Abstract
Redox-regulated proteins play fundamentally important roles not only during the defense of organisms against oxidative stress conditions but also as targets of cellular signaling events. This realization has spurred the development of proteomic techniques geared towards characterizing the redoxome; proteins with highly reactive cysteine residues, whose thiol oxidation state controls the function of the proteins, and by extension, the pathways they are part of. We will here summarize the most recent advances made in the field of redox proteomic analyses, aimed to elucidate the cellular redox networks that appear to control pro- and eukaryotic organisms.
Introduction
Cellular redox conditions set the pace for many processes, ranging from gene expression and protein translation to metabolism, proteostasis, cell signaling and apoptosis [1]. Thus it is not surprising that perturbations in redox homeostasis are associated with many different disease conditions, including cancer, type 2 diabetes, cardiovascular and neurodegenerative diseases, and aging [2,3]. Alterations in the cellular redox status or in the levels of specific reactive oxygen species (ROS) or nitrogen species (RNS) are most often sensed by proteins with redox-reactive cysteine residues, whose thiol oxidation states exert control over the activity of the proteins [4-6]. High levels of ROS are produced by cells of the innate immune system in response to bacterial infections, are found at sites of chronic inflammation, or are generated during many disease conditions, such as ischemic stroke [7-9]. High ROS levels trigger rapid oxidative activation of anti-oxidant transcription factors, including OxyR, Yap1p and Nrf2 [10], activate redox-regulated chaperones, such as Hsp33 and peroxiredoxin [11,12], and alter the function of many redox-senitive metabolic enzymes, like GAPDH, to re-route metabolic fluxes important for the restoration of redox homeostasis [6]. Low levels of ROS are instead continuously produced by NADPH oxidases, by proteins of the mitochondrial electron transport chain and during oxidative protein folding in the endoplasmic reticulum [13]. At this low level, ROS act as signaling molecules to mediate localized signaling events via the oxidative modification of redox sensitive key players, including phosphatases (e.g., phosphatase and tensin homologue, PTEN) and kinases (e.g., JNK, PKC) [13-15].
The types of oxidative thiol modifications that affect redox-sensitive cysteines depend on the local protein environment of the thiol groups and the prevailing redox conditions. An increase in the ratio of oxidized to reduced glutathione (GSSG to GSH), for instance, leads most often to S-glutathionylation (-S-S-G). Presence of hydrogen peroxide (H2O2), peroxynitrite (ONOO-) or hypochlorous acid (HOCl) commonly causes sulfenic (-SOH) acid formation [14,16]. Although stable sulfenic acid formation has been demonstrated to occur in a number of redox sensitive proteins (e.g., NADH peroxidase, GAPDH), their highly reactive nature often leads to further oxidation reactions with nearby thiols to form disulfide bonds (S-S) [17]. Alternatively, sulfenic acids are further oxidized to sulfinic (-SO2H) or sulfonic acids (-SO3H) (Fig. 1). Due to the largely irreversible nature of these overoxidation reactions, however, these thiol modifications usually play only minor roles in controlling redox-regulated proteins and serve as biomarkers for oxidative protein damage [18]. Presence of RNS has been shown to lead to S-nitrosylation (S-NO) in a large subset of redox-sensitive proteins [16,19] (Fig. 1).
Fig. 1. Select detection methods for oxidative cysteine modifications.
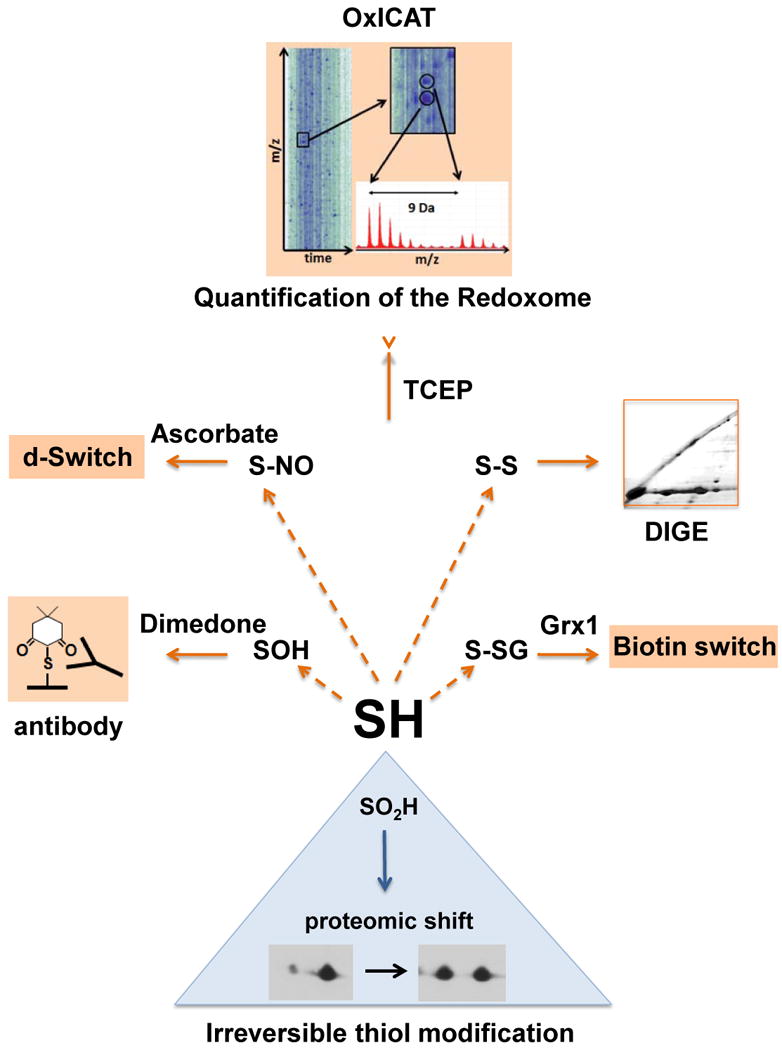
Reversible oxidative thiol modifications include cysteine sulfenic acid (-SOH), which can be specifically labeled with dimedone or dimedone derivatives, and either visualized using dimedone specific antibodies, or enriched and subsequently identified [27,29]. Proteins containing disulfide bonds (-S-S-) can be visualized using diagonal gel electrophoresis (DIGE), in which proteins are first separated under non-reducing conditions, followed by a separation under reducing conditions in the second dimension [55]. The biotin switch assay (see Fig. 2 for details) is commonly used for the detection of S-nitrosylated (-S-NO) and S-glutathionylated (-S-SG) proteins [16]. Very recently, a quantitative mass spectrometric biotin switch assay termed “d-Switch” was introduced, which quantifies S-nitrosylations using mass spectrometry [56]. OxICAT, a quantitative mass spectrometric method, monitors the absolute oxidation status of hundreds of different protein thiols in a single experiment [48]. Irreversible oxidation of cysteines to sulfinic acids (-SO2H) can be visualized and quantified as shifts in the isoelectric point of proteins on 2D gels.
Most oxidative thiol modifications are fully reversible in vivo and dedicated, highly conserved systems exist, which control the redox status of proteins and maintain the cellular redox homeostasis. Members of the thioredoxin family facilitate the reduction of sulfenated, nitrosylated and disulfide-bonded proteins [20], while glutaredoxins, such as Grx-1 reduce mixed disulfides with glutathione (Fig. 1) [21]. Recently, a highly conserved class of sulfinic acid reductases (i.e., sulfiredoxins) has been identified, which reduce overoxidized type 2 peroxiredoxins [22] and moonlight as de-glutathionylation enzymes [23]. This ability to rapidly and reversibly modify structurally and/or functionally important cysteine thiols makes redox regulation an excellent mechanism to regulate protein activity on a posttranslational level. With the development of highly quantitative redox proteomic methods, which we will summarize below, we have now the possibility to globally identify these redox-regulated proteins, collectively called “the redoxome” [24], and further characterize the many redox-sensitive networks that appear to exist in the cell.
Detection and in vivo relevance of sulfenic acids
Sulfenic acids are highly reactive precursors for most known stable oxidative thiol modifications, and thus play a central role in redox-regulatory processes [25]. In recent years, a number of innovative techniques have been developed to detect and identify proteins that show increased sulfenic acid formation [25]. Some of the methods rely on the ex vivo reduction of sulfenic acids with arsenite, a variation of the biotin switch assay (BSA) that is outlined in detail below [16]. Other methods make use of fluorescent- or affinity-tagged dimedone (5,5-dimethyl-1,3-cyclohexanedione) derivatives, which specifically alkylate and thereby tag sulfenic acids. Subsequent enrichment of the tagged proteins from complex samples allows their mass spectrometric identification [26]. Another very powerful approach has recently been introduced by the Carroll lab, who developed DAz-2 (4-(3-azidopropyl)cyclohexane-1,3-dione), a highly membrane permeable dimedone derivative, which can be used to trap sulfenic acids as they form in the cell [27]. DAz-2 labeled proteins were conjugated to phosphine-biotin via their azide group, purified and identified using LC-MS/MS. By using this method in HeLa cells, nearly all previously known sulfenic acid containing proteins were confirmed and more than 170 new candidate proteins were identified [27]. In contrast to these methods, which rely on the interaction of protein sulfenic acids with small alkylating reagents, a protein-based assay was recently introduced as well [28]. This assay makes use of a small His-tagged peptide, derived from the oxidative stress transcription factor Yap1, which contains one highly reactive cysteine. This cysteine traps sulfenic acids in other proteins by forming intermolecular disulfides. Ni-affinity chromatography serves to purify and enrich the proteins, which are identified upon DTT-mediated release from the Yap1-peptide. Using targeted expression of the Yap1p-peptide, this approach opens up the possibility to detect protein sulfenic acids even in distinct sub-cellular compartments [28]. In addition, a sulfenic acid-specific antibody is now available, which allows the detection and profiling of sulfenic acids using protein microarrays and the direct visualization of sulfenic acids as they develop in the cell using microscopy [29].
Taken together, detection of sulfenic acid-containing proteins has come a long way. However, a few major limitations still remain; the main concern is the instability of sulfenic acids, which makes them hard to trap. Identification of sulfenic acid containing proteins often requires their enrichment, which obscures any information about the extent to which a protein is actually sulfenated in the cell. Thus, the development of quantitative differential sulfenic acid trapping techniques is needed to distinguish between reduced, sulfenated or stably oxidatively modified cysteines within a protein population. Only then will we be able to differentiate between proteins that use sulfenic acid formation as final oxidation product, and proteins that use it as transient intermediate on their way to more stable oxidative thiol modifications. This distinction is crucial for our understanding of what sequence or structural features in proteins mediate the redox sensitivity of cysteine thiols and support the formation of stable oxidative modifications.
Biotin Switch Assay – Recent Advances to a Powerful Approach
Although semi-quantitative by nature, the biotin switch assay has greatly contributed to the elucidation of many redox-sensitive pathways in pro- and eukaryotic cells [16,19]. Countless S-nitrosylated, sulfenated proteins and S-glutathionylated proteins have been pulled out of complex protein mixtures by this approach. Originally introduced by Jaffrey and Snyder to identify S-nitrosylated proteins [30], and subsequently modified and optimized by numerous groups (see below), the principle behind this method is surprisingly simple (Fig. 2); free cysteines in complex protein mixtures are first alkylated using the reversible, thiol-specific alkylation reagent methyl methanethiosulfonate (MMTS) [30]. Existing oxidative thiol modifications are then reduced and switched for the stable biotin, hence the name. The crucial step lies in the choice of reducing agents, which mediates the biotin switch; incubation with ascorbate/CuCl2 [31] or siapinic acid, an apparently more S-nitrosylation-selective reductant [32], is used to specifically reduce and subsequently label S-nitrosylated proteins. Arsenite has been shown to specifically liberate sulfenated cysteines [33] while incubation of the alkylated protein mixture with glutaredoxin (Grx) triggers the specific reduction of S-glutathionylated cysteines [34] (Fig. 2). The biotin-tagged proteins are then either visualized by western blot and/or enriched for by affinity purification and subsequently identified [16].
Fig. 2. Detection of specific cysteine modifications - the biotin switch assay.
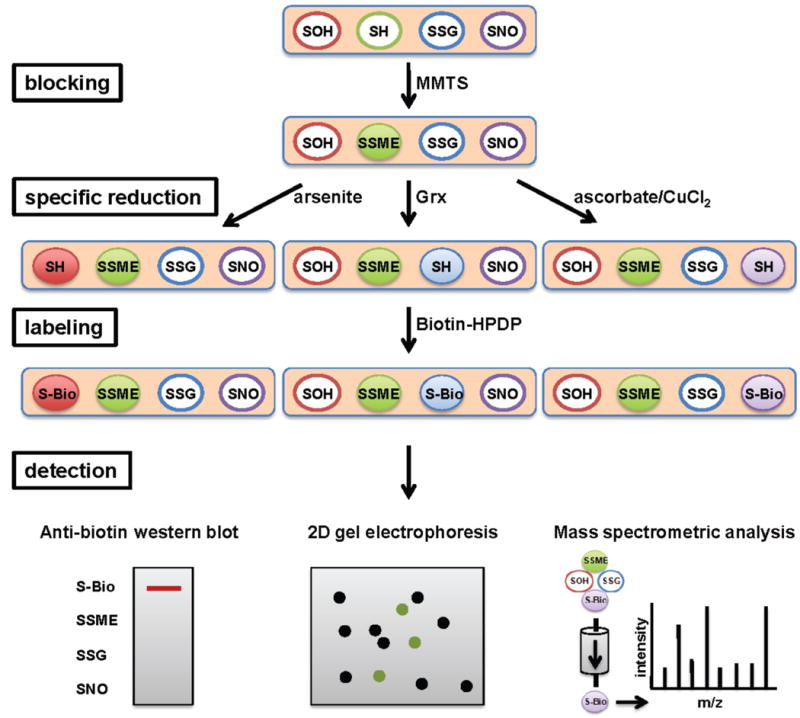
To detect sulfenated, nitrosated or glutathionylated proteins in cell lysates, samples are first incubated with the reversible thiol-alkylating reagent MMTS to block all free thiols. Then, depending on the type of cysteine modification present, proteins are either incubated with arsenite to reduce sulfenic acids, with ascorbate to reduce S-nitrosylations or with the small protein glutaredoxin (Grx) to reduce S-glutathionylated cysteines. These cysteine thiols are subsequently labeled with biotin-HPDP, thus tagging only those proteins, which originally contained the thiol modification. Proteins are then separated on 1D or 2D gels and biotinylated proteins are detected by western blot analysis using antibodies against biotin, or enriched by affinity chromatography and identified by MS analysis.
The realization that numerous important proteins and cellular pathways are regulated by S-nitrosylation drove the development of many variations on the biotin switch assay theme, some simply to increase sensitivity and detection of S-nitrosylated proteins, others to aid in the identification of the affected cysteines and the quantification of S-nitrosylation. The fluorescence-switch method, for instance, uses fluorescein-5-maleimide instead of biotin to label, visualize and identify nitrosylated proteins in endothelial cells using 2D gels [35]. The irreversible biotinylation procedure (i.e., IDP) introduced the use of the irreversible isotope-coded affinity tag (ICAT) labeling to identify S-nitrosylated cysteines using LC-MS/MS [36]. In SNO-SID (i.e., S-NO identification), a proteolytic cleavage step was introduced to enrich for only those peptides that previously carried the modification, an advance that led to the identification of nearly 70 nitrosylated peptides in rat cerebellum lysates [37]. The related SNO-RAC technique went a step further by incorporating quantitative iTRAQ tags to the purified peptides, which allowed the first quantitative comparison of nitrosylation profiles in various samples [38]. Finally, in 2010, the d-Switch method was introduced, which allows the quantification of S-nitrosylation at distinct sites in proteins using a differential thiol trapping approach combined with quantitative mass spectrometry (see below) [39].
Although the biotin switch assay has been instrumental in identifying many oxidatively modified proteins in complex protein mixtures, its development has seen its share of challenges. Ascorbate, for instance, appears to reduce not only S-nitrosylations but also “soft” cysteine modifications, including sulfenic acids and even disulfide bonds [40], and was found to require CuCl2 to increase its sensitivity [31]. The reversible thiol blocking reagent MMTS has been suggested to not only alkylate cysteines but to potentially even introduce oxidative thiol modifications into proteins [41]. These concerns illustrate how methods, as simple as they initially seem, require constant re-evaluation and improvement, and warrant the careful verification of identified oxidative thiol modifications by alternative strategies. Despite these labor pains, however, it is now clear that S-nitrosylated and S-glutathionylated proteins are involved in dozens of different cell signaling pathways, a realization that would not have been possible without this powerful method.
The Quantitative Redoxome – Determining the Thiol Oxidation State in vivo
One major advance in determining the in vivo thiol oxidation status of proteins came with the introduction of acid-trapping techniques, where cells are lysed with acids like trichloroacetic acid [42]. Since rates of thiol-disulfide exchange reactions decrease 10-fold for each pH unit below the cysteine's pKa value, low pH incubation effectively “freezes” the in vivo thiol oxidation status and prevents oxidation artifacts during cell lysis. Subsequent modification of all accessible thiol groups and comparison with the thiol oxidation status of proteins in control samples has contributed significantly to the view that proteins can undergo reversible thiol modifications in vivo. However, many of these initial studies failed to identify the affected cysteine(s), and, even more importantly, none were able to determine the absolute oxidation status of the cysteines. Yet this information is crucial to evaluate how oxidative thiol modification might affect the structure or function of the respective protein, and to assess whether a large enough population of any given protein is affected to make a physiological impact in the cell. One major advance came with the establishment of differential thiol labeling techniques, in which all reduced cysteines are first labeled with one alkylating agent, while all oxidized cysteines are, after their reduction, labeled with either another variant of the same reagent or a different alkylating reagent [43,44] (Fig. 3). In combination with 2D gels, this strategy provided information about the relative amount of oxidized protein over total protein, and redox changes could be expressed as fold-changes in oxidation status. This semi-quantitative technology has been successfully used to identify many redox sensitive proteins in bacteria, yeast and higher organisms, to determine the substrate proteins of many pro- and eukaryotic oxidoreductases, and to monitor changes in the redox status of proteins in response to disease conditions and aging [43,45-47].
Fig. 3. OxICAT – Identification of the redoxome.
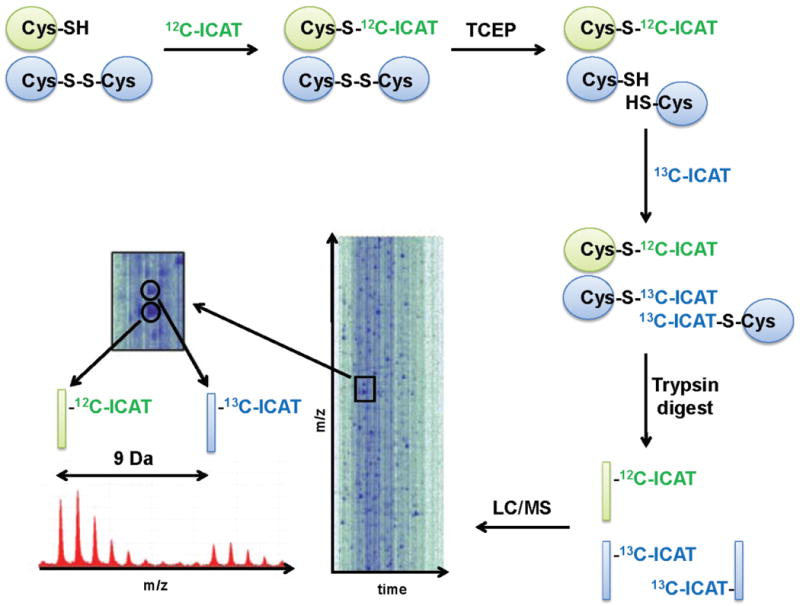
Cells or tissues are harvested on TCA to maintain the in vivo thiol redox status of the proteins. Protein samples are incubated with light 12C-ICAT under protein-denaturing conditions to label all reduced protein thiols. Light ICAT is removed, and, after reduction with the general reductant TCEP, all previously oxidized proteins are labeled with the 9 Da heavier 13C-ICAT. The differentially labeled proteins are digested with trypsin, enriched by affinity chromatography, separated by HPLC (LC) and identified by MS and tandem MS analysis. A graphical representation of an LC/MS run is shown. The cysteine-containing peptide of a protein that was partially oxidized in vivo co-elutes as an ICAT pair from the HPLC (inset). Analysis of the mass spectrum reveals two mass peaks, which are precisely 9 Da apart, corresponding to the light and heavy-labeled version of the peptide. Because the ICAT-labeled peptides are chemically identical, their behavior in the mass spec is identical and the relative signal intensity of the two peaks can be used to determine the absolute oxidation status of the cysteine in vivo. Tandem MS analysis is then used to identify the peptide and the cysteine(s) affected.
An important recent advance came with the realization by us and others that the highly quantitative ICAT, which comes in an isotopically light 12C-ICAT form and a 9 Da heavier 13C-ICAT form, is a thiol specific-trapping reagent and thus suitable for a quantitative redox proteomic approach [48,49] (Fig. 3). In this method, which we termed OxICAT, reduced cysteines are labeled with light ICAT, while all previously oxidized cysteines are upon reduction labeled with heavy ICAT. This differential labeling causes a mass difference of 9 Da or multiples of 9 Da depending on the number of oxidized cysteines. The biotin-moiety of the ICAT reagent allows the enrichment of only cysteine-containing peptides, which are then separated by HPLC and identified using MS and tandem MS analysis (Fig. 3). Mass spectrometric analysis not only provides the identification of the peptide and of the redox sensitive cysteine, but at the same time, delivers the exact oxidation status of the cysteine-containing peptides. Because both light and heavy labeled peptides are chemically identical, they co-elute from the HPLC and fly to the same extent in the mass spectrometer. Thus, mass peaks that are precisely 9 Da or multiples of 9 Da apart correspond to the reduced and oxidized species of the peptide, with the respective peak areas reflecting their true in vivo distribution (Fig. 3). By using the OxICAT method [48], the absolute oxidation status of thousands of individual thiols can be monitored and quantified in a single experiment. OxICAT is widely applicable and has been used to determine the steady-state redox status of high abundance proteins in the nucleus, to identify oxidation-sensitive proteins in uni- and multicellular organisms (e.g., E. coli, C. elegans), and to characterize and determine the redox sensitivity of defined proteins both in vitro and in vivo [48] [49-51]. While this method is very powerful and provides the exact oxidation states for thousands of peptides at the same time, many of which can be identified by MS/MS analysis, its analysis is time-consuming. An slightly less quantitative alternative is the comparative ICAT-analysis, which assesses the relative oxidation status of protein thiols between samples [53-55]. Reduced cysteines from one sample are labeled with light ICAT, while the reduced cysteines of the other sample are labeled with heavy ICAT. Like in other quantitative proteomics approaches, the samples are then mixed in equal ratios and any deviation from the expected equal signal intensity of ICAT pairs is interpreted as a potential change in the oxidation status of the corresponding peptide. However, since alterations in the steady-state peptide concentration lead to similar results, additional control experiments are necessary to confirm the redox sensitivity of the identified proteins.
Conclusions
Over the last decade, tremendous progress in the field of redox proteomics has been made. Hundreds of proteins have been identified, which use reversible thiol modifications to adjust their functions to the prevailing redox conditions and ROS levels in the cell, coining the term “Redoxome” [24]. By developing methods for the identification and quantification of oxidative cysteine modifications, many new insights in the field of redox-regulation have been gained. It is now clear that complex cellular pathways can be controlled by the redox status of as little as one single cysteine (e.g., active site cysteine in GAPDH), and that disturbances of the cellular redox homeostasis have severe physiological consequences, associated with an increasing number of pathological conditions. By applying these new quantitative tools to model systems and disease models, we have now the opportunity to ask more sophisticated questions about cause and effects of redox alterations, aimed to improve antioxidant therapies and intervention strategies with the ultimate goal to maintain cellular redox homeostasis.
Acknowledgments
We thank Dr. James Bardwell for critically reading this manuscript. We thank Dr. Jean-Francois Collet for providing us with the DIGE image shown in Fig.1. This work was supported by the National Institute of Aging Grant AG027349.
Abbreviations
- BSA
biotin switch assay
- DTT
dithiothreitol
- GapDH
Glyceraldehyde 3-phosphate dehydrogenase
- Grx
glutaredoxin
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- IAM
iodoacetamide
- IBP
irreversible biotinylation procedure
- ICAT
Isotope-Coded Affinity Tag
- iTRAQ
isobaric tag for relative and absolute quantitation
- MMTS
methanethiosulfonate
- MS
mass spectrometry
- NADH
Nicotinamide adenine dinucleotide
- NADPH
Nicotinamide adenine dinucleotide phosphate
- NEM
N-ethylmaleimide
- PTEN
Phosphatase and tensin homologue
- ROS
reactive oxygen species
- TCEP
tris(2-carboxyethyl)phosphine
References
*of special interest
** of outstanding interest
- 1.Halliwell B, Gutteridge JMC. Free radicals in biology and medicine. 4th. Oxford; New York: Oxford University Press; 2007. [Google Scholar]
- 2.Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- *3.Stadtman ER. Protein oxidation in aging and age-related diseases. Ann N Y Acad Sci. 2001;928:22–38. doi: 10.1111/j.1749-6632.2001.tb05632.x. [DOI] [PubMed] [Google Scholar]
- *4.Paulsen CE, Carroll KS. Orchestrating redox signaling networks through regulatory cysteine switches. ACS Chem Biol. 2010;5:47–62. doi: 10.1021/cb900258z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klomsiri C, Karplus PA, Poole LB. Cysteine-Based Redox Switches in Enzymes. Antioxid Redox Signal. 2010 doi: 10.1089/ars.2010.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandes N, Schmitt S, Jakob U. Thiol-based redox switches in eukaryotic proteins. Antioxid Redox Signal. 2009;11:997–1014. doi: 10.1089/ars.2008.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller RA, Britigan BE. Role of oxidants in microbial pathophysiology. Clin Microbiol Rev. 1997;10:1–18. doi: 10.1128/cmr.10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen CL, Bayraktutan U. Oxidative stress and its role in the pathogenesis of ischaemic stroke. Int J Stroke. 2009;4:461–470. doi: 10.1111/j.1747-4949.2009.00387.x. [DOI] [PubMed] [Google Scholar]
- 9.Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic Biol Med. 2010 doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antelmann H, Helmann JD. Thiol-based redox switches and gene regulation. Antioxid Redox Signal. 2010 doi: 10.1089/ars.2010.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winter J, Ilbert M, Graf PC, Ozcelik D, Jakob U. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell. 2008;135:691–701. doi: 10.1016/j.cell.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jang HH, Lee KO, Chi YH, Jung BG, Park SK, Park JH, Lee JR, Lee SS, Moon JC, Yun JW, et al. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell. 2004;117:625–635. doi: 10.1016/j.cell.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Veal EA, Day AM, Morgan BA. Hydrogen peroxide sensing and signaling. Mol Cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- *14.Dalle-Donne I, Rossi R, Colombo G, Giustarini D, Milzani A. Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem Sci. 2009;34:85–96. doi: 10.1016/j.tibs.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 15.Tonks NK. Redox redux: revisiting PTPs and the control of cell signaling. Cell. 2005;121:667–670. doi: 10.1016/j.cell.2005.05.016. [DOI] [PubMed] [Google Scholar]
- **16.Burgoyne JR, Eaton P. A rapid approach for the detection, quantification, and discovery of novel sulfenic acid or S-nitrosothiol modified proteins using a biotin-switch method. Methods Enzymol. 2010;473:281–303. doi: 10.1016/S0076-6879(10)73015-9. [DOI] [PubMed] [Google Scholar]; ** An excellent review about the application of the biotin switch method.
- 17.Yeh JI, Claiborne A, Hol WG. Structure of the native cysteine-sulfenic acid redox center of enterococcal NADH peroxidase refined at 2.8 A resolution. Biochemistry. 1996;35:9951–9957. doi: 10.1021/bi961037s. [DOI] [PubMed] [Google Scholar]
- 18.Dalle-Donne I, Scaloni A, Giustarini D, Cavarra E, Tell G, Lungarella G, Colombo R, Rossi R, Milzani A. Proteins as biomarkers of oxidative/nitrosative stress in diseases: the contribution of redox proteomics. Mass Spectrom Rev. 2005;24:55–99. doi: 10.1002/mas.20006. [DOI] [PubMed] [Google Scholar]
- *19.Foster MW, Hess DT, Stamler JS. Protein S-nitrosylation in health and disease: a current perspective. Trends Mol Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koharyova M, Kolarova M. Oxidative stress and thioredoxin system. Gen Physiol Biophys. 2008;27:71–84. [PubMed] [Google Scholar]
- 21.Gallogly MM, Mieyal JJ. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr Opin Pharmacol. 2007;7:381–391. doi: 10.1016/j.coph.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Rhee SG, Jeong W, Chang TS, Woo HA. Sulfiredoxin, the cysteine sulfinic acid reductase specific to 2-Cys peroxiredoxin: its discovery, mechanism of action, and biological significance. Kidney Int Suppl. 2007:S3–8. doi: 10.1038/sj.ki.5002380. [DOI] [PubMed] [Google Scholar]
- 23.Findlay VJ, Townsend DM, Morris TE, Fraser JP, He L, Tew KD. A novel role for human sulfiredoxin in the reversal of glutathionylation. Cancer Res. 2006;66:6800–6806. doi: 10.1158/0008-5472.CAN-06-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *24.Chiappetta G, Ndiaye S, Igbaria A, Kumar C, Vinh J, Toledano MB. Proteome screens for Cys residues oxidation: the redoxome. Methods Enzymol. 2010;473:199–216. doi: 10.1016/S0076-6879(10)73010-X. [DOI] [PubMed] [Google Scholar]
- 25.Kettenhofen NJ, Wood MJ. Formation, Reactivity, and Detection of Protein Sulfenic Acids. Chem Res Toxicol. 2010 doi: 10.1021/tx100237w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson KJ, Klomsiri C, Codreanu SG, Soito L, Liebler DC, Rogers LC, Daniel LW, Poole LB. Use of dimedone-based chemical probes for sulfenic acid detection methods to visualize and identify labeled proteins. Methods Enzymol. 2010;473:95–115. doi: 10.1016/S0076-6879(10)73004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **27.Leonard SE, Reddie KG, Carroll KS. Mining the thiol proteome for sulfenic acid modifications reveals new targets for oxidation in cells. ACS Chem Biol. 2009;4:783–799. doi: 10.1021/cb900105q. [DOI] [PubMed] [Google Scholar]; ** This paper describes an innovative approach to trap and identify sulfenic acids as they develop in the cell.
- 28.Takanishi CL, Ma LH, Wood MJ. A genetically encoded probe for cysteine sulfenic acid protein modification in vivo. Biochemistry. 2007;46:14725–14732. doi: 10.1021/bi701625s. [DOI] [PubMed] [Google Scholar]
- 29.Seo YH, Carroll KS. Profiling protein thiol oxidation in tumor cells using sulfenic acid-specific antibodies. Proc Natl Acad Sci U S A. 2009;106:16163–16168. doi: 10.1073/pnas.0903015106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *30.Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE. 2001;2001:pl1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Kettenhofen NJ, Shiva S, Hogg N, Gladwin MT. Copper dependence of the biotin switch assay: modified assay for measuring cellular and blood nitrosated proteins. Free Radic Biol Med. 2008;44:1362–1372. doi: 10.1016/j.freeradbiomed.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kallakunta VM, Staruch A, Mutus B. Sinapinic acid can replace ascorbate in the biotin switch assay. Biochim Biophys Acta. 1800:23–30. doi: 10.1016/j.bbagen.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 33.Saurin AT, Neubert H, Brennan JP, Eaton P. Widespread sulfenic acid formation in tissues in response to hydrogen peroxide. Proc Natl Acad Sci U S A. 2004;101:17982–17987. doi: 10.1073/pnas.0404762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shelton MD, Chock PB, Mieyal JJ. Glutaredoxin: role in reversible protein s-glutathionylation and regulation of redox signal transduction and protein translocation. Antioxid Redox Signal. 2005;7:348–366. doi: 10.1089/ars.2005.7.348. [DOI] [PubMed] [Google Scholar]
- 35.Tello D, Tarin C, Ahicart P, Breton-Romero R, Lamas S, Martinez-Ruiz A. A “fluorescence switch” technique increases the sensitivity of proteomic detection and identification of S-nitrosylated proteins. Proteomics. 2009;9:5359–5370. doi: 10.1002/pmic.200900070. [DOI] [PubMed] [Google Scholar]
- 36.Huang B, Chen C. Detection of protein S-nitrosation using irreversible biotinylation procedures (IBP) Free Radic Biol Med. 49:447–456. doi: 10.1016/j.freeradbiomed.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 37.Hao G, Derakhshan B, Shi L, Campagne F, Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc Natl Acad Sci U S A. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA, Stamler JS. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat Biotechnol. 2009;27:557–559. doi: 10.1038/nbt.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **39.Sinha V, Wijewickrama GT, Chandrasena RE, Xu H, Edirisinghe PD, Schiefer IT, Thatcher GR. Proteomic and mass spectroscopic quantitation of protein S-nitrosation differentiates NO-donors. ACS Chem Biol. 5:667–680. doi: 10.1021/cb100054m. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper introduces the first quantitative method to detect S-nitrosylations in vivo.
- 40.Giustarini D, Dalle-Donne I, Colombo R, Milzani A, Rossi R. Is ascorbate able to reduce disulfide bridges? A cautionary note. Nitric Oxide. 2008;19:252–258. doi: 10.1016/j.niox.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 41.Karala AR, Ruddock LW. Does s-methyl methanethiosulfonate trap the thiol-disulfide state of proteins? Antioxid Redox Signal. 2007;9:527–531. doi: 10.1089/ars.2006.1473. [DOI] [PubMed] [Google Scholar]
- **42.Hansen RE, Winther JR. An introduction to methods for analyzing thiols and disulfides: Reactions, reagents, and practical considerations. Anal Biochem. 2009;394:147–158. doi: 10.1016/j.ab.2009.07.051. [DOI] [PubMed] [Google Scholar]; A must-read guideline how to work with redox-sensitive proteins.
- 43.Leichert LI, Jakob U. Protein thiol modifications visualized in vivo. PLoS Biol. 2004;2:e333. doi: 10.1371/journal.pbio.0020333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riederer IM, Riederer BM. Differential protein labeling with thiol-reactive infrared DY-680 and DY-780 maleimides and analysis by two-dimensional gel electrophoresis. Proteomics. 2007;7:1753–1756. doi: 10.1002/pmic.200601007. [DOI] [PubMed] [Google Scholar]
- 45.Le Moan N, Clement G, Le Maout S, Tacnet F, Toledano MB. The Saccharomyces cerevisiae proteome of oxidized protein thiols: contrasted functions for the thioredoxin and glutathione pathways. J Biol Chem. 2006;281:10420–10430. doi: 10.1074/jbc.M513346200. [DOI] [PubMed] [Google Scholar]
- 46.Perez VI, Pierce A, de Waal EM, Ward WF, Bokov A, Chaudhuri A, Richardson A. Detection and quantification of protein disulfides in biological tissues a fluorescence-based proteomic approach. Methods Enzymol. 2010;473:161–177. doi: 10.1016/S0076-6879(10)73008-1. [DOI] [PubMed] [Google Scholar]
- 47.Marino SM, Li Y, Fomenko DE, Agisheva N, Cerny RL, Gladyshev VN. Characterization of surface-exposed reactive cysteine residues in Saccharomyces cerevisiae. Biochemistry. 2010;49:7709–7721. doi: 10.1021/bi100677a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **48.Leichert LI, Gehrke F, Gudiseva HV, Blackwell T, Ilbert M, Walker AK, Strahler JR, Andrews PC, Jakob U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci U S A. 2008;105:8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper describes OxICAT, the first method that quantifies the absolute oxidation status of proteins in vivo and in vitro.
- 49.Go YM, Pohl J, Jones DP. Quantification of redox conditions in the nucleus. Methods Mol Biol. 2009;464:303–317. doi: 10.1007/978-1-60327-461-6_17. [DOI] [PubMed] [Google Scholar]
- 50.Kumsta C, Thamsen M, Jakob U. Effects of Oxidative Stress on Behavior, Physiology, and the Redox Thiol Proteome of Caenorhabditis elegans. Antioxid Redox Signal. 2010 doi: 10.1089/ars.2010.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yi L, Jenkins PM, Leichert LI, Jakob U, Martens JR, Ragsdale SW. Heme regulatory motifs in heme oxygenase-2 form a thiol/disulfide redox switch that responds to the cellular redox state. J Biol Chem. 2009;284:20556–20561. doi: 10.1074/jbc.M109.015651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sethuraman M, McComb ME, Huang H, Huang S, Heibeck T, Costello CE, Cohen RA. Isotope-coded affinity tag (ICAT) approach to redox proteomics: identification and quantitation of oxidant-sensitive cysteine thiols in complex protein mixtures. J Proteome Res. 2004;3:1228–1233. doi: 10.1021/pr049887e. [DOI] [PubMed] [Google Scholar]
- 53.Hagglund P, Bunkenborg J, Maeda K, Svensson B. Identification of thioredoxin disulfide targets using a quantitative proteomics approach based on isotope-coded affinity tags. J Proteome Res. 2008;7:5270–5276. doi: 10.1021/pr800633y. [DOI] [PubMed] [Google Scholar]
- 54.Fu C, Hu J, Liu T, Ago T, Sadoshima J, Li H. Quantitative analysis of redox-sensitive proteome with DIGE and ICAT. J Proteome Res. 2008;7:3789–3802. doi: 10.1021/pr800233r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leichert LI, Jakob U. Global methods to monitor the thiol-disulfide state of proteins in vivo. Antioxid Redox Signal. 2006;8:763–772. doi: 10.1089/ars.2006.8.763. [DOI] [PubMed] [Google Scholar]
- 56.Sinha V, Wijewickrama GT, Chandrasena RE, Xu H, Edirisinghe PD, Schiefer IT, Thatcher GR. Proteomic and mass spectroscopic quantitation of protein S-nitrosation differentiates NO-donors. ACS Chem Biol. 2010;5:667–680. doi: 10.1021/cb100054m. [DOI] [PMC free article] [PubMed] [Google Scholar]


