Abstract
Saccharomycescerevisiae antizyme (AZ) resembles mammalian AZ in its mode of synthesis by translational frameshifting and its ability to inhibit and facilitate the degradation of ornithine decarboxylase (ODC). Despite many studies on the interaction of AZ and ODC, the ODC: AZ complex has not been purified from any source and thus clear information about the stoichiometry of the complex is still lacking. In this study we have studied the yeast antizyme protein and the ODC: AZ complex. The far UV CD spectrum of the full-length antizyme shows that the yeast protein consists of 51% β-sheet, 19% α-helix, and 24% coils. Surface plasmon resonance analyses show that the association constant (KA) between yeast AZ and yeast ODC is 6× 107 (M−1). Using purified His-tagged AZ as a binding partner, we have purified the ODC:AZ inhibitory complex. The isolated complex has no ODC activity. The molecular weight of the complex is 90 kDa, which indicates a one to one stoichiometric binding of AZ and ODC in vitro. Comparison of the circular dichroism (CD) spectra of the two individual proteins and of the ODC: AZ complex shows a change in the secondary structure in the complex.
Keywords: affinity constants, antizyme-ODC complex, circular dichroism, protein-protein interaction, surface plasmon resonance
Introduction
Polyamines are aliphatic charged molecules that play an important role in various cellular functions (1–4). Ornithine decarboxylase (ODC) is the first rate-limiting enzyme in polyamine biosynthetic pathway in yeast and other eukaryotes. It catalyzes the decarboxylation of ornithine to putrescine (3). The expression of ODC is tightly regulated by feedback inhibition by its end products spermidine and spermine. In addition, an efficient unique, auto-regulatory circuit controlling both the synthesis and the uptake of polyamines is mediated by a polyamine-induced protein termed antizyme (AZ). AZ acts as a polyamine sensor and is synthesized from two open overlapping reading frames via an unusual, polyamine-stimulated ribosomal frameshifting(5–9). Recently, an orthologue of mammalian antizyme was identified in budding yeast (Saccharomycescerevisiae) and cloned (10). The yeast AZ resembles mammalian AZ in its mode of synthesis by translational frameshifting and its ability to facilitate the degradation of ODC. Interaction between ODC and AZ results in inhibition of ODC activity and the ODC protein is targeted to be degraded by the 26 S proteasome without ubiquitination (11–13).
Although there are many studies on the interaction and inhibition of ODC by AZ, so far the inactive ODC: AZ complex has not been purified from any source. Hence, clear information are still lacking about the stoichiometry of the complex and the structural changes upon interaction of the two proteins. In the present study we have purified the yeast antizyme from E. coli expression systems and have used it to trap ornithine decarboxylase to purify the resulting ODC: AZ complex. We also determined the binding affinities, change in secondary structure upon interaction and stoichiometry of the ODC: AZ complex.
Materials and Methods
Purification of yeast antizyme
The yeast antizyme coding sequence (10) was amplified by PCR and cloned in Champion pET100 vector (Invitrogen, Carlsbad, CA) with a N-terminal 6XHis-tag. The primers used in making the construct were- forward 5′-CACATGTATGAAGTAATACAGAAAAGG-3′ and reverse 5′-TTAGCATTCAAACTCTAAAATAACAAAG-3′. The frameshift site was deleted (ΔT) from the sequence to make an inframe construct by site directed mutagenesis using the primer pairs: forward 5′-GGTGCGCGGACATCCCTCTA-3′ and reverse 5′-TAGAGGGATGTCCGCGCACC-3′ following the site directed mutagenesis protocol (Stratagene, La Jolla, CA). All the constructs and mutations were confirmed by DNA sequencing. The resulting plasmid was transformed into E. coli strain BL21 (DE3) and selected on LB plates containing ampicillin. A selected clone was inoculated in LB plus ampicillin (100 µg/ml) and the overnight culture was diluted into a 50-liter culture with a starting OD600 of 0.1 in a bio-fermentor and incubated at 37°C. When the OD reached 1.0, 0.5 mM, IPTG was added and the culture was incubated further for another three hours at 25°C, to induce the protein. Under these conditions the antizyme protein was mostly in the soluble fraction. In each batch of purification, 40 grams of E. coli cells were extracted with TALON extractor buffer (Clontech), in presence of protease inhibitor cocktail (Pierce, Rockford, IL), 1 mM PMSF, 20 µg/ml lysozyme and 10 µg/ml DNase I at room temperature, and spun at 10,000 RPM for 30 min. Super flow cobalt resin (TALON, Clontech, Mountain View, CA) was equilibrated with equilibration/wash buffer (50 mM sodium phosphate, 300 mM NaCl, pH 7.0) and the supernatant from the E. coli extract was mixed with the metal affinity (Cobalt) resin equilibrated with above buffer. The cobalt column containing the protein was washed three times with equilibrium/wash buffer, with a final wash using 10 mM imidazole in the same buffer. The protein was eluted with 100 mM imidazole, 400 mM NaCl, 10% glycerol, in 50 mM sodium phosphate buffer (pH 7.5). The purified protein is stable and soluble in the presence of 50 mM sodium phosphate, 300–400 mM NaCl, 1 mM DTT, 10% glycerol, at pH 7.5. Using these conditions the purified protein can be stored for a longer period of time (3 months) at −80°C without any precipitation or loss of activity.
Purification of antizyme:ODC complex
Yeast ODC was over-expressed in E. coli polyamine mutant [HT252 (Δ (speAspeB) Δ (speCglc) Δ (speED)] and purified according to Gupta et al(14). Briefly, yeast ODC was subcloned from pSPE1U containing the SPE1 gene of Saccharomyces cerevisiae(15) into the modified phagemid pTZ19U (16) into the NdeI-SmaI sites. The resulting plasmid was used to transform E. coli polyamine mutant (HT252). Another plasmid pGP1-2 (which contain the T7 RNA polymerase gene under temperature sensitive control) was also used to transform the above E. coli strain to construct a temperature inducible yeast ODC overexpression strain of E. coli. The protein was induced by incubation at 43° C and purified using three sequential chromatographic systems (Accell QMA- ionexchange, Waters, Milford, MA; Butyl-sepharose- hydrophobic and Mono-Q- ionexchange, GE Healthcare, Biosciences, Uppsala, Sweden) to near homogeneity. Antizyme and ODC protein at a 1:2 molar ratio were mixed on ice for 10 min in binding buffer (50 mM sodium phosphate, 50 µM pyridoxal phosphate and 1 mM β- mercaptoethanol, 100 mM NaCl, pH7.5) and then mixed with TALON resin equilibrated with the same buffer. The mixture was spun at 5,000 rpm and the supernatant was removed. The resin was washed four times with the binding buffer and a final wash with the same buffer containing 10 mM imidazole. The bound protein complex was eluted with the binding buffer containing 100 mM imidazole.
Biophysical techniques
Antizyme and the ODC: AZ complex were characterized separately via size exclusion chromatography on a Superdex 200 10/300 GL column (GE Healthcare, Biosciences) coupled to a DAWN-EOS multiangle laser light scattering detector (Wyatt Technology, Santa Barbara, CA) and a Milton Roy SM3100 variable-wavelength UV–visible absorbance detector (Thermo Finnegan, West Palm Beach, FL) or a refractive index detector (Optilab Rex, Wyatt Technology, Santa Barbara, CA) installed in parallel. The eluent was pumped at 0.5 ml/min. Prior to measurement each protein was filtered using a 0.1 µm Anotop filter (Whatman). Data were collected using ASTRA 4.9 data collector and molecular mass calculations were performed by ASTRA software (Wyatt Technology) (17, 18).
Circular dichroism (CD) spectra were recorded on a Jasco J-715 spectropolarimeter. The spectra were recorded with a 0.1 cm path length cuvettee using a scan rate of 10 nm/min and a time constant of 10 s at far-UV (from 260–190 nm). All data were averaged over a minimum of 4 scans. Ellipticities were calculated by following the CONTIN method (19):
-
{Θ}M = (100 . Θobs)/(d . C)
{Θ}MRE = (100 . Θobs)/[d . C . (n - 1)]
Where {Θ}M is the calculated molar ellipticity (expressed in degrees per square centimeter per decimole), {Θ}MRE is the calculated mean residue ellipticity (expressed as degrees square centimeter per decimole), Θobs is the observed ellipticity (expressed in degrees), d is the path length (in centimeter), C is the molar protein concentration, and n is the total number of amino acids in protein or protein-protein complex. Near UV (from 250–390 nm) and visible spectra were measured using a 1 cm path-length cuvette (protein concentration= 1mg/ml).
Surface plasmon resonance of antizyme and ODC interaction was studied in a Biacore 2000 optical biosensor at 25 °C (GE Healthcare Biosciences). At first 900 response unit (RU) of antizyme was attached by amine coupling to the surface of a CH5 sensor chip (research grade) using N-hydroxysuccinimide and N-ethyl- N dimethylaminopropyl carbodiimide activation. Ethanolamine was used to quench the unreacted N-hydroxysuccinimide groups. Corresponding reference surfaces were prepared in the absence of antizyme. The mobile phase buffer (HBS-EP, 10 mM HEPES, pH 7.4, 150 mM NaCl, 3.4 mM EDTA, and 0.005% Tween-20) was filtered (0.2 µm) and degassed. The binding of different concentrations of ODC (analyte) to antizyme (ligand) was assayed, and the bound analytes were removed by washing with the HBS-EP buffer. Each assay was performed to find an association and dissociation curve, and each curve was corrected for non-specific binding by subtracting the signal obtained from the blank reference flow cell. Kinetic constants from association and dissociation curves were derived from linear transformation of the data obtained from at least three different concentrations of analyte. The kinetic parameters obtained were compared with those estimated by fitting the data to the simple 1:1 Langmuir interaction model by using BIA evaluation software (version 3.1).
Assays of ornithine decarboxylase, antizyme and protein
Ornithine decarboxylase was assayed as described earlier (20). Antizyme activity was measured on the basis of inhibition of ornithine decarboxylase in the same assay mixture as ODC. Antizyme protein was incubated with ornithine decarboxylase on ice or at room temperature for 10 min and then the substrate 14C ornithine mixture (400 nCi/ml of L-[1–14C] ornithine, 100 µM cold ornithine) was added and the reaction mixture was incubated further for 30 min at 37 °C. The antizyme activity was calculated by subtracting the ornithine decarboxylase activity in the presence of antizyme from the ornithine decarboxylase activity in the absence of antizyme. Protein content was measured by the Bradford method (21) using Biorad reagent (Biorad, Hercules, CA).
Results
Purification of yeast antizyme
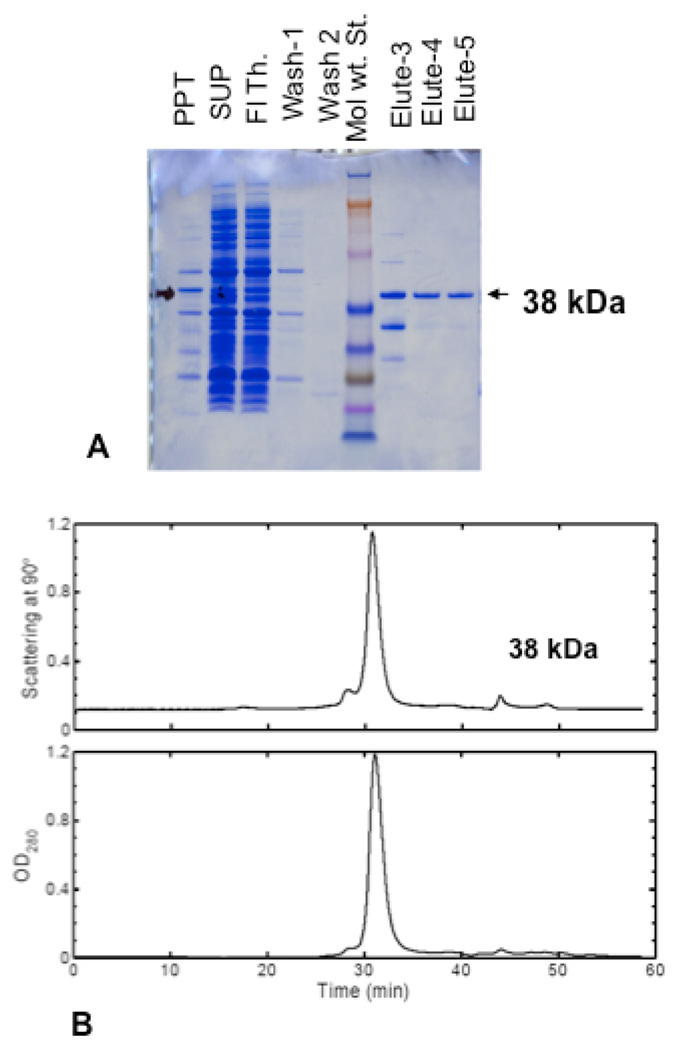
The purified 6Xhis-tagged protein eluted from a cobalt column was nearly homogeneous as shown by SDS-PAGE assays under reducing condition (Fig. 1A). To determine the native molecular weight, the same protein was analyzed by size exclusion chromatography (Superdex 200) followed by light scattering (SEC-light scattering) as shown in Fig. 1B. The results from these studies show the yeast AZ is a monomer of 38 kDa (with 3kDa for the N-terminal His-Tag).
Fig. 1. Purification of yeast antizyme protein.
(A) SDS-PAGE showing the purification of his-tagged antizyme. The yeast AZ protein was overexpressed in E. coli and purified using cobalt column (TALON) as mentioned in Materials and Methods. Aliquots of samples from different steps were run on SDS-PAGE together with protein standard. (B) The native molecular weight of his-tagged antizyme protein was determined by size exclusion chromatography followed by light scattering. Purified antizyme protein (250 µl) from cobalt column at 1mg/ml concentration was loaded into a superdex-200 gel filtration column (GL 30/200) equilibrated with phosphate base saline and was run at a flow rate of 0.5 ml/min. The top panel shows the light scattering at 90° angles and the bottom panel shows optical density at 280 nm. The average molecular weight was determined by the measurement of scattered light in comparison to the protein concentration. The system was calibrated using bovine serum albumin and ovalbumin as standards.
In vitro inhibition of ODC by AZ and equilibrium constants
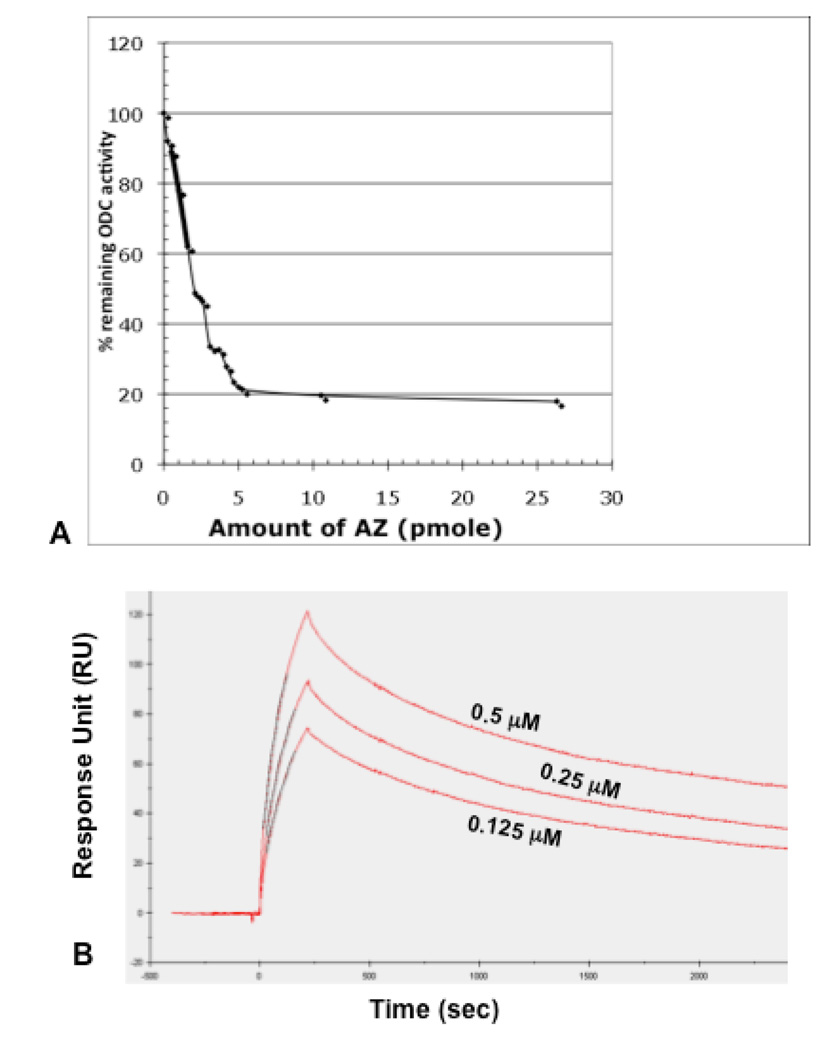
As shown in Fig. 2A, ODC was inhibited by purified AZ. The assay was performed in the presence of a constant amount of purified ODC (2 pmoles/ assay) with varying amount of antizyme. The inhibition was linear with different AZ amount up to 75% of inhibition of ODC. We have also studied the antizyme ODC interaction by surface plasmon resonance (SPR) analyses as shown in Fig. 2B. Purified antizyme was immobilized on the surface of a CM5 chip as ligand, and a solution of different concentration of ODC was passed as analyte over the chip surface. Real-time binding to ligand antizyme was assayed by using a Biacore 2000 instrument. As a control, bovine serum albumin at 5–10 µM was also passed over the chip (data not shown). SPR studies were carried out as a function of concentration of ODC to obtain values of rate constants for association and dissociation (ka and kd), and the equilibrium dissociation (KD) and association constants (KA). The values are ka (M−1 s−1)= 3.14× 104; kd (s−1)= 4.9× 10−4; KA (M−1)= 6.2× 107 and KD (M)= 1.73× 10−8, which indicate a tight binding of AZ to ODC protein.
Fig. 2. Inhibition of ornithine decarboxylaseby yeast antizyme and affinity constants of protein-protein interaction.
(A) 2 pmole of ODC was incubated in the presence and absence of antizyme, and the ODC activity remaining after incubation was determined by 14CO2 trapped in Ba(OH)2 soaked filter paper. Assays were performed in triplicates and the average data are presented here. (B) Sensogram showing the binding of different concentration of ODC protein (0.125, 0.25 and 0.5 µM) to the surface immobilized antizyme protein, as obtained from surface plasmon resonance analyses in a Biacore 2000 as described in Materials and Methods.
Ornithine decarboxylase forms a 1:1 stoichiometric complex with antizyme in vitro
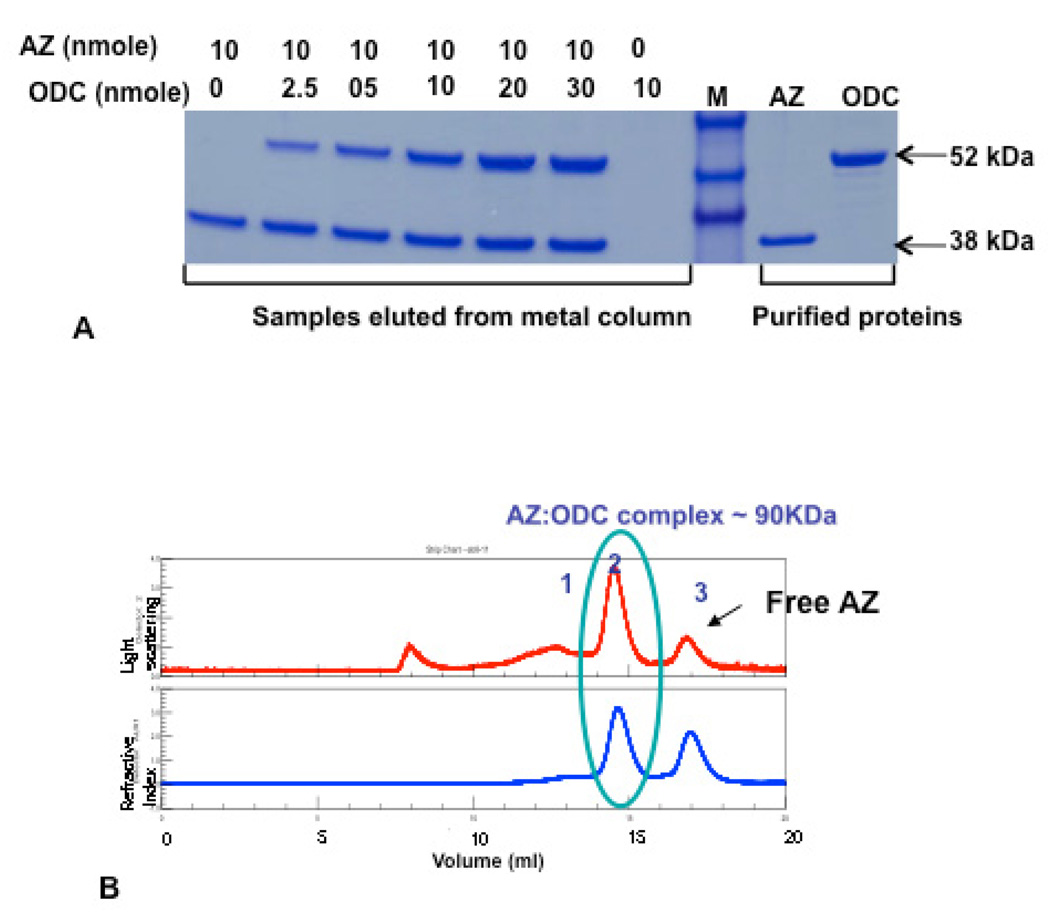
In order to determine the stoichiometry, we purified the ODC:AZ complex. The non-covalent interaction of ODC and AZ forms a ODC: AZ complex, which is very tight and has no ODC activity. As shown in Fig. 3A the inactive complex can be separated on SDS-PAGE, so there is no covalent modification of ODC by AZ binding. The complex was subjected to further analysis by SEC-light scattering analyses to show a molecular weight of 90 kDa, which corresponds to one monomer of ODC binding to one monomer of AZ (Fig 3B).
Fig. 3. Stoichiometry of yeast ODC:AZ complex.
(A) Purified ODC was incubated overnight with His-tagged purified antizyme in different molar ratios as indicated in the figure. The resulting complex was trapped on cobalt (TALON) column and purified as described in Materials and Methods. An aliquot of the purified sample was run on SDS-PAGE together with protein standard. (B). The purified ODC: AZ complex was assayed on SEC-lightscattering to determine the native molecular weight of the ODC: AZ complex. The top panel is showing the dynamic (multi angle) light scattering, the bottom panel is showing the refractive index for protein concentration determination as determined by a refractive index detector (Optilab Rex).
Circular dichroism spectra show change in secondary structure in ODC: AZ complex
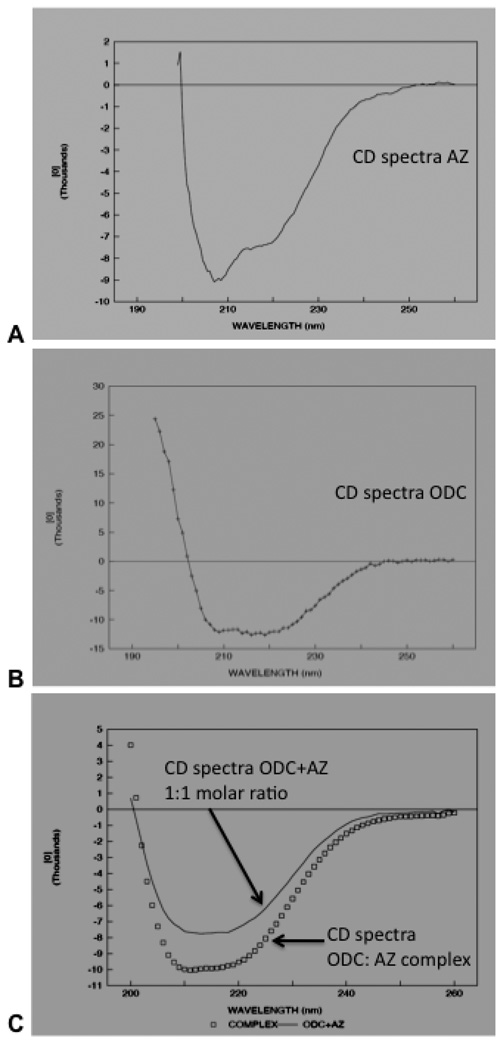
We next studied the change in secondary structure of ODC upon binding to AZ by CD analyses. The secondary structure of purified AZ, ODC and ODC: AZ complex proteins were analyzed by far UV circular dichroism spectroscopy (from 260–190 nm) (Fig. 4). The antizyme protein consists of β-sheet (51±2.5%), α-helix (19±1.2%), coil (24±2%) and turn (6±2.%) (Fig 4A). Far UV-CD spectroscopy shows that the secondary structure of ODC protein consists of α-helix (39±1%) and β-sheets (32±2%), remainder (20±1%) and turn (9±2%) (Fig. 4B). As shown in Fig 4C, the far UV CD spectra revealed asignificant change in the secondary structure of either ODC and/or AZ upon binding to each other. When the purified ODC: AZ CD spectra are compared with the overall sum of AZ and ODC CD spectra in 1:1 molar ratio, there was a significant change in secondary structure. The CONTIN analyses results show increase in β-sheet in ODC: AZ complex (49%) as compared to the sum of molar mixture of ODC and AZ (41%β-sheet). However, the near ultra violet and visible CD spectra of the complex (arising from aromatic side chains and pyridoxal phosphate, respectively) were the sum of those of the ODC and AZ 1:1 molar mixture, within experimental error (data not shown).
Fig. 4. CD spectra of purified AZ, ODC and ODC: AZ complex show a change in secondary structure in the complex.
Far UV (260–190 nm) CD spectra of purified antizyme (A) and ornithine decarboxylase (B) proteins. Antizyme or ornithine decarboxylase proteins at 0.1 mg/ml were run on a CD (JASCO) and the mean ellipticity of the molecule was determined. The data was analyzed to calculate the percentage of α-helices and β-sheets present in antizyme protein as mentioned in the text. (C). Far UV (260–190 nm) CD spectra of purified ODC: AZ complex (0.1 mg/ml) is shown here. The data were calculated and compared with the sum of the CD spectra of 1:1 molar ratios two molecules.
Discussion
The regulation of ornithine decarboxylase by antizyme is a classic phenomenon of protein-protein interaction. The enzymatically active ODC dimer is inhibited by antizyme (22) and a number of studies have shown that antizyme binds to ODC and forms an inactive complex that facilitate degradation of ODC by the 26S proteasome (22, 23). However, definitive evidence of the stoichiometry of the complex has been lacking, because the ODC: AZ complex has not been purified in these studies. For example, Heller et al (1976) (5) and Fujita et al (1982) (23) have observed that the mammalian ODC:AZ complex was eluted before active ODC and hence the molecular weight was higher than dimeric ODC. These results suggested that a dimeric ODC formed a complex with monomeric antizyme. On the other hand, Mamroud-Kidron et al (1994) (22) based on mouse antizyme-ODC interaction studies and cross-linking experiments postulated that monomeric ODC was tightly associated with antizyme. An earlier attempt to purify ODC:antizyme complex by using an affinity column via binding ornithine decarboxylase in a sepharose-pyridoxamine phosphate column failed (6).
In this study, we have purified the ODC: AZ complex by using an antizyme-cobalt column. We have determined the molecular weight of the complex by size exclusion chromatography and light scattering as 90,000 (Fig. 3), which corresponds to a 1:1 stoichiometric complex comprising of a monomer of ODC (52 kDa) and a monomer of antizyme (38 kDa, including 3 kDa as His tag). Comparison of the CD spectra of AZ, ODC and the purified complex shows a change in the secondary structure in the complex (Fig. 4). Thus, the calculated sum of CD spectra of 1:1 molar mixture of ODC and AZ is different when compared to the spectrum of the purified ODC:AZ complex (Fig. 4C). This inactive complex results from an ionic interaction of ODC and antizyme, as increasing salt concentration reduces the binding of antizyme to ODC and also the complex can be partially dissociated by increasing NaCl concentration above 150 mM (data not shown). The ODC inhibition by antizyme can also be reversed by α-difluromethylornithine (DFMO) inactivated-ODC (data not shown). Thus the inactivation of yeast ODC by AZ is reversible.
We have also purified and characterized the yeast antizyme protein. The full-length yeast antizyme protein is a monomer of 38 kDa (with 3 kDa as N-terminal 6X his tag) (Fig 1). It is more soluble than the mammalian protein and can be maintained as a soluble protein without precipitation for a long time (more than three months) in presence of 300–400 mM NaCl containing buffer (pH7.5) at −80 °C (data not shown). In contrast, the full-length rat AZ protein was insufficiently soluble at low concentration; even the AZ1 Δ69 (rat antizyme 1 deleted in N-terminal 69 amino acid residues) showed limited solubility and aggregation at 1mg/ml concentration (24). The full-length secondary structure of yeast antizyme was determined (Fig. 4A), which mainly consists of β-sheet (51±2.5%), α-helix (19±1.2%) and coil (24±2%).
We determined the binding affinities between purified yeast antizyme and ODC protein by surface plasmon resonance (SPR) analyses (Fig. 3B). The binding affinity of yeast antizyme to yeast ODC is ~1/1000th as compared to mammalian antizyme-ODC (6, 25). This could to due to presence of fewer residues in the interaction site of yeast ODC and antizyme.
Although Saccharomyces cerevisiae antizyme performs the same function of ODC regulation as its mammalian counterpart, it is different from the mammalian antizyme in several aspects, such as i) the yeast antizyme molecular weight is ~1.4 times higher than the mammalian antizyme; ii) yeast antizyme has only minimal effect on polyamine transport (26); iii) yeast antizyme binds to yeast ODC and is degraded by yeast proteasome, but not by mammalian proteasome in an in vitro system (26). Although mammalian and yeast ornithine decarboxylases show a comparable high degree of identity (~35%), yeast and mammalian antizymes are remarkably divergent (<10% identity).
Despite the above differences, the similarity in the conserved domain and its regulation by frame shifting indicate a common evolutionary origin of S. cerevisiae antizyme (10, 27). As definitively shown here antizyme forms a heterodimer with ODC consisting of one molecule each of ODC and antizyme; it is possible that antizyme binds more efficiently to ODC monomer and dissociates ODC from the dimer to monomer form. Crystal structure of mouse ODC showed that the protein is ahomodimer (28), however, ODC:AZ interaction is possible due to rapid equilibrium between ODC dimer and monomer (29, 30, 22). Therefore, our result support the postulations made by other researchers using mammalian ODC and antizyme proteins (12, 22). The CD spectra showed a change in secondary structure in the yeast ODC: AZ complex, which also supports the finding that the mouse antizyme caused a conformationalchange in mouse ODC, which made the C terminus of ODC more accessible for binding (31). Thus, all these observations and our results support a general conclusion on regulation of ornithine decarboxylase by antizyme in various eukaryotic organisms.
Acknowledgement
This research was supported by the Intramural Research Program of the NIH (National Institute of Diabetes, Digestive and Kidney Diseases). The authors would like to thank Dr. Herbert Tabor and Dr. Allen P. Minton for supporting this research and critical reviewing of the manuscript. We would also like to thank Dr. BegoñaMonterroso for helping in dynamic light scattering experiment of ODC-AZ complex.
Abbreviation
- AZ
antizyme
- CD
circular dichroism
- ODC
ornithine decarboxylase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tabor CW, Tabor H. Polyamines. Annu Rev Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- 2.Pegg AE. Recent advances in the biochemistry of polyamines in eukaryotes. Biochem J. 1986;234:249–262. doi: 10.1042/bj2340249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen SS. A Guide to the Polyamines. New York: Oxford Univ. Press; 1998. pp. 1–595. [Google Scholar]
- 4.Wallace HM, Fraser AV, Hughes A. A perspective of polyamine metabolism. Biochem J. 2003;376:1–14. doi: 10.1042/BJ20031327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heller JS, Fong WF, Canellakis ES. Induction of a protein inhibitor to ornithine decarboxylase by the end products of its reaction. Proc Natl Acad Sci U S A. 1976;73:1858–1862. doi: 10.1073/pnas.73.6.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitani T, Fujisawa H. Purification and some properties of a protein inhibitor (antizyme) of ornithine decarboxylase from rat liver. J Biol Chem. 1984;259:10036–10040. [PubMed] [Google Scholar]
- 7.Matsufuji S, Matsufuji T, Miyazaki Y, Murakami Y, Atkins JF, Gesteland RF, Hayashi S. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell. 1995;80:51–60. doi: 10.1016/0092-8674(95)90450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rom E, Kahana C. Polyamines regulate the expression of ornithine decarboxylase antizyme in vitro by inducing ribosomal frame-shifting. Proc Natl Acad Sci U S A. 1994;91:3959–3963. doi: 10.1073/pnas.91.9.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell JL, Judd GG, Bareyal-Leyser A, Ling SY. Feedback repression of polyamine transport is mediated by antizyme in mammalian tissue-culture cells. Biochem J. 1994;299:19–22. doi: 10.1042/bj2990019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palanimurugan R, Scheel H, Hofmann K, Dohmen RJ. Polyamines regulate their synthesis by inducing expression and blocking degradation of ODC antizyme. EMBO J. 2004;23:4857–4867. doi: 10.1038/sj.emboj.7600473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murakami Y, Tanaka K, Matsufuji S, Miyazaki Y, Hayashi S. Antizyme, a protein induced by polyamines, accelerates the degradation of ornithine decarboxylase in Chinese-hamster ovary-cell extracts. Biochem J. 1992;283:661–664. doi: 10.1042/bj2830661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, Coffino P. Regulated degradation of ornithine decarboxylase requires interaction with the polyamine-inducible protein antizyme. Mol Cell Biol. 1992;12:3556–3562. doi: 10.1128/mcb.12.8.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murakami Y, Matsufuji S, Hayashi S, Tanahashi N, Tanaka K. Degradation of ornithine decarboxylase by the 26S proteasome. Biochem Biophys Res Commun. 2000;267:1–6. doi: 10.1006/bbrc.1999.1706. [DOI] [PubMed] [Google Scholar]
- 14.Gupta R, Hamasaki-Katagiri N, White Tabor C, Tabor H. Effect of spermidine on the in vivo degradation of ornithine decarboxylase in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2001;98:10620–10623. doi: 10.1073/pnas.181341298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie QW, Tabor CW, Tabor H. Ornithine decarboxylase in Saccharomyces cerevisiae: chromosomal assignment and genetic mapping of the SPE1 gene. Yeast. 1990;6:455–460. doi: 10.1002/yea.320060602. [DOI] [PubMed] [Google Scholar]
- 16.Tabor S, Richardson CC. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987;84:4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Attri AK, Minton AP. New methods for measuring macromolecular interactions in solution via static light scattering: basic methodology and application to nonassociating and self-associating proteins. Anal Biochem. 2005;337:103–110. doi: 10.1016/j.ab.2004.09.045. [DOI] [PubMed] [Google Scholar]
- 18.Attri AK, Minton AP. Composition gradient static light scattering: a new technique for rapid detection and quantitative characterization of reversible macromolecular hetero-associations in solution. Anal Biochem. 2005;346:132–138. doi: 10.1016/j.ab.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 19.Provencher SW, Glockner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- 20.Chattopadhyay MK, Tabor CW, Tabor H. Studies on the regulation of ornithine decarboxylase in yeast: effect of deletion in the MEU1 gene. Proc Natl Acad Sci U S A. 2005;102:16158–16163. doi: 10.1073/pnas.0507299102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 22.Mamroud-Kidron E, Omer-Itsicovich M, Bercovich Z, Tobias KE, Rom E, Kahana C. A unified pathway for the degradation of ornithine decarboxylase in reticulocyte lysate requires interaction with the polyamine-induced protein, ornithine decarboxylase antizyme. Eur J Biochem. 1994;226:547–554. doi: 10.1111/j.1432-1033.1994.tb20079.x. [DOI] [PubMed] [Google Scholar]
- 23.Fujita K, Murakami Y, Hayashi S. A macromolecular inhibitor of the antizyme to ornithine decarboxylase. Biochem J. 1982;204:647–652. doi: 10.1042/bj2040647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffman DW, Carroll D, Martinez N, Hackert ML. Solution structure of a conserved domain of antizyme: a protein regulator of polyamines. Biochemistry. 2005;44:11777–11785. doi: 10.1021/bi051081k. [DOI] [PubMed] [Google Scholar]
- 25.Cohavi O, Tobi D, Schreiber G. Docking of antizyme to ornithine decarboxylase and antizyme inhibitor using experimental mutant and double-mutant cycle data. J Mol Biol. 2009;390:503–515. doi: 10.1016/j.jmb.2009.05.029. [DOI] [PubMed] [Google Scholar]
- 26.Porat Z, Landau G, Bercovich Z, Krutauz D, Glickman M, Kahana C. Yeast antizyme mediates degradation of yeast ornithine decarboxylase by yeast but not by mammalian proteasome: new insights on yeast antizyme. J Biol Chem. 2008;283:4528–4534. doi: 10.1074/jbc.M708088200. [DOI] [PubMed] [Google Scholar]
- 27.Ivanov IP, Atkins JF. Ribosomal frameshifting in decoding antizyme mRNAs from yeast and protists to humans: close to 300 cases reveal remarkable diversity despite underlying conservation. Nucleic Acids Res. 2007;35:1842–1858. doi: 10.1093/nar/gkm035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Almrud JJ, Oliveira MA, Kern AD, Grishin NV, Phillips MA, Hackert ML. Crystal structure of human ornithine decarboxylase at 2.1 A resolution: structural insights to antizyme binding. J Mol Biol. 2000;295:7–16. doi: 10.1006/jmbi.1999.3331. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell JL, Chen HJ. Conformational changes in ornithine decarboxylase enable recognition by antizyme. Biochim Biophys Acta. 1990;1037:115–121. doi: 10.1016/0167-4838(90)90109-s. [DOI] [PubMed] [Google Scholar]
- 30.Coleman CS, Stanley BA, Viswanath R, Pegg AE. Rapid exchange of subunits of mammalian ornithine decarboxylase. J Biol Chem. 1994;269:3155–3158. [PubMed] [Google Scholar]
- 31.Li X, Coffino P. Degradation of ornithine decarboxylase: exposure of the C-terminal target by a polyamine-inducible inhibitory protein. Mol Cell Biol. 1993;13:2377–2383. doi: 10.1128/mcb.13.4.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]