used to perform searches against a database compiled from the SwissProt (release 54.8) and Trembl (release 37.8) databases, using the following settings: taxonomy: human; mass tolerances: 10 ppm (MS) and 0.8 Da (MS/MS); enzyme: Trypsin/P; Variable modifications: N-Ac-protein, methionine oxidation and dioxidation, cystein trioxidation and proline hydroxylation; allowed miscleavages: 2. The resulting peptide identifications were automatically filtered using the home-made software IRMa and consolidated into an MS identification database (msiDB). The criteria used to filter identifications were: Rank = 1, peptide score >identity threshold (p<0.05). The target-decoy approach was used to estimate false positive identifications rates. The average false discovery rate received during this analysis was below 2%.
CE-MS sample preparation, analysis and data processing
The urine samples were prepared as described [20]. Shortly, 0.7-mL aliquots were diluted 1:1 with an aqueous solution containing 2 M urea, 10 mM NH4OH and 0.02% SDS. For removal of high-molecular mass proteins, ultrafiltration using Centrisart ultracentrifugation filter devices (20 kDa molecular mass cutoff; Sartorius, Goettingen, Germany) was used. Centrifugation was carried out at 3,000 × g until 1.1 mL of filtrate was obtained. After desalting on PD-10 columns (GE Healthcare, Munich, Germany) pre-equilibrated with 0.01% NH4OH, samples were lyophilized and stored at 4°C. Before CE-MS analysis, samples were reconstituted in 9 μL water (plus, if applicable, 1 μL external standard stock solution; see below) irrespective of protein content to keep reference standards constant.
CE-MS analysis was performed in two different laboratories (Hannover and Glasgow) essentially as described [21]. A P/ACE MDQ capillary electrophoresis system (Beckman Coulter, Fullerton, CA, USA) was coupled to either a micrOTOF MS (Bruker Daltronic, Bremen, Germany) in Hannover, Germany, or to a micrOTOF-Q MS/MS (Bruker Daltronic, Bremen, Germany) in Glasgow, UK. Twenty % acetonitrile (Sigma-Aldrich, Taufkirchen, Germany) in HPLC-grade water (Roth, Karlsruhe, Germany) supplemented with 0.94% formic acid (Sigma-Aldrich) was used as running buffer. For CE-MS analysis, the electrospray ionization interface from Agilent Technologies (Palo Alto, CA, USA) was used. Spectra were recorded over an m/z-range of 350 to 3,000 and accumulated every 3 seconds. The MosaiquesVisu software was used to deconvolute mass spectral ion peaks from the same molecule at different charge states into a single mass [22]. To achieve high mass accuracy, deconvoluted TOF signals were calibrated based on FT-ICR-derived accurate masses (mass deviation <1 ppm) as described [23]. After calibration, mass deviation of TOF-MS data was found to be 2 ± 8 ppm. A probabilistic clustering process was implemented in parallel for normalization of CE-retention times and calibration of signal intensities. This was accomplished by local regression analysis of each analysis to specific urinary calibrants that serve as internal standards [11]. All detected peptides were deposited, matched and annotated in a Microsoft SQL database.
The analytical characteristics of the CE-MS system were extensively investigated and described by Theodorescu et al. [20]; and Haubitz et al. [10]. Briefly, the average recovery of the sample after preparation was 85%. The limit of detection was found to be approx. 1 fmol. Resolution >8,000 ensured the detection of monoisotopic mass signals for z≤6.
Quantification using stable isotope labelled external standards by CEMS analysis
For absolute quantification stable isotope-labelled peptides were used as described [11]. Samples (9 μL) were spiked with 1 μL of a stable isotope labelled external standard solution (for concentration of the different peptides see [11), and 290 nL of this sample was injected into the CE, resulting in between 0,09 – 20 ng of each synthetic peptide being injected. Resulting MS signal (ion counts) were correlated with the known concentration of the standard peptides. Amount of endogenous peptide was assessed using the calibration curves previously established. The analysis was repeated 15 times, and the mean and standard deviation of the results are reported in Table 1.
LC-MS sample preparation, analysis and data processing
LC-MS analysis of endogeneous peptides was performed with an Ultimate3000 system (Dionex, Amsterdam, Netherlands) coupled to an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). High-resolution MS scans were acquired in the Orbitrap analyzer while MS/MS scans were triggered in parallel in the LTQ ion trap. Urine samples were prepared as described above (see section `CE-MS sample preparation, analysis and data processing'). Lyophilized peptides were reconstituted in 14 μL of 2% acetonitrile, 0.05% trifluoroacetic acid, and an aliquot of 5 μL (approximatively 7 μg total peptide) was loaded onto a C18 precolumn (300 μm ID × 5 mm, Dionex) at 20 μL/min in 2% acetonitrile, 0.05% trifluoroacetic acid. After desalting, the precolumn was switched online with the analytical column (75 μm ID × 15 cm PepMap C18, Dionex) and equilibrated in solvent A (5% acetonitrile, 0.2% formic acid). Peptides were eluted using a 0 to 60 % gradient of solvent B (80% acetonitrile, 0.2% formic acid) during 60 min at 300 nL/min flow rate. The LTQ-Orbitrap was operated in data dependent acquisition mode. Survey full scan MS spectra (m/z mass range 300–2000) were acquired in the Orbitrap with the resolution set to a value of 60 000 at m/z 400 (target value of 1 000 000 charges in the linear ion trap). The five most intense ions per survey scan (excluding singly-charged) were selected for MS/MS fragmentation using collisionally induced dissociation (CID) and the resulting fragments were analyzed in the linear ion trap (parallel mode, target value 10 000). Collision energy was set to 35% for MS/MS. Dynamic exclusion was employed within 60 seconds to prevent repetitive selection of the same peptide. For each peptide mixture (male and female), seven replicate LC-MS runs were performed. RAW files were converted into mzxml files with the use of ReAdW (version 4.2.0). For further comparisons this file format was used with msInspect (version 2.0) (http://proteomics.fhcrc.org/CPL/msinspect/) to generate a peaklist of the MS data. For matching of the various LC-MS data, these were calibrated with respect to abundance based on the 29 internal standards and using the same procedure as described above for CE-MS.
Database search from MS/MS data acquired with the LTQ-Orbitrap was performed using the Mascot Daemon software (version 2.2.03, Matrix Science, London, UK). The following parameters were set for creation of the peak lists: parent ions in the mass range 400–4500, no grouping of MS/MS scans, and threshold at 1000. Data were searched against human entries in the Uniprot database (release 14.0, 72400 sequences, compiled from the SwissProt (release 56.0) and Trembl (release 39.0) databases), without any enzyme specificity. No fixed modification was selected, and oxidation of methionine and proline were set as variable modifications. The mass tolerance was set to 10 ppm and 0.8 Da for MS and MS/MS respectively. Mascot results were parsed with the in-house developed software MFPaQ version 4.0 (http://mfpaq.sourceforge.net/) and top ranking peptides with a Mascot score of more than 35 were automatically selected. In order to calculate the False Discovery Rate (FDR), the search was performed using the “decoy” option in Mascot, and MFPaQ used the same threshold to validate decoy and target hits. The FDR was calculated for each database search at the peptide level (FDR = number of validated decoy hits / (number of validated target hits + number of validated decoy hits) × 100), and the average value for all samples analysed was 2.8 % for a Mascot score of 35. In addition, criteria related to the mass accuracy and the frequency of detection in all replicate samples were also used to validate peptides reported in the final list. Deviation between the experimental m/z and the theoretical m/z of the identified sequence was calculated for all peptides, boxplot analysis was performed to detect the outliers. The median mass shift was 2.2 ppm, peptides identified with a mass deviation >2.5 ppm from this value were removed from the list. The MS signal intensity of the sequenced peptides was extracted from each replicate run of male and female samples using MFPaQ. Only peptides for which MS signal was found in at least 4 of the 7 analysis for each sample were provided in the list of sequenced peptides.
In addition, several peptides were also previously sequenced using LC-MS/MS analysis in other laboratories, as recently described in detail [23]. Here, the average false discovery rate received during these analyses was below 2%.
To enable comparison of CE- and LC-MS data, the identity of peptides in CE- and LC-MS was established on the basis of sequence, mass and CE- migration time. In the absence of sequence information, peptides were considered identical if the mass deviation was less than 30 ppm and the difference in abundance was less than 4-fold.
Results
In a multi-institutional effort we have analyzed the urinary proteome and peptidome of two “standard” human urine samples, representing pooled urine from healthy human male and female. The depth of the urinary proteome/peptidome covered by this multi-institutional analysis is reflected in Figure 1. Analyses were performed in different laboratories using nearly all available state-of the art proteomics technologies and instruments, including 2DE- in combination with MALDI TOF MS, LC-MS and -MS/MS using an LTQ-Orbitrap MS, 1D gel in combination with nano-LC MS/MS using a hybrid linear ion trap Fourier transform MS (LTQ-FT Ultra; this method will be abbreviated as GeLC-MS) as well as capillary electrophoresis coupled to mass-spectrometry (CE-MS). In all cases, participating laboratories applied their established protocols for sample preparation, analysis, and data processing (Summarized in Table 1B).
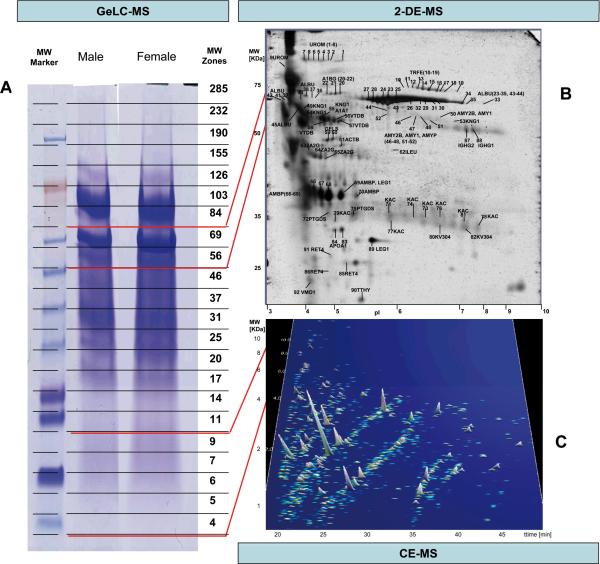
Figure 1. Representative examples of analysis of the human standard urine sample by different proteomics technologies.
A) 1D gel used for fractionation in the GeLC-MS enables separation by mass, and allows for ample material in the subsequent LC-MS/MS analysis of the respective tryptic fragments. The respective molecular weight zones that were excised are indicated on the right. B) 2DE-MS of the female sample, due to its high resolution, enables separation of isoforms/different post-translational modifications. At the same time, representation of the entire gel in the subsequent MS/MS analysis, as in the 1DE, is generally not preformed, due to the size of the gel and the consequently high number of LC-MS/MS analyses required. C) CE-MS (shown is the female sample) enables analysis of the lower molecular weight portion of the urinary proteome that cannot be addressed adequately by SDS-gel based approaches. However, the evidently very high resolution can only be utilized in the analysis of molecules at lower mass, in general < 15 kDa.
Table 1B.
Summary of methods and technologies employed for the analysis of the standard urine samples.
| Technique | Sample preparation | Separation | Digestion | MS | Identification | Quantification | ||
|---|---|---|---|---|---|---|---|---|
| Engine | Modifications | Accuracy | ||||||
| 2DE | UF (3000MWCO) TCA/NLS precipitation | IEF3–10, 11%PAGE | trypsin | MALDI TOF TOF | Mascot 2.2.01 | Fixed modification: carbamidomethylation (Cys); Variable: Oxidation (Met)* | 25ppm (MS) | N/A |
|
| ||||||||
| GeLCMS | UF(5000MWCO) | 4–12%Bis-Tris PAGE and Ultimate 3000 coupled to MS | oxidation trypsin | LTQ-FT Ultra | Mascot 2.2.03 | Variable modifications: N-Ac-protein, methionine oxidation and dioxidation, cystein trioxidation and proline hydroxylation | 10ppm (MS), 0.8 Da (MS/MS) | N/A |
|
| ||||||||
| CE-MS | UF (20000MWCO), desalting (PD-10), lyophilization | P/ACE MDQ CE coupled to MS | none | micrOTOF MS (H)or micrOTOF-Q MS/MS (G) | Mascot Internet version, OMSSA 2.1.4 | Variable modifications: oxidation of methionine, lysine, and proline | 0.5 Da (MS), 0.7 Da (MS/MS) | Internal standards and spiked isotope labeled peptides |
|
| ||||||||
| LC-MS and MS/MS | UF (20000MWCO), desalting (PD-10), lyophilization | Ultimate3000 coupled to MS | none | LTQ-Orbitrap | Mascot 2.2.03 | Variable modfications: oxidation of methionine, lysine and proline | 10ppm (MS), 0.8 Da (MS/MS) | Internal standards |
UF: ultrafiltration; MWCO: Molecular weight cut-off; N/A: not applicable; H: Hannover; G: Glasgow;
an alternative search method with multiple variable modifications was also tested as described in Material-Methods (2DE analysis)
Clinical characterization of samples
In order to provide commonly used clinical reference points, volunteers were characterized for various clinico-pathological characteristics as shown in Table 1A. In addition, the urine samples were characterized by conventional immunological assays (ELISA) for certain disease-specific biomarkers used in routine clinical laboratory testing. Specifically, biomarkers associated with IgA-Nephropathy (CD14, IgA and galactose-deficient IgA1) were examined by routine clinical ELISAs and results are provided in Table 1A.
2DE
The identification score, peptide coverage and peptides for each of the identified protein spots (Figure 1) are provided in Supplementary `Detailed Tables', spreadsheet `2DE Master'. In addition, identification and predicted modification data received from the analysis of the various peptides in comparison to those of the respective peptides identified during the GeLC-MS analysis and in a similar mass zone are provided (Figure 2 and Supplementary `Detailed Tables', spreadsheet `2DE Master'). A total of 92 protein spots were identified using PMF, whose position in the gel and identity are in general agreement to published urinary data from normal individuals using similar sample preparation and electrophoretic protocols (3, 17, 24, 25). Further, CE-MS analysis of tryptic peptides from spots identified by MALDI as albumin and serotransferin isoforms provided increased sequence coverage and predicted the presence of selected modifications per isoform (Supplementary Figure 1 a–h and Supplementary `Detailed Tables', spreadsheet `2DEspots-CEMS Analysis').
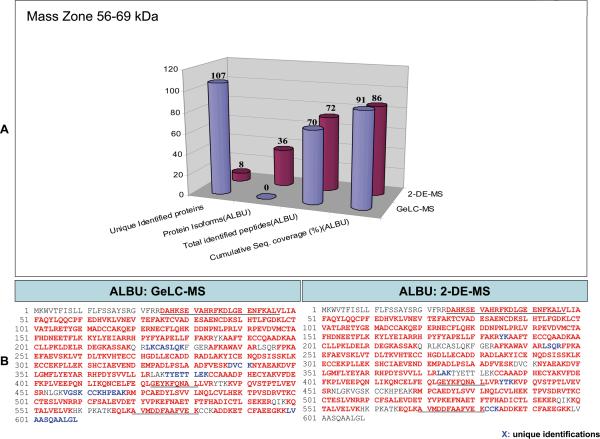
Figure 2. Comparison of the identification results from GeLC-MS and 2DE.
A) Graphical representation of the results from the analysis of the 56-69kDa band zone by GeLC-MS and 2DE referring to the number of unique protein identifications provided by each technique; using albumin as an example, a comparison of the number of detected isoforms, peptides, and respective cumulative sequence coverage (%) provided by each technique is also made. B) The sequences of the identified peptides using GeLC-MS and 2DE are also shown. Red characters indicate the identified peptides of the protein and blue characters indicate unique identifications by one approach. For comparison, regions of the protein detected as native peptides by CE-MS and/or LC-MS are underlined.
GeLC-MS
Analyses of the 44 gel bands (see Figure 1) corresponding to male and female urines resulted in ~10,000 identifications pointing to ~2,450 protein accessions (including same and sub-sets). Results were compiled by sample and by band and proteins grouped in a similar way as Mascot does. However, the protein representative of each group was selected on the basis of its emergence in the list of identifications. If two proteins shared the same set of peptides in a given analysis, the representative protein would be the one which appeared most frequently as master in all the analyses. The results by sample and by band are presented in Supplementary `Detailed Tables', spreadsheets: `GeLC-MSMS Identification List' and `GeLC-MSMS Heat Map' respectively. A table summarizing the different protein groups, their representatives and the list of proteins belonging to each group, is also presented. (Supplementary `Detailed Tables', spreadsheet `GeLC-MSMS Protein Groups').
CE-MS
CE-MS analysis of male and female standard sample was repeated 25 times in Hannover and 7 times in Glasgow to ensure high confidence in the detected peptides. Only peptides that could be detected in at least 30% of these analyses (in either sample, using either instrumental setup) were accepted. A graphical depiction of the data is shown in Figure 3A. The calibrated and annotated CE-MS data are available in Supplementary `Detailed Tables', spreadsheet: `CE-(ESI)-MS male, female'. In the resulting CE-MS profiles, each peptide is identifiable by its mass and CE migration time, and ion signal intensity is used as measure for relative abundance. The sequences of 292 peptides (including PTMs) identified by LC-MS/MS are given. Identified peptides originate from 31 different protein precursors. Absolute quantification of some peptides was done using external stable isotope-labelled peptide standards. These results are provided in Table 1.
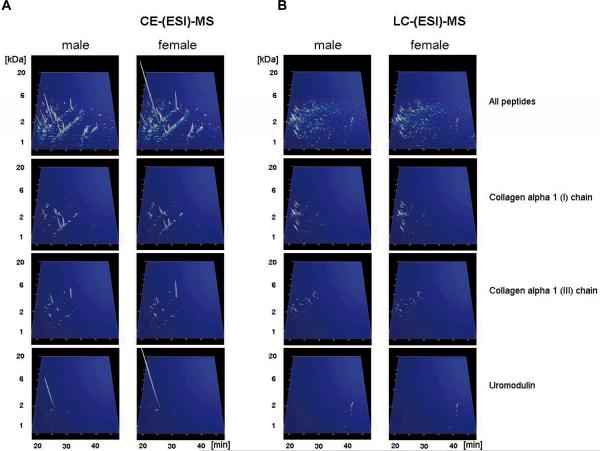
Figure 3. Display of the low molecular weight proteome of the human urine standard.
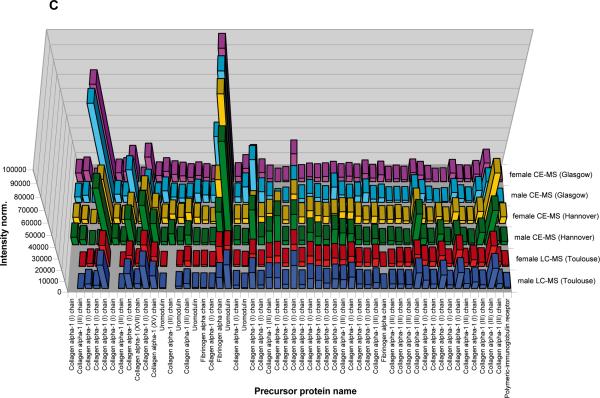
A) Peptides and proteins detected using CE-MS: shown are compiled patterns of the individual analyses. The molecular mass on a logarithmic scale (0.8 – 20 kDa, indicated on the left) is plotted against normalized migration time (18–48 min, indicated on the bottom). Average signal intensity is encoded by peak height and color. Upper panel: all detected signals (corresponding to Supplementary `Detailed Tables', spreadsheet: `CE-(ESI)-MS male, female'). Lower panels: Distribution of the collagen alpha 1 (I), the collagen alpha 1 (III), and the uromodulin peptides, as indicated on the left. B) Peptides and proteins detected using LC-MS. Shown are compiled patterns of the individual analyses. The molecular mass on a logarithmic scale (0.8 – 20 kDa, indicated on the left) is plotted against retention time (18–48 min, indicated on the bottom). Average signal intensity is encoded by peak height and color. Upper panel: all detected signals (corresponding to Supplementary `Detailed Tables', spreadsheet: `LC-(ESI)-MS female, male'). Lower panels: Distribution of the collagen alpha 1 (I), the collagen alpha 1 (III), and the uromodulin peptides, as indicated on the right. C) Inter-laboratory and inter-platform comparisons of the 40 most abundant sequenced peptides (based on ion counting) in the standard male and the female samples. These peptides account for ~40% of the total signal. The majority of these peptides represent degradation products from collagen, uromodulin, and fibrinogen, as indicated on the bottom. As expected, a very high similarity between the data obtained on the two samples on an identical instrument can be observed, with most of these peptides being among the 40 most abundant peptides in both samples. When comparing data obtained on similar, yet different instruments (comparing upper two panels from CE-TOF with the middle 2 panels from CE-Q-TOF), good comparability can be observed, and most peptides can be detected with similar abundance. Analysis of the same samples on a completely different platform (LC-coupled Orbitrap) results in similar data. Most noticeable: the absence of very small and very large peptides in the LC-MS analysis. This is likely due to small and highly charged peptides not binding to the reversed-phase material, while large peptides likely precipitating on the column, and thereby not eluting.
To evaluate comparability between the male and the female sample, peptides that are among the 40 most abundant (based on ion-counting) peaks from each sample were compared after calibration of the ion-counts. Most of these 40 peaks were among the 40 most abundant in both samples. In total 47 peptides (33 of the 40 were found among the 40 most abundant in both samples) were compared. Further, and as evident from the comparison shown in Figure 3C, most peaks were recorded with similar intensity in the male and the female samples, indicating the high quality of sample material. The intervariability between the labs is displayed by an overlap of 94.4% using the most abundant peptides, which demonstrated approximately 40% of the sample amount).
Notably, as these samples serve as “healthy human urine standards”, we also assessed their scoring versus previously established biomarker patterns for the following diseases: diabetes [26], diabetic nephropathy [27], chronic kidney disease [26], IgA nephropathy [28], ANCA-associated vasculitis [10], bladder cancer [20] and coronary artery disease [29], using CE-MS. In all of these biomarker models, both samples scored as normal healthy controls, confirming that they represent adequate baseline controls.
LC-MS and MS/MS
LC-MS profiling of male and female standard sample was performed on an LTQ-Orbitrap, and was repeated 7 times to ensure high confidence in the detected peptides. The results of this analysis are shown in Figure 3B and provided in Supplementary `Detailed Tables', spreadsheet: `LC-(ESI)-MS male, female'. As in the case of CE-MS analysis, we compared male and female datasets by aligning the 40 most abundant peaks from each sample. Most of these 40 peaks were the same for the two samples and in total 46 peptides were compared and showed very similar intensity profiles (data not shown).
In parallel, MS/MS sequencing of the most abundant peptide ions was performed in the LTQ ion-trap. 282 unique peptides (detected in at least 4 of the 7 analyses for each sample) were identified, with validation criteria based on Mascot scoring, with a median mass shift of 2.2 ppm and a maximum standard mass deviation of ± 2.5 ppm from this value. These sequenced peptides originate from 31 different protein precursors, among them are highly abundant collagens, but also other proteins already known to produce proteolytic fragments in urine. Matching of identified sequences to LC-MS peaks extracted with msInspect was performed based on accurate mass and elution time, as shown in Supplementary `Detailed Tables', spreadsheet: `LC-(ESI)-MS male, female'.
Application of standard urine in inter-lab and platform comparability
To evaluate comparability between different instrumental setups in different laboratories, the 40 most abundant peaks in the individual samples analyzed using CE-TOF (Hannover) and CE-Q-TOF (Glasgow) were identified and compared. Again, most of these 40 peaks were among the 40 most abundant in all of the datasets, resulting in a total of only 66 peptides. These 40 peaks account for approximately 40% of the total signal (see Supplementary `Detailed Tables', spreadsheet: `CE-(ESI)-MS male, female').
As a proof of principle for the application of these standard samples in estimating comparability of different MS platforms, we further compared datasets obtained on the two CE-MS platforms (see above) with those obtained using an Ultimate3000 nanoLC system coupled to an LTQ-Orbitrap mass spectrometer, sited in Toulouse (France). When comparing the 40 most abundant sequenced peptides detected at the different sites, high consistency of the datasets was apparent (overlap: male 87%, female 89%), as well as some expected differences (see Supplementary `Detailed Tables', spreadsheet: `Comparison CE-MS vs. LC-MS' and Figure 3C). For example, several low molecular weight peptides detected by CE-MS were missing in the LC-MS data, likely due to their inability to bind to the LC column materials. Additionally, several high molecular weight peptides were absent in the LC-MS data, likely because of precipitation on the column material. This comparison highlights one advantage of CE in comparison to LC: selective loss of peptides is generally not observed. These results show that on the basis of a common standard a comparative analysis between datasets generated on different platforms and at different sites is possible after appropriate adjustment and recalibration of the data.
Data integration from different techniques
Data generated by different proteomics technologies are not always comparable and thereby combination of observations results in some information loss. For example, while GeLC-MS provides identification of tryptic peptides via sequence annotation, it generally does not provide a measure of abundance. In contrast, CE- or LC-MS gives measures of abundance, but not immediate sequence identification. In an effort to integrate the acquired data in an informative way, two tables were generated compiling respectively the proteomics (“bottom-up” approaches: 2DE, GeLC-MS) and peptidomics data (LC-MS and CE-MS, “top-down” approaches; Supplementary `Compiled Tables'). A detailed (within the limitations of each technique) description of the physicochemical properties of the identified proteins and peptides is provided. Specifically, in the case of the proteomics data (Supplementary `Compiled Tables', spreadsheet `GeLC-MS and 2DE combined'), the observed MW and pI values of the proteins, as applicable, are shown. It should be noted that no homogenization of MW calculations of the two techniques was made since rules for this were not always evident. For example, by 2DE distinct isoforms of albumin were observed at 70, 68, 65 kDa, which could all correspond to the 69 kDa Albumin detected by GeLC-MS. It should also be noted that same identifications at adjacent MW zones may not correspond to distinct isoforms but rather to cross-contamination. Since distinction of the 2 possibilities is impossible, we show all available information. In the case of the low molecular weight proteome and to display all data available, we combined the CE- and the LC-MS data (Supplementary `Compiled Tables', spreadsheet `CE-MS and LC-MS combined'). While this was possible with high confidence for the sequenced peptides, to do so in the absence of sequence was challenging. The second parameters that are used for identification, CE migration time or LC retention time, cannot be correlated to each other. This is also evident from Figure 3, where no correlation can be found for the entire CE- and LC-MS analyses, or the data that show distribution of the peptides from collagen alpha 1 (I), collagen alpha 1 (III), or uromodulin. As a compromise aiming at avoiding artificial duplication (a single peptide being listed as two different peptides), but at the same time avoiding reporting two different peptides as identical, we have considered peptides identical, if the mass deviation between the LC- and the CE-MS data was less than 30 ppm, and the difference between the normalized signal amplitude was below 4-fold. The combined data are given in Supplementary `Compiled Tables', spreadsheet: `CE-MS and LC-MS combined'.
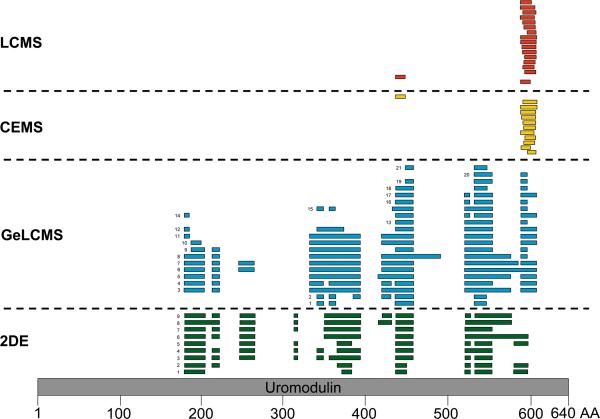
Combination of the proteomics with peptidomics data in one table is impossible due to the completely different data representation. Nevertheless, an effort to combine sequence information received from the different techniques may be made: For example, mapping of the identified peptides of uromodulin (uromodulin spots detected by 2DE are shown in Figure 1, CEMS uromodulin fragments detected by CE-MS and LC-MS are shown in Figure 3) on the protein sequence, reveals that the N- and C-termini of the protein have not been detected by any of the applied techniques (Figure 4). The “native” peptides identified by the peptidomics approaches are located close to the C-terminus of the protein. In the case of albumin, even though very extensive sequence coverage is received by the combination of techniques, a peptide of the N-terminus of the protein remained undetected in the vast majority of applied techniques (Figure 2B) with the only exception being the analysis of tryptic digests of 2DE spot 24 by CE-MS (see Supplementary Figure 1e). These observations, even though not conclusive at this point, particularly in view of the expected continuous data accumulation from this sample, may form the basis for hypothesis-driven research questions relating to expected native peptides, “hot spots” for protein modifications, specific proteolytic sites etc.
Figure 4. Comparison of the identification results from the different techniques.
Position of the uromodulin peptides detected by the employed techniques (2DE, GeLC-MS, CE-MS, and LC-MS).on the protein sequence. The numeric labeling is based on the protein theoretical primary sequence (grey bar). The numbers in front of the bars correspond to the spot numbers of the 2DE master gel (see Supplementary `Detailed Tables', spreadsheet: `2DEMaster') or to the gel band in the case of GeLC-MS (see Supplementary `Detailed Tables', spreadsheet: `GeLC-MSMS Identification List'). The GeLC-MS female and male data are merged. In the case of the 2DE and GeLC-MS approaches the detected peptides correspond to tryptic digests of the respective spots or bands. In the case of CE- and LCMS analysis the naturally occurring peptides are shown, as identified following tandem mass spectrometry.
Discussion
We have generated two human urine samples that are available in large quantities, likely represent of the currently best characterized urine specimens, and can be used as “standards” in future urine proteomics analysis to enable inter-laboratory comparison of datasets.
In contrast to previous reports, analysis involved a definition of the actual biochemical entities, i.e. proteins/peptides associated with molecular mass and detected posttranslational modifications (within the limitation of the technologies employed), and the relative abundance of these compounds.
Proteins and peptides in urine generally are present in several different forms due to different posttranslational processing, including specific proteolysis [23]. This is also evident from the data presented here, where we describe several different and distinct proteins and peptides that originate from the same parental protein. It is essential to accurately define these entities, as only some are biomarkers for disease (e.g. different, specific degradation products in renal pathology [30], and these different forms must be clearly distinguished). The combination of different technologies allowed coverage of a wide mass range from the protein down to the peptide level, revealing the advantages of the different technologies, and their complementarities. Specifically, and as expected, the various platforms and technologies yielded differences in the number of detected and identified peptides and proteins. However, both, extensive overlaps and extensive complementarities in the information provided by different datasets were observed. For example, while the uromodulin protein was detectable in both, 2DE and GeLC-MS experiments, naturally existing uromodulin peptides could only be identified by CE-MS or LC-MS analysis (Figure 3). In addition, the presence of different uromodulin isoforms could be observed only on 2D gels.
Due to the age of the participating volunteers (Table 1A), and the increasing evidence supporting the presence of age-associated protein changes to urine [31], the reported proteome is considered more representative of younger age groups. In addition, while the two samples have been exhaustively analyzed by current state-of-the art methods, they were not analyzed “to completion”, i.e., not all of their compounds are currently known and quantified. Notably, the sample collection protocol that was followed is not compatible with the recommended procedure for the study of exosomal or other urine solid phase proteins [4, 9]. In addition the collection of more quantitative data is undoubtedly needed and is currently under way. Nevertheless due to the combinatorial application of techniques, these samples likely represent of the most extensively and comprehensively investigated urine samples reported.
These multi-institutional and multi-technology based datasets of urinary proteins and peptides from two distinct standard samples are the foundation for the comprehensive compilation of the urine proteome. To facilitate this effort, the full list of identified peptides and proteins and pertinent separation and MS data are also available at the European Kidney and Urine Proteomics Action website (www.eurokup.org; 32) and Human ProteinPedia (accession number HuPA_00668) Importantly future datasets that are obtained using these two samples will be also made freely available via the EuroKUP website. The development of unified systems for protocol and data reporting, processing, comparison and mining for clinical urine proteomics is evident and mandatory for achieving clinical impact. This is under development in the EuroKUP consortium and will result in a continuous increase of the depth and breath of the characterization of these standard samples. Thus, they will be a resource of increasing value to the community and an invitation to optimize and complete the knowledge on the human urinary proteome/peptidome. Investigators interested in acquiring aliquots of the standard urine sample are invited to contact Harald Mischak (mischak@mosaiques.de).
Supplementary Material
Acknowledgement
This work was funded in part by grants from the European Union through InGenious HyperCare (grant LSHM-C7-2006-037093) to HM and the EuroKUP COST Action (BM0702). AV, MA, MM, JZ and JG, CM, MC acknowledge financial support from FP7 DECanBio (201333). In addition, DB, CL, AGP, BM and JPS acknowledge financial support from the Agence Nationale pour la Recherche (ANR-07-PHYSIO-004-01). JN and HS acknowledge support by NIH grants DK075868 and DK078244.
Abbreviations
- EuroKUP
European Kidney and Urine Proteomics COST Action
- GeLC-MS
1D gel analysis in combination with nano-LC MS/MS (using LTQ-FT Ultra)
- HUKPP
Human Urine and Kidney Proteome Project
- HAA-IgA
Helix aspersa agglutinin reactive IgA
- NGAL
neutrophil gelatinase-associated lipocalin
Footnotes
Present address for Dr Kolch: Systems Biology Ireland, University College Dublin
Conflict of Interest Statement There is no conflict of interest for any of the authors with the exception of HM who is a founder of Mosaiques-Diagnostics and developed CE-MS for proteomics clinical applications.
References
- [1].Davis MT, Spahr CS, McGinley MD, Robinson JH, et al. Towards defining the urinary proteome using liquid chromatography-tandem mass spectrometry. II. Limitations of complex mixture analyses. Proteomics. 2001;1:108–117. doi: 10.1002/1615-9861(200101)1:1<108::AID-PROT108>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- [2].Pang JX, Ginanni N, Dongre AR, Hefta SA, Opitek GJ. Biomarker discovery in urine by proteomics. J. Proteome. Res. 2002;1:161–169. doi: 10.1021/pr015518w. [DOI] [PubMed] [Google Scholar]
- [3].Thongboonkerd V, McLeish KR, Arthur JM, Klein JB. Proteomic analysis of normal human urinary proteins isolated by acetone precipitation or ultracentrifugation. Kidney Int. 2002;62:1461–1469. doi: 10.1111/j.1523-1755.2002.kid565.x. [DOI] [PubMed] [Google Scholar]
- [4].Pisitkun T, Johnstone R, Knepper MA. Discovery of Urinary Biomarkers. Mol. Cell Proteomics. 2006;5:1760–1771. doi: 10.1074/mcp.R600004-MCP200. [DOI] [PubMed] [Google Scholar]
- [5].Decramer S, Gonzalez de Peredo A, Breuil B, Mischak H, Monsarrat B, Bascands JL, Schanstra JP. Urine in clinical proteomics. Mol Cell Proteomics. 2008;7(10):1850–62. doi: 10.1074/mcp.R800001-MCP200. [DOI] [PubMed] [Google Scholar]
- [6].Weissinger EM, Schiffer E, Hertenstein B, Ferrara JL, et al. Proteomic patterns predict acute graft-versus-host disease after allogeneic hematopoietic stem cell transplantation. Blood. 2007;109:5511–5519. doi: 10.1182/blood-2007-01-069757. [DOI] [PubMed] [Google Scholar]
- [7].Ein-Dor L, Zuk O, Domany E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5923–5928. doi: 10.1073/pnas.0601231103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. The human urinary proteome contains more than 1500 proteins including a large proportion of membranes proteins. Genome Biol. 2006;7:R80.1–R80.16. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gonzales PA, Pisitkun T, Hoffert JD, Tchapyjnikov D, et al. Large-scale proteomics and phosphoproteomics of urinary exosomes. J Am Soc Nephrol. 2009;20:363–379. doi: 10.1681/ASN.2008040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Haubitz M, Good DM, Woywodt A, Haller H, et al. Identification and Validation of Urinary Biomarkers for Differential Diagnosis and Evaluation of Therapeutic Intervention in ANCA associated Vasculitis. Molecular & Cellular Proteomics. 2009 doi: 10.1074/mcp.M800529-MCP200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jantos-Siwy J, Schiffer E, Brand K, Schumann G, et al. Quantitative Urinary Proteome Analysis for Biomarker Evaluation in Chronic Kidney Disease. J. Proteome. Res. 2009;8:268–281. doi: 10.1021/pr800401m. [DOI] [PubMed] [Google Scholar]
- [12].Kistler AD, Mischak H, Poster D, Dakna M, et al. Identification of a unique urinary biomarker profile in patients with autosomal dominant polycystic kidney disease. Kidney Int. 2009 doi: 10.1038/ki.2009.93. [DOI] [PubMed] [Google Scholar]
- [13].Havanapan PO, Thongboonkerd V. Are protease inhibitors required for gel-based proteomics of kidney and urine? J Proteome Res. 2009;8:3109–3117. doi: 10.1021/pr900015q. [DOI] [PubMed] [Google Scholar]
- [14].Thongboonkerd V, Mungdee S, Chiangjong W. Should urine pH be adjusted prior to gel-based proteome analysis? J Proteome Res. 2009;8:3206–3211. doi: 10.1021/pr900127x. [DOI] [PubMed] [Google Scholar]
- [15].Schaub S, Wilkins J, Weiler T, Sangster K, et al. Urine protein profiling with surface-enhanced laser-desorption/ionization time-of-flight mass spectrometry. Kidney Int. 2004;65:323–332. doi: 10.1111/j.1523-1755.2004.00352.x. [DOI] [PubMed] [Google Scholar]
- [16].Chevallet M, Diemer H, Van DA, Villiers C, Rabilloud T. Toward a better analysis of secreted proteins: the example of the myeloid cells secretome. Proteomics. 2007;7:1757–1770. doi: 10.1002/pmic.200601024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zerefos PG, Vougas K, Dimitraki P, Kossida S, et al. Characterization of the human urine proteome by preparative electrophoresis in combination with 2-DE. Proteomics. 2006;6:4346–4355. doi: 10.1002/pmic.200500671. [DOI] [PubMed] [Google Scholar]
- [18].Chevallet M, Luche S, Rabilloud T. Silver staining of proteins in polyacrylamide gels. Nat. Protoc. 2006;1:1852–1858. doi: 10.1038/nprot.2006.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Suzuki H, Moldoveanu Z, Hall S, Brown R, et al. IgA1-secreting cell lines from patients with IgA nephropathy produce aberrantly glycosylated IgA1. J Clin. Invest. 2008;118:629–639. doi: 10.1172/JCI33189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Theodorescu D, Wittke S, Ross MM, Walden M, et al. Discovery and validation of new protein biomarkers for urothelial cancer: a prospective analysis. Lancet Oncol. 2006;7:230–240. doi: 10.1016/S1470-2045(06)70584-8. [DOI] [PubMed] [Google Scholar]
- [21].Wittke S, Mischak H, Walden M, Kolch W, et al. Discovery of biomarkers in human urine and cerebrospinal fluid by capillary electrophoresis coupled to mass spectrometry: towards new diagnostic and therapeutic approaches. Electrophoresis. 2005;26:1476–1487. doi: 10.1002/elps.200410140. [DOI] [PubMed] [Google Scholar]
- [22].Neuhoff N, Kaiser T, Wittke S, Krebs R, et al. Mass spectrometry for the detection of differentially expressed proteins: a comparison of surface-enhanced laser desorption/ionization and capillary electrophoresis/mass spectrometry. Rapid Communications in Mass Spectrometry. 2004;18:149–156. doi: 10.1002/rcm.1294. [DOI] [PubMed] [Google Scholar]
- [23].Coon JJ, Zürbig P, Dakna M, Dominiczak AF, et al. CE-MS analysis of the human urinary proteome for biomarker discovery and disease diagnostics. Proteomics Clin. Appl. 2008;2:964–973. doi: 10.1002/prca.200800024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Thongboonkerd V, Chutipongtanate S, Kanlaya R. Systematic evaluation of sample preparation methods for gel-based human urinary proteomics: quantity, quality, and variability. J. Proteome. Res. 2006;5:183–191. doi: 10.1021/pr0502525. [DOI] [PubMed] [Google Scholar]
- [25].Oh J, Pyo JH, Jo EH, Hwang SI, et al. Establishment of a near-standard two-dimensional human urine proteomic map. Proteomics. 2004;4:3485–3497. doi: 10.1002/pmic.200401018. [DOI] [PubMed] [Google Scholar]
- [26].Snell-Bergeon JK, Maahs DM, Ogden LG, Kinney GL, et al. Evaluation of urinary biomarkers for coronary artery disease, diabetes, and diabetic kidney disease. Diabetes Technol. Ther. 2009;11:1–9. doi: 10.1089/dia.2008.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Rossing K, Mischak H, Dakna M, Zürbig P, et al. Urinary Proteomics in Diabetes and CKD. J Am Soc Nephrol. 2008;19:1283–1290. doi: 10.1681/ASN.2007091025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Julian BA, Wittke S, Novak J, Good DM, et al. Electrophoretic methods for analysis of urinary polypeptides in IgA-associated renal diseases. Electrophoresis. 2007;28:4469–4483. doi: 10.1002/elps.200700237. [DOI] [PubMed] [Google Scholar]
- [29].Zimmerli LU, Schiffer E, Zürbig P, Kellmann M, et al. Urinary proteomics biomarkers in coronary artery disease. Mol Cell Proteomics. 2008;7:290–298. doi: 10.1074/mcp.M700394-MCP200. [DOI] [PubMed] [Google Scholar]
- [30].Candiano G, Musante L, Bruschi M, Petretto A, et al. Repetitive fragmentation products of albumin and alpha1-antitrypsin in glomerular diseases associated with nephrotic syndrome. J. Am. Soc. Nephrol. 2006;17:3139–3148. doi: 10.1681/ASN.2006050486. [DOI] [PubMed] [Google Scholar]
- [31].Zürbig P, Decramer S, Dakna M, et al. The human urinary proteome reveals high similarity between kidney aging and chronic kidney disease. Proteomics. 2009;9(8):2108–17. doi: 10.1002/pmic.200800560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Vlahou A, Schanstra J, Frokiaer J, El NM, Spasovski G, Mischak H, Domon B, Allmaier G, Bongcam-Rudloff E, Attwood T. Establishment of a European Network for Urine and Kidney Proteomics. J Proteomics. 2008;71:490–492. doi: 10.1016/j.jprot.2008.06.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.