Abstract
The tyrosine kinase receptor, HER2 is a crucial prognostic marker and therapeutic target for breast cancer; however, the downstream targets and biological effectors of HER2 remain unclear. We investigated the relationship between HER2 and the transcription factor FoxM1 in breast cancer. HER2 and FoxM1 expression levels were compared in breast carcinoma cell lines, paraffin embedded breast cancer patient samples and at the mRNA level in purified breast epithelial cells. To further examine the relationship between HER2 and FoxM1 expression, we either overexpressed or siRNA-mediated depleted endogenous HER2 in breast cancer cell lines. Additionally, a mammary epithelium-targeted HER2 (neu) transgenic mouse model was also used to assess the effect of HER2 on FoxM1 levels. Furthermore, the effect of the HER2-tyrosine kinase inhibitor lapatinib on FoxM1 in HER2 positive breast cancer cells was investigated. HER2 protein levels directly correlated with FoxM1 expression in both breast carcinoma cell lines and paraffin embedded breast cancer patient samples. Moreover, in purified breast epithelial cells, overexpression of HER2 was associated with high levels of FoxM1 mRNA, suggesting that the upregulation of FoxM1 expression is at least partially mediated transcriptionally. Furthermore, overexpression or ablation of endogenous HER2 resulted in parallel changes in FoxM1 expression. Critically, mammary epithelium-targeted HER2 mouse tumours also resulted in increased FoxM1 expression, suggesting that HER2 directed FoxM1 expression occurs in vivo and may be a critical downstream effector of HER2-targeting therapies. Indeed, treatment of breast cancer cells with lapatinib reduced FoxM1 expression at protein, mRNA and gene promoter levels. Moreover, analysis of normal and breast cancer patient samples revealed that elevated FoxM1 expression at protein and mRNA levels correlated with breast cancer development, but not significantly with cancer progression and survival. Our results indicate that the HER2 receptor regulates the expression of the FoxM1 transcription factor, which has a role in breast cancer development.
Introduction
Human epidermal growth factor receptor-2 (HER2/neu also known as ErbB2) is a member of the epidermal growth factor receptor (EGFR) family of receptor tyrosine kinases (RTKs), which also include EGFR (ErbB1), HER3 (ErbB3), and HER4 (ErbB4). EGFR family members are typically activated in a ligand-dependent manner, and ligand-binding promotes receptor homo- or hetero-dimerisation and subsequent downstream signalling. HER2 does not bind ligands directly, but can be transactivated by the other EGFRs, leading to an amplification of signal transduction (1). EGFR family members are established regulators of cell proliferation and survival, and emerging evidence suggests that aberrant signalling of EGFR family members is a common feature of many tumour types (1, 2). In particular, overexpression of HER2 is a common feature of solid tumours and is believed to be a primary transformation event (1). Moreover, the amplification of HER2 in primary breast tumours correlates with poor prognosis and disease progression (3–5), whilst blocking HER2 activity using humanised antibodies or small molecules has proved to be an effective strategy in the treatment of breast cancer. A HER2 blocking antibody, trastuzumab (also known as Herceptin), has been demonstrated to significantly reduce the risk of breast cancer recurrence in HER2 positive patients compared to standard adjuvant therapy alone (6). Lapatinib (GW572016) is a small molecule dual tyrosine kinase inhibitor (TKI) for HER2 that acts through competitive inhibition of ATP-binding to the receptor tyrosine kinase domain (7–9). Lapatinib has also been shown to cause growth delay and cell death in breast cancer cell lines and human tumour xenografts expressing high levels of EGFR and/or HER2. Recent phase II/III clinical studies also demonstrated that lapatinib was well tolerated and provided anti-tumour activity in patients with breast as well as with other types of cancer when used as a monotherapy or in combination with other anti-cancer treatments (10, 11). However, whilst enhanced EGFR expression may have been shown to function primarily through two pathways the ERK1/2 MAP kinase and phosphatidylinositol 3-kinase (PI3-K)/protein kinase B (PKB also called Akt) signalling cascades (1, 2, 11), a complete understanding of the mechanism by which HER2 promotes tumourgenesis remains lacking.
Key features of HER2 deregulation are chromosome instability, disruption of mitotic checkpoints, deregulation of the cell death programmes, and enhanced cell cycle progression (12, 13). Recent research has highlighted the FoxM1 transcription factor, a member of the Forkhead box family, as a ubiquitously expressed key transcriptional regulator of the cell cycle (14, 15). Moreover, FoxM1 exhibits cell cycle-dependent expression, which increases at the beginning of S-phase, and peaks in G2- and M-phases (16). Consistent with FoxM1 being involved in cell cycle control, knock-out mice are embryonic leathal and display abnormalities in the heart and liver, with many cells exhibiting polyploidy (17). Moreover, FoxM1 is induced in liver regeneration and tissue repair mouse models, which correlates with expression of numerous mitotic promoting genes (18–20). Depletion of FoxM1 with siRNA, results in a delay in G2, mitotic spindle defects, chromosome missegregation and mitotic catastrophe (21–23). Evidence from microarray and other molecular analysis has revealed that FoxM1 regulates these functions by regulating genes essential for DNA synthesis, faithful chromosome regulation and mitosis, including p21Cip1, p27Kip1, Skp2, Cks1, cdc25b, Aurora B kinase, survivin, polo like kinase-1 (PLK1), centromere protein A (CENPA), CENPB, Nek-2 and KIF20A (14). Current understanding of the mechanisms by which FoxM1 is regulated is largely limited to post-translational modifications; FoxM1 is activated by cyclin-CDK and ERK mediated phosphorylation (14, 24, 25) which promotes nuclear localization, and expression of cell cycle regulators such as cdc25b, survivin, and polo like kinase-1 (PLK1) (14). Previous studies have shown that FoxM1 is overexpressed in breast cancer and that elevated FoxM1 promotes tumour progression in glioma cells, colorectal cancer, prostate carcinomas and lung adenocarcinomas (22, 26–29). Moreover, ectopic expression of FoxM1 promoted the development and growth of prostate cancer in mouse models (26). Conversely, reduced FoxM1 levels can decrease tumourogenity and lung tumour mass in mouse models (27–29). Given the overlapping roles of FoxM1 and HER2 in cell proliferation, survival, G2/M checkpoints, chromosome instability and breast cancer development, it is intriguing to suggest that HER2 may in part regulate FoxM1 expression in breast cancer transformation and thereby promote cell cycle progression and deregulation of the mitotic checkpoints. We sought to test this hypothesis by studying the relationship between FoxM1 and HER2 expression in breast cancer.
Materials and Methods
Cell culture and mice
The human breast carcinoma cell lines BT474, SKBR3, MCF-7, MDA-MB-231 and MDA-MB-453 were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 2 mM glutamine, and 100 units/ml penicillin/streptomycin, in a humidified incubator in an atmosphere of 10% or 5% CO2 at 37 °C. Lapatinib was obtained from GlaxoSmithKline, dissolved in DMSO and used at a final concentration of 1 µM. Wild-type and MMTV-c-neu mice described previously (30) were maintained at Dana-Faber Cancer Institute facilities, Harvard Medical School, Boston USA.
Plasmids and transfections
The human FoxM1 promoter construct is a gift from Professor Rene H. Medema (University Medical Center Utrecht, the Netherlands), and has previously been described (31). The human HER2 expression vector pCDNA3.1-HER2 cDNA was a generous gift from Professor William J Gullick, (the University of Kent, Canterbury, Kent, UK). For transfections, cells were seeded to a confluency of about 50% and incubated for 6 h with a master mix of transfection reagents containing Fugene-6 (Qiagen, Crawley, UK) and the plasmid DNA in a ratio of 3:1 diluted in Optimem. Fresh media was then added to replace the transfection reagent and cells were allowed to grow overnight before use.
Western Blotting
Western blotting was performed on whole cell extracts prepared by lysing cells in Nonidet P-40 lysis buffer (1% Nonidet P-40, 100 mM NaCl and 20 mM Tris-HCl, pH 7.4) or 200 mM NaCl, 50 mM Tris-HCl, pH 7.4, 2mM EDTA, 1% Triton X-100, 10% glycerol both with addition of 10 mM NaF, 1 mM sodium orthovanadate, 30 mM Na β-glycerophosphate, and protease inhibitors (“Complete” protease inhibitor mixture, as instructed by the manufacturer, Roche Applied Science, Lewes, UK) on ice for 15 min. Insoluble material was removed by centrifugation, and protein concentration was determined by Bio-Rad Dc protein assay (Bio-Rad, Hemel Hempstead, UK). 20 µg of protein was size-fractionated using SDS-PAGE, and electro-transferred onto Protran nitrocellulose membranes (Schliecher and Schuell, Dassel, Germany). Membranes were blocked in 5% BSA or 5% milk in Tris-buffered saline plus 0.5% Tween for 30 min at RT and then incubated with specific antibodies. Antibodies used were phospho-HER2 (PN2A; Biosource; Invitrogen), cdc25b (DCS-162; Abcam, Cambridge, UK), HER2 (C-18), Cyclin B (H-433), PLK (F8), tubulin (H235), FoxM1 (C-20) and actin (I-19) were obtained from Santa Cruz Biotechnology (Autogen Bioclear, Wiltshire, UK). Primary antibodies were detected using horseradish peroxidase-linked anti-mouse or anti-rabbit conjugates as appropriate (DAKO, Ely, UK), and visualized using the ECL detection system (Amersham Biosciences, Amersham UK).
Luciferase reporter assay
Cells were transfected with the human FoxM1 promoter and Renilla (pRL-TK; Promega, Southampton, UK) as internal transfection control using Fugene-6 (Qiagen, Crawley, UK). For promoter analysis, 24 h after transfection, cells were then collected, washed twice in PBS, and harvested for firefly/Renilla luciferase assays using the Dual-Glo™ Luciferase reporter assay system (Promega) according to manufacturer’s instruction. Luminescence was then read using the 9904 TOPCOUNT Perkin Elmer (Beaconsfield, UK) plate reader.
Real-time quantitative PCR (RTQ-PCR)
Frozen samples from patients, who had undergone surgery at Charing Cross Hospital (London, UK), were used for RNA extraction. All of the samples were frozen in liquid nitrogen immediately after resection and stored at −80°C until use. Total RNA (2 µg) isolated using the RNeasy Mini kit (Qiagen, Crawley, UK) was reverse transcribed using the Superscript III reverse transcriptase and random primers (Invitrogen, Paisley, UK), and the resulting first strand cDNA was used as template in the real-time PCR. All samples were performed in triplicates. The following gene-specific primer pairs were designed using the ABI Primer Express software: FoxM1-sense: 5’-TGCAGCTAGGGATGTGAATCTTC-3’ and FoxM1-antisense: 5’-GGAGCCCAGTCCATCAGAACT-3’; L19-sense 5’-GCGGAAGGGTACAGCCAAT-3’ and L19-antisense GCAGCCGGCGCAAA-3’. Specificity of each primer was determined using NCBI BLAST module. Real time PCR was performed with ABI PRISM 7700 Sequence Detection System using SYBR Green Mastermix (Applied Biosystems, Brackley, UK). FoxM1 and L19 transcript levels were quantified using the standard curve method. L19, a non-regulated ribosomal housekeeping gene was used as an internal control to normalise input cDNA.
Gene Silencing with Small Interfering RNAs (siRNAs)
For gene silencing, cells were transiently transfected with 50 nM of the following siRNA reagents purchased from Dharmacon (Lafayette, CO): HER2 siGENOME SMARTpool or siCONTROL non-targeting siRNA. BT474 or SKBR3 cells were cultured in six-well plates until 60% confluent and transfected with oligonucleotides using OligofectAMINE (Invitrogen) according to manufacturer’s instructions. Cell lysates were collected for Western blot analysis 48 h post transfection.
Immunohistochemical analysis (IHC) and Samples
The patient materials used in this study are retrospective samples collected between 1980 and 2000. The ethical approval of this study has been granted by the Riverside Research Ethics Committee, Hammersmith London-reference number: 05/Q0411/57. The mouse paraffin-embedded sections originated from mice kept at the animal facilities at Dana-Farber Cancer Institute, Harvard Medical School, Boston, USA. For immunohistochemical staining, formalin fixed paraffin-embedded samples from patients, who had undergone surgery at Charing Cross Hospital between 1981 and 2003, were retrieved from the files of the Hammersmith Hospital, London, United Kingdom. The majority of this series have already been characterized for tumour type and grade, and the main breast cancer markers, including HER2 and ER. For mouse study, mammary glands isolated from wild type or MMTV-c-neu mice were formalin fixed, paraffin wax embedded before being used for IHC staining. Immunohistochemistry was performed with a FoxM1 antibody (clone C-20, dilution 1:450, Santa Cruz) using the streptavidin-biotin-peroxidase technique. Heat-induced antigen retrieval using the Citrate Buffer (Sodium Citrate) was performed in a microwave for 14 min with 5 min prewarming. Cytospin samples were incubated with 0.2 % Triton X-100 for 10 min before performing the streptavidin-biotin-peroxidase procedure. HER2 status was determined by staining using the anti-HER-2/neu (4B5 or CB11, Santa Cruz) rabbit monoclonal antibody on the Ventana Benchmark XT according to the manufacturer’s protocol (Ventana Medical Systems, Inc, AZ, USA). Counterstaining was in haematoxylin. A semi-quantitative assessment of FoxM1 and HER2 expression was performed for all human and mouse invasive tumours, according to the following criteria: For FoxM1 assessment, − and + cases were classified as negative cases and cases with ++/+++ expression intensity were categorised as positive cases. For HER2 classification, all cases − and + were classed as negative according with pathological established criteria for HER2 amplification and only +++ HER2 expression were considered positive. HER2 FISH analyses are conducted for ++ cases.
Statistical analysis
Kaplan–Meier survival analysis and a non-parametric log-rank test (Mantel-Cox test) were used to test the survival distributions of the two sample groups and considered significant at p<0.05. Pearson’s correlation analysis was used to test the relationship between FoxM1 status and tumour grade, or FoxM1 status and HER2 expression, classified according to the above description, and was considered significant at p<0.05. To test for differences between tumour grade and FoxM1/L19 mRNA one-way-analysis of variance (ANOVA) was performed followed by Dunnett’s t-test, and the mean difference considered significant at the p<0.05 level. All statistical analysis was performed SPSS v.15.
Purification of malignant epithelial cells from primary tumours
The tissue was minced and digested at 37°C with type IA collagenase (1 mg/ml) in RPMI-1640 plus 5% foetal calf serum (FCS), 2 mM L-glutamine, 100 U/ml penicillin, 0.1 mg/ml streptomycin, 50 U/ml polymixin B and 2.5 mg/ml amphotericin B. The digestion was continued until a predominantly single-cell suspension was achieved (usually 2–5 h). Undigested material was removed using a 50 mm pore nylon mesh. Tissue remaining on the mesh was subjected to further collagenase digestion (1–3 h) and again filtered through a 50 mm pore nylon mesh. The two filtrates were refiltered with progressively smaller filter sizes (50–28 mm). The tumour epithelial cells were immunoaffinity purified using superparamagnetic, polystyrene beads (Dynal Ltd, New Ferry, Wirral, UK) coated with a mouse IgG1 monoclonal antibody (MAb Ber-EP4) specific for two (34 and 39 kDa) glycopolypeptide membrane antigens (32). Cells were then cytospun for 5 min, air dried and stored at −20 °C. This method has previously been characterised, and tumour cells isolated using this protocol commonly exhibit a high degree of purity (>95%) (33, 34). As with the immunohistochemical stained samples, HER2 status of the purified epithelial cells from primary tumours were determined by fluorescent in situ hybridisation (FISH). Validation of the purified tumour cells was performed by staining with hematoxylin/eosin (H&E) or cytokeratins 8 and 18 (MAb CAM 5.2, Becton Dickinson, Oxford, UK) (32).
Cell Cycle Analysis
Cell cycle analysis was performed by propidium iodide (PI) staining. Subconfluent cells with or without drug treatment were trypsinized, collected by centrifugation, and resuspended in PBS, before fixing in 90% ethanol. The fixed cells were incubated with 5 µg/ml propidium iodide, 0.1 mg/ml RNase A, 0.1% Nonidet P-40, and 0.1% trisodium citrate for 30 min prior to analysis using a Becton Dickinson FACScanto analyzer. The cell cycle profile was analyzed using Cell Diva software (Becton Dickinson UK Ltd).
Results
HER2 Correlates with FoxM1 Expression in Breast Cancer Cell Lines
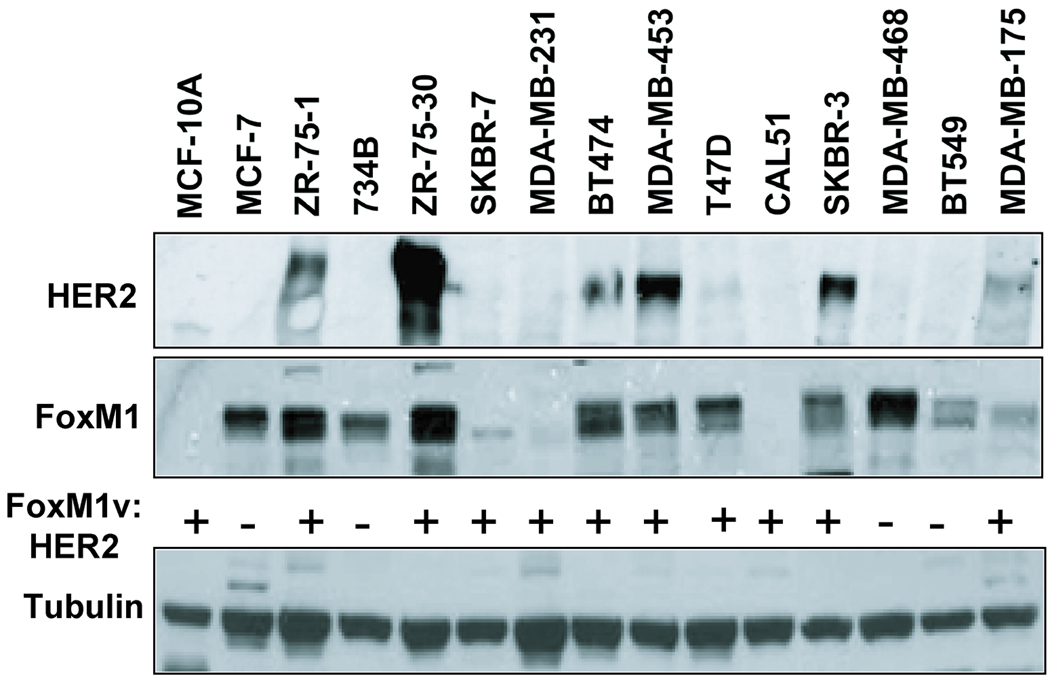
To examine whether FoxM1 expression correlates with HER2 expression levels, we analyzed HER2 and FoxM1 expression in a panel of 15 different breast cell lines by Western blotting. Consistent with our hypothesis that HER2 may regulate FoxM1 expression, 7 breast carcinoma cell lines expressing high levels of HER2 were also positive for FoxM1 expression (Fig. 1), whilst 4 cell lines that had relatively low HER2 also displayed low levels of FoxM1 expression. Therefore, 11 of the 15 breast cancer cell lines examined demonstrated a positive correlation between the levels of HER2 and FoxM1 (Fig. 1). Four of the cell lines do not possess a correlation, where it is possible that other mechanisms may influence their FoxM1 levels, therefore negating the effect of HER2. For example, EGFR or other growth factor receptors may play a more predominant role in regulating FoxM1 in these cell lines. Moreover, these cell lines may have been affected by continuous culturing, which may in turn affect their cell cycle and subsequently FoxM1 levels. Yet, importantly FoxM1 is expressed at high levels in all cell lines where HER2 is over-expressed, suggesting HER2 overexpression drives FoxM1 expression in breast cancer cells.
Figure 1. HER2 correlates with FoxM1 protein expression in breast carcinoma cell lines.
Protein lysates were prepared from fifteen different cancer cell lines, as indicated, and 20 µg was analyzed by Western blotting using specific antibodies against HER2 (top panel is a low exposure and bottom panel is a short exposure), FoxM1 and tubulin. + indicates positive correlation, and - denotes no correlation for each cell line.
FoxM1 expression levels are elevated in HER2 positive breast cancer patient tumour samples
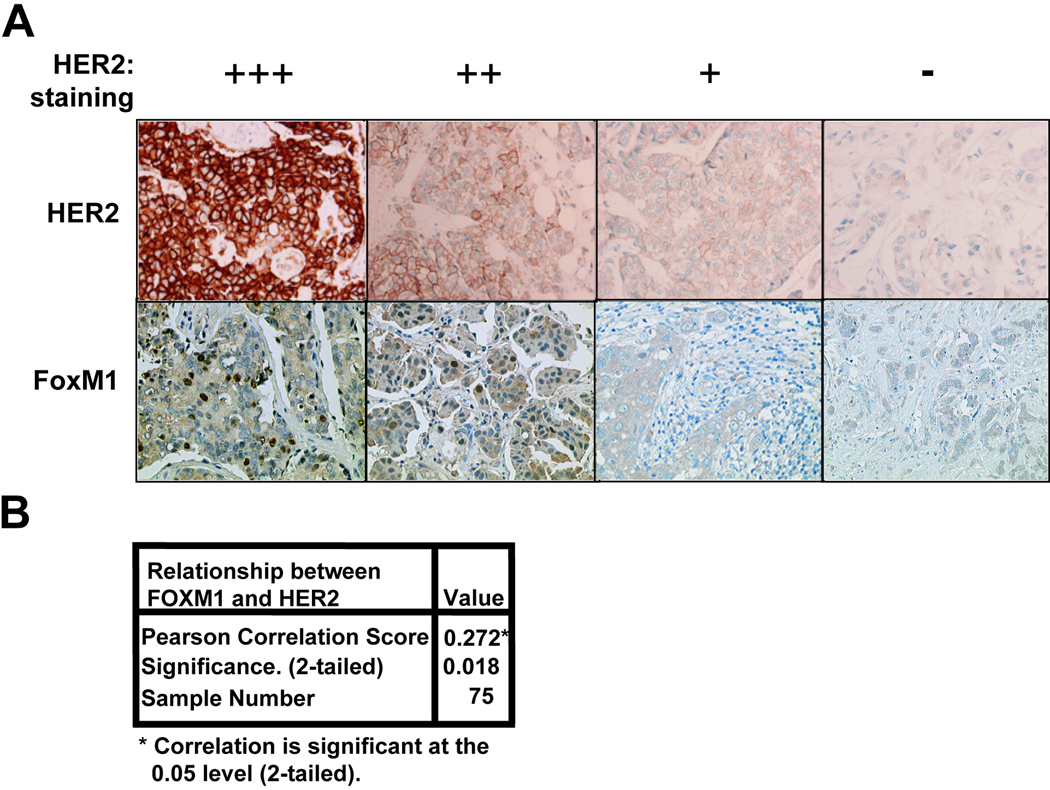
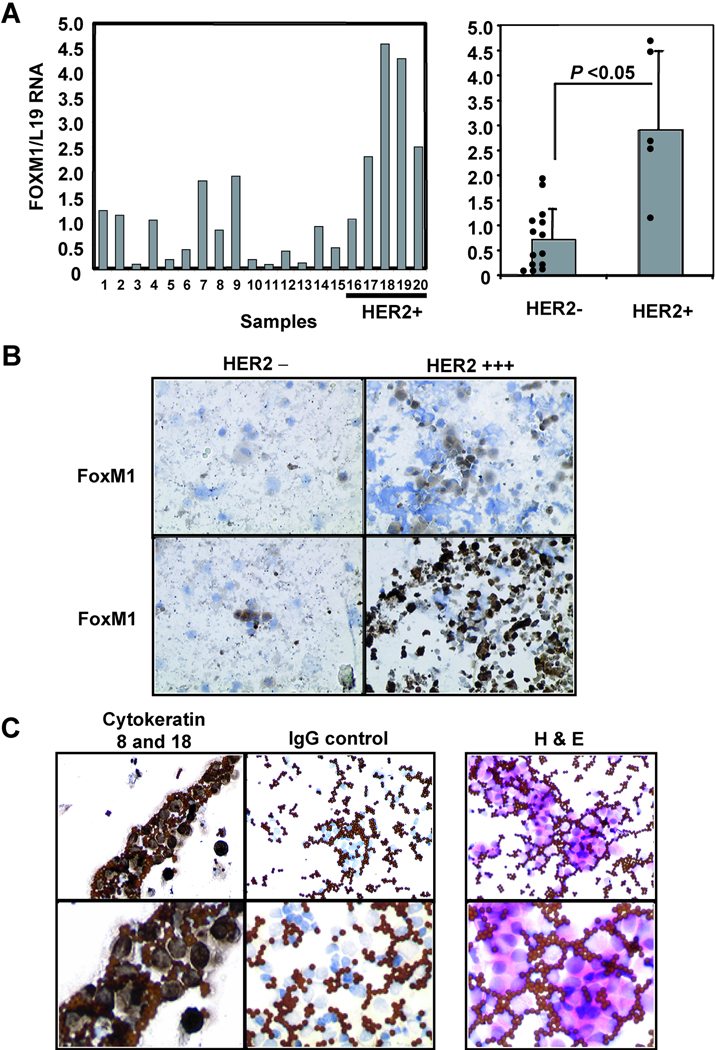
To confirm that the correlation between FoxM1 and HER2 expression is also valid in tumour tissue, a series of 112 paraffin-embedded breast cancer patient samples were immunohistochemically stained for HER2 and FoxM1 (Table 1). As controls HER2 and FoxM1 antibodies were used to perform immunohistochemical staining on a HER2 positive cell line, BT474, and a HER2 negative cell line MDA-MB-231, which revealed similar expression to that detected by Western blotting (data not shown). As expected, in the breast cancer patient samples, HER2 staining is concentrated around the plasma membrane of the cells (Fig. 2A), whilst FoxM1 was located predominantly in the nucleus. Notably, significant FoxM1 expression could only be detected in the epithelial but not the stromal cells of the breast tissue, further indicating that the FoxM1 staining is specific. Conversely, breast tissue samples devoid of HER2 staining also lacked FoxM1 expression. Statistical analysis showed that there is a significant and positive association between FoxM1 and HER2 expression (Pearson correlation=0.272, two-tailed p<0.005), but that no significant association between FoxM1 expression and tumour grade, or patient survival could be found (Fig. 2B and data not shown). To investigate whether HER2 overexpression was associated with increased levels of FoxM1 expression at transcript level, we examined FoxM1 mRNA levels by RTQ-PCR in epithelial cells purified from breast cancer tumour biopsies. Consistent with our previous data, RTQ-PCR results indicated that when patient samples were categorised by their HER2 status, it was found that the HER2 positive samples had significantly higher levels of FoxM1 transcripts (p<0.05) (Fig. 3A). The protein expression levels of HER2 and FoxM1 in these purified cells was further confirmed by immunohistochemical staining following cytospin (Fig 3B).
Table 1.
Information on patient age, nodal status, pathological diagnosis, tumour size, differentiation grade recorded, and ER, HER2 and FoxM1 status
| FOXMI negative | FOXMI positive | ||||
|---|---|---|---|---|---|
| n | (%) | n | (%) | ||
| Grade | 0 | 6 | 5.5 | 3 | 2.8 |
| I | 7 | 6.4 | 3 | 2.8 | |
| II | 37 | 33.9 | 21 | 19.3 | |
| III | 19 | 17.4 | 13 | 11.9 | |
| Size | 3.1 (2.5)a | 2.4 (13.6)b | 2.6 (2.0)a | 1.3 (4.8)b | |
| Nodal Status | No | 2.2 | 25.6 | 9 | 10.5 |
| Yes | 34 | 39.5 | 21 | 24.4 | |
| Estrogen Receptor | Negative | 40 | 53.3 | 20 | 26.7 |
| positive | 5 | 6.7 | 10 | 13.3 | |
| Her2 status | Negative | 40 | 53.3 | 20 | 26.7 |
| Positive | 5 | 6.7 | 10 | 13.3 | |
| Age | 57.1 (58.0)a | 11.7 (54.0)b | 58.5 (59.5)a | 13.8 (63.0)b | |
Mean (Median).
SD. (range).
Figure 2. HER2 protein expression correlates with FoxM1 expression in breast cancer patient samples.
(A) Tumour tissue samples obtained from breast cancer patients that had been formalin fixed paraffin-embedded were immunohistochemically stained with HER2 and FoxM1 antibodies using the streptavidin-biotin-peroxidase technique. Examples of four paired samples stained with antibodies against HER2 (left column) and FoxM1 (right column) are shown (X200 magnification). (B) FoxM1 status and HER2 expression was determined by immunohistochemistry as in (A) and categorised as described in material and methods. 112 samples were analysed of which 75 were FoxM1 and HER2 positive, 109 were FoxM1 positive, and 78 HER2 positive. The correlation between FoxM1 status and HER2 expression levels was analysed using a 2-tailed Pearson’s correlation test, and was considered significant at p<0.05.
Figure 3. HER2 expression correlates with FoxM1 expression in cells isolated from breast cancer patients.
(A) RNA was isolated from epithelial cells purified from primary tumours and subjected to RTQ-PCR with FoxM1 and L19 primers. The left-hand graph shows the FoxM1/L19 ratio of the 20 tumour samples examined, with the cases positive for HER2 (as determined by FISH) indicated on the right. The right-hand graph illustrates the FoxM1/L19 ratio of the samples after categorising into either HER2 negative or positive. The correlation shown to be significant using the students t-test; errors bars indicated standard deviation, p<0.05. (B) The purified epithelial tumour cells were cytospun and subsequently stained for HER2 and FoxM1 as described above. Examples of staining are shown (X400 magnification). (C) Isolated epithelial tumour cells were validated for purity by staining with hematoxylin/eosin (H&E) and for cytokeratins 8 and 18. Upper panel shows 20X and lower panel shows 40X magnification.
Altering the HER2 Expression modulates FoxM1 expression in breast cancer cell lines
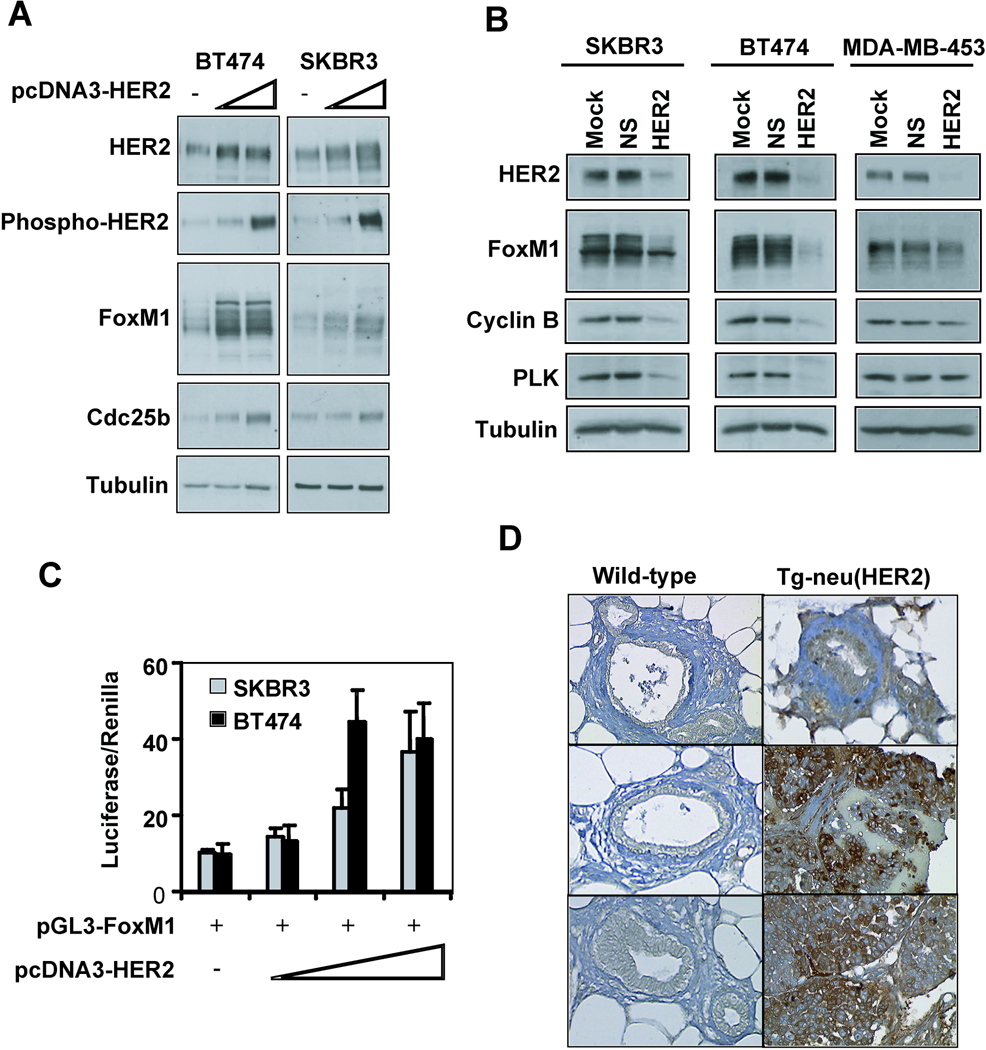
To determine whether HER2 regulates FoxM1 expression, SKBR3, BT474 and MDA-MB- 453 cells were transfected with HER2 expression vectors. Western blotting revealed that the overexpressed HER2 was active at the plasma membrane, as revealed by increased phosphorylated HER2 (Fig 4A). In SKBR3 and BT474 the higher HER2 levels and resultant increase in phosphorylated HER2 coincided with higher FoxM1 levels. Additionally, the FoxM1 transcriptional target cdc25b increased in protein levels, suggesting that there is a functional significance to the HER2-mediated increase in FoxM1.
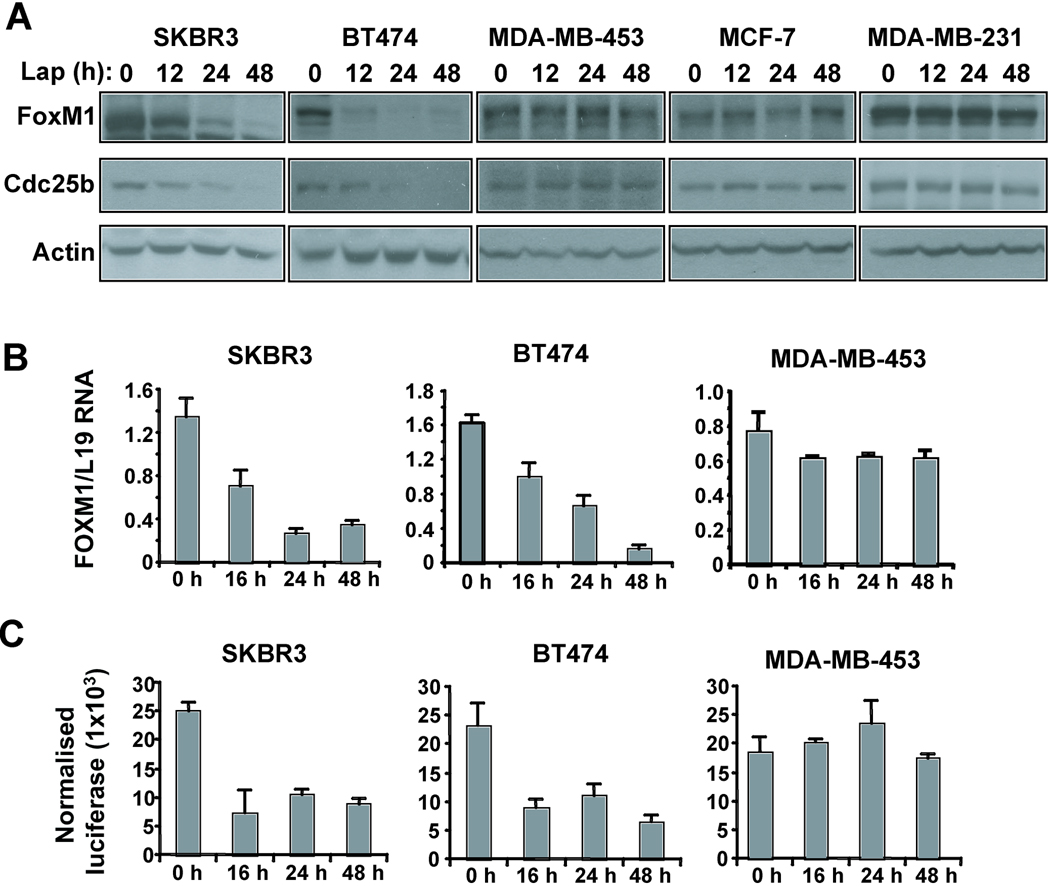
Figure 4. Altering the HER2 expression levels modulates FoxM1 expression breast cancer cells.
(A) SKBR3 or BT474 cells were transfected with either 3 µg or 5 µg of pCDNA3-HER2 or empty vector for 48 hours before protein lysate was prepared. Lysate (20 µg) was subjected to Western blotting with the indicated antibodies. (B) SKBR3, BT474 or MDA-MB-453 cells were transfected with mock (transfection reagent alone), 50 nM non-specific siRNA or 50 nM HER2 siRNA for 48 h. Subsequently cells were lysed and proteins (20 µg) analysed by Western blotting with the specified antibodies. (C) SKBR3 or BT474 cells were transfected with 20 ng of the FoxM1 promoter and increasing amounts (0.1, 0.2, 0.25 ng) of the HER2 expression plasmid for 24 hours before cells were collected for luciferase reporter assays. The relative luciferase activity values are corrected for co-transfected renilla activity. All data shown represent the average of three independent experiments, and the error bars show the standard deviation (mean ± SD). (D) Mouse epithelial tissue from either wild-type mammary glands or mammary tissue-overexpressing HER2 was isolated and stained for FoxM1. A total of at least six controls and six MMTV-neu samples were examined and staining from three controls and MMTV-neu samples were shown (X200 magnification).
To further validate FoxM1 as a downstream target of HER2 the effect of HER2 knockdown on FoxM1 expression was determined in SKBR3, BT474, MDA-MB-453 and MCF7 cells. Ablation of HER2 protein in SKBR3, BT474 and MDA-MB-453 resulted in reduced FoxM1 expression and concomitant decrease in target gene expression (Fig. 4B). Furthermore, increasing levels of the HER2 expression activated a FoxM1 promoter/reporter construct in SKBR3 and BT474 cells (Fig. 4C), suggesting that HER2 regulates FoxM1 expression at the promoter level. To assess the effect of increased HER2 levels on FoxM1 expression during tumourigenesis, mouse mammary glands were isolated from either wild-type mice (n=6) or mouse mammary tumour virus (MMTV)-neu transgenic mice (n=6) and stained for FoxM1 expression. Epithelial cells from wild-type mammary glands had low FoxM1 expression, while a large proportion of the cells from mice expressing mammary epithelium-targeted HER2 had high levels of FoxM1 (Fig. 4D).
The HER2 inhibitor lapatinib reduces FoxM1 expression in sensitive cell lines
To further confirm that FoxM1 is a downstream signalling target of HER2 in breast cancer, we next studied the effects of the anti-HER2 tyrosine kinase inhibitor (TKI) on FoxM1 expression in sensitive and resistant breast carcinoma cell lines. To this end, the lapatinib sensitive cells lines SKBR3 and BT474, as well as the resistant MDA-MB-453, MCF-7 and MDA-MB-231 cells were treated with 1 µM lapatinib for the indicated time points before protein lysate was subjected to Western blotting. Treatment with lapatinib led to a decrease in FoxM1 protein levels by 24 h in SKBR3 and by 12 h in the more sensitive BT474 (Fig. 5A). This coincided with a decrease in cdc25b, a FoxM1 target gene. However, FoxM1 and cdc25b levels did not change with lapatinib treatment of the resistant cell lines. Furthermore, RTQ-PCR analysis demonstrated that lapatinib reduced FoxM1 transcripts in the sensitive, but not in the resistant cell lines (Fig 5B). Transient transfection of the sensitive cell lines with the FoxM1 promoter reporter construct showed a decrease in promoter activity when treated with lapatinib, whilst there was no change in promoter activity in the MDA-MB-453 cells (Fig. 5C). Cell cycle assays showed that significant lapatinib-induced cell cycle arrest only occurs after 48 h, whilst FoxM1 protein levels decrease by 12 h lapatinib treatment, suggesting the changes in FoxM1 are not as a result of changes in the cell cycle (data not shown). Nevertheless, subsequent down-regulation of FoxM1 expression could be due to the accumulation of cells at G1 after prolonged lapatinib treatment, as FoxM1 expression has been shown to be induced during the cell cycle at S, G2/M phases (15).
Figure 5. Treatment of sensitive cells with lapatinib results in reduced FoxM1 expression levels.
(A) SKBR3, BT474, MDA-MB-453, MCF-7 or MDA-MB-231 cells were treated with 1µM lapatinib for the indicated time points before protein lysate was prepared. Lysate (20 µg) was subjected to Western blotting with the indicated antibodies. (B) SKBR3, BT474 or MDA-MB-453 cells were treated with 1µM lapatinib for the indicated time points before cells were collected and total RNA isolated. FoxM1 mRNA levels were analysed by RTQ-PCR normalised with L19 mRNA levels. (C) SKBR3, BT474 or MDA-MB-453 cells were transfected with 10 ng of the FoxM1 promoter for 24 h, treated with 1µM lapatinib for the indicated time points before cells were collected for luciferase reporter assays. The relative luciferase activity values are corrected for co-transfected renilla activity. All data shown represent the average of three independent experiments, and the error bars show the standard deviation (mean ± SD).
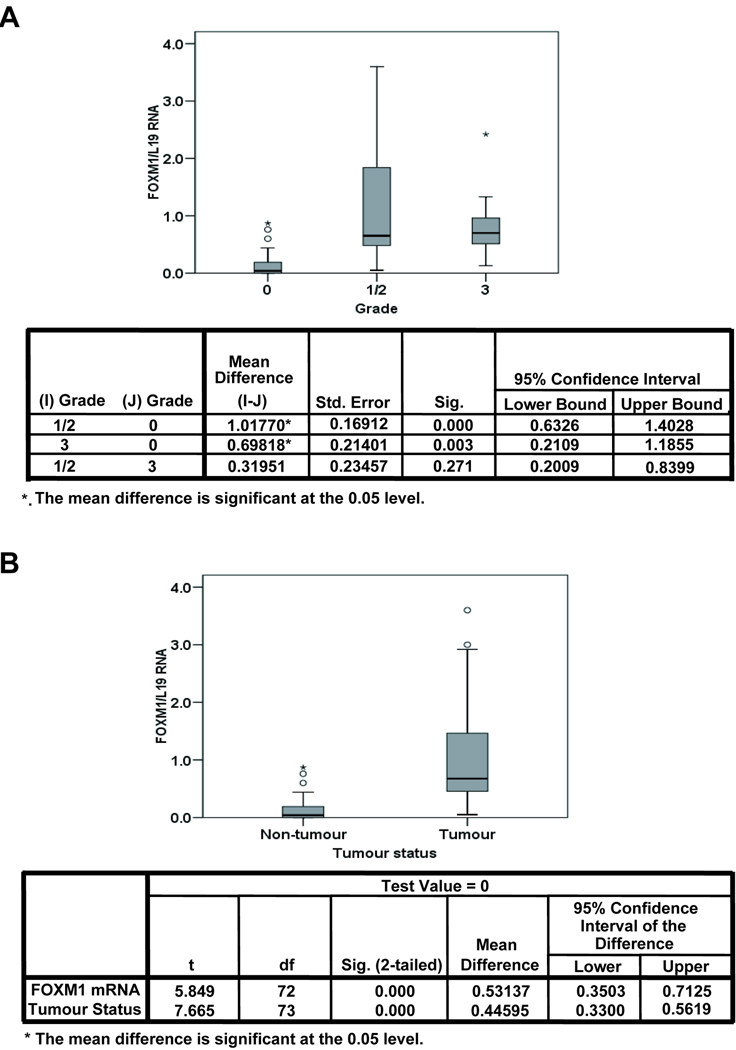
FoxM1 expression correlates with tumourigenesis
In order to investigate the relevance of FoxM1 expression in breast cancer development and progression, the relationship between FoxM1 mRNA level and tumour grade was examined. To this end, the FoxM1 mRNA was examined in 75 individual non-matched patient samples, of which 41 were non-tumour tissue, 21 samples were from grade 1/2 classified tumours, and 12 from grade 3 classified tumours. Analysis of variance (ANOVA) showed there were no significant variations in FoxM1 mRNA level between grade 1/2 and 3 patients (p=0.271), but very significant differences were found between non-tumour versus grade 1/2 and grade 3, (p<0.01 and <0.01, respectively) (Fig 6A). Further analysis revealed that differences in FoxM1 mRNA level between non-tumour and tumour tissues were also significant (p<0.001) (Fig 6B), indicating that overexpression of FoxM1 has a role in breast cancer tumourigenesis.
Figure 6. Relationship between FoxM1 mRNA and tumour grade in breast cancer.
The relationship between FoxM1 mRNA and tumour grade was examined. FoxM1 was quantified using RTQ-PCR and normalized using the housekeeping gene L19. 75 individual non-matched patient samples were analysed, of which 41 were non-tumour tissue, 21 samples were from grade 2 classified tumours, and 12 from grade 3 classified tumours. FoxM1/L19 mRNA for grades (A) and for non-tumour versus tumour tissue (B) are shown using Box-and-Whisker plots illustrating the median values, first and third quartile, and smallest and largest values of FoxM1/L19 mRNA at each grade. To test for differences between tumour grade and FoxM1/L19 mRNA one-way-analysis of variance (ANOVA) was performed followed by Dunnett’s t-test, and the mean difference considered significant at the p<0.05 level (A).
Discussion
In this study we have demonstrated a strong association between HER2 and FoxM1 expression in breast cancer, and that the manipulation of HER2 levels resulted in parallel changes in FoxM1 expression. The expression levels of FoxM1 were consistently associated with HER2 in both breast carcinoma cell lines and paraffin embedded breast cancer patient samples. Moreover, using purified breast epithelial tumour cells, we also established that overexpression of HER2 correlated with high levels of FoxM1 mRNA expression, suggesting that the HER2-associated up-regulation of FoxM1 expression is at least partially mediated at the mRNA level. Additionally, overexpression or siRNA-mediated depletion of HER2 in breast cancer cell lines resulted in parallel changes in FoxM1 protein levels, indicating that FoxM1 expression levels were dependent upon HER2 expression levels. This result is further confirmed in vivo using a transgenic mouse model, where mammary epithelium-targeted HER2 expression led to FoxM1 overexpression. Furthermore, treatment of HER2 overexpessing breast cancer cells with the HER2-tyrosine kinase inhibitor lapatinib also produced a reduction in FoxM1 expression at the protein, mRNA level and a decrease in gene promoter activity. Consistent with this, titration of HER2 increased FoxM1 reporter activity, suggesting that the up-regulation of FoxM1 expression is at least partially mediated at the mRNA level through regulation of the FoxM1 promoter. Moreover, lapatinib-induced cell cycle arrest only after FoxM1 protein levels decreased, suggesting the changes in FoxM1 are not predominantly a result of the cell cycle. In addition, although there was no difference in FoxM1 mRNA levels between tumour grades, there was a significant difference between non-tumour and tumour tissue, suggesting that FoxM1 mRNA level is an important marker for breast cancer development.
Elevated HER2 activity is thought to be a primary transformation event during cancer development, and overexpression of the HER2 receptor and amplification of the gene occurs in up to 25% of breast cancer patients. This is associated with a more aggressive disease outcome and HER2 positivity has become one of the major prognostic indicators in early breast cancer (1, 3–5). Consequently, HER2 has become a target for therapeutic intervention and drugs such as Herceptin, a monoclonal antibody which targets the extracellular domain of the HER2 receptor, play a key role in the management of early stage disease. One year of adjuvant treatment with herceptin has been shown in the HERA trial to have an overall survival benefit after adjuvant chemotherapy, reducing the risk of recurrence and preventing the development of metastatic disease (35, 36). This makes HER2 and its downstream targets, such as FoxM1, attractive therapeutic targets for the prevention and treatment of breast cancer. However, to date the efficacy of the HER2-directed therapies have been limited by the development of de novo and acquired resistance, the mechanism of which is currently poorly understood (11, 37–40). Thus, despite the well-established role of HER2 in breast cancer development, a complete understanding of the downstream effectors of HER2 inhibition remain elusive. In this study, our data suggest for the first time that FoxM1 is a primary cellular target of HER2-dependant signalling in breast cancer cells, and that HER2 signals activate FoxM1 expression at the protein, mRNA and gene promoter levels. Several studies have implicated FoxM1 in tumourogenesis and as a marker for poor prognosis (22, 26–29, 41) for example, ectopic expression of FoxM1 induces the formation and growth of prostate cancer mice (26). We have recent unpublished data showing very significant differences in FoxM1 between non-tumour versus grade 1/2 and grade 3, but no significant variations in FoxM1 level between grade 1/2 and 3 patients; indicating that overexpression of FoxM1 has a role in breast cancer tumourogenesis. However, the mechanisms involved FoxM1 deregulation and its role in cancer formation remain largely unknown. Therefore, elucidating these factors will be beneficial in identifying better biomarkers and potential drug targets.
Moreover, we have established that FoxM1 is a target of lapatinib, and may therefore play an important role in the mode of action of this therapeutic drug. Given the critical role of FoxM1 as a regulator of cell cycle progression, mitotic defects and the G2/M cell cycle checkpoint, it is possible that a reduction in FoxM1 expression is a critical event in lapatinib-induced toxicity in breast cancer cells. In support of this, we found that FoxM1 decreased prior to cell cycle arrest in lapatinib-treated SKBR3 cells. Indeed, previous studies have supported the direct targeting of FoxM1 as an effective anti-cancer strategy. For example, a recent cell based screen has identified the antibiotic, Siomycin A, as an inhibitor of FoxM1 (42). Siomycin A was able to reduce transforming ability and induce apoptosis in transformed but not normal cells, through down-regulating FoxM1 expression and its transcriptional activity (42). Another anti-FoxM1 therapeutic strategy that originated from findings in cancer cell lines was that the transcription activity of FoxM1 can be inhibited by the tumour suppressor p14ARF (p19ARF in mice), through a region mapped to residues 26–46 of the protein (43). In addition, administration of a cell-penetrating ARF26–44 peptide to mice diminished FoxM1 function in vivo, causing selective apoptosis and reduced proliferation and angiogenesis in hepatocellular carcinomas (HCC) (44). Our data point to the intriguing possibility that the concomitant targeting of FoxM1 and HER2 may provide additional levels of cell death, delay or circumvent the development of resistance, whilst retaining tumour-selectivity.
The mechanism by which HER2 regulates FoxM1 expression levels may also be critical to understanding the mechanism of acquired resistance to HER2-targeted therapies. Previous studies have shown that HER2 and its co-receptor, EGFR signal through the PI3-K-Akt pathway to repress FoxO3a (45–47), a transcription factor involved in lapatinib-mediated cell cycle arrest and apoptosis (36, 45). Recent work has also implicated FoxO3a in the regulation of FoxM1 (48), and thus the PI3-K-Akt-FoxO3a signalling axis may provide the link between HER2 and FoxM1 expression. Work is currently underway in our lab to further delineate the signalling events connecting HER2 with FoxM1. Our results have established a strong link between the expression of HER2 and FoxM1 in breast cancer. Our data also indicates that FoxM1 is a crucial downstream signalling target of HER2, and that HER2 signalling regulates FoxM1 expression at protein, mRNA and promoter levels in breast cancer, thus explaining the overlapping functions of HER2 and FoxM1. Our results suggest that FoxM1 is an important diagnostic marker for breast cancer development, which may play a critical role in tumourigenesis and is therefore a potential cellular target for therapeutic intervention, especially in HER2-targeted therapy resistant cancers.
Acknowledgements
Grant support: Cancer Research UK (R. E. Francis, S. S. Myatt, R. C. Coombes, E.W-F. Lam, A. Filipovic, C. Palmieri), Association for International Cancer Research (R. E Francis, E. W.-F. Lam) Breast Cancer Research Trust (J. Krol, E. W.-F. Lam), Biotechnology and Biological Sciences Research Council (B. Peck, E. W.-F. Lam)
Abbreviations
- HER2
Human epidermal growth factor receptor-2
- EGFR
growth factor receptor
- RTK
receptor tyrosine kinases
- PI3-K
phosphatidylinositol 3-kinase
- PKB
protein kinase B
- PLK1
polo like kinase-1
- CENPA
centromere protein A
- IHC
immunohistochemical
- FCS
foetal calf serum
- FISH
fluorescent in situ hybridisation
- H&E
hematoxylin/eosin
- PI
propidium iodide
- MMTV
mammary tumour virus
References
- 1.Zhang H, Berezov A, Wang Q, Zhang G, Drebin J, Murali R, Greene MI. ErbB receptors: from oncogenes to targeted cancer therapies. J Clin Invest. 2007;117:2051–2058. doi: 10.1172/JCI32278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 3.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 4.van de Vijver M, van de Bersselaar R, Devilee P, Cornelisse C, Peterse J, Nusse R. Amplification of the neu (c-erbB-2) oncogene in human mammmary tumors is relatively frequent and is often accompanied by amplification of the linked c-erbA oncogene. Mol Cell Biol. 1987;7:2019–2023. doi: 10.1128/mcb.7.5.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varley JM, Swallow JE, Brammar WJ, Whittaker JL, Walker RA. Alterations to either c-erbB-2(neu) or c-myc proto-oncogenes in breast carcinomas correlate with poor short-term prognosis. Oncogene. 1987;1:423–430. [PubMed] [Google Scholar]
- 6.Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE, Jr, Davidson NE, Tan-Chiu E, Martino S, Paik S, Kaufman PA, Swain SM, Pisansky TM, Fehrenbacher L, Kutteh LA, Vogel VG, Visscher DW, Yothers G, Jenkins RB, Brown AM, Dakhil SR, Mamounas EP, Lingle WL, Klein PM, Ingle JN, Wolmark N. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 7.Ciardiello F, Caputo R, Bianco R, Damiano V, Fontanini G, Cuccato S, De Placido S, Bianco AR, Tortora G. Inhibition of growth factor production and angiogenesis in human cancer cells by ZD1839 (Iressa), a selective epidermal growth factor receptor tyrosine kinase inhibitor. Clin Cancer Res. 2001;7:1459–1465. [PubMed] [Google Scholar]
- 8.Wakeling AE, Guy SP, Woodburn JR, Ashton SE, Curry BJ, Barker AJ, Gibson KH. ZD1839 (Iressa): an orally active inhibitor of epidermal growth factor signaling with potential for cancer therapy. Cancer Res. 2002;62:5749–5754. [PubMed] [Google Scholar]
- 9.Nelson MH, Dolder CR. Lapatinib: a novel dual tyrosine kinase inhibitor with activity in solid tumors. Ann Pharmacother. 2006;40:261–269. doi: 10.1345/aph.1G387. [DOI] [PubMed] [Google Scholar]
- 10.Ciardiello F. Epidermal growth factor receptor inhibitors in cancer treatment. Future Oncol. 2005;1:221–234. doi: 10.1517/14796694.1.2.221. [DOI] [PubMed] [Google Scholar]
- 11.Montemurro F, Valabrega G, Aglietta M. Lapatinib: a dual inhibitor of EGFR and HER2 tyrosine kinase activity. Expert Opin Biol Ther. 2007;7:257–268. doi: 10.1517/14712598.7.2.257. [DOI] [PubMed] [Google Scholar]
- 12.Asanuma H, Torigoe T, Kamiguchi K, Hirohashi Y, Ohmura T, Hirata K, Sato M, Sato N. Survivin expression is regulated by coexpression of human epidermal growth factor receptor 2 and epidermal growth factor receptor via phosphatidylinositol 3-kinase/AKT signaling pathway in breast cancer cells. Cancer Res. 2005;65:11018–11025. doi: 10.1158/0008-5472.CAN-05-0491. [DOI] [PubMed] [Google Scholar]
- 13.Jeruss JS, Liu NX, Chung Y, Magrane G, Waldman F, Edgerton S, Yang X, Thor AD. Characterization and chromosomal instability of novel derived cell lines from a wt-erbB-2 transgenic mouse model. Carcinogenesis. 2003;24:659–664. doi: 10.1093/carcin/bgg001. [DOI] [PubMed] [Google Scholar]
- 14.Laoukili J, Stahl M, Medema RH. FoxM1: at the crossroads of ageing and cancer. Biochim Biophys Acta. 2007;1775:92–102. doi: 10.1016/j.bbcan.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 16.Korver W, Roose J, Clevers H. The winged-helix transcription factor Trident is expressed in cycling cells. Nucleic Acids Res. 1997;25:1715–1719. doi: 10.1093/nar/25.9.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korver W, Schilham MW, Moerer P, van den Hoff MJ, Dam K, Lamers WH, Medema RH, Clevers H. Uncoupling of S phase and mitosis in cardiomyocytes and hepatocytes lacking the winged-helix transcription factor Trident. Curr Biol. 1998;8:1327–1330. doi: 10.1016/s0960-9822(07)00563-5. [DOI] [PubMed] [Google Scholar]
- 18.Ye H, Kelly TF, Samadani U, Lim L, Rubio S, Overdier DG, Roebuck KA, Costa RH. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol Cell Biol. 1997;17:1626–1641. doi: 10.1128/mcb.17.3.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye H, Holterman AX, Yoo KW, Franks RR, Costa RH. Premature expression of the winged helix transcription factor HFH-11B in regenerating mouse liver accelerates hepatocyte entry into S phase. Mol Cell Biol. 1999;19:8570–8580. doi: 10.1128/mcb.19.12.8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Hung NJ, Costa RH. Earlier expression of the transcription factor HFH-11B diminishes induction of p21(CIP1/WAF1) levels and accelerates mouse hepatocyte entry into S-phase following carbon tetrachloride liver injury. Hepatology. 2001;33:1404–1414. doi: 10.1053/jhep.2001.24666. [DOI] [PubMed] [Google Scholar]
- 21.Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, Medema RH. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 22.Wonsey DR, Follettie MT. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res. 2005;65:5181–5189. doi: 10.1158/0008-5472.CAN-04-4059. [DOI] [PubMed] [Google Scholar]
- 23.Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, Tan Y, Ackerson T, Costa RH. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25:10875–10894. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luscher-Firzlaff JM, Lilischkis R, Luscher B. Regulation of the transcription factor FOXM1c by Cyclin E/CDK2. FEBS Lett. 2006;580:1716–1722. doi: 10.1016/j.febslet.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 25.Major ML, Lepe R, Costa RH. Forkhead box M1B transcriptional activity requires binding of Cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol Cell Biol. 2004;24:2649–2661. doi: 10.1128/MCB.24.7.2649-2661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalin TV, Wang IC, Ackerson TJ, Major ML, Detrisac CJ, Kalinichenko VV, Lyubimov A, Costa RH. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 2006;66:1712–1720. doi: 10.1158/0008-5472.CAN-05-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim IM, Ackerson T, Ramakrishna S, Tretiakova M, Wang IC, Kalin TV, Major ML, Gusarova GA, Yoder HM, Costa RH, Kalinichenko VV. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 2006;66:2153–2161. doi: 10.1158/0008-5472.CAN-05-3003. [DOI] [PubMed] [Google Scholar]
- 28.Liu M, Dai B, Kang SH, Ban K, Huang FJ, Lang FF, Aldape KD, Xie TX, Pelloski CE, Xie K, Sawaya R, Huang S. FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 2006;66:3593–3602. doi: 10.1158/0008-5472.CAN-05-2912. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida Y, Wang IC, Yoder HM, Davidson NO, Costa RH. The forkhead box M1 transcription factor contributes to the development and growth of mouse colorectal cancer. Gastroenterology. 2007;132:1420–1431. doi: 10.1053/j.gastro.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 30.Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411:1017–1021. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- 31.Korver W, Roose J, Heinen K, Weghuis DO, de Bruijn D, van Kessel AG, Clevers H. The human TRIDENT/HFH-11/FKHL16 gene: structure, localization, and promoter characterization. Genomics. 1997;46:435–442. doi: 10.1006/geno.1997.5065. [DOI] [PubMed] [Google Scholar]
- 32.Kothari MS, Ali S, Buluwela L, Livni N, Shousha S, Sinnett HD, Vashisht R, Thorpe P, Van Noorden S, Coombes RC, Slade MJ. Purified malignant mammary epithelial cells maintain hormone responsiveness in culture. Br J Cancer. 2003;88:1071–1076. doi: 10.1038/sj.bjc.6600866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diaz-Arias AA, Loy TS, Bickel JT, Chapman RK. Utility of BER-EP4 in the diagnosis of adenocarcinoma in effusions: an immunocytochemical study of 232 cases. Diagn Cytopathol. 1993;9:516–521. doi: 10.1002/dc.2840090509. [DOI] [PubMed] [Google Scholar]
- 34.Fellowes VS, Husebekk A, Gress RE, Vance BA. Minimal residual disease detection in breast cancer: improved sensitivity using cytokeratin 19 and epidermal growth factor receptor RT-PCR. Int J Oncol. 2004;24:861–867. [PubMed] [Google Scholar]
- 35.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, Smith I, Gianni L, Baselga J, Bell R, Jackisch C, Cameron D, Dowsett M, Barrios CH, Steger G, Huang CS, Andersson M, Inbar M, Lichinitser M, Lang I, Nitz U, Iwata H, Thomssen C, Lohrisch C, Suter TM, Ruschoff J, Suto T, Greatorex V, Ward C, Straehle C, McFadden E, Dolci MS, Gelber RD. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353:1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 36.Hegde PS, Rusnak D, Bertiaux M, Alligood K, Strum J, Gagnon R, Gilmer TM. Delineation of molecular mechanisms of sensitivity to lapatinib in breast cancer cell lines using global gene expression profiles. Mol Cancer Ther. 2007;6:1629–1640. doi: 10.1158/1535-7163.MCT-05-0399. [DOI] [PubMed] [Google Scholar]
- 37.Johnston SR, Head J, Pancholi S, Detre S, Martin LA, Smith IE, Dowsett M. Integration of signal transduction inhibitors with endocrine therapy: an approach to overcoming hormone resistance in breast cancer. Clin Cancer Res. 2003;9:524S–532S. [PubMed] [Google Scholar]
- 38.Ranson M, Hammond LA, Ferry D, Kris M, Tullo A, Murray PI, Miller V, Averbuch S, Ochs J, Morris C, Feyereislova A, Swaisland H, Rowinsky EK. ZD1839, a selective oral epidermal growth factor receptor-tyrosine kinase inhibitor, is well tolerated and active in patients with solid, malignant tumors: results of a phase I trial. J Clin Oncol. 2002;20:2240–2250. doi: 10.1200/JCO.2002.10.112. [DOI] [PubMed] [Google Scholar]
- 39.Metro G, Finocchiaro G, Cappuzzo F. Anti-cancer therapy with EGFR inhibitors: factors of prognostic and predictive significance. Ann Oncol. 2006;17 Suppl 2:ii42–ii45. doi: 10.1093/annonc/mdj920. [DOI] [PubMed] [Google Scholar]
- 40.Cappuzzo F, Finocchiaro G, Metro G, Bartolini S, Magrini E, Cancellieri A, Trisolini R, Castaldini L, Tallini G, Crino L. Clinical experience with gefitinib: an update. Crit Rev Oncol Hematol. 2006;58:31–45. doi: 10.1016/j.critrevonc.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 41.Martin KJ, Patrick DR, Bissell MJ, Fournier MV. Prognostic breast cancer signature identified from 3D culture model accurately predicts clinical outcome across independent datasets. PLoS ONE. 2008;3:e2994. doi: 10.1371/journal.pone.0002994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radhakrishnan SK, Bhat UG, Hughes DE, Wang IC, Costa RH, Gartel AL. Identification of a chemical inhibitor of the oncogenic transcription factor forkhead box M1. Cancer Res. 2006;66:9731–9735. doi: 10.1158/0008-5472.CAN-06-1576. [DOI] [PubMed] [Google Scholar]
- 43.Kalinichenko VV, Major ML, Wang X, Petrovic V, Kuechle J, Yoder HM, Dennewitz MB, Shin B, Datta A, Raychaudhuri P, Costa RH. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–850. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gusarova GA, Wang IC, Major ML, Kalinichenko VV, Ackerson T, Petrovic V, Costa RH. A cell-penetrating ARF peptide inhibitor of FoxM1 in mouse hepatocellular carcinoma treatment. J Clin Invest. 2007;117:99–111. doi: 10.1172/JCI27527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krol J, Francis RE, Albergaria A, Sunters A, Polychronis A, Coombes RC, Lam EW. The transcription factor FOXO3a is a crucial cellular target of gefitinib (Iressa) in breast cancer cells. Mol Cancer Ther. 2007;6:3169–3179. doi: 10.1158/1535-7163.MCT-07-0507. [DOI] [PubMed] [Google Scholar]
- 46.Guo S, Sonenshein GE. Forkhead box transcription factor FOXO3a regulates estrogen receptor alpha expression and is repressed by the Her-2/neu/phosphatidylinositol 3-kinase/Akt signaling pathway. Mol Cell Biol. 2004;24:8681–8690. doi: 10.1128/MCB.24.19.8681-8690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jackson JG, Kreisberg JI, Koterba AP, Yee D, Brattain MG. Phosphorylation and nuclear exclusion of the forkhead transcription factor FKHR after epidermal growth factor treatment in human breast cancer cells. Oncogene. 2000;19:4574–4581. doi: 10.1038/sj.onc.1203825. [DOI] [PubMed] [Google Scholar]
- 48.Delpuech O, Griffiths B, East P, Essafi A, Lam EW, Burgering B, Downward J, Schulze A. Induction of Mxi1-SR alpha by FOXO3a contributes to repression of Myc-dependent gene expression. Mol Cell Biol. 2007;27:4917–4930. doi: 10.1128/MCB.01789-06. [DOI] [PMC free article] [PubMed] [Google Scholar]