Abstract
Background
Nucleic-acid-testing (NAT) to diagnose HIV infection in children under age 18 months provides a barrier to HIV-testing in exposed children from resource-constrained settings. The ultrasensitive HIV- p24- antigen (Up24) assay is cheaper and easier to perform and is sensitive (84–98%) and specific (98–100%). The cut-point optical density (OD) selected for discriminating between positive and negative samples may need assessment due to regional differences in mother-to-child HIV-transmission rates.
Objectives
We used receiver operator characteristics (ROC) curves and logistic regression analyses to assess the effect of various cut-points on the diagnostic performance of Up24 for HIV-infection status among HIV-exposed children. Positive and negative predictive values at different rates of disease prevalence were also estimated.
Study design
A study of Up24 testing on dried blood spot (DBS) samples collected from 278 HIV-exposed Haitian children, 3–24-months of age, in whom HIV-infection status was determined by NAT on the same DBS card.
Results
The sensitivity and specificity of Up24 varied by the cut-point-OD value selected. At a cut-point-OD of 8-fold the standard deviation of the negative control (NCSD), sensitivity and specificity of Up24 were maximized [87.8% (95% CI, 83.9–91.6) and 92% (95% CI, 88.8–95.2), respectively]. In lower prevalence settings (5%), positive and negative predictive values of Up24 were maximal (75.9% and 98.8%, respectively) at a cut-point-OD that was 15-fold the NCSD.
Conclusions
In low prevalence settings, a high degree of specificity can be achieved with Up24 testing of HIV-exposed children when a higher cut-point OD is used; a feature that may facilitate more frequent use of Up24 antigen testing for HIV-exposed children.
1. Background
The World Health Organization recommends highly active antiretroviral therapy (HAART) for all children diagnosed with HIV infection at two years of age and younger.1 HAART for HIV-infected children in resource-constrained settings (RCS) is hindered by limited access to nucleic acid testing (NAT) required for diagnosis. Frequent HIV-testing of infants with ongoing exposure to HIV through breast-feeding is also deterred. Evaluation of ultrasensitive p24 (Up24) antigen assays on liquid or dried plasma2–8 or blood spots9–13, that is more feasible for RCS, yielded sensitivities of 84–98% and specificities of 98–100%. More recently, Mwapasa et. al.11 showed that the performance characteristics of a simplified Up24 assay had a sensitivity of 84% and 98%, respectively, and the positive and negative predictive values was influenced by the cutoff OD selected to distinguish positive and negative samples, and by HIV prevalence.
2. Objective
We used receiver operator characteristics (ROC) curves and logistic regression analyses to assess the effect of various cut-points on the diagnostic performance of Up24 to correctly classify HIV-infection status among HIV-exposed children. Positive and negative predictive values at different rates of disease prevalence were also estimated.
3. Study Design
As part of the JHPIEGO-UNICEF and Caris, Early Infant Diagnosis (EID) program in Haiti, DBS (Whatman # 903 cards; Fischer Scientific, MA) were collected on 278 HIV-exposed, Haitian children between 0.1 to 24 months of age for testing by HIV DNA polymerase chain reaction (HIV-DNA PCR; AMPLICOR HIV-1 DNA test version 1.5;Roche Diagnostics GMBH, Germany). Testing was done in a laboratory in Kenya accredited for infant HIV testing. This laboratory also participates in an external quality assurance program. DBS were labeled with a unique code without identifiers, collection date, and age before stored at room temperature in a polythene bag with desiccant, but without humidity indicator. The results of HIVNDA PCR testing were reported to clinical sites with a median turnaround time of 1.8 months (IQR: 0.6 – 2.4 months). Only the clinical program staff maintained the ability for using the unique codes to connect results back to patient records. The residual DBS were shipped to the U.S. for testing by Up24 antigen to assess potential for implementation in Haiti. DBS were stored at 4° C until tested. Laboratory personnel performing the Up24 antigen assays were blinded to the PCR results. The study was approved by the Institutional Review Board at Johns Hopkins University with waiver of consent, and also by the Haitian Ministry of Health and the CARIS Foundation, the EID implementing partner.
Whole blood was eluted from two whole spots from the DBS card using slight modifications of published methods12,13. The modifications included increasing the volume (250 μl) of SNCR lysis buffer14 and the addition of a quick spin (two minutes in a microfuge, Centrifuge 5424, Eppendorf, Germany) over a column (Cat # 28704, Qiagen Inc., CA) to remove coagulated blood generated during the heat dissociation step. These additional steps enriched the yield of the final elution volume without compromising assay sensitivity or specificity (data not shown); 200 μl of eluate was used for Up24 antigen testing with ELAST amplification according to the manufacturer’s directions (Perkin Elmer, MA). DBS samples from HIV-seronegative persons were also run with each assay.
The mean optical density (OD) of four replicate negative-controls from HIV-negative serum (kit reagent) was used to establish the mean negative control OD value. This value was subtracted from all samples to yield an adjusted OD value that was used in the subsequent analyses. The standard deviation of OD values from the four replicate negative controls (termed NCSD henceforth) was calculated. Multiples (0–15) of these NCSDs were used as a range of cut-point or to assess sensitivity and specificity. Analyses of sensitivity and specificity and area under the ROC curve (AUC)15 were performed for multiples of the NCSDs as the OD cut-point value. A logistic regression analysis was performed with the HIV-DNA PCR test results and the undichotomized, adjusted OD values. Descriptive statistics were calculated for age at sample collection, and duration of storage. Median characteristics of subgroups were compared using the Wilcoxon Rank-Sum. Comparisons of the sensitivity, specificity and AUC between subgroups determined by the median age and storage duration were performed using an optimized cut-point OD value (multiple of the NCSD).15 Statistical analyses were performed using STATA v 10.1 (STATA Statistical Software: College Station, TX: STATACorp LP, 2009).
4. Results
The cohort of 278 children were at a median age of four months (IQR: 2.0 – 7.7 months) at the time of DBS collection. Samples were stored for a median of 14 months (IQR: 11.4 – 15.7 months; range 7–26 months) before testing with the Up24 assay. The HIV- infection prevalence was 14.7% by HIV DNA PCR testing. The median age of the 41 HIV- infected infants was four months (IQR: 2.2 – 7.4 months).
Figure 1 shows the distribution of adjusted OD values for HIV-exposed children who tested positive or negative by HIV-DNA PCR, and for the HIV-seronegative controls. The median OD for children identified as infected by HIV-DNA PCR was significantly higher (p < 0.001) as compared to those who tested negative by HIV-DNA PCR (2.24 [IQR, 0.58–3.16] versus 0.05 [IQR, −0.023–0.11]), respectively (Fig. 1). One HIV-DNA PCR negative sample seemed to be an outlier, but repeat testing based on Up24 results were not planned.
Fig. 1.
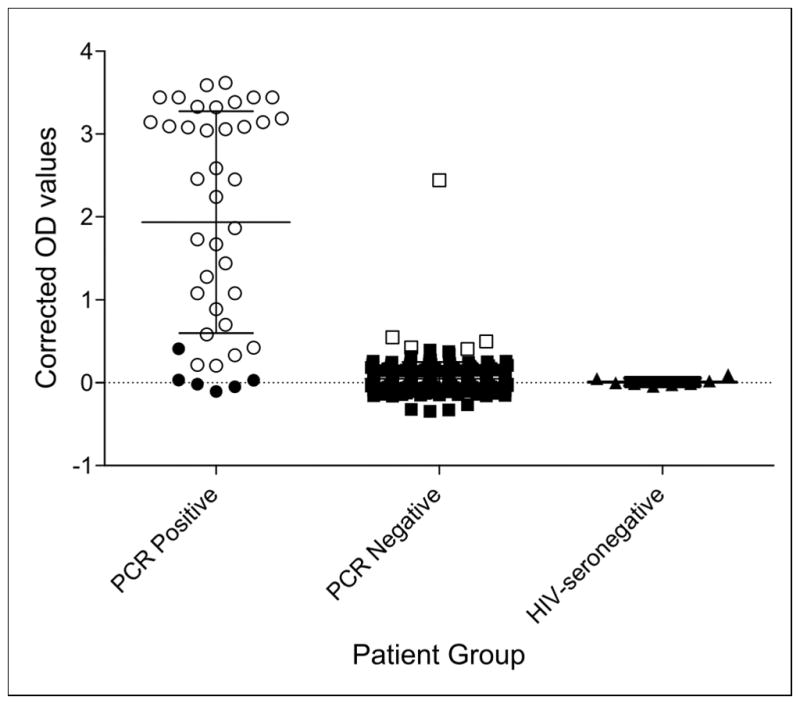
Optical Density (OD) values measured at 490nm with reference at 630nm adjusted by subtracting OD of the average of the negative controls by Ultrasensitive p24 (Up24) antigen testing on dried blood spots (DBS) from HIV-exposed children with HIV-DNA PCR confirmed HIV infection status. Open circles (black) are samples that tested positive by both HIV-DNA PCR and Up24, filled circles (black) represent the samples that were positive by HIV-DNA but negative by Up24. Filled squares (black) represent the samples that tested negative by both HIV-DNA PCR and Up24, and open squares (black) represent HIV-DNA PCR negative samples that tested positive by Up24. Black triangles are DBS samples from HIV-seronegative persons. The median OD for children identified as infected by HIV-DNA PCR was significantly higher (p < 0.001) as compared to those who tested negative by HIV-DNA PCR (2.24 [IQR, 0.58–3.16] versus 0.05 [IQR, −0.023–0.11]), respectively. The median OD for DBS from eight HIV-seronegative controls was −0.006 (IQR, −0.02 – 0.04).
Previous studies on the use of Up24 antigen testing to diagnose HIV infection in children have used various approaches to discriminate between positive and negative samples; a cut-point OD value equal to or greater than five times the NCSD is often used.2,4,7,8,10,12,13 Using this criterion, the overall sensitivity of Up24 antigen assay was 87.8% and the specificity was 86.5% (Table 1) with 86.7% of 278 HIV-exposed infants correctly classified. Five of 41 (12%) infected children were misclassified as uninfected, and 32 of 237 uninfected children (13.5%) were misclassified as infected.
Table 1.
Sensitivity and Specificity of Up24 for Infant HIV diagnosis using Receiver Operator Characteristics Curves (N=278)
| Multiples of NCSDa for Cut-Point OD | Sensitivity (%)(95% CI) | Specificity (%)(95% CI) | AUCb | Correct Classification (%) |
|---|---|---|---|---|
| 0 | 92.7(89.6–95.7) | 46.8(41.0–52.7) | 0.69 | 53.6 |
| 1 | 92.7(89.6–95.7) | 58.2(52.4–64.0) | 0.75 | 63.3 |
| 2 | 92.7(89.6–95.7) | 65.4(59.8–70.9) | 0.79 | 69.4 |
| 3 | 87.8(89.6–95.7) | 74.3(69.1–79.4) | 0.81 | 76.2 |
| 4 | 87.8(83.9–91.6) | 81.4(76.8–86.0) | 0.85 | 82.4 |
| 5 | 87.8(83.9–91.6) | 86.5(82.5–90.5) | 0.87 | 86.7 |
| 6 | 87.8(83.9–91.6) | 89.9(86.3–93.4) | 0.89 | 89.6 |
| 7 | 87.8(83.9–91.6) | 91.6(88.3–94.8) | 0.90 | 91.0 |
| 8c | 87.8(83.9–91.6) | 92.0(88.8–95.2) | 0.90 | 91.4 |
| 9 | 80.5(75.8–85.1) | 92.8(89.8–95.9) | 0.87 | 91.0 |
| 10 | 80.5(75.8–85.1) | 93.7(90.8–96.5) | 0.87 | 91.7 |
| 11 | 80.5(75.8–85.1) | 96.2(93.9–98.5) | 0.88 | 93.9 |
| 12 | 80.5(75.8–85.1) | 97.0(95.0–99.0) | 0.89 | 94.6 |
| 13 | 80.5(75.8–85.1) | 97.9(96.2–99.6) | 0.88 | 95.0 |
| 14 | 78.0(73.2–82.9) | 98.3(96.8–99.8) | 0.88 | 95.3 |
| 15 | 78.0(73.2–82.9) | 98.7(97.4–100.0) | 0.88 | 95.7 |
NCSD – Standard deviation of negative controls per assay
AUC – Area under the receiver operating characteristic curve
Multiple of NCSD with maximum sensitivity and specificity
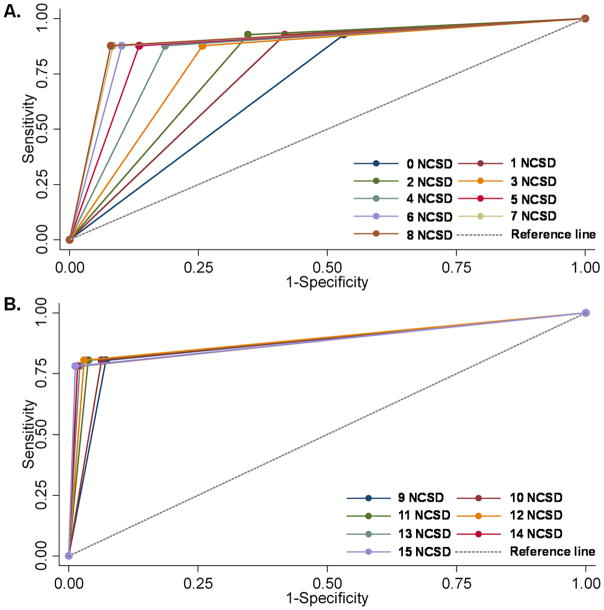
The sensitivity and specificity of Up24 for identifying HIV-infection in children was examined using ROC curves over a range of cut-point OD values (Table 1). With increasing multiples of the NCSD, sensitivity decreased and specificity increased. The Youden index16,17, the cut-point at which sensitivity (87.8%) and specificity (92%); were optimized was identified at 8-fold the NCSD; with 91.4% of children correctly classified. A cut-point OD that was 15 times the NCSD resulted in correct classification of 95.7% of the children’s infection status.
Another approach to evaluate the discriminatory capabilities of Up24 is the AUC derived from the ROC curve (Table 1, Fig. 2).18 The AUC, a summary statistic for the ROC curve, is the probability that the measure used to classify samples will rank a randomly chosen positive sample higher than a randomly chosen negative sample.18 The AUC was lowest (0.69) at zero times the NCSD and highest between 7 to 15 times NCSD (range: 0.88–0.90); these comparisons were statistically different across the 15 NCSD multiples (p < 0.001). Using logistic regression analysis, the OD cut-point that optimized sensitivity (87.8%) and specificity (92%) was similarly identified as eight times the NCSD with 91.4% of the samples correctly classified.
Fig. 2.
Receiver Operator Characteristics curves of the Up24 antigen data generated at the different cut-point optical density tested between 0–8-fold NCSD [Panel A] and 8–15-fold NCSD [Panel B]. The diagonal or chance line shown in grey is the reference line use to calculate the area under curve (AUC) for each ROC curve.18,19
Since age of testing and duration of storage of DBS may also affect the performance characteristics of Up24 antigen detection in DBS9,13, we examined, using a logistic regression model with 8-fold NCSD as the cut-point, their effect on the sensitivity and specificity of Up24 antigen assay. No significant difference in the median age (p = 0.87) or duration of storage (p=0.246) between the samples with negative and positive PCR results were found.
Table 2 summarizes the positive and negative predictive values for Up24 antigen assay at different HIV prevalence levels and when multiples of 5, 8 and 15-fold the NCSD are used for the cut-point. In areas where the expected prevalence of true infection is approximately 5%, the positive and negative predictive values of Up24 at 8- and 15-fold the NCSD were 36.6 and 99.3%, and 75.9 and 98.8%, respectively.
Table 2.
Effect of Varying Population Prevalence of HIV Infection on Test Performance Characteristics (PPVa and NPVa) at Different ODa Cut-Point Values
| OD Cut-off Value (Sensitivity, Specificity) | 5* NCSDb (87.8%, 86.5%) | 8* NCSD (87.8%, 92.0%) | 15* NCSD (78.0%, 98.7%) | |||
|---|---|---|---|---|---|---|
| Prevalence of True Infection | PPV(%) | NPV (%) | PPV(%) | NPV(%) | PPV(%) | NPV(%) |
| 1% | 6.2 | 99.9 | 10 | 100 | 37.7 | 100 |
| 5% | 25.5 | 99.3 | 36.6 | 99.3 | 75.9 | 98.8 |
| 10% | 41.9 | 98.5 | 54.9 | 98.5 | 86.9 | 97.6 |
| 15% | 53.4 | 97.6 | 65.9 | 97.7 | 91.3 | 96.2 |
PPV-Positive predictive value; NPV-Negative predictive value
NCSD – Standard deviation of negative controls by assay
Fold of NCSD
5. Discussion
In this study of 278 HIV-exposed children for whom the prevalence of HIV infection was determined at 14.5% by HIV-DNA PCR, we found that the sensitivity and specificity of Up24 testing on DBS samples varied significantly based on the cut-point selected. When the Youden index (8-fold NCSD) was used, sensitivity and specificity of Up24 was 88% and 92%, respectively. The cut-point identified by ROC curves was similar to that identified using AUC and logistic regression analyses. By increasing the cut-point OD to 11- to 15-times the standard deviation of the negative controls, specificities between 96–98% was achieved. In countries where PMTCT programs are established, it is expected that a high proportion (nearly 93–99%) of HIV-exposed children will not acquire infection.19,20 Therefore, a technically non-demanding test with high specificity is likely to be beneficial for classifying infection status of HIV-exposed children.
Our study is limited by the prolonged storage of samples (median 14-months) before testing, which may explain the lower sensitivity of Up24 in this study (88%) compared with other studies (84–98%) done on DBS samples9. In addition, Up24 testing was not performed in a resource-constrained setting, yet the assay required slight modifications from published methods to achieve optimal performance, highlighting the need for standardized protocols for Up24 antigen testing. A recent study by Mwapasa showed a similar degree of specificity (>96%) of Up24 testing for infant HIV diagnosis with a simplified Up24 test when tested across the different HIV prevalence levels as assessed in our stuy11. Together, these findings suggest that in low prevalence settings, a high degree of specificity can be achieved with Up24 testing of HIV-exposed children when a higher cut-point OD is used; a feature that may facilitate more frequent testing of HIV-exposed children.
Acknowledgments
We thank the study participants, the Haitian Ministry of Health, and the CARIS Foundation for making this study possible.
Sources of financial support: This work was funded by ICTR award (D.P.) made possible through Grant Number UL1 RR 025005 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH″. The work was also supported the National Institutes of Health grant # R01HD057784 (D.P.).
Footnotes
Conflicts of interest: The authors do not have an association that might pose a conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Antiretroviral therapy for HIV infection in infants and children: Towards universal access. WHO/UNAIDS; 2010. [PubMed] [Google Scholar]
- 2.De Baets AJ, Edidi BS, Kasali MJ, Beelaert G, Schrooten W, Litzroth A, et al. Pediatric human immunodeficiency virus screening in an African district hospital. Clin Diagn Lab Immunol. 2005 Jan;12(1):86–92. doi: 10.1128/CDLI.12.1.86-92.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fiscus SA, Wiener J, Abrams EJ, Bulterys M, Cachafeiro A, Respess RA. Ultrasensitive p24 antigen assay for diagnosis of perinatal human immunodeficiency virus type 1 infection. J Clin Microbiol. 2007 Jul;45(7):2274–7. doi: 10.1128/JCM.00813-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.George E, Beauharnais CA, Brignoli E, Noel F, Bois G, De Matteis RP, et al. Potential of a simplified p24 assay for early diagnosis of infant human immunodeficiency virus type 1 infection in Haiti. J Clin Microbiol. 2007 Oct;45(10):3416–8. doi: 10.1128/JCM.01314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knuchel MC, Tomasik Z, Speck RF, Luthy R, Schupbach J. Ultrasensitive quantitative HIV-1 p24 antigen assay adapted to dried plasma spots to improve treatment monitoring in low-resource settings. J Clin Virol. 2006 May;36(1):64–7. doi: 10.1016/j.jcv.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 6.Respess RA, Cachafeiro A, Withum D, Fiscus SA, Newman D, Branson B, et al. Evaluation of an ultrasensitive p24 antigen assay as a potential alternative to human immunodeficiency virus type 1 RNA viral load assay in resource-limited settings. J Clin Microbiol. 2005 Jan;43(1):506–8. doi: 10.1128/JCM.43.1.506-508.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schupbach J, Boni J, Tomasik Z, Jendis J, Seger R, Kind C. Sensitive detection and early prognostic significance of p24 antigen in heat-denatured plasma of human immunodeficiency virus type 1-infected infants. Swiss Neonatal HIV Study Group. J Infect Dis. 1994 Aug;170(2):318–24. doi: 10.1093/infdis/170.2.318. [DOI] [PubMed] [Google Scholar]
- 8.Sutthent R, Gaudart N, Chokpaibulkit K, Tanliang N, Kanoksinsombath C, Chaisilwatana P. p24 Antigen detection assay modified with a booster step for diagnosis and monitoring of human immunodeficiency virus type 1 infection. J Clin Microbiol. 2003 Mar;41(3):1016–22. doi: 10.1128/JCM.41.3.1016-1022.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cachafeiro A, Sherman GG, Sohn AH, Beck-Sague C, Fiscus SA. Diagnosis of human immunodeficiency virus type 1 infection in infants by use of dried blood spots and an ultrasensitive p24 antigen assay. J Clin Microbiol. 2009 Feb;47(2):459–62. doi: 10.1128/JCM.01181-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knuchel MC, Jullu B, Shah C, Tomasik Z, Stoeckle MP, Speck RF, et al. Adaptation of the ultrasensitive HIV-1 p24 antigen assay to dried blood spot testing. J Acquir Immune Defic Syndr. 2007 Mar 1;44(3):247–53. doi: 10.1097/QAI.0b013e31802c3e67. [DOI] [PubMed] [Google Scholar]
- 11.Mwapasa V, Cachafeiro A, Makuta Y, Beckstead DJ, Pennell ML, Chilima B, et al. Using a simplified human immunodeficiency virus type 1 p24 antigen assay to diagnose pediatric HIV-infection in Malawi. J Clin Virol. 2010 Dec;49(4):299–302. doi: 10.1016/j.jcv.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patton JC, Sherman GG, Coovadia AH, Stevens WS, Meyers TM. Ultrasensitive human immunodeficiency virus type 1 p24 antigen assay modified for use on dried whole-blood spots as a reliable, affordable test for infant diagnosis. Clin Vaccine Immunol. 2006 Jan;13(1):152–5. doi: 10.1128/CVI.13.1.152-155.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patton JC, Coovadia AH, Meyers TM, Sherman GG. Evaluation of the ultrasensitive human immunodeficiency virus type 1 (HIV-1) p24 antigen assay performed on dried blood spots for diagnosis of HIV-1 infection in infants. Clin Vaccine Immunol. 2008 Feb;15(2):388–91. doi: 10.1128/CVI.00265-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schupbach J, Boni J, Bisset LR, Tomasik Z, Fischer M, Gunthard HF, et al. HIV-1 p24 antigen is a significant inverse correlate of CD4 T-cell change in patients with suppressed viremia under long-term antiretroviral therapy. J Acquir Immune Defic Syndr. 2003 Jul 1;33(3):292–9. doi: 10.1097/00126334-200307010-00002. [DOI] [PubMed] [Google Scholar]
- 15.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982 Apr;143(1):29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 16.Youden WJ. Index for rating diagnostic tests. Cancer. 1950 Jan;3(1):32–5. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 17.Schisterman EF, Perkins NJ, Liu A, Bondell H. Optimal cut-point and its corresponding Youden Index to discriminate individuals using pooled blood samples. Epidemiology. 2005 Jan;16(1):73–81. doi: 10.1097/01.ede.0000147512.81966.ba. [DOI] [PubMed] [Google Scholar]
- 18.Fawcett T. An introduction to ROC analysis. Pattern Recognition Letters. 2006;(27):861–74. [Google Scholar]
- 19.Chasela CS, Hudgens MG, Jamieson DJ, Kayira D, Hosseinipour MC, Kourtis AP, et al. Maternal or infant antiretroviral drugs to reduce HIV-1 transmission. N Engl J Med. 2010 Jun 17;362(24):2271–81. doi: 10.1056/NEJMoa0911486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mofenson LM. Protecting the next generation--eliminating perinatal HIV-1 infection. N Engl J Med. 2010 Jun 17;362(24):2316–8. doi: 10.1056/NEJMe1004406. [DOI] [PubMed] [Google Scholar]