Abstract
GATA family transcription factors are core components of the vertebrate heart gene network. GATA factors also contribute to heart formation indirectly through regulation of endoderm morphogenesis. However, the precise impact of GATA factors on vertebrate cardiogenesis is masked by functional redundancy within multiple lineages. Early heart specification in the invertebrate chordate Ciona intestinalis is similar to that of vertebrates but only one GATA factor, Ci-GATAa, is expressed in the heart progenitor cells and adjacent endoderm. Here we delineate precise, tissue specific contributions of GATAa to heart formation. Targeted repression of GATAa activity in the heart progenitors perturbs their transcriptional identity. Targeted repression of endodermal GATAa function disrupts endoderm morphogenesis. Subsequently, the bilateral heart progenitors fail to fuse at the ventral midline. The resulting phenotype is strikingly similar to cardia bifida, as observed in vertebrate embryos when endoderm morphogenesis is disturbed. These findings indicate that GATAa recapitulates cell-autonomous and non-cell-autonomous roles performed by multiple, redundant GATA factors in vertebrate cardiogenesis.
Keywords: Cardiac morphogenesis, Ascidian, Cell Migration, Cardia Bifida, Endoderm
Introduction
The GATA family of zinc finger transcription factors plays a central role in vertebrate heart formation (Peterkin et al., 2005; Srivastava, 2006). Members of the GATA, Nkx2.5, Tbx and Hand gene families are integrated by highly conserved, reciprocal interactions to form a cardiac regulatory “kernel” (Davidson and Erwin, 2006; Olson, 2006; Peterkin et al., 2005). Of the six vertebrate GATA paralogues, three (GATA4, 5 and 6) are expressed in developing cardiomyocytes (Molkentin, 2000; Patient and McGhee, 2002; Peterkin et al., 2005). Congenital mutations in GATA4 are linked to human heart defects (Garg et al., 2003; McFadden and Olson, 2002; Posch et al., 2008; Rajagopal et al., 2007; Wolf and Basson, 2010). Additionally, experimental studies demonstrate that GATA factors help establish the myocardiocyte lineage in developing embryos. (Kikuchi et al., 2010; Olson and Schneider, 2003; Singh et al., 2010; Takeuchi and Bruneau, 2009).
GATA4,5 and 6 function cell-autonomously to direct myocardioblast specification and differentiation. GATA factors bind upstream of heart gene promoters to directly regulate their expression (Dodou et al., 2004). Simultaneous knockdown of GATA4,5 and 6 prevents expression of myocardial marker genes (Peterkin et al., 2007; Zhao et al., 2008). In contrast, disruption of GATA function outside of the cardiac mesoderm does not affect myocardial gene expression (Gannon and Bader, 1995; Peterkin et al., 2009; Reecy et al., 1999). Intriguingly, a recent study has shown that GATA factors also re-establish the cardiomyocyte lineage during regeneration (Kikuchi et al., 2010).
GATA factors expressed in the endoderm and other non-cardiac lineages instruct vertebrate myocardioblast position. In wild type embryos, bilateral fields of myocardial precursors converge at the ventral midline, fusing to form a single progenitor field. Disruption of endoderm specification or morphogenesis blocks myocardioblast convergence (Alexander et al., 1999; Kikuchi et al., 2000; Schier et al., 1997). The resulting bilateral heart fields continue to differentiate into distinct beating clusters of heart tissue, a phenotype known as cardia bifida. Similarly, embryos with disrupted GATA function have deformed endoderm and develop cardia bifida (Haworth et al., 2008; Kuo et al., 1997; Molkentin et al., 1997; Peterkin et al., 2009; Reiter et al., 1999).
Functional overlap between GATA 4,5 and 6 in both the pre-cardiac and adjacent lineages has made it difficult to dissect their precise, tissue specific roles. Recent studies have confirmed a high degree of functional overlap between GATA 4,5 and 6 (Holtzinger and Evans, 2007; Peterkin et al., 2007; Singh et al., 2010; Zhao et al., 2008). Additionally, each GATA factor has a unique, dynamic expression domain encompassing the pre-cardiac mesoderm, endoderm, and additional adjacent lineages such as the zebrafish yolk syncytial layer. Examination of non-cell autonomous contributions by GATA factors in the endoderm and other adjacent lineages has proven particularly difficult due to the cellular complexity of vertebrate embryos. Therefore, we have initiated a functional study of GATA in the simple chordate Ciona intestinalis.
Ciona has proven to be an excellent model for dissecting conserved aspects of chordate heart development (Davidson, 2006). Ciona is a member of the tunicates, a group of organisms that diverged just prior to two rounds of genome duplication within their sister clade, the vertebrate chordates. Thus Ciona has a single copy of many essential heart genes, including single orthologues to the vertebrate GATA4,5,6, Nkx2.5 and Hand gene families (GATAa, Nkx and Hand respectively). Additionally, low embryonic cell numbers has facilitated comprehensive mapping of the Ciona heart lineage (Davidson and Levine, 2003; Satou et al., 2004). Ciona heart tissue is derived from four B7.5 lineage blastomeres in the gastrulating embryo. These cells are demarcated by expression of the conserved pre-cardiac specification factor, Mesp (Davidson et al., 2005; Saga et al., 1999; Satou et al., 2004). By the end of neurulation, the B7.5 blastomeres divide to generate two distinct daughter cell lineages, the trunk ventral cells (TVCs) and the anterior tail muscle precursors (ATMs). All Ciona heart cells are derived from the TVCs (Satou et al., 2004). The initial establishment of TVC identity is driven by FGF mediated activation of the Ets1/2 transcription factor (Davidson et al., 2006). Activated Ets1/2 promotes the expression of two primary TVC transcription factors, Hand-like (also referred to as Notrlc, (Satou et al., 2004)) and FoxF (Beh et al., 2007, Davidson et al. 2006). Bilateral pairs of TVCs migrate along the endoderm towards the ventral midline where they meet to form a single cardiac progenitor pool (Christiaen et al., 2008; Davidson, 2006; Stolfi et al., 2010).. The bilateral fusion of Ciona TVCs resembles the midline convergence of vertebrate myocardial precursors (Olson and Schneider, 2003). Subsequently, a subset of TVCs migrate to form pharyngeal mesoderm, expressing islet and other orthologues to vertebrate secondary heart field markers (Stolfi et al., 2010). Thus, Ciona cardiogenesis represents a conserved blueprint for deciphering related but more complex processes underlying vertebrate heart development.
In this study, we have exploited the genomic and cellular simplicity of Ciona embryos to delineate distinct roles for GATAa in the endoderm and cardiac mesoderm.
Materials and Methods
Embryo isolation and manipulation
Ciona intestinalis adults were purchased from M-REP (San Marcos, CA). Protocols for fertilization, electroporation and culturing (in artificial sea water, Crystal Sea Marine Mix) are as described previously (Corbo et al., 1997). Embryos were staged according to an established developmental timeline (Hotta et al., 2007).
Molecular cloning
The Ttf-1 enhancer, also known as Titf-1 (Ristoratore et al., 1999; Shi and Levine, 2008) was amplified from genomic DNA with the primers: Ttf-enh-NcoI-5’and Ttf-enh-NotI-3’(all primer sequences in Supplemental Table 1) and cloned upstream of lacZ in the pCES vector (Harafuji et al., 2002) to make the Ttf-lacZ construct. To create the Ttf-GFP-strabismus construct, the strabismus coding region was amplified from the Ciona Gene Collection 1 library clone GC01o06 (Satou et al., 2002) with the primers StrabGFP-NheI-5’and StrabGFP-EcoRI-3’ and then cloned into a modified form of the Mesp-EtsVp16 construct (Davidson et al., 2006) in which gfp replaced the nuclear localization sequence upstream of the EtsVp16 domain using a NotI/NheI restriction digest. The full length strabismus coding region was then swapped for the EtsVp16 domain by a NheI/EcoRI restriction digest to create the gfp-strabismus fusion gene and then cloned into the Ttf-lacZ vector by a NotI/BlpI restriction digest, replacing the lacZ gene. The coding sequence for the GATAa DNA binding domain (GATAa-DBD) was amplified from the Ciona Gene Collection 1 library clone GC02d03 (Satou et al., 2002) using the primers GATAa-NheI-5’ and GATAa-SpeI-3’ and cloned downstream of the Mesp enhancer and nuclear localization signal and upstream of the WRPW sequence using NheI and SpeI sites in the Mesp-EtsWRPW construct (Davidson et al., 2006). Both the GATAa-DBD and the GATAa-WRPW domains were then cloned downstream of the Hand-like (Davidson and Levine, 2003) and Ttf enhancers using NotI/BlpI restriction digests. The Mesp-lacZ and Mesp-GFP constructs were described elsewhere (Davidson et al., 2005). The esconsin-3xgfp coding region was a gift from François Robin (Roure et al., 2007) and was amplified using the primers Esconsin-NotI-5’ and Esconsin-EcoRI-3’ and reamplified using nested PCR, then swapped for lacZ in the Mesp-lacZ construct using a NotI/EcoRI digest, to create the Mesp-esc-3xGFP construct (referred to as Mesp-esc-GFP in the paper).
Histochemistry and imaging
X-gal staining and fluorescent in situ hybridizations were performed as described previously (Beh et al., 2007) with the following exceptions for the in situ protocol. Following hybridization, embryos were washed with TNT (0.1M Tris-HCl pH 7.4, 0.15M NaCl, 0.1% Tween-20), blocked for 1 hour in TNB (0.1M Tris-HCl pH 7.4, 0.15M NaCl, 1% BSA) and then incubated overnight at 4°C in primary antibodies: 1:1000 POD anti-DIG (Roche) and 1:1000 mouse anti-β-galactosidase or 1:1000 rabbit anti-GFP. The embryos were washed with TNT and then incubated for 5 minutes in FITC-tyramide working solution (Perkin Elmer) in order to visualize antisense RNA probes for Hand-like, FoxF, GATAa, BMP2/4 and Nkx. Embryos were washed with TNT, blocked for 1 hour in TND (0.1M Tris-HCl pH 7.4, 0.15M NaCl, 2% Natural Donkey Serum), and incubated overnight at 4°C in secondary antibodies (1:1000 Donkey anti-mouse Alexa Fluor 647, 1:1000 Donkey anti-rabbit Alexa Fluor 555). Embryos were washed in TNT and mounted with 100% glycerol. The antisense RNA probes for GATAa and BMP2/4 were created from Gene Collection 1 library clones, GC02d03 and GC15c08 respectively (Satou et al., 2002) using T7, T3 and GATAa-probe-5’ (sequence in Supplemental Table 1) primers. Antisense RNA probes for Hand-like, FoxF and Nkx were previously described (Beh et al., 2007; Davidson and Levine, 2003).
Immuno-staining was conducted in accordance with the protocols detailed in (Veeman et al., 2008) or (Dong et al., 2009). All antibodies were purchased from Invitrogen apart from the mouse anti-β-galactosidase (Promega, Z378A) and were used in 1:1000 dilution. Embryos were mounted in 100% glycerol and imaged with a Zeiss LSM 510 META NLO laser scanning confocal microscope equipped with 40× oil immersion objective (numerical aperture 1.3). Z-stacks were taken at intervals of 2µm and reconstructed using the Zeiss LSM image software and Imaris Bitplane 7.0. Further image processing was performed with Adobe PhotoShop. Images of immuno-stained embryos are representative of highly consistent phenotypes seen in at least 10 individuals for each condition including at least two independent trials.
Results
Comparative analysis of GATAa expression
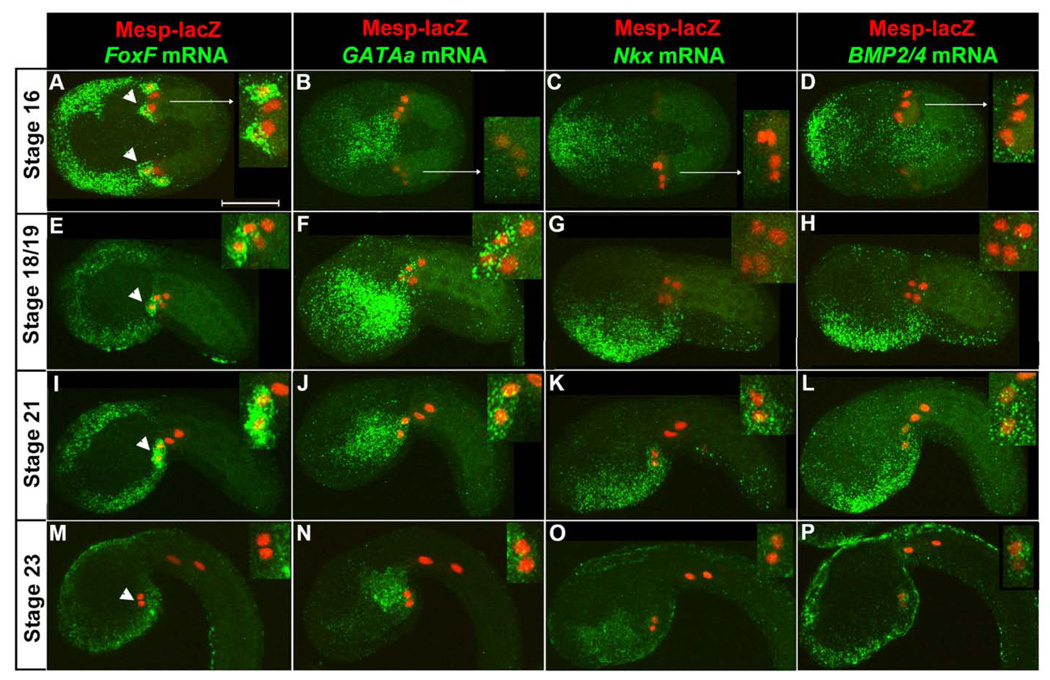
To compare the expression patterns of GATAa and other early heart progenitor (TVC) genes (FoxF, Nkx and BMP2/4) (Beh et al., 2007; Davidson, 2006), we marked the B7.5 lineage using Mesp-lacZ and performed fluorescent in situ hybridizations (Beh et al., 2007). We collected embryos at 60 minute intervals encompassing initial TVC specification, stage 16, through completion of TVC midline fusion, stage 23 (Hotta et al., 2007). We then examined all marker genes in parallel, using a single staged sample for each trial.
At stage 16, 8 hours post fertilization (hpf), FoxF was robustly expressed in newly emerged TVCs (arrowheads, Fig. 1A) and trunk ectoderm. In contrast, GATAa, Nkx and Bmp2/4 did not appear to be expressed in the TVCs although they were expressed in neighboring lineages (Fig. 1B–D). In particular, GATAa was strongly expressed in the posterior endoderm while Nkx and BMP2/4 were expressed in an overlapping region of the anterior/ventral epidermis and Nkx was expressed in portions of the ventral endoderm. Between stages 18 and 19 (9 hpf), we began to detect robust expression of GATAa in the TVCs along with continued expression in the posterior endoderm and variable, weak expression in the anterior endoderm (Fig. 1F). In contrast, the expression patterns of FoxF, Nkx and BMP2/4 remained unaltered (Fig. 1E,G,H). Most critically, the TVCs did not appear to express either Nkx or BMP2/4 during these stages (Fig. 1G,H). By stage 21 (10 hpf), TVCs initiated migration into the trunk region (Fig. 1I–L). Nkx and BMP2/4 expression in the TVCs rose to detectable levels (Fig. 1K,L) while FoxF and GATAa expression domains were unchanged (Fig. 1I,J). As the TVCs completed midline convergence (stage 23, 12 hpf, Fig. 1M–P), GATAa expression persisted in the TVCs (Fig. 1N). Endodermal GATAa displayed a sharpened boundary, as expression in the anterior domain diminished (Fig. 1N). The BMP2/4 expression domain appeared unchanged (Fig. 1P), while Nkx expression in the TVCs appeared to be greatly reduced (Fig. 1O). Interestingly, FoxF was no longer detected in the TVC lineage (Fig. 1M).
Figure 1. Temporal analysis of GATAa expression relative to other heart progenitor genes.
Representative expression patterns from stage 16 to stage 23 as indicated (visualized transcripts denoted above each column in green, stages to the left, n=20–30 embryos for each condition). All embryos shown anterior to the left in this and all subsequent figures. Embryos in (A–D) were imaged from the ventral side while embryos in (E–P) were imaged laterally, dorsal side up. Nuclei of TVCs and ATMs are marked by β-gal antibody staining (red). Inset panels display magnified views of the B7.5 lineage. Note that the TVCs are the more anterior lineage, as marked by arrowheads in the first column. The weak β-gal staining on one side of the embryos in (C, D) represents mosaic incorporation of transgenic markers. Scale bar: 50 µm.
In summary, our in situ expression studies demonstrate that GATAa is expressed in the TVCs after initial heart progenitor markers (such as FoxF) and before Nkx and BMP2/4. The apparent centrality of GATAa expression in the temporal framework of heart gene expression may reflect a central functional role in the heart regulatory network. To explore this possibility, we began a series of studies focused on disrupting GATAa function.
Cell-autonomous GATAa activity is required for heart progenitor migration and proliferation
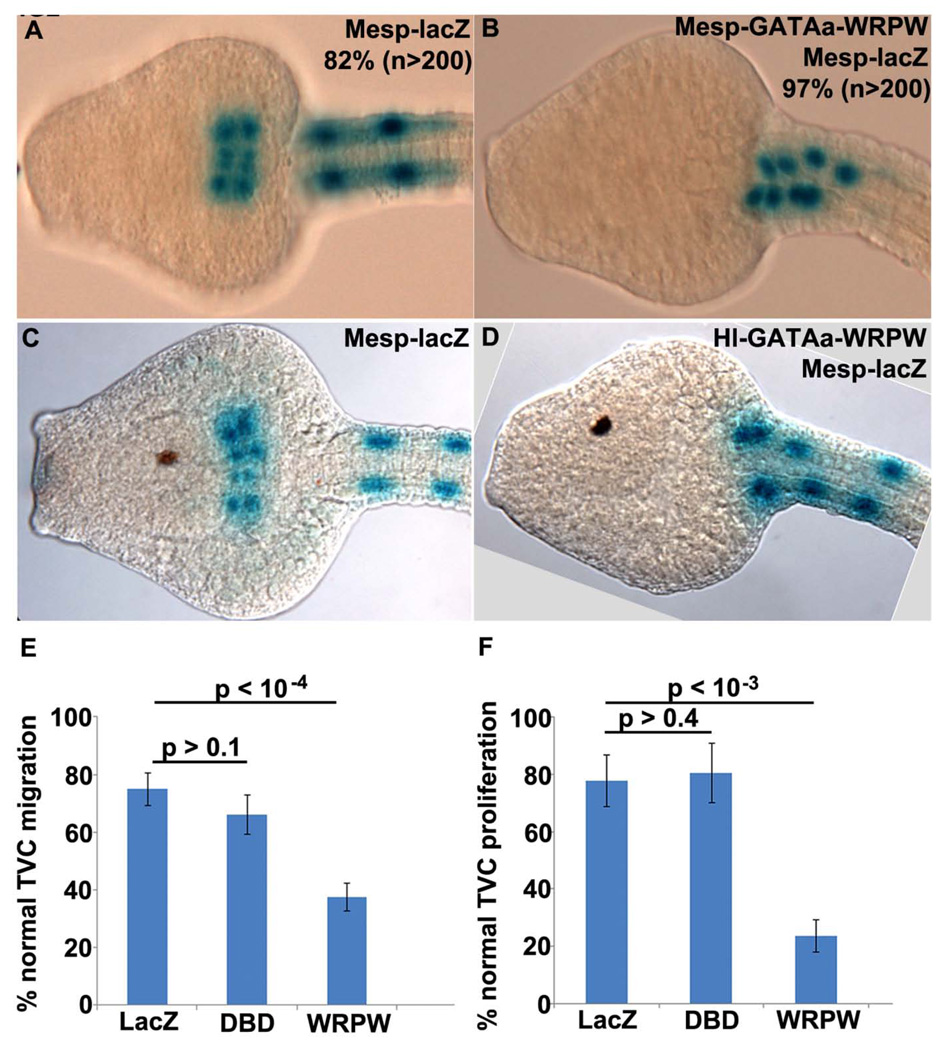
We first examined the cell-autonomous function of GATAa by disrupting its activity in the TVC lineage. For this purpose we fused the GATAa DNA binding domain to the Drosophila hairy WRPW repressor motif (Barolo and Levine, 1997; Fisher et al., 1996). A similar strategy has been successfully employed to disrupt FoxF and Ets1/2 activity in the B7.5 lineage (Beh et al., 2007; Christiaen et al., 2008; Davidson et al., 2006). . Initially, we used the Mesp enhancer to express this dominant repressor fusion protein in the B7.5 lineage (Mesp-GATAa-WRPW). In stage 24 control embryos, labeled TVCs consistently migrated into the trunk, fused at the midline and underwent a single round of division (Fig. 2A). In Mesp-GATAa-WRPW transgenic embryos, the TVCs failed to detach, migrate or proliferate (Fig. 2B). These results suggest that GATAa activity plays a critical role in modulating trunk ventral cell behavior.
Figure 2. Disruption of GATAa activity in the TVCs affects their migration and proliferation.
(A–D) Representative ventral views of stage 24 embryos (13.5 hpf) expressing Mesp-lacZ alone (A, C) or co-electroporated with Mesp-GATAa-WRWP (B) or Hl-GATAa-WRPW (D). Occurrence of highly consistent phenotypes are included numerically in panels (A,B) while more variable phenotypes observed in (C,D) are represented graphically in the following panels. (E, F) Plots showing percentage of stage 24 embryos with normal TVC migration (E) and proliferation (F). In both graphs, LacZ = Hl-lacZ / Mesp-lacZ embryos, DBD = Hl-GATAa-DBD / Mesp-lacZ embryos, WRPW = Hl-GATAa-WRPW / Mesp-lacZ embryos. (E) LacZ (n=813), DBD (n=478), WRPW (n=804). (F) LacZ (n=314), DBD (n=361), WRPW (n=543). In this and subsequent graphs each sample includes at least two independent trials, p values were calculated with a two sample t-test assuming unequal variances and error bars correspond to Standard Error of the Mean (SEM).
We were concerned, however, that early expression of GATAa-WPRW under the Mesp driver might enhance potential off-target effects. The Mesp enhancer is predicted to drive expression of GATAa-WRPW during early gastrula stages, approximately four hours prior to endogenous GATAa expression in the TVC lineage (Fig. 1F, (Davidson et al., 2005)). Moreover, the Mesp enhancer drives GATAa-WRPW expression in the entire B7.5 lineage (both TVCs and ATMs) while endogenous GATAa expression is restricted to the TVCs. To address these concerns, we conducted additional studies using an enhancer element located upstream of Hand-like to regulate expression of GATAa fusion constructs (Davidson and Levine, 2003). This Hand-like enhancer (Hl) activates expression in the trunk ventral cells immediately following FGF mediated specification, as seen for the endogenous Hand-like transcript (Stacie Ilchena, in preparation). Thus Hl-GATAa-WRPW is predicted to perturb GATAa activity in a spatiotemporal pattern that closely parallels endogenous GATAa expression.
We examined the impact of Hl-GATAa-WRPW on both TVC migration and proliferation (Fig. 2C–F). For a more rigorous control, we included embryos in which the GATAa DNA binding domain alone was expressed in the TVCs (Hl-GATAa-DBD). In stage 24 controls, there was no significant variation in either TVC migration or proliferation (Fig. 2C,E,F). In Hl-GATAa-WRPW transgenic embryos, there was a clear, significant disruption of TVC migration and proliferation (Fig. 2D–F).
In summary, targeted disruption of GATAa activity perturbs TVC migration and proliferation. These results suggest that GATAa plays a conserved, central role in the Ciona heart gene network, regulating target genes involved in TVC behavior.
Targeted repression of GATAa activity in the TVCs disrupts heart progenitor gene expression
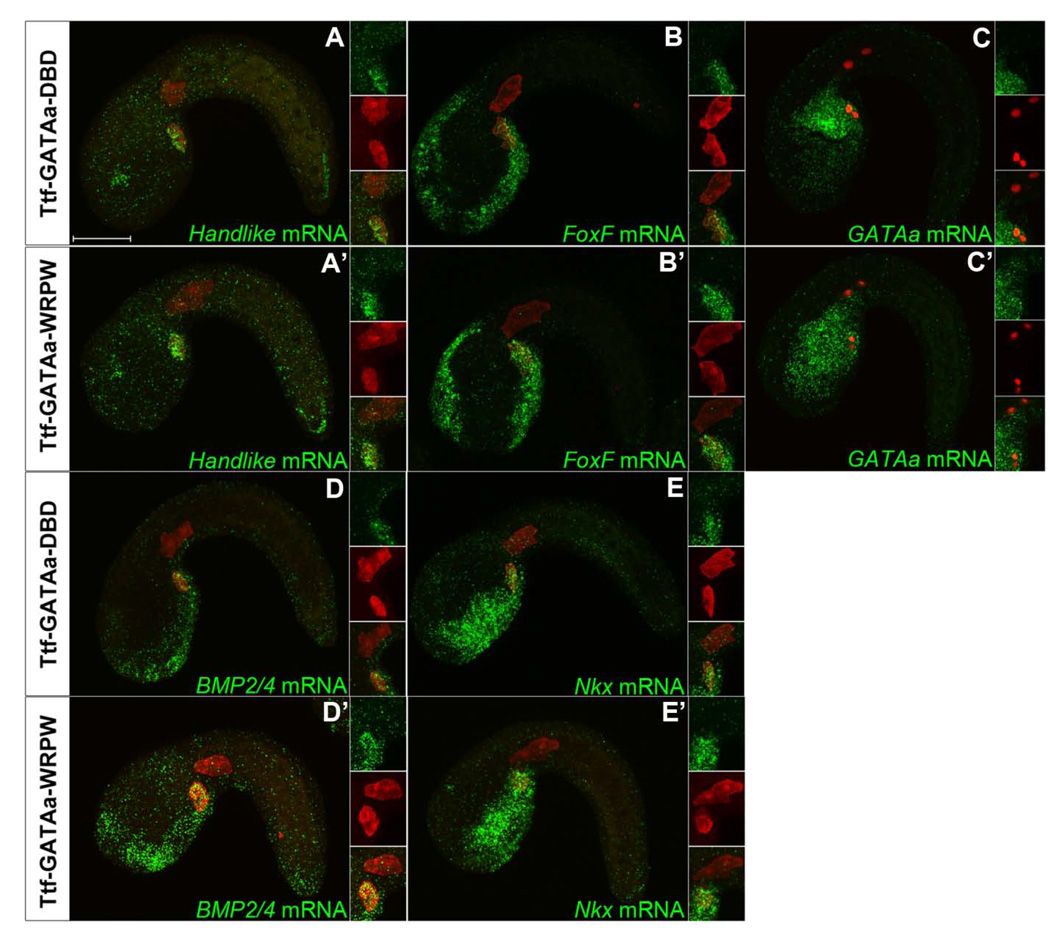
We next examined the impact of Hl-GATAa-WRPW on TVC gene expression (Fig. 3). In both Hl-lacZ (data not shown) and Hl-GATAa-DBD control embryos, Hand-like, FoxF, Nkx, BMP2/4 and GATAa were consistently expressed in the trunk ventral cells (Fig. 3A–D and data not shown). In Hl-GATAa-WRPW embryos (Fig. 3A’–D’) TVC expression of FoxF, Nkx BMP2/4 and GATAa itself was either eliminated or significantly reduced while expression in adjacent lineages was not affected. Among the embryos showing reduced expression, marker gene transcript was often present in the more anterior “leader” cell within bilateral TVC pairs (data not shown). In contrast, Hl-GATAa-WRPW did not substantially down-regulate Hand-like expression (Stacie Ilchena, in preparation). Thus, the Hl-GATAa-WRPW construct does not indiscriminately down-regulate all TVC genes. Moreover, expression of the Hl-GATAa-WRPW construct is not subject to auto-repression. Furthermore, due to the impact of Hl-GATAa-WRPW on GATAa expression, the observed perturbations of TVC behavior and gene expression in these transgenic embryos are likely to reflect a robust down-regulation of GATAa targets, without competition from endogenous GATAa.
Figure 3. Disruption of GATAa activity in the TVCs disrupts cardiac gene expression.
(A–D) Representative images of stages 22 (11–11.5 hpf) embryos co-electroporated with Mesp-lacZ (red) and either Hl-GATAa-DBD (A–D) or Hl-GATAa-WRPW (A’–D’) and examined for expression of FoxF, Nkx, BMP2/4 and GATAa. Number of embryos showing the displayed expression profile vs. total number examined is shown on the upper right. (A’–D’) Embryos represent the predominant staining pattern, no expression in the TVCs. However, additional embryos showed reduced expression (staining in one ‘leader’ TVC) for each of these probes as follows; (A’) Another 4/29 embryos showed reduced FoxF expression in the TVCs (23/29 reduced or eliminated); (B’) Another 5/18 embryos showed reduced Nkx expression in the TVCs (14/18 reduced or eliminated); (C’) Another 4/22 embryos showed reduced BMP2/4 expression in the TVCs (20/22 reduced or eliminated). The insets show magnified views of the TVCs and include individual and merged channels. Scale bar: 50 µm.
In summary, in situ expression assays indicate that GATAa plays a central role in TVC transcriptional identity, participating in three distinct regulatory functions; maintaining expression of a subset of primary TVC genes (FoxF but not Hand-like); perpetuating its own expression and; helping to initiate the expression of ensuing transcription factors including Nkx, the sole orthologue to the vertebrate heart kernel gene Nkx2.5.
Endodermal GATAa activity is required for heart progenitor midline convergence
We next asked whether disruption of GATAa function in the endoderm impacts heart development. For this purpose, we disrupted GATAa activity by expressing the dominant negative repressor GATAa-WRPW using the characterized pan-endoderm enhancer for thyroid transcription factor Ttf-1, also known as Titf-1 (Ristoratore et al., 1999; Shi and Levine, 2008). We observed the effect of this manipulation on transgenically labeled TVCs (Fig. 4). In Ttf-GATAa-WRPW embryos, TVCs detached and migrated anteriorly but failed to converge at the midline (Fig. 4B). Thus, at stage 24, Ttf-GATAa-WRPW embryos contained two distinct bilateral groups of heart progenitor cells. We also examined TVC number at stage 24 and found that GATAa-WRPW had a modest but significant impact on TVC proliferation (Fig. 4D).
Figure 4. Disruption of GATAa activity in the endoderm affects heart progenitor convergence at the midline.
(A, B) Representative ventral views of stage 24 (13.5 hpf) embryos co-electroporated with Mesp-lacZ along with Ttf-GATAa-DBD or Ttf-GATAa-WRPW as indicated. (C, D) Plots showing percentage of stage 24 embryos with normal TVC convergence (C) and proliferation (D). In both graphs, LacZ = Ttf-lacZ/Mesp-lacZ embryos, DBD = Ttf-GATAa-DBD/Mesp-lacZ embryos, WRPW = Ttf-GATAa-WRPW/Mesp-lacZ embryos. (C) LacZ (n=108), DBD (n=170), WRPW (n=439). (D) LacZ (n=112), DBD (n=214), WRPW (n=674). (E–H) Ventral views of stage 24 embryos co-electroporated with Mesp-esc-gfp along with Ttf-GATAa-DBD or Ttf-GATAa-WRPW. Embryos were stained with phalloidin to label F-actin (grey). (E, F) A single confocal plane of the embryo shows the architecture of the endodermal tissue underlying the TVCs. (G, H) Same embryos as in (E, F) but showing a superimposed projection of the labeled TVCs (green). Red line in (G) and (H) highlights the shape of the gut lumen. Scale bar: 10 µm.
To assess the effect of the Ttf-GATAa-WRPW construct on endoderm morphology, we examined the shape of the developing gut cavity in stage 24 embryos (Fig. 4E–H). In ventral views of control embryos, endodermal epithelia appeared to fold around a T-shaped lumen (Fig. 4E, red dotted line in 4G). In Ttf-GATAa-WRPW embryos, the anterior endoderm still appeared to form an intact epithelium but the enclosed lumen was often widened and did not appear to extend posteriorly (Fig. 4F, red dotted line in 4H).
In summary, we have shown that repression of endodermal GATAa activity perturbs endoderm morphogenesis and disrupts medial migration of the TVCs. This leads to a phenotype (TVC bifida) remarkably similar to the cardia bifida phenotype associated with perturbation of endodermal GATA in vertebrate embryos (Haworth et al., 2008; Kuo et al., 1997; Molkentin et al., 1997; Peterkin et al., 2009; Reiter et al., 1999).
Targeted disruption of GATAa activity in the endoderm does not disrupt TVC marker gene expression
We next examined expression of TVC marker genes (Hand-like, FoxF, GATAa, BMP2/4 and Nkx) in Ttf-lacZ, Ttf-GATAa-DBD and Ttf-GATAa-WRPW transgenic embryos at stage 22 (Fig. 5) and stage 23 (data not shown). There was no detectable difference in the TVC expression of these markers between Ttf-GATAa-WRPW and control embryos. There was also no discernible difference in TVC detachment and initial anterior migration. These results suggest that loss of GATAa activity in the endoderm does not disrupt the transcriptional network required to establish TVC identity. We also noted that Ttf-GATAa-WRPW does not eliminate endogenous GATAa expression in the posterior endoderm (Fig.5C’). Indeed, it appears that Ttf-GATAa-WRPW generated ectopic expression of GATAa in the anterior endoderm (Fig. 5C’). Thus, it appears that GATAa auto-regulates through distinct feedback loops in the endoderm and TVC lineages.
Figure 5. Disruption of endodermal GATAa activity affects endodermal GATAa expression but has no discernable effect on TVC gene expression.
Representative stage 22 (11–11.5 hpf) embryos co-electroporated with Mesp-lacZ (red) and either Ttf-GATAa-DBD (A–E), or Ttf-GATAa-WRPW (A’–E’) and probed for expression of Hand-like, FoxF, GATAa, BMP2/4 and Nkx as indicated. The displayed patterns were highly consistent in all embryos examined (n=20–30 embryos for each condition). The insets show magnified views of the TVC region including both single and merged channels. Scale bar: 50 µm.
Our results suggest that heart progenitors in Ciona and vertebrate embryos converge on the midline through conserved interactions with the underlying endoderm. In both vertebrate and Ciona embryos, bifida phenotypes resulting from loss of endodermal GATAa function do not appear to involve disruption of heart progenitor transcriptional identity (Fig. 5) and (Peterkin et al., 2009; Reiter et al., 1999). Instead, cardia and TVC bifida reflect disruptions in endoderm morphogenesis (Fig. 4). The cellular impact of GATA disruption on vertebrate endoderm morphogenesis has not been evaluated. We have therefore begun to explore this process in Ciona.
Disruption of GATAa activity interferes with folding of the posterior endoderm
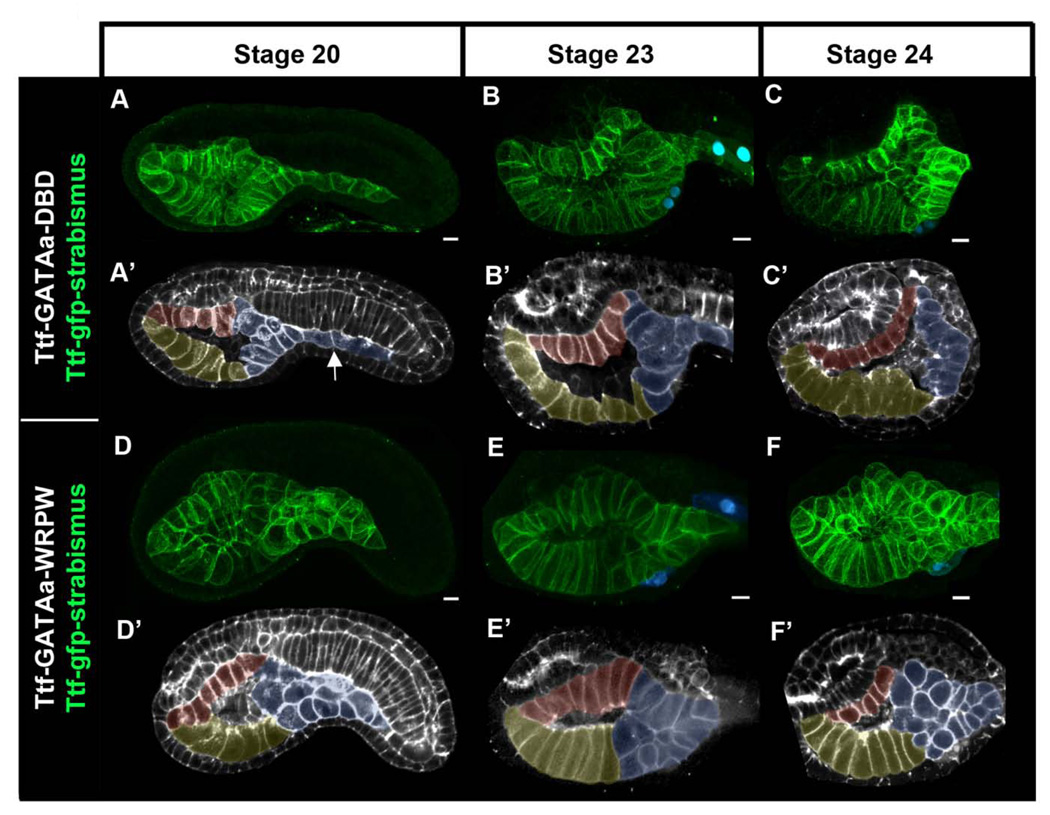
We exploited the cellular simplicity of Ciona embryos to examine how disruption of GATAa activity affects endoderm morphogenesis. We first examined endoderm morphogenesis in control embryos (Fig. 6A–C’). For this purpose we labeled endoderm cell membranes by transgenic expression of a GFP-strabismus fusion protein under the Ttf enhancer (Ttf-GFP-strabismus). We fixed Ttf- GFP-strabismus / Ttf-GATAa-DBD embryos at hourly intervals beginning at gastrulation (stage 10) and continuing until stage 24 when TVC fusion was complete. Endoderm development in Ttf-GATAa-DBD embryos was indistinguishable from that in wild type labeled embryos (Fig. 6A–C’ and data not shown). By the end of neurulation (stage 16), endoderm precursor cells formed a three layered rudiment just ventral to the developing notochord (data not shown). Approximately two hours later (stage 20), the trunk endoderm formed a trapezoidal sac lined by three epithelial surfaces; a dorsal roof, a ventral floor and a posterior wall (Fig. 6A, pseudo-colored as red, yellow and blue respectively in Fig. 6A’). In the tail region, multiple layers of posterior endoderm had converged and elongated to form the endodermal strand (Fig. 6A’ white arrow). Over the next four hours (stages 22–24, Fig. 6B–C’) the roof and floor of the gut cavity lengthened along the anterior/posterior axis while the posterior wall narrowed and elongated to form a single layer.
Figure 6. Targeted disruption of GATAa activity perturbs posterior endoderm morphogenesis.
(A–F) Partial lateral projections of representative embryos co-electroporated with Mesp-lacZ to mark the heart progenitors (blue), Ttf-GFP-strabismus to mark the endoderm (green), and either Ttf-GATAa-DBD (A–C’) or Ttf-GATAa-WRPW (D–F’). Embryos were fixed at the stage indicated at the top of the figure. Note that GFP-strabismus was enriched basolateraly in polarized epithelia as observed previously (Gravel et al., 2010). (A’–F’) To clarify endoderm morphology, schematic representations of corresponding embryos (A–F) were created using single optical sections including F-actin staining (grey) with the GFP-strabismus expressing endoderm cells pseudo-colored (yellow = ventral floor, red posterior endoderm). White arrow indicates the endodermal strand. Scale bar: 10 µm.
Based on our observations of control embryos, we evaluated the effect of Ttf-GATAa-WRPW on endoderm morphogenesis. During gastrulation and neurulation, morphogenesis of the endoderm rudiment is highly dynamic, making it difficult to discern consistent differences between wild-type and Ttf-GATAa-WRPW embryos (data not shown). We first observed consistent disruption of endoderm morphology in Ttf-GATAa-WRPW embryos at stage 20 (Fig. 6D, D’). This disruption was particularly evident in the posterior endoderm (blue, Fig. 6D’). In this domain, endoderm cells coalesced into a disorganized group, 1–4 layers wide and 1–2 layers thick. The endodermal strand did not elongate posteriorly into the nascent tail and the posterior wall of the gut cavity did not appear to form (compare Fig. 6B,C to 6E,F). Strikingly, the anterior gut often appeared relatively normal with a well-delineated ventral floor and dorsal roof. This disparity in anterior and posterior development was maintained at later stages (Fig. 6E–F’).
In summary, these results indicate that endodermal GATAa activity is required for morphogenesis of the posterior endoderm. In particular, it appears that GATAa activity mediates the coordinated convergence of endoderm precursors as they form the endodermal strand and rear wall of the nascent gut.
Discussion
GATAa acts cell-autonomously to regulate the heart progenitor gene network
Our results indicate that GATAa plays a conserved, cell-autonomous role establishing cardioblast transcriptional identity. Through sequential expression studies, we show that GATAa occupies a central position in the Ciona cardioblast gene network. Through targeted repression assays, we demonstrate that GATAa functions cell-autonomously in the TVCs to regulate this gene network. We have demonstrated that GATAa either directly or indirectly participates in three core nodes of the TVC network; maintenance of prior network components (FoxF), regulation of its own expression and; expression of new components, including the heart kernel gene Nkx. Cell-autonomous GATAa activity thereby promotes the defining behaviors of the TVC lineage, including detachment, migration and proliferation. Thus, Ciona GATAa appears to perform a suite of cell-autonomous functions fulfilled by GATA 4,5 and 6 in vertebrate cardiomyocytes (Peterkin et al., 2005). However, our understanding of the precise role of GATAa in these conserved regulatory networks is far from complete. Future experiments will focus on a more comprehensive characterization of GATAa-regulated TVC genes. In particular, these efforts will discriminate between direct vs. indirect GATAa regulation of cardiac network components. Additionally, our repression based assays are designed to investigate the contribution of GATAa mediated target gene activation. Future studies will investigate the potential contribution of GATAa mediated repression in the heart network.
The position of GATAa in the Ciona heart regulatory kernel may reflect the ancestral chordate network. The regulatory architecture of the vertebrate heart network appears to have undergone significant divergence. In some vertebrate embryos, GATA factors play a primary role in establishing heart kernel gene expression, while in others they function downstream of Nkx2.5 (Peterkin et al., 2005). We have found that GATAa is expressed prior to Nkx and that GATAa activity is required for Nkx expression. Thus our data suggest that GATAa functions upstream of Nkx. This may represent the ancestral chordate pattern. However, substantial evaluation of chordate heart network evolution requires further research on the precise regulatory relationships between cardiac genes in Ciona, cephalochordates and basal vertebrates.
GATAa exhibits differential modes of auto-regulation in the TVC and endoderm lineages
Through examination of endogenous GATAa expression in response to disruptions of GATAa function, we have revealed distinct cell-lineage specific modes of GATAa auto-regulation. In the TVCs, it appears that GATAa activity is required for GATAa expression, supporting the presence of a direct or indirect positive feedback loop. In the posterior endoderm, it appears that GATAa activity is not required for GATAa expression, indicating the absence or weakening of this positive feedback. More surprisingly, in the anterior endoderm it appears that GATAa activity is required to suppress GATAa expression, indicating the presence of a tissue specific, indirect negative-feedback loop.
GATAa feedback loops in the heart gene network may involve mutually supportive regulatory interactions with FoxF. Previous studies in Ciona indicate that TVC GATAa expression is partially dependent on FoxF activity (Beh et al., 2007; Christiaen et al., 2008) and here we show that FoxF expression is dependent on GATAa activity. Studies of GATA4 regulation in mouse embryos have identified a critical role for FoxF and GATA auto-regulatory binding sites in an early lateral plate mesoderm enhancer (Rojas et al., 2005). These observations warrant further, more rigorous studies of presumed recursive regulatory interactions between FoxF and GATAa in Ciona. In particular, it will be important to determine whether there are direct or indirect regulatory interactions between these two transcription factors and whether these interactions represent a conserved node in chordate heart or lateral plate gene networks.
The presence of lineage specific GATAa auto-regulatory loops in Ciona has fascinating implications for vertebrate GATA function. Previous studies have suggested that feedback loops between vertebrate GATA factors ensure that loss of a single GATA family leads to the compensatory up-regulation of the other family members (Kuo et al., 1997). However, the regulatory mechanisms underlying GATA feedback loops and their potential contributions to vertebrate cardiogenesis and heart disease remain poorly characterized. In particular, the possibility that these loops along with associated auto-regulatory loops may vary in a lineage specific fashion has not been explored. Further studies of the rudimentary Ciona GATAa regulatory network have the potential to disentangle this critical aspect of chordate heart evolution and development.
GATAa acts non-cell-autonomously to direct midline convergence of heart progenitor cells
This study indicates that GATAa plays a conserved, non-cell-autonomous role in the convergence of bilateral heart progenitor fields. Through sequential in situ hybridizations, we delineate persistent GATAa expression in the posterior endoderm. Lineage specific functional assays demonstrate that endodermal GATAa activity is required for morphogenesis of posterior gut tissues. In accordance with studies in vertebrates, our results suggest that proper endoderm morphogenesis in Ciona is required for convergence of bilateral heart progenitors but is not required for the establishment of cardioblast transcriptional identity (Peterkin et al., 2009; Reiter et al., 1999). This interpretation is supported by the normal detachment and anterior migration of TVCs in Ttf-GATAa-WRPW embryos. However, we have only examined a subset of early TVC marker genes. Future work will focus on elucidating the impact of endodermal GATAa activity on comprehensive TVC gene expression. It will also be critical to examine the impact of endoderm GATAa activity on TVC expression at later stages. We are particularly interested in determining whether endodermal GATAa activity or endoderm morphogenesis is required for the late stage demarcation between heart and pharyngeal muscle progenitors within the TVC lineage (Stolfi et al., 2010).
GATAa and endoderm morphogenesis
The complex architecture of embryonic vertebrate endoderm has made it difficult to distinguish the precise contribution of GATA to endoderm morphogenesis. We have therefore begun to exploit the cellular simplicity of Ciona to observe the precise impact of GATA function on endoderm formation. Confocal imaging of fluorescently labeled endoderm has permitted a three dimensional reconstruction of the developing gut. Based on this analysis, prior lineage data (Nishida, 1987) and additional unpublished data (K. Ragkousi, in preparation), we have subdivided the Ciona gut primordium into three presumptive morphogenetic regions; 1) the dorsal roof of the anterior gut cavity; 2) the ventral floor of the anterior gut cavity; 3) the posterior endoderm, including the posterior wall of the gut cavity and the endodermal strand. Targeted manipulations of endodermal GATAa activity have a differential impact on posterior endoderm morphogenesis in accordance with GATAa expression in this lineage. In particular, we find that GATAa activity is required for the formation of the posterior gut wall and the elongation of the endodermal strand.
Endoderm morphogenesis and heart progenitor convergence in chordate embryos
The remarkable similarity of the bifid phenotypes observed in Ciona and vertebrate embryos may reflect a conserved, chordate role for the endoderm in instructing heart progenitor convergence. In vertebrate embryos, convergence of heart progenitors requires convergence of the underlying foregut epithelia. However, the nature of foregut convergence is variable and reflects fundamental differences in embryonic development. In zebrafish embryos, a flat sheet of endoderm converges towards the midline to form a thickened rod (Warga and Nusslein-Volhard, 1999). Endoderm convergence in Xenopus is less well-characterized but appears to involve the movement of bifid foregut precursors over a layer of deep endoderm (Li et al., 2008). In contrast, mouse and chick foregut convergence is associated with a deep invagination of the foregut pocket (Tremblay and Zaret, 2005). These studies suggest that instructive interactions between the converging foregut and heart progenitors may have been maintained despite gross shifts in foregut morphogenesis. These conserved instructive interactions may reflect an ancestral vertebrate mechanism for heart progenitor convergence. Alternatively, they may reflect a more basal chordate program or they may have arisen independently in multiple clades. A precise characterization of endoderm/heart precursor interactions within vertebrate and invertebrate chordates is required to distinguish between these possibilities.
Interactions between heart progenitors and the endoderm epithelia
The nature of endoderm/heart progenitor interactions in vertebrate embryos remains poorly characterized. The associated convergence of the vertebrate foregut and heart fields imply a mechanical link that drags the heart progenitors into position. Alternatively, the foregut may provide signals that facilitate heart progenitor migration. Our preliminary observations suggest that the Ciona gut rudiment does not undergo midline convergence. Instead, the ventral floor of the endoderm appears to broaden laterally as the adjacent TVCs converge (data not shown). Time-lapse imaging analysis is required to discern whether individual endoderm cells converge in association with the TVCs despite the apparent lack of overall endoderm convergence. We anticipate that further research in Ciona will provide valuable insights into the precise nature of conserved endoderm/heart progenitor interactions.
Conclusions
In this study we demonstrate that GATAa is a functional orthologue of the vertebrate GATA factors, directing cardiomyocyte specification and positioning. Remarkably, disruption of GATA function in the endoderm leads to bifida of the heart progenitors. Thus, despite extremely low cell numbers in the Ciona embryo, it appears that conserved interactions with the endoderm promote heart progenitor convergence. Further studies in Ciona will permit high resolution analysis of endoderm/heart precursor interactions and help us formulate new testable hypotheses regarding the role of endoderm in vertebrate cardiogenesis.
Supplementary Material
Acknowledgments
We are grateful to Mike Levine, Shota Chiba, Michael Veeman, Ute Rothbächer, Lionel Christiaen, Bill Smith and Patrick Lemaire for training and advice as well as François Robin for the esconsin-3xgfp construct. We also thank Tom Bunch and Antony Jose for their critical comments on the manuscript. This work was funded by NIH grant (HL091027) to B.D. and an American Heart Association postdoctoral fellowship (0920117G) to K.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander J, Rothenberg M, Henry GL, Stainier DYR. casanova plays an early and essential role in endoderm formation in zebrafish. Developmental Biology. 1999;215:343–357. doi: 10.1006/dbio.1999.9441. [DOI] [PubMed] [Google Scholar]
- Barolo S, Levine M. hairy mediates dominant repression in the Drosophila embryo. Embo J. 1997;16:2883–2891. doi: 10.1093/emboj/16.10.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beh J, Shi W, Levine M, Davidson B, Christiaen L. FoxF is essential for FGF-induced migration of heart progenitor cells in the ascidian Ciona intestinalis. Development. 2007;134:3297–3305. doi: 10.1242/dev.010140. [DOI] [PubMed] [Google Scholar]
- Christiaen L, Davidson B, Kawashima T, Powell W, Nolla H, Vranizan K, Levine M. The transcription/migration interface in heart precursors of Ciona intestinalis. Science. 2008;320:1349–1352. doi: 10.1126/science.1158170. [DOI] [PubMed] [Google Scholar]
- Corbo JC, Levine M, Zeller RW. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development. 1997;124:589–602. doi: 10.1242/dev.124.3.589. [DOI] [PubMed] [Google Scholar]
- Davidson B. Ciona intestinalis as a model for cardiac development. Semin Cell Dev Biol. 2006 doi: 10.1016/j.semcdb.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B, Levine M. Evolutionary origins of the vertebrate heart: Specification of the cardiac lineage in Ciona intestinalis. Proc Natl Acad Sci U S A. 2003;100:11469–11473. doi: 10.1073/pnas.1634991100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B, Shi W, Beh J, Christiaen L, Levine M. FGF signaling delineates the cardiac progenitor field in the simple chordate, Ciona intestinalis. Genes Dev. 2006;20:2728–2738. doi: 10.1101/gad.1467706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B, Shi W, Levine M. Uncoupling heart cell specification and migration in the simple chordate Ciona intestinalis. Development. 2005;132:4811–4818. doi: 10.1242/dev.02051. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- Dodou E, Verzi MP, Anderson JP, Xu S-M, Black BL. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development. 2004;131:3931–3942. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- Dong B, Horie T, Denker E, Kusakabe T, Tsuda M, Smith WC, Jiang D. Tube formation by complex cellular processes in Ciona intestinalis notochord. Developmental Biology. 2009;330:237–249. doi: 10.1016/j.ydbio.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher A, Ohsako S, Caudy M. The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein- protein interaction domain. Mol. Cell. Biol. 1996;16:2670–2677. doi: 10.1128/mcb.16.6.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon M, Bader D. Initiation of cardiac differentiation occurs in the absence of anterior endoderm. Development. 1995;121:2439–2450. doi: 10.1242/dev.121.8.2439. [DOI] [PubMed] [Google Scholar]
- Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, Matsuoka R, Cohen JC, Srivastava D. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- Gravel M, Iliescu A, Horth C, Apuzzo S, Gros P. Molecular and cellular mechanisms underlying neural tube defects in the loop-tail mutant mouse. Biochemistry. 2010;49:3445–3455. doi: 10.1021/bi902180m. [DOI] [PubMed] [Google Scholar]
- Harafuji N, Keys DN, Levine M. Genome-wide identification of tissue-specific enhancers in the Ciona tadpole. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:6802–6805. doi: 10.1073/pnas.052024999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haworth K, Kotecha S, Mohun T, Latinkic B. GATA4 and GATA5 are essential for heart and liver development in Xenopus embryos. BMC Developmental Biology. 2008;8:74. doi: 10.1186/1471-213X-8-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzinger A, Evans T. Gata5 and Gata6 are functionally redundant in zebrafish for specification of cardiomyocytes. Developmental Biology. 2007;312:613–622. doi: 10.1016/j.ydbio.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta K, Mitsuhara K, Takahashi H, Inaba K, Oka K, Gojobori T, Ikeo K. A web-based interactive developmental table for the ascidian Ciona intestinalis, including 3D real-image embryo reconstructions: I. From fertilized egg to hatching larva. Developmental Dynamics. 2007;236:1790–1805. doi: 10.1002/dvdy.21188. [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, MacRae CA, Stainier DYR, Poss KD. Primary contribution to zebrafish heart regeneration by gata4+ cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Trinh LA, Reiter JF, Alexander J, Yelon D, Stainier DYR. The zebrafish bonnie and clyde gene encodes a Mix family homeodomain protein that regulates the generation of endodermal precursors. Genes & Development. 2000;14:1279–1289. [PMC free article] [PubMed] [Google Scholar]
- Kuo C, Morrisey E, Anandappa R, Sigrist K, Lu M, Parmacek M, Soudais C, Leiden J. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- Li Y, Rankin SA, Sinner D, Kenny AP, Krieg PA, Zorn AM. Sfrp5 coordinates foregut specification and morphogenesis by antagonizing both canonical and noncanonical Wnt11 signaling. Genes & Development. 2008;22:3050–3063. doi: 10.1101/gad.1687308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden DG, Olson EN. Heart development: learning from mistakes. Current Opinion in Genetics & Development. 2002;12:328–335. doi: 10.1016/s0959-437x(02)00306-4. [DOI] [PubMed] [Google Scholar]
- Molkentin J. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem. 2000;275:38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- Molkentin J, Lin Q, Duncan S, Olson E. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- Nishida H. Cell lineage analysis in ascidian embryos by intracellular injection of a tracer enzyme. III. Up to the tissue restricted stage. Dev Biol. 1987;121:526–541. doi: 10.1016/0012-1606(87)90188-6. [DOI] [PubMed] [Google Scholar]
- Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN, Schneider MD. Sizing up the heart: development redux in disease. Genes & Development. 2003;17:1937–1956. doi: 10.1101/gad.1110103. [DOI] [PubMed] [Google Scholar]
- Patient R, McGhee J. The GATA family (vertebrates and invertebrates) Curr Opin Genet Dev. 2002;12:416–422. doi: 10.1016/s0959-437x(02)00319-2. [DOI] [PubMed] [Google Scholar]
- Peterkin T, Gibson A, Loose M, Patient R. The roles of GATA-4, -5 and -6 in vertebrate heart development. Semin Cell Dev Biol. 2005;16:83–94. doi: 10.1016/j.semcdb.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Peterkin T, Gibson A, Patient R. Redundancy and evolution of GATA factor requirements in development of the myocardium. Dev Biol. 2007;311:623–635. doi: 10.1016/j.ydbio.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterkin T, Gibson A, Patient R. Common genetic control of haemangioblast and cardiac development in zebrafish. Development. 2009;136:1465–1474. doi: 10.1242/dev.032748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posch MG, Perrot A, Schmitt K, Mittelhaus S, Esenwein E-M, Stiller B, Geier C, Dietz R, Geβner R, Özcelik C, Berger F. Mutations in GATA4, NKX2.5, CRELD1, and BMP4 are infrequently found in patients with congenital cardiac septal defects. American Journal of Medical Genetics Part A. 2008;146A:251–253. doi: 10.1002/ajmg.a.32042. [DOI] [PubMed] [Google Scholar]
- Rajagopal SK, Ma Q, Obler D, Shen J, Manichaikul A, Tomita-Mitchell A, Boardman K, Briggs C, Garg V, Srivastava D, Goldmuntz E, Broman KW, Woodrow Benson D, Smoot LB, Pu WT. Spectrum of heart disease associated with murine and human GATA4 mutation. Journal of Molecular and Cellular Cardiology. 2007;43:677–685. doi: 10.1016/j.yjmcc.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reecy JM, Li X, Yamada M, DeMayo FJ, Newman CS, Harvey RP, Schwartz RJ. Identification of upstream regulatory regions in the heart-expressed homeobox gene Nkx2-5. Development. 1999;126:839–849. doi: 10.1242/dev.126.4.839. [DOI] [PubMed] [Google Scholar]
- Reiter J, Alexander J, Rodaway A, Yelon D, Patient R, Holder N, Stainier D. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes Dev. 1999;13:2983–2995. doi: 10.1101/gad.13.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristoratore F, Spagnuolo A, Aniello F, Branno M, Fabbrini F, Di Lauro R. Expression and functional analysis of Cititf1, an ascidian NK-2 class gene, suggest its role in endoderm development. Development. 1999;126:5149–5159. doi: 10.1242/dev.126.22.5149. [DOI] [PubMed] [Google Scholar]
- Rojas A, De Val S, Heidt AB, Xu S-M, Bristow J, Black BL. Gata4 expression in lateral mesoderm is downstream of BMP4 and is activated directly by Forkhead and GATA transcription factors through a distal enhancer element. Development. 2005;132:3405–3417. doi: 10.1242/dev.01913. [DOI] [PubMed] [Google Scholar]
- Roure A, Rothbächer U, Robin F, Kalmar E, Ferone G, Lamy C, Missero C, Mueller F, Lemaire P. A multicassette gateway vector set for high throughput and comparative analyses in Ciona and vertebrate embryos. PLoS ONE. 2007;2:e916. doi: 10.1371/journal.pone.0000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki J, Inoue T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- Satou Y, Imai KS, Satoh N. The ascidian Mesp gene specifies heart precursor cells. Development. 2004;131:2533–2541. doi: 10.1242/dev.01145. [DOI] [PubMed] [Google Scholar]
- Satou Y, Yamada L, Mochizuki Y, Takatori N, Kawashima T, Sasaki A, Hamaguchi M, Awazu S, Yagi K, Sasakura Y, Nakayama A, Ishikawa H, Inaba K, Satoh N. A cDNA resource from the basal chordate Ciona intestinalis. Genesis. 2002;33:153–154. doi: 10.1002/gene.10119. [DOI] [PubMed] [Google Scholar]
- Schier AF, Neuhauss SC, Helde KA, Talbot WS, Driever W. The one-eyed pinhead gene functions in mesoderm and endoderm formation in zebrafish and interacts with no tail. Development. 1997;124:327–342. doi: 10.1242/dev.124.2.327. [DOI] [PubMed] [Google Scholar]
- Shi W, Levine M. Ephrin signaling establishes asymmetric cell fates in an endomesoderm lineage of the Ciona embryo. Development. 2008;135:931–940. doi: 10.1242/dev.011940. [DOI] [PubMed] [Google Scholar]
- Singh MK, Li Y, Li S, Cobb RM, Zhou D, Lu MM, Epstein JA, Morrisey EE, Gruber PJ. Gata4 and Gata5 cooperatively regulate cardiac myocyte proliferation in mice. Journal of Biological Chemistry. 2010;285:1765–1772. doi: 10.1074/jbc.M109.038539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Stolfi A, Gainous TB, Young JJ, Mori A, Levine M, Christiaen L. Early chordate origins of the vertebrate second heart field. Science. 2010;329:565–568. doi: 10.1126/science.1190181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay KD, Zaret KS. Distinct populations of endoderm cells converge to generate the embryonic liver bud and ventral foregut tissues. Developmental Biology. 2005;280:87–99. doi: 10.1016/j.ydbio.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Veeman MT, Nakatani Y, Hendrickson C, Ericson V, Lin C, Smith WC. chongmague reveals an essential role for laminin-mediated boundary formation in chordate convergence and extension movements. Development. 2008;135:33–41. doi: 10.1242/dev.010892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warga RM, Nusslein-Volhard C. Origin and development of the zebrafish endoderm. Development. 1999;126:827–838. doi: 10.1242/dev.126.4.827. [DOI] [PubMed] [Google Scholar]
- Wolf M, Basson CT. The molecular genetics of congenital heart disease: a review of recent developments. Current Opinion in Cardiology. 2010;25:192–197. doi: 10.1097/HCO.0b013e328337b4ce. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Watt AJ, Battle MA, Li J, Bondow BJ, Duncan SA. Loss of both GATA4 and GATA6 blocks cardiac myocyte differentiation and results in acardia in mice. Developmental Biology. 2008;317:614–619. doi: 10.1016/j.ydbio.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.