Abstract
Background
Tyrosine hydroxylase (TH) is the rate-limiting enzyme in catecholamine biosynthesis. Common genetic variation at the human TH promoter predicts alterations in autonomic activity and blood pressure, but how such variation influences human traits, and specifically whether such variation affects transcription, is not yet known.
Methods and Results
Pair-wise LD across the TH locus indicated that common promoter variants (C-824T, G-801C, A-581G, and G-494A) were located in a single 5′ LD block in white, black, Hispanic and Asian populations. Polymorphisms C-824T and A-581G were located in highly conserved regions, and were predicted to disrupt known transcriptional control motifs: MEF2, SRY, and FOXD1 at C-824T; and G/C-rich binding factors (such as SP1, AP2, and EGR1) at A-581G. At C-824T and A-581G, promoter/luciferase reporter plasmids indicated differential allele strength (T>C at C-824T; G>A at A-581G) under both basal circumstances and secretory stimulation. C-824T and A-581G displayed the most pronounced effects on both transcription in cella and catecholamine secretion in vivo. We further probed the functional significance of C-824T and A-581G by co-transfection of trans-activating factors in cella; MEF2, SRY, and FOXD1 differentially activated C-824T, while the G/C-rich binding factors SP1, AP2, and EGR1 differentially activated A-581G. At C-824T, factor MEF2 acted in a directionally coordinate fashion (at T>C) to explain the in vivo trait associations, while at A-581G factors SP1/AP2/EGR1 displayed similar differential actions (at G>A). Finally, chromatin immunoprecipitation (ChIP) demonstrated that the endogenous factors bound to the motifs in cella.
Conclusion
We conclude that common genetic variants in the proximal TH promoter, especially at C-824T and A-581G, are functional in cella, and alter transcription so as to explain promoter marker-on-trait associations in vivo. MEF2, FOXD1, and SRY contribute to functional differences in C-824T expression, while SP1, AP2, and EGR1 mediate those of A-581G. The SRY effect on TH transcription suggests a mechanism whereby males and females may differ in sympathetic activity and hence BP. These results point to new strategies for diagnostic and therapeutic intervention into disorders of human autonomic function, and their cardiovascular consequences.
Keywords: Tyrosine hydroxylase, hypertension, gene expression, catecholamine, genetics, autonomic, nervous system
INTRODUCTION
The enzyme tyrosine hydroxylase (tyrosine 3-monooxygenase; TH; EC-1.14.16.2) catalyzes the rate-limiting step in catecholamine biosynthesis: conversion of the amino acid L-tyrosine to L-dihydroxyphenylalanine (DOPA).
In previous clinical studies1, 2, we found that common allelic variation within the TH locus exerts a heritable effect on autonomic control of the circulation; changes in catecholamine secretion and the BP response to environmental stress suggested consequences for later development of cardiovascular disease traits. Common variants in the promoter region2, especially C-824T and A-581G, predicted heritable alterations in multiple autonomic traits, biochemical and physiological, and ultimately to the disease trait of hypertension. However, it was not clear whether such variants were in themselves functional, or simply exerted a statistical association with autonomic traits by virtue of close proximity to an actual causative locus (i.e., by linkage disequilibrium [or LD]),
In this study, we probed the functional consequences of variation in the proximal promoter region of TH, beginning with promoter haplotype/luciferase reporter plasmids. We went on to computational approaches, site-directed mutagenesis, and characterization of likely trans-acting factors, both endogenous (by chromatin immunoprecipitation, or ChIP) and exogenous (by co-transfection). We thereby developed evidence that variation in the promoter, especially at C-824T and A-581G, disrupts particular motifs (MEF2, SRY, and FOXD1 at C-824T; G/C-rich binding factors at A-582G) to alter transcriptional activity.
METHODS
Linkage Disequilibrium (LD)
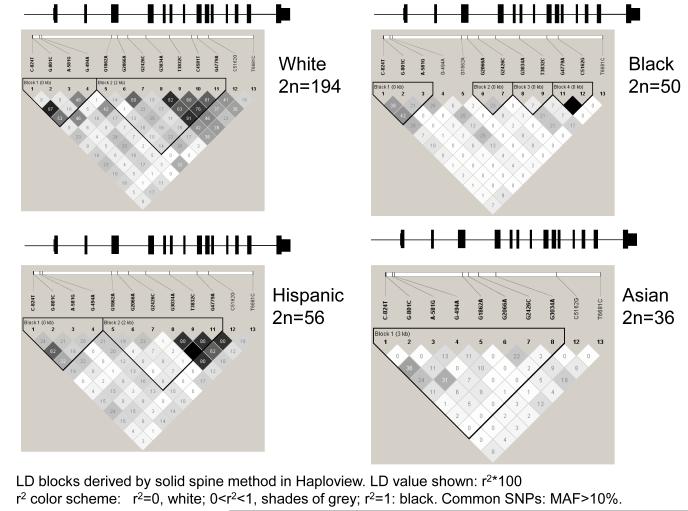
Patterns of linkage disequilibrium (LD) were analyzed and visualized by the software Haploview3. LD blocks were derived by the solid spline criterion in Haploview3, from unphased diploid genotypes of subjects from 4 diverse biogeographic ancestry groups systematically sequenced across the TH locus: white (European ancestry, 2n=194 chromosomes), black (sub-Saharan African ancestry, 2n=50), Hispanic (Mexican American, 2n=56), and east Asian (2n=36).
Statistical analyses
Statistical analyses were carried out with SPSS-11.5 (Chicago, IL). Parametric, general linear model (typically, 2-way ANOVA) analyses was used to test the associations of haplotypes and luciferase reporter activity for each construct. Interactions of promoter variants (haplotypes or SNPs) with stimuli (e.g., nicotine) was carried out in the univariate tests of the general linear model, wherein there was a single dependent variable (e.g., luciferase activity), and the interactive (multiplicative) effects of the two independent variables (e.g., haplotype and drug) could be modeled. Each factor (independent variable, or the interaction between two independent variables) was assessed for significance using F-tests, with significance defined as p≤0.05. Effects co-transfected transcription factors were also analyzed by both 2-tailed T-test and univariate tests of the general linear model.
Biochemical phenotyping: Catecholamines
Samples for measurement of plasma and urine catecholamines were quickly frozen at −70°C, prior to a sensitive radioenzymatic assay based on catechol-O-methylation 4. The assay uses a pre-concentration step that increases sensitivity by ~10-fold over other catechol-O-methyltransferase-based assays and ~20-fold over many HPLC assays, permitting accurate measurement of basal plasma epinephrine levels, which are at the limit of sensitivity for HPLC assays. Urine catecholamine values were normalized to creatinine excretion in the same sample.
Computational prediction of transcription factor binding motifs overlying TH promoter common variants
Multiple sequence alignments were performed by Clustal-W. Potential transcription factors binding motifs were predicted beginning with the web-based “phylogenetic footprinting” methods at Consite <http://asp.ii.uib.no:8090/cgi-bin/CONSITE/consite/>5 and Conreal <http://conreal.niob.knaw.nl/> 6, focusing on areas of conserved sequence across species. We also studied potential motifs with MacVector7, evaluating both alleles of each biallelic variant.
Exogenous/co-transfected transcription factors
Plasmids containing cDNAs encoding transcription factors MEF2 and FOXD1 were from Open Biosystems (Huntsville, AL). SP1, AP2α, and EGR-1 were from Stratagene (La Jolla, CA). MEF2 and its dominant-negative mutant mMEF2 8 were from John C. McDermott (York University, Toronto, Canada). cDNAs were either obtained in pCMV expression plasmids, or subcloned into pcDNA-3.1 (Invitrogen, Carlsbad, CA). The SRY full-length open reading frame was amplified from human male genomic DNA and subcloned into pcDNA3.1 vector between KpnI and XhoI sites.
TH promoter haplotype/luciferase reporter activity assays
Human TH haplotype-specific promoter fragments, corresponding to TH -944/-1 bp in TH isoform B (source clones NM_000360, NT_009237.17, and NP_000351), were PCR-amplified from genomic DNA of known homozygotes (after resequencing), and cloned into the promoter-less firefly luciferase reporter plasmid pGL3-Basic (Promega, Madison, WI). Promoter positions are numbered upstream (−) of the start codon. Common haplotypes (>1-5% frequency) of the TH promoter were generated by site-directed mutagenesis from a known haplotype, verified by sequencing, and numbered as previously described2. Supercoiled plasmids were purified on columns (Qiagen, Valencia, CA) prior to transfection.
Luciferase reporter assays of TH promoter variants
PC12 pheochromocytoma cells were transfected (at 50-60% confluence, 1 day after 1:4 splitting) with 1 μg of supercoiled promoter haplotype-firefly luciferase reporter plasmid and 10 ng of the Renilla luciferase expression plasmid pRL-TK (Promega Inc., Madison, WI) as an internal control per well, by the liposome method (Superfect; Qiagen, Valencia, CA). The firefly and Renilla luciferase activities in cell lysates were measured 16-24 hours after transfection, using the Dual Luciferase® reporter assay system (Promega, Madison, WI), and the results were expressed as either the ratio of firefly/Renilla luciferase activity, or firefly activity/total protein in the lysate9. Each experiment included four replicates. Results were expressed as mean ± SEM. Statistical significance was calculated using Student’s t-test or one-way ANOVA, and significance was established at the p <0.05 level.
Sympathochromaffin secretory stimulation: Nicotine and pituitary adenylate cyclase-activating peptide (PACAP) effects on TH promoter haplotype/luciferase reporter activity
PC12 cells were treated with nicotine (1 mM) or PACAP (200 nM), versus mock, in normal growth media after transfection. Luciferase activity was assayed with D-luciferin sodium (Sigma, St. Louis, MO), and normalized to total protein. Results are shown as the fold-stimulation by treatment.
Trans-activation: Co-transfection of transcription factors with TH promoter haplotype/reporters
50 or 100 ng of each transcription factor plasmid, or pcDNA-3.1 empty vector, were co-transfected into PC12 cells, along with 1 μg of TH promoter haplotypes differing by particular variants. Luciferase activities were assayed as above and normalized by total protein. Reaction of the TH promoter to trans-activation was expressed as fold-change of the mean value of luciferase activity between the transcription factor-transfected group and the mock-transfected (empty vector, pcDNA-3.1) group.
Chromatin Immunoprecipitation (ChIP)
PC12 cells were transfected with particular TH promoter haplotype/reporter to get the different allele for each variant. ChIP assays were carried out using the kit from Upstate Biotechnology (Lake Placid, NY, USA). Cells (5-10×106) were cross-linked in 1% formaldehyde for 10 min at 37°C and washed with ice-cold PBS containing a protease inhibitor mixture (Sigma P8340; 10 μg/ml), then resuspended in cell lysis buffer (0.1% Triton X-100, 10 mM KCl, and 10 mM Tris, pH 8). After sonication (Branson) and centrifugation, samples were pre-cleared with a salmon sperm DNA/protein A agarose 50% slurry to reduce non-specific background, and incubated with factor-specific antibodies (Santa Cruz Biotechnology [SCBT]; or pre-immune IgG as a negative control) at 4°C overnight with rotation. Characteristics of the SCBT antibodies were as follows: MEF2, sc-313X, rabbit polyclonal IgG; Foxd1, sc-47585X, goat polyclonal IgG; SRY, sc-8233X, goat polyclonal IgG; Sp1, sc-420X, mouse monoclonal IgG; Ap2, sc-184X, rabbit polyclonal IgG; Egr-1, sc-110X, rabbit polyclonal IgG. The immune complex was captured with a salmon sperm DNA/protein A slurry after incubation at 4°C for 1 hour. The immune complex was pelleted, washed 4 times and eluted by 1%SDS/0.1 M NaHCO3, then cross-links were reversed by heating in 0.2 M NaCl at 65°C for 4 hours, followed by proteinase K digestion at 45°C for 1 hour. The DNA was subsequently extracted and purified with QIAquick PCR kits (Qiagen).
Immunoprecipitated nucleosomal DNA samples were analyzed by PCR using primers forming an amplicon that encompassed the C-824T (-908 to -754 bp, sense: 5′-CTGGTGGGGTAGAGGAGAAA-3′, antisense: 5′-ACCCCACTCAGCCCCTCTAT-3′) or A-581G (-671 to -512 bp, sense: 5′-GCGTGGCGTCTCCTTAGA-3′, antisense: 5′-GAGTGCCATCTGCCCACA-3′) sites in the human TH promoter. Extracted DNA from the chromatin fractions before antibody precipitation was used as a positive control (“input DNA”). To ensure that the PCR amplification was in the linear range, reactions with different amounts of input DNA samples were carried out for various (typically 15-40) cycle numbers; a linear range of amplification typically occurred at ~25 cycles. After amplification, PCR products were separated on 1.5% agarose gels.
Chromatin isolation and PCR after the transfection of TH promoter into PC12 cells
TH promoter haplotype-1 (6 μg in 6 μl) was transfected into 10-cm plates of PC12 cells (~107 cells/plate) at ~80-90% confluence, as described above. Chromatin was isolated by differential centrifugation, as outlined below10, with a protocol from <http://www.lamondlab.com/pdf/chromatinisolation.pdf>. Briefly, 24 hours post-transfection, cells were lysed in an isotonic buffer containing 0.1% Triton-X100, then centrifuged at low speed (1300G at 4°C for 5 min), to obtain a low-speed pellet (P1) and low-speed supernate (S1), after which the low-speed supernate (S1) was centrifuged at high speed (20000G at 4°C for 5 min) to obtain a soluble “cytosolic” fraction (S2). The low-speed pellet (P1) was then subjected to hypotonic nuclear lysis and centrifuged at low speed (1700G at 4°C for 5 min) to obtain a soluble nuclear fraction (S3) and a chromatin-enriched fraction (P3). P3 was re-suspended in 100 μl of distilled water. Aliquots of equal amounts (2 μg protein each) of the S2, S3 and P3 fractions were subjected to PCR amplification with the same primers flanking the C-824T polymorphism, as were used for ChIP, as described above. After amplification or 15, 20, or 25 cycles, PCR products were separated on 1.5% agarose/TBE gels, followed by ethidium bromide (0.2 mg/L) staining.
RESULTS
Linkage disequilibrium (LD) patterns in the TH promoter region
We scored 13 common variants (MAF>10%) spanning the locus in subjects of European, African, Hispanic, or Asian ancestry. To visualize patterns of SNP associations, pair-wise correlations among the 13 common SNPs were quantified as linkage disequilibrium (LD) parameter r2 by the solid spine criterion in Haploview3 across the TH locus. In these subjects, a block of LD in the promoter region was maintained in subjects of European, Hispanic and Asian ancestry, though in blacks the promoter block was somewhat truncated, and in Asians the 5′-block extended into exon-4 (Figure 1A). TH promoter allele and haplotype frequencies differed across the 4 biogeographic ancestry groups (On-line supplementary Table1).
Figure 1A. Genetic variation at the human TH locus.
LD blocks across the locus in 4 biogeographic ancestry groups. Common (minor allele frequency >10%) SNPs used to demonstrate LD (numbering from the ATG translational start codon): C-824T, G-801C, A-581G, G-494A, G1862A, G2066A, G2426C, G3034A, T3832C, C4581T, G4779A, C5162G, and T6681C. LD blocks were assigned by the solid spline method in Haploview. The TH exon/intron structure is superimposed above each panel; there were no common variants in the final (most 3′) exon.
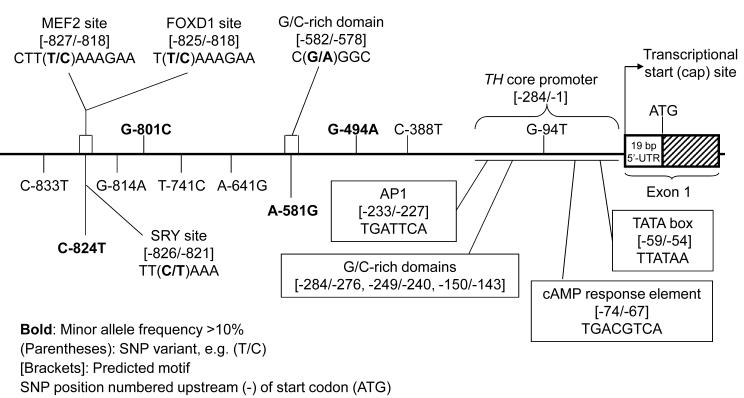
Genetic variation in the proximal human TH promoter: Motifs
Numbering upstream (5′, - [minus]) from the translational start codon (ATG), motifs identified included: -59/-54 TATA box (TTATAA), -74/-67 cAMP response element (TGACGTCA), and -233/-227 AP1 site (TGATTCA). Several G/C rich regions (-284/-276, -249/-240, -150/-143) were also noted in the proximal promoter. By resequencing across the TH locus2, we identified 10 polymorphisms in the promoter, 4 of which were common (MAF>10%): C-824T (rs10770141, 38.7%), G-801C (rs10840490, 11.6%), A-581G (rs10770140, 38.1%), and G-494A (rs11042962, 43.1%). Of note, the very proximal “core” promoter (-284/-1 bp) was devoid of common variation. Using prediction algorithms, we identified motifs likely to be disrupted by the promoter variants (Figure 1B). Among these, C-824T may alter the binding potential for factors MEF2, FOXD1 and SRY, while A-581G is located in a G/C rich region, a likely binding site for such factors as SP1, AP2 and EGR1. We did not identify motifs altered by common variants G-801C or G-494A.
Figure 1B. Genetic variation at the human TH locus.
Genetic variation in the proximal TH promoter: Effect on domains and motifs. The C-824T polymorphism interrupts motifs for transcription factors MEF2, FOXD1 and SRY, while A-581G disrupts a G/C-rich domain, likely altering binding of transcription factors such as SP1, AP2 and EGR1. Functional elements in the core TH promoter are derived from previous reports in the literature, largely documenting sites in the rat promoter11-13.
Effect of TH promoter variants on transcriptional activity
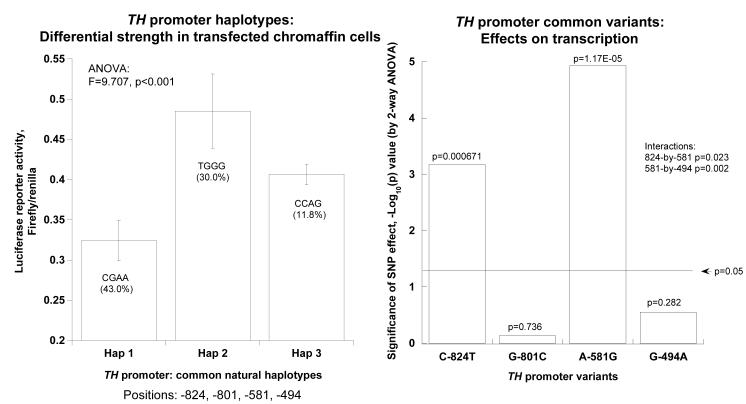
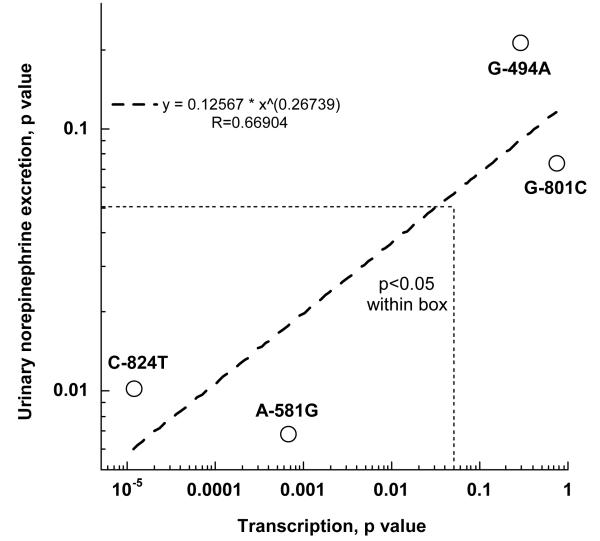
Twenty-four hours after transfection into PC12 cells, human TH promoter haplotypes showed substantial differences in luciferase reporter activity (at p<0.001, Figure 2A, left panel). Among the common haplotypes, haplotype-2 (at 30% of chromosomes) was more active (p<0.001) than haplotype-1 (at 43%). Information on the haplotypes and their frequencies in different biogeographic ancestry groups is provided in on-line supplemental Table 1. By 2-way ANOVA, 2 of the 4 common variants (C-824T, A-581G) altered transcriptional activity (each at p<0.0001, Figure 2A, right panel), though the other 2 had no independent effects. Common variants C-824T and A-581G displayed interactive effects on transcription (p=0.00067), while variants A-581G and G-494A also interacted (p=1.17E-05). The interactions indicate that the effect of any particular SNP is highly dependent on genetic background (i.e., other SNPs in the same promoter haplotype). Comparison of these in cella results with the effects of the same 4 variants in vivo (Figure 2B) indicates that the same variants (C-824T and A-581G) that exert the greatest effect on TH transcription in cella also have the most pronounced effect on human catecholamine excretion in vivo 14.
Figure 2A. TH promoter haplotype activities. Statistical analyses were based on results from 8 naturally occurring haplotypes (haplotypes 1, 2, 3, 7, 8, 9, 11, and 13; on-line supplemental Table 1).
Basal activity.
Left panel: Haplotype effects. Differential luciferase reporter activity in transfected pheochromocytoma (PC12) cells. Here only the 3 most common haplotypes (frequency ≥10%) are shown, based on the 4 common promoter variants: C-824T, G-801C, A-581G, and G-494A. ANOVA is shown for haplotype number as the independent variable. Results of ANOVA are shown for the haplotype as the independent variable, and luciferase activity as the dependent variable.Right panel: Individual SNP effects. Results of ANOVA are shown for the individual SNP as the independent variable, with luciferase activity as the dependent variable. A −log[10]p value plot is shown for significance of each of the 4 common variants.
Figure 2B. TH promoter haplotype activities. Statistical analyses were based on results from 8 naturally occurring haplotypes (haplotypes 1, 2, 3, 7, 8, 9, 11, and 13; on-line supplemental Table 1).
TH promoter common variants: Effects on gene expression in cella and in vivo. The common variants C-824T and A-581G that exert the greatest effect on TH transcription (horizontal axis) also have the most pronounced effect on human catecholamine excretion in vivo (vertical axis). The horizontal axis is derived from p values obtained by ANOVA with the individual SNP as the independent variable, and luciferase activity as the dependent variable (Figure 2A, right panel). Results in vivo (vertical axis) were obtained in catecholamine-phenotyped subjects of self-identified European ancestry, in analyses wherein the independent variable was the promoter SNP, and the dependent variable was renal norepinephrine excretion. Thus, both analyses utilized individual SNPs (rather than haplotypes) as the independent variables.
Effect of secretory stimulation (nicotine and PACAP) on transcriptional efficiency of TH promoter variants
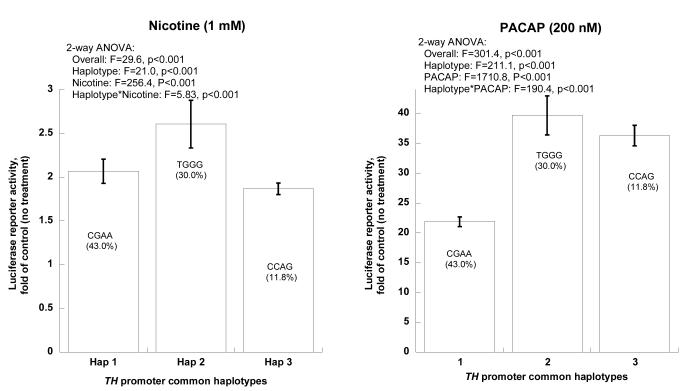
TH promoter/luciferase reporters exhibited uniformly greater activity when treated by the secretory stimuli nicotine (1 mM) or PACAP (200 nM), with an especially marked effect of PACAP. Consistently, haplotype-2 displayed greater responses to both nicotine (~2.6-fold) and PACAP (~39.7-fold) than did haplotype-1 (~2.07-fold for nicotine, ~21.9-fold for PACAP; Figure 2C).
Figure 2C. TH promoter haplotype activities. Statistical analyses were based on results from 8 naturally occurring haplotypes (haplotypes 1, 2, 3, 7, 8, 9, 11, and 13; on-line supplemental Table 1).
Secretory stimulation. Effects of nicotine or PACAP on common TH promoter haplotype transcriptional activities. Bars show the fold-increase of luciferase reporter activity after stimulation, for each common haplotype.
We then considered the individual variants, on balanced haplotype backgrounds (Figures 2D, 2E). Information on the haplotypes used in this study is given in on-line supplemental Table 2.
Figure 2D. TH promoter haplotype activities. Statistical analyses were based on results from 8 naturally occurring haplotypes (haplotypes 1, 2, 3, 7, 8, 9, 11, and 13; on-line supplemental Table 1).
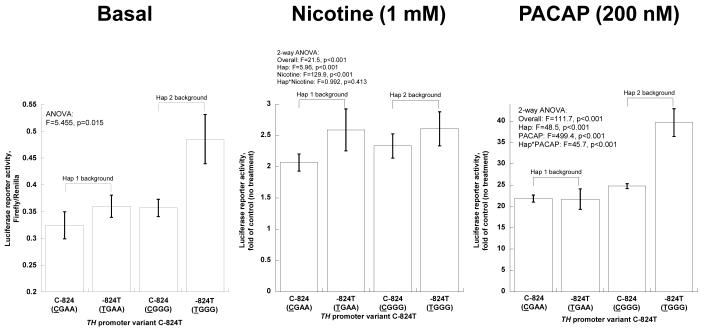
TH promoter common variant C-824T: Functional variation under basal and stimulated circumstances. Experiments were conducted on two haplotype backgrounds. Bars show mean ± SEM luciferase activity for each state. C-824 (CGAA) is from haplotype-1, -824T (TGAA) from haplotype-13, C-824 (CGGG) from haplotype-8, and -824T (TGGG) from haplotype-2 (on-line supplemental Table 2). Results are shown as fold of basal (no drug treatment).
Figure 2E. TH promoter haplotype activities. Statistical analyses were based on results from 8 naturally occurring haplotypes (haplotypes 1, 2, 3, 7, 8, 9, 11, and 13; on-line supplemental Table 1).
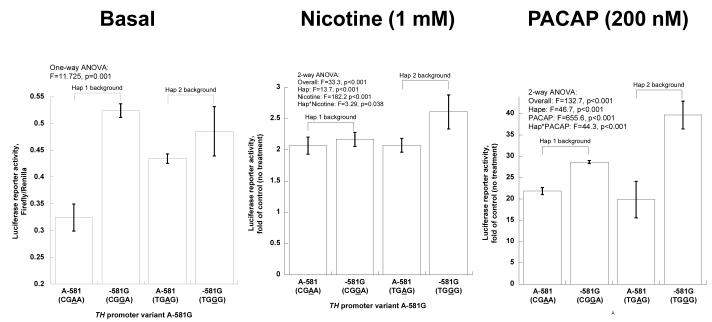
TH promoter common variant A-581G: Functional variation under basal and stimulated circumstances. Experiments were conducted on two haplotype backgrounds. Bars show mean ± SEM luciferase activity for each state. A-581 (CGAA) is from haplotype-1, -581G (CGGA) from site-directed (artificial) mutation 1M of haplotype-1, A-581 (TGAG) from haplotype-11, and -581G (TGGG) from haplotype-2 (on-line supplemental Table 2). Results are shown as fold of basal (no drug treatment).
C-824T
Under basal circumstances (Figure 2D, left), greater activity of the T allele was most apparent on the hapotype-2 background. When stimulated by nicotine, C-824T responded differentially, with an increased effect of the drug on the T allele, on either Hap-1 or Hap-2 backgrounds (Figure 2D, middle). When stimulated by PACAP, the increased response of the T allele was most apparent on the hap-2 background (Figure 2D, right),
A-581G
In the basal state (Figure 2E, left), greater activity of the G allele was especially apparent on the hap-1 background. During nicotine stimulation, greater activity of the G allele was most apparent on the hap-2 background (Figure 2E, middle). In response to PACAP, the G allele was more responsive on either background, though most prominently on hap-2 (Figure 2E, right).
TH C-824T polymorphism: Role of transcription factor MEF2
Sequence conservation/alignment
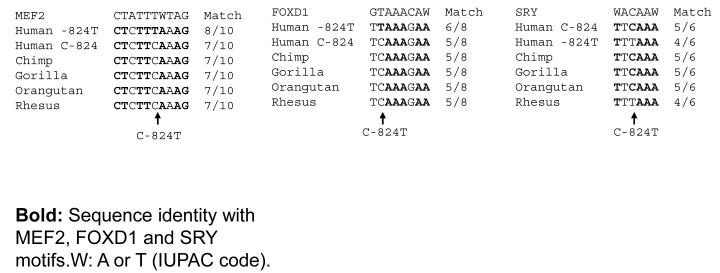
C-824T is located in a region highly conserved across sequenced primates (on-line complementary Figure 1), with the C allele ancestral in the primate lineage. In this local region, there is a partial consensus match for an MEF2 site (Figure 3A), with an improved match for the T allele (8/10 bp) over the C allele (7/10 bp).
Figure 3A. TH promoter variant C-824T.
Consensus motifs match for MEF2, FOXD1 and SRY at C-824T across primate species.
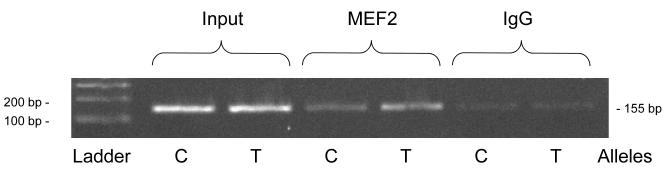
Endogenous MEF2 (by ChIP) (Figure 3B)
Figure 3B. TH promoter variant C-824T.
Endogenous MEF2: ChIP. The C allele (CGAA) is from haplotype-1, and the T allele (TGAA) from haplotype-13 (on-line supplemental Table 2).
We probed the potential interaction of endogenous MEF2 with C-824T by ChIP: we found that both the C and T alleles were indirectly captured by an antibody directed toward MEF2, with a suggestion of increased binding by the T allele.
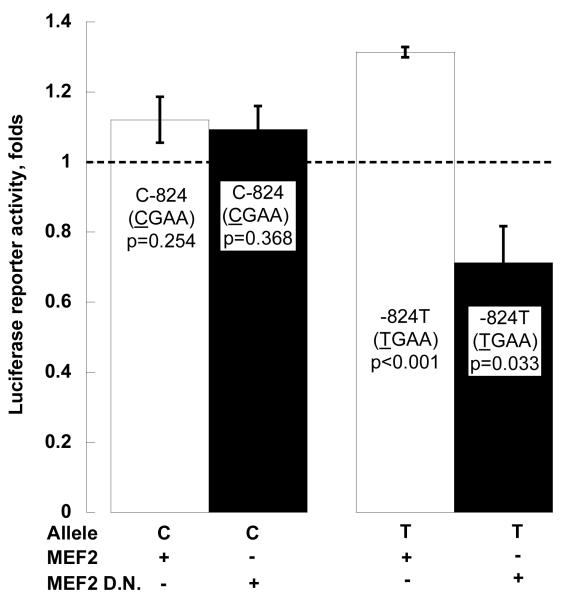
Exogenous (transfected) MEF2 (Figure 3C)
Figure 3C. TH promoter variant C-824T.
Exogenous MEF2: Co-transfection of an expression plasmid for MEF2, or its dominant negative (D.N.) mutant, mMEF2. The C allele (CGAA) is from haplotype-1, and the T allele (TGAA) from haplotype-13 (on-line supplemental Table 2). Results are shown as fold of basal (co-transfection with empty pCMV vector, shown as the dashed line).
Co-transfected human MEF2 increased TH promoter expression to 131.3% on the T allele (p<0.001), but had little effect on C allele (p=0.254). MEF2 dominant negative mutant mMEF2 decreased TH promoter expression down to 71.3% on the T allele (p=0.033), but had little effect on the C allele (p=0.368). Two-way ANOVA documented the substantial effects of MEF2 and its dominant negative mutant on TH transcription (F=11.931, p=0.001).
TH C-824T polymorphism: Role of transcription factors FOXD1 and SRY
Sequence homology/alignment
C-824T also displayed partial consensus matches for transcription factors FOXD1 and SRY (Figure 3A). At FOXD1, the T allele was a better match than C (6/8 versus 5/8 bp), while at SRY, the C allele displayed a better match than T (5/6 versus 4/6 bp, Figure 3A).
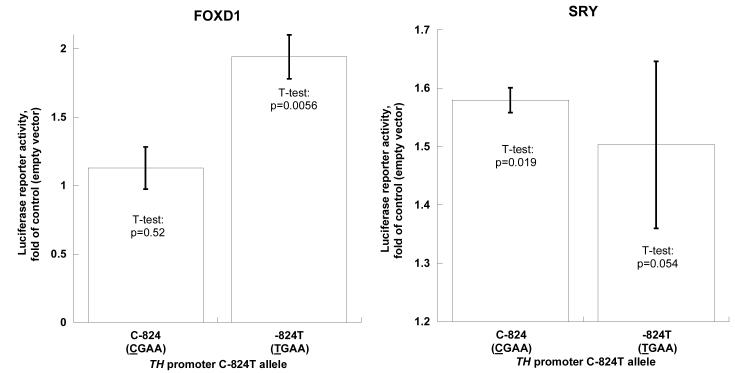
Exogenous (transfected) factors
When co-transfected with FOXD1 into PC12 cells, the T allele was substantially stimulated (1.94-fold, p=0.0056), while the C allele had little response (1.12-fold, p=0.52). When co-transfected with SRY, the C allele displayed only a slightly greater response (1.57-fold, p=0.019) than the T allele (1.50-fold, p=0.054, Figure 4A). Two-way ANOVA showed that both FOXD1 (F=11.369, p=0.006) and SRY (F=22.183, p=0.001) had significant effects on TH promoter transcription (Figure 4A).
Figure 4A. TH promoter variant C-824T: FOXD1 and SRY motifs.
Exogenous factors: Co-transfection of an expression plasmid for FOXD1 or SRY. The C allele (CGAA) is from haplotype-1, and the T allele (TGAA) from haplotype-13 (on-line supplemental Table 2). Results are shown as fold of basal (co-transfection with empty pCMV vector).
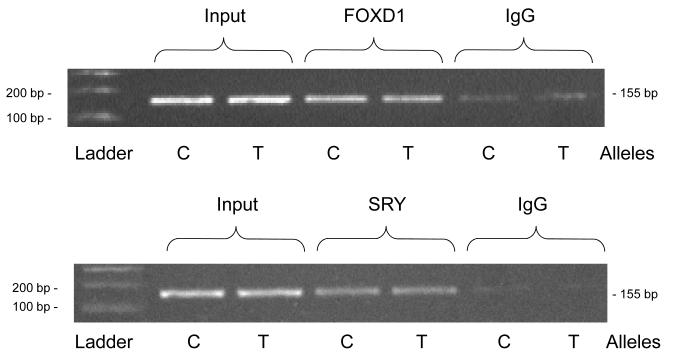
Endogenous factors (by ChIP)
During ChIP, both the C and T alleles could be captured by anti-FOXD1 and anti-SRY antibodies (Figure 4B), with little difference between the two alleles in quantity of PCR product.
Figure 4B. TH promoter variant C-824T: FOXD1 and SRY motifs.
Endogenous FOXD1 or SRY: by ChIP. The C allele (CGAA) is from haplotype-1, and the T allele (TGAA) from haplotype-13 (on-line supplemental Table 2).
TH A-581G polymorphism: Effect of transcription factors SP1, AP2 and EGR1
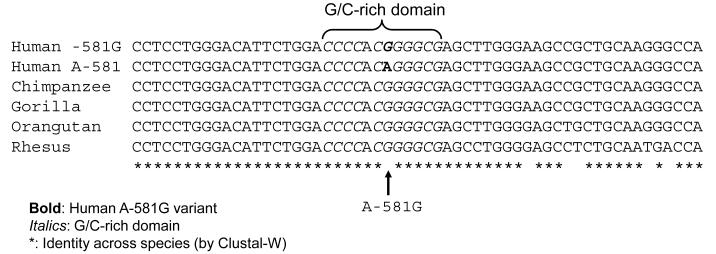
Sequence homology/alignment (Figure 5A)
Figure 5A. TH promoter variant A-581G: G/C-rich motif.
Inter-species primate sequence homology in the region of A-581T.
TH A-581G is located in conserved a G/C-rich region, CCCCAC[G/A]GGGCG, with the G allele ancestral in the primate lineage. Computationally, the G allele is predicted to exhibit enhanced binding to transcription factors SP1, AP2 and EGR1.
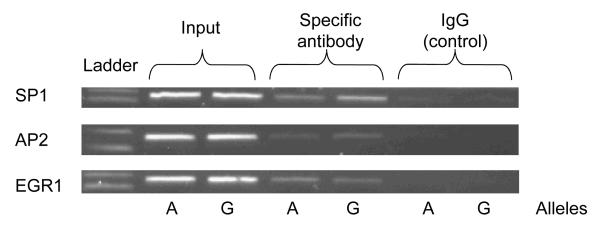
Endogenous factors (by ChIP)
During ChIP, each of transcription factors (SP1, AP2 and EGR1) on the target promoter could be captured by specific antibodies, with a greater signal than IgG control serum, though the two alleles did not differ substantially in binding (Figure 5B).
Figure 5B. TH promoter variant A-581G: G/C-rich motif.
Endogenous factors by ChIP: SP1, AP2, EGR1. The A allele (TGAG) is from haplotype-11, and the G allele (TGGG) from haplotype-2 (on-line supplemental Table 2).
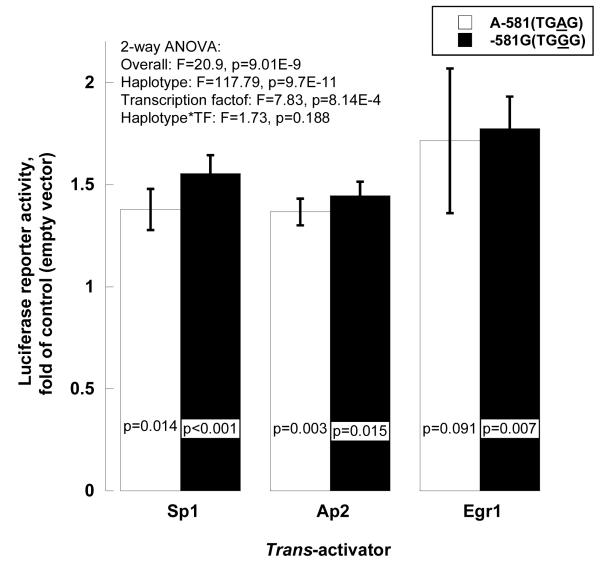
Exogenous (transfected) factors
Each of the three transcription factors (SP1, AP2 and EGR1) increased gene expression, with a greater response for the G than the A allele (Figure 5C).
Figure 5C. TH promoter variant A-581G: G/C-rich motif.
Exogenous factors by co-transfection: SP1, AP2, EGR1. The A allele (TGAG) is from haplotype-11, and the G allele (TGGG) from haplotype-2 (on-line supplemental Table 2). Results are shown as fold of basal (co-transfection with empty pCMV vector).
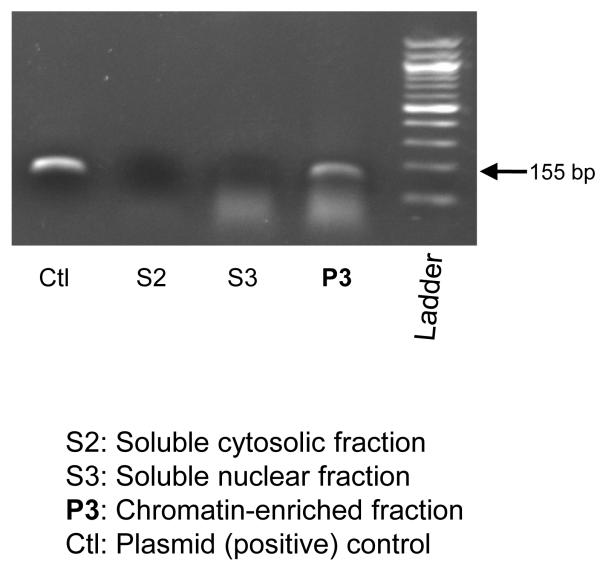
Presence of the transfected TH promoter plasmid in the chromatin fraction of PC12 cells
To explore whether the ChIP technique is appropriate for transfected plasmids, we studied the subcellular localization of the human TH promoter/luciferase reporter plasmid after transfection/expression in PC12 cells. When different fractions were analyzed by PCR, using primers flanking the C-824T site, the plasmid was localized to the P3 (chromatin) fraction of the cell (Figure 6); the plasmid was not observed in either the cytosolic or the soluble nuclear fractions of the cell.
Figure 6. Chromatin isolation from human TH promoter-transfected PC12 cells: Presence of the transfected plasmid in the chromatin fraction of the cell.
One day after transfection of the human TH-haplotype-1 promoter/luciferase reporter plasmid, subcellular fractionation was performed, and equal amounts (2.18 μg protein) of each subcellular fraction were used as template for PCR, over 15, 20 or 25 cycles, using an amplicon spanning TH promoter polymorphism C-824T. Results from 25 PCR cycles are shown. The PCR positive control plasmid was the human TH-haplotype-1 promoter/luciferase reporter (10 ng transfected). The chromatin isolation protocol was from the literature10.
DISCUSSION
Overview
Human tyrosine hydroxylase (TH) represents a rate-limiting point for control of catecholamine biosynthesis and potentially blood pressure. Our previous work showed that TH promoter common haplotypes (across C-824T→G-801C→A-581G→G-494A) exhibited pleiotropic effects on autonomic function, increasing both norepinephrine excretion and blood pressure during stress2. In those studies, individual variation at C-824T and A-581G were especially predictive of autonomic function, with the (minor) -824T and -581G alleles associated with increased catecholamine secretion and stress BP increments in twin pairs2. In this study, we explored how common genetic variation in the TH proximal promoter influences gene expression. We present evidence from several approaches (bioinformatic, expression activity, and binding) in which we found that both C-824T and A-581G conferred functional changes upon the TH promoter, and that particular transcription factors were implicated: at C-824T, MEF2, FOXD1, and SRY; and at A-581G, G/C-rich domain recognition factors SP1, AP2, and EGR1.
TH promoter variant C-824T
T allele with increased transcriptional activity
In the previous clinical study2, we discovered that the TH promoter C-824T minor (T) allele was associated with both higher urinary catecholamine excretion and greater changes in BP response to cold stress2. Here, we used transfected TH promoter haplotype/luciferase reporters to establish that that T allele confers increased transcriptional activity in cella (~1.36-fold greater than the C allele, p=0.0082; haplotypes-2 and -8). In addition, the T allele displayed an augmented response to typical chromaffin cell secretory stimuli such as nicotine or PACAP (T versus C: ~2.59- versus ~2.07-fold for nicotine, haplotypes-13 and -1; ~39.7- versus ~24.8-fold for PACAP, haplotypes-2 and -8).
Role of MEF2: Endogenous and exogenous
The T allele was predicted to have increased binding to transcription factor MEF2, and this increment was confirmed by ChIP, indicating that endogenous MEF2 can interact with the motif. Co-transfection of MEF2 yielded a greater increase in TH promoter/luciferase activity in the T than the C versions, and co-transfected dominant negative mMEF2 8 decreased TH promoter/luciferase reporter activity at the T but not the C version. Each of these in cella results suggests a differential role for MEF2 at the T allele in vivo.
C-824T is located in an A/T-rich region constituting a binding motif for the transcription factor MEF215. The MEF2/MADS family of transcription factors are widespread in expression, and involved in the regulation of multiple cellular processes including muscle differentiation, immune cell apoptosis, and the survival of neurons16, 17. Preferential activation of the T allele by MEF2 in cella may begin to explain the exaggerated in vivo effects of -824T on catecholamine secretion and blood pressure in the population14.
Role of SRY: Endogenous and exogenous
Sex-determining region Y (SRY), a transcription factor from the High Mobility Group family, initiates male sex determination in mammals 18. Previous reports indicated that an Sry expression plasmid trans-activated a Th promoter/reporter gene, although it was not clear whether the effect was direct19. Sry gene delivery to the rodent adrenal medulla increased not only Th enzymatic activity, but also plasma norepinephrine and ultimately blood pressure20; this finding, coupled with interaction of Sry with the tyrosine hydroxylase promoter, implicates the Sry allele as a hypertensive component of the SHR (spontaneously hypertensive rat) Y chromosome21. Our data showing that SRY binds to the C-824T motif, and increases TH transcription, suggest that SRY may play a role in maintenance of catecholamine synthesis and hence BP, and thus might contribute to well-documented male/female differences in population BP 2.
Role of FOXD1: Endogenous and exogenous
FOXD1 (FOrkhead boX D1; also known as FREAC4 or FKHL8) is a member of the forkhead/winged helix family of transcription factors, which are characterized by a conserved monomeric DNA-binding domain called the winged helix22. Although FOXD1 has been implicated in neuronal induction and differentiation, this is the initial report about its potential role in control of catecholamine biosynthesis.
Multiple factors
At C-824T, we found that the interaction of multiple factors (MEF2, SRY, and FOXD1) with their target motifs was likely to be disrupted by the polymorphic base; the evidence arose from bioinformatic, functional, and binding analyses. How such factors might interact at the motif in vivo, whether additively or synergistically, remains unexplored.
TH promoter A-581G
Transcriptional activity
In the previous clinical study 2, we found that the -581G allele predicted increased catecholamine secretion and stress BP changes in twin pairs. Here we found that the G allele promoted increased basal gene expression (luciferase activity) in cella, with augmented responses to the secretory stimuli nicotine and PACAP.
Role of G/C-rich binding factors
A-581G occurred in a relatively G/C rich region, which presented the potential for binding by such transcription factors as SP123, AP224 or EGR125. Here we found that co-transfection of either SP1, AP2, or EGR1 preferentially trans-activated the G allele. The effects of endogenous SP1, AP2 and EGR1 in cella were captured by ChIP. How might changes in the effects of such factors influence human catecholaminergic responses? EGR1 (Early Growth Response 1), a transcription factor with three zinc fingers of the Cys2His2 class26, is rapidly induced by stimuli modeling stress effects in experimental animal models27, 28, thereby altering transcription of genes encoding the catecholamine biosynthetic enzymes TH29, PNMT30, 31, and DBH32. This report represents an initial finding that naturally occurring genetic polymorphism in the catecholaminergic pathway may alter the binding patterns of G/C-rich motif recognition factors, such as EGR1, to affect autonomic responses in human populations.
Conclusions and perspectives
We conclude that TH promoter common polymorphisms C-824T and G-581A are not only statistical predictors of catecholamine secretory and stress BP traits in vivo2, but also causally responsible for alterations in transcriptional efficiency of the TH gene. These conclusions emerge from a convergence of computational and experimental approaches, in which experiments documented the effects of particular factors, both endogenous and exogenous. The pleiotropic effects of the two polymorphisms seem to arise from differential actions of specific transcription factors at the TH promoter: MEF2/SRY/FOXD1 at C-824T, and SP1/AP2/EGR1 at A-581G. The SRY effect on TH transcription suggests a mechanism whereby males and females may differ in sympathetic activity and hence BP. The results may augment our understanding of molecular events underlying inter-individual variations in autonomic function, and the genetic predisposition to such cardiovascular disease states as hypertension.
Stress exposure activates the sympathoneural system, resulting in catecholamine release mediating the “fight or flight” response. Chronic stress is associated with the development of numerous disorders including cardiovascular diseases such as hypertension. Probing genetic factors underlying stress responses may be important for prevention and treatment of such disorders. Tyrosine hydroxylase (TH) is the rate-limiting enzyme in biosynthesis of catecholamines. Here we studied two common genetic variants in the promoter (transcriptional control) region of the TH gene. Changes in gene expression seemed to arise from differential actions of particular transcription factors at the two variable sites. The results may augment our understanding of molecular events underlying inter-individual variations in autonomic function, and the genetic predisposition to such cardiovascular disease states as hypertension.
Supplementary Material
Acknowledgments
Funding Sources: National Institutes of Health, Department of Veterans Affairs.
Abbreviations
- AP2
Transcription factor, Activating enhancer-binding Protein 2.
- ChIP
Chromatin immunoprecipitation.
- EGR-1
Transcription factor, Early Growth Response protein 1.
- EMSA
Electrophoretic Mobility Shift assays.
- FOXD1
Transcription factor, Forkhead bOX D1.
- LD
Linkage disequilibrium
- MEF2
Transcription factor, Myocyte Enhancer Factor-2.
- SNP
Single Nucleotide Polymorphism.
- SP1
Transcription factor, Specificity Protein 1.
- SRY
Transcription factor, Sex-determining Region Y.
Footnotes
Conflict of Interest Disclosures: None.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Zhang L, Rao F, Wessel J, Kennedy BP, Rana BK, Taupenot L, Lillie EO, Cockburn M, Schork NJ, Ziegler MG, O’Connor DT. Functional allelic heterogeneity and pleiotropy of a repeat polymorphism in tyrosine hydroxylase: prediction of catecholamines and response to stress in twins. Physiol Genomics. 2004;19:277–291. doi: 10.1152/physiolgenomics.00151.2004. [DOI] [PubMed] [Google Scholar]
- 2.Rao F, Zhang L, Wessel J, Zhang K, Wen G, Kennedy BP, Rana BK, Das M, Rodriguez-Flores JL, Smith DW, Cadman PE, Salem RM, Mahata SK, Schork NJ, Taupenot L, Ziegler MG, O’Connor DT. Tyrosine hydroxylase, the rate-limiting enzyme in catecholamine biosynthesis: discovery of common human genetic variants governing transcription, autonomic activity, and blood pressure in vivo. Circulation. 2007;116:993–1006. doi: 10.1161/CIRCULATIONAHA.106.682302. [DOI] [PubMed] [Google Scholar]
- 3.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 4.Kennedy B, Ziegler MG. A more sensitive and specific radioenzymatic assay for catecholamines. Life Sci. 1990;47:2143–2153. doi: 10.1016/0024-3205(90)90314-h. [DOI] [PubMed] [Google Scholar]
- 5.Sandelin AWW, Lenhard B. ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 2004;32 doi: 10.1093/nar/gkh372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berezikov E, Guryev V, Cuppen E. CONREAL web server: identification and visualization of conserved transcription factor binding sites. Nucleic Acids Res. 2005;33:W447–450. doi: 10.1093/nar/gki378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rastogi PA. MacVector. Integrated sequence analysis for the Macintosh. Methods Mol Biol. 2000;132:47–69. doi: 10.1385/1-59259-192-2:47. [DOI] [PubMed] [Google Scholar]
- 8.Ornatsky OI, Andreucci JJ, McDermott JC. A dominant-negative form of transcription factor MEF2 inhibits myogenesis. J Biol Chem. 1997;272:33271–33278. doi: 10.1074/jbc.272.52.33271. [DOI] [PubMed] [Google Scholar]
- 9.Mahapatra NR, Mahata M, Ghosh S, Gayen JR, O’Connor DT, Mahata SK. Molecular basis of neuroendocrine cell type-specific expression of the chromogranin B gene: Crucial role of the transcription factors CREB, AP-2, Egr-1 and Sp1. J Neurochem. 2006;99:119–133. doi: 10.1111/j.1471-4159.2006.04128.x. [DOI] [PubMed] [Google Scholar]
- 10.Wysocka J, Reilly PT, Herr W. Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol Cell Biol. 2001;21:3820–3829. doi: 10.1128/MCB.21.11.3820-3829.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong DL, Tank AW. Stress-induced catecholaminergic function: transcriptional and post-transcriptional control. Stress. 2007;10:121–130. doi: 10.1080/10253890701393529. [DOI] [PubMed] [Google Scholar]
- 12.Lewis-Tuffin LJ, Quinn PG, Chikaraishi DM. Tyrosine hydroxylase transcription depends primarily on cAMP response element activity, regardless of the type of inducing stimulus. Mol Cell Neurosci. 2004;25:536–547. doi: 10.1016/j.mcn.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 13.Nagamoto-Combs K, Piech KM, Best JA, Sun B, Tank AW. Tyrosine hydroxylase gene promoter activity is regulated by both cyclic AMP-responsive element and AP1 sites following calcium influx. Evidence for cyclic amp-responsive element binding protein-independent regulation. J Biol Chem. 1997;272:6051–6058. doi: 10.1074/jbc.272.9.6051. [DOI] [PubMed] [Google Scholar]
- 14.Rao F, Wen G, Gayen JR, Das M, Vaingankar SM, Rana BK, Mahata M, Kennedy BP, Salem RM, Stridsberg M, Abel K, Smith DW, Eskin E, Schork NJ, Hamilton BA, Ziegler MG, Mahata SK, O’Connor DT. Catecholamine release-inhibitory peptide catestatin (chromogranin A(352-372)): naturally occurring amino acid variant Gly364Ser causes profound changes in human autonomic activity and alters risk for hypertension. Circulation. 2007;115:2271–2281. doi: 10.1161/CIRCULATIONAHA.106.628859. [DOI] [PubMed] [Google Scholar]
- 15.Karasseva N, Tsika G, Ji J, Zhang A, Mao X, Tsika R. Transcription enhancer factor 1 binds multiple muscle MEF2 and A/T-rich elements during fast-to-slow skeletal muscle fiber type transitions. Mol Cell Biol. 2003;23:5143–5164. doi: 10.1128/MCB.23.15.5143-5164.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao Z, Bonni A, Xia F, Nadal-Vicens M, Greenberg ME. Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science. 1999;286:785–790. doi: 10.1126/science.286.5440.785. [DOI] [PubMed] [Google Scholar]
- 17.Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA, Olson EN. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- 18.Dubin RA, Ostrer H. Sry is a transcriptional activator. Mol Endocrinol. 1994;8:1182–1192. doi: 10.1210/mend.8.9.7838151. [DOI] [PubMed] [Google Scholar]
- 19.Milsted A, Serova L, Sabban EL, Dunphy G, Turner ME, Ely DL. Regulation of tyrosine hydroxylase gene transcription by Sry. Neurosci Lett. 2004;369:203–207. doi: 10.1016/j.neulet.2004.07.052. [DOI] [PubMed] [Google Scholar]
- 20.Ely D, Milsted A, Bertram J, Ciotti M, Dunphy G, Turner ME. Sry delivery to the adrenal medulla increases blood pressure and adrenal medullary tyrosine hydroxylase of normotensive WKY rats. BMC Cardiovasc Disord. 2007;7:6. doi: 10.1186/1471-2261-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turner ME, Farkas J, Dunmire J, Ely D, Milsted A. Which Sry Locus Is the Hypertensive Y Chromosome Locus? Hypertension. 2008 doi: 10.1161/HYPERTENSIONAHA.108.124131. [DOI] [PubMed] [Google Scholar]
- 22.Pierrou S, Hellqvist M, Samuelsson L, Enerback S, Carlsson P. Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. Embo J. 1994;13:5002–5012. doi: 10.1002/j.1460-2075.1994.tb06827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferrari N, Desmarais D, Royal A. Transcriptional activation of the neuronal peripherin-encoding gene depends on a G + C-rich element that binds Sp1 in vitro and in vivo. Gene. 1995;159:159–165. doi: 10.1016/0378-1119(95)00140-2. [DOI] [PubMed] [Google Scholar]
- 24.Williams T, Tjian R. Characterization of a dimerization motif in AP-2 and its function in heterologous DNA-binding proteins. Science. 1991;251:1067–1071. doi: 10.1126/science.1998122. [DOI] [PubMed] [Google Scholar]
- 25.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 26.O’Donovan KJ, Tourtellotte WG, Millbrandt J, Baraban JM. The EGR family of transcription-regulatory factors: progress at the interface of molecular and systems neuroscience. Trends Neurosci. 1999;22:167–173. doi: 10.1016/s0166-2236(98)01343-5. [DOI] [PubMed] [Google Scholar]
- 27.Papanikolaou NA, Sabban EL. Sp1/Egr1 motif: a new candidate in the regulation of rat tyrosine hydroxylase gene transcription by immobilization stress. J Neurochem. 1999;73:433–436. doi: 10.1046/j.1471-4159.1999.0730433.x. [DOI] [PubMed] [Google Scholar]
- 28.Liu X, Kvetnansky R, Serova L, Sollas A, Sabban EL. Increased susceptibility to transcriptional changes with novel stressor in adrenal medulla of rats exposed to prolonged cold stress. Brain Res Mol Brain Res. 2005;141:19–29. doi: 10.1016/j.molbrainres.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 29.Nakashima A, Ota A, Sabban EL. Interactions between Egr1 and AP1 factors in regulation of tyrosine hydroxylase transcription. Brain Res Mol Brain Res. 2003;112:61–69. doi: 10.1016/s0169-328x(03)00047-0. [DOI] [PubMed] [Google Scholar]
- 30.Wong DL, Siddall BJ, Ebert SN, Bell RA, Her S. Phenylethanolamine N-methyltransferase gene expression: synergistic activation by Egr-1, AP-2 and the glucocorticoid receptor. Brain Res Mol Brain Res. 1998;61:154–161. doi: 10.1016/s0169-328x(98)00225-3. [DOI] [PubMed] [Google Scholar]
- 31.Kvetnansky R, Kubovcakova L, Tillinger A, Micutkova L, Krizanova O, Sabban EL. Gene expression of phenylethanolamine N-methyltransferase in corticotropin-releasing hormone knockout mice during stress exposure. Cell Mol Neurobiol. 2006;26:735–754. doi: 10.1007/s10571-006-9063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng SY, Serova LI, Glazkova D, Sabban EL. Regulation of rat dopamine beta-hydroxylase gene transcription by early growth response gene 1 (Egr1) Brain Res. 2008;1193:1–11. doi: 10.1016/j.brainres.2007.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.