Abstract
Background/Aims
Few reports have described the association between mutations in the entire X gene of the hepatitis B virus (HBV) and the clinical status of HBV-infected patients. We studied the association between HBV X gene mutations and the disease status of patients infected with HBV genotype C.
Methods
Mutations in the HBV X genes of 194 patients were determined by direct sequencing. The subject population consisted of patients with chronic hepatitis (n=60), liver cirrhosis (n=65), and hepatocellular carcinoma (HCC) (n=69). The sequencing results of these 3 groups were compared.
Results
Each of the mutations G1386M, C1485T, C1653T, T1753V, A1762T, and G1764A was significantly associated with the patient's clinical status. The T1753V (p<0.001) and A1762T/G1764A (p<0.001) mutations were found more frequently in Hepatitis B e antigen (HBeAg)-negative than in HBeAg-positive patients. Specific X gene mutations (G1386M, C1653T, and A1762T/G1764A) were more prevalent in patients with liver cirrhosis and HCC than in chronic hepatitis patients (p<0.005 for all). In addition, the T1753V (p<0.001) and C1485T (p<0.001) mutations were significantly more prevalent in HCC patients than in chronic hepatitis patients. Only the prevalence of the T1753V mutation increased as the HBV infection progressed from liver cirrhosis to HCC (p=0.023).
Conclusions
Our findings show a difference in the pattern of X gene mutations that were associated with the clinical status of patients with chronic HBV infection.
Keywords: Hepatitis B virus, Hepatocellular carcinoma, Cirrhosis, Liver disease, X mutation
INTRODUCTION
Hepatitis B virus (HBV) infection is a worldwide health concern, and more than 350 million people are chronic carriers of this virus.1 HBV is the prototype member of the family Hepadnaviridae, with a compact genome of approximately 3.2 kb arranged in a circular, partially double-stranded DNA molecule.2 The viral genome contains 4 open reading frames encoding its own proteins with 4 promoters and 2 enhancers.3
The clinical patterns of hepatitis B are diverse, ranging from natural recovery to acute hepatitis, chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (HCC). The different clinical patterns are influenced by various factors that include the natural history of HBV infection, host susceptibility, duration of the infection, environmental factors, and viral factors.4-8
Regarding viral factors, the core promoter/precore mutations are known to be associated with the severity of liver disease, such as fulminant hepatitis, chronic active hepatitis, and HCC.9-11 Specific mutations in the regions where the X gene and enhancer II/core promoter regions overlap are especially associated with the development of HCC.12-16
The HBV X protein (HBx) is associated with hepatocarcinogenesis through a multistep process, which includes induction of reactive oxygen species, alteration of mitochondrial function and physiology, and DNA damage.17-19 In hepatic inflammation, HBx induces many pro-inflammatory cytokines at the transcriptional level.20,21 The protease inhibitor-like structure of HBx may also cause accumulation of toxic factors, thereby resulting in severe hepatocellular injury.22 The mutations in HBx cause enhancement of various HBx effects.18,23 These observations provide evidence that HBx is involved in the progression of liver disease. However, very few reports have described the association of X gene mutations with disease progression in chronic HBV infection.24-26 Thus, the aim of our study was to evaluate the association of X gene mutations with the different clinical status of patients with HBV genotype C infection.
MATERIALS AND METHODS
1. Patients
We collected serum from 194 HBV-infected patients (chronic hepatitis, 60; liver cirrhosis, 65; HCC, 69) who visited the outpatient department at the Wonkwang University Hospital from January 2005 to December 2008. The following exclusion criteria were employed for the study patients: acute hepatitis B infection, history of previous antiviral therapeutic trials, hepatitis C or D virus infection, alcohol abuse, or other causes of liver disease, such as autoimmune hepatitis, Wilson's disease, or hemochromatosis. Chronic hepatitis is defined by persistent or intermittent elevation of alanine transaminase (ALT) levels for 6 months and such elevations are not observed in cirrhosis and/or HCC.
The diagnosis of liver cirrhosis was based on imaging findings acquired by ultrasonography, computed tomography (CT), magnetic resonance imaging (MRI) the clinical and laboratory findings. HCC was diagnosed by histology or a combination of α-fetoprotein levels and imaging findings (hepatic angiography, CT, or MRI). The study protocol was approved by the Ethics Committee of our institution, and informed consent was obtained from all the patients. All collected serum samples were stored at -20℃ until the analysis.
2. Serological assays for HBV markers and viral load
The hepatitis B markers hepatitis B surface antigen (HBsAg), hepatitis B surface antibody (HBsAb), hepatitis B e antigen (HBeAg), and anti-hepatitis B e antibody (HBeAb) were tested using electrochemiluminescence assay kits (Roche, Rotkreuz, Switzerland). The HBV viral load was assayed using the Artus HBV LC PCR Kit (Qiagen, Hamburg, Germany), which has a detection limit of 200 copies/mL.
3. Extraction of HBV DNA, amplification, and sequencing of X genes
HBV DNA was extracted from serum (200 µL) by using a commercial kit (QIAamp DNA Blood Mini Kit; Qiagen, Hilden, Germany) following the manufacturer's protocol. The first round of PCR was performed using the following primer pair: sense primer S1 (5'-GAACCTTTGTGGCTCCTCTG-3' [nt: 1241-1260]) and antisense primer AS1 (5'-AAAAAGAGAGTAACTCCACAG-3' [nt: 1935-1955]). The second nested PCR was performed with the sense primer S1 (5'-GCAGCCGGTCTGGAGCGAAA-3' [nt: 1301-1320]) and antisense primer AS1 (5'-ACAGCTTGGAGGCTTGAACA-3' [nt: 1861-1880]).
PCR products were purified on 2% agarose gel by using a commercial kit (QIAquick Gel Extraction Kit; Qiagen, Hilden, Germany), and residual primers and dNTPs were removed. The purified PCR products were directly sequenced by the dideoxy method with a BigDye® Terminator V3.1 ready reaction cycle sequencing kit and analyzed using an ABI PRISM 377 DNA Sequencer (Applied Biosystems, Foster City, CA, USA).
4. HBV genotyping
The HBV genomic sequences were multiple aligned using CLUSTAL W/X software. The genetic distances were estimated using Kimura's two-parameter model, and phylogenetic trees were constructed using the neighbor-joining method.27,28
5. Determination of nucleotide variations
The nucleotide sequences from position 1374 to 1838, located in the region of the X gene, were examined, and the sequences were verified in both directions. The nucleotide variations were defined by differences in the consensus sequences. Mutations with a frequency >10% were selected.
6. Statistical analysis
The data were analyzed using the SPSS version 11.5 statistical software package (SPSS Inc., Chicago, IL, USA). For comparisons of the collected data, the chi-square test, Fisher exact test, Student's t-test, and multivariate logistic regression analysis were performed. The differences between the groups with p values <0.05 were considered statistically significant.
RESULTS
1. Baseline clinical characteristics, nucleotide variability, and mutational patterns of HBV X gene according to the clinical status
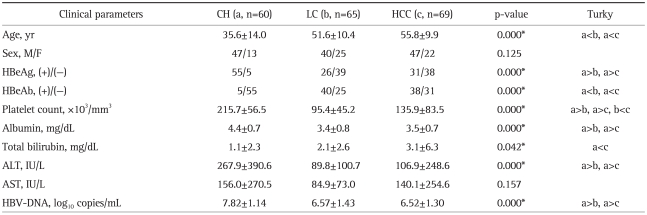
The subject population consisted of patients who had the following 3 diseases: chronic hepatitis (n=60), liver cirrhosis (n=65), and HCC (n=69). All patients were infected with HBV genotype C. Table 1 shows serum HBV DNA concentrations, age, sex, liver function, and HBeAg serostatus in the study population. Chronic hepatitis patients showed a younger age, higher albumin, higher ALT, higher platelet count, higher HBV DNA, higher HBeAg positive rate, and lower HBeAb positive rate than liver cirrhosis and HCC patients. However, liver cirrhosis patients and HCC patients were not significantly different from each other except in the platelet count.
Table 1.
Clinical Characteristics of Patients in the Study
Data are presented as number or mean±SD. The significance of differences (p-value) in the indicated group is shown.
CH, chronic hepatitis; LC, liver cirrhosis; HCC, hepatocelluar carcinoma; HBeAg, hepatitis B e antigen; HBeAb, hepatitis B e antibody; HBV-DNA, hepatitis B virus-DNA.
*p<0.05.
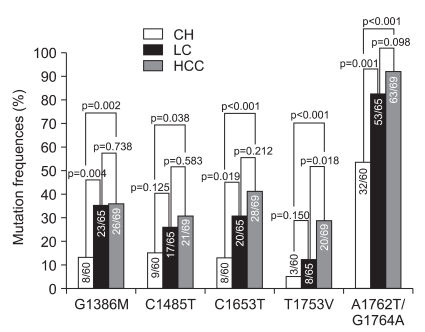
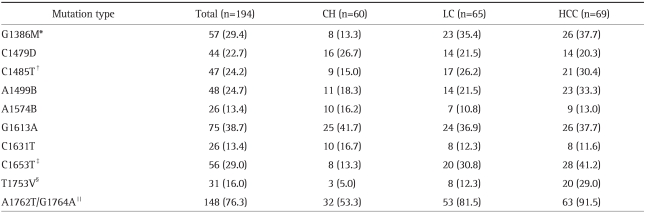
All subjects (n=194) showed 10 mutations with a frequency >10%. Six of the ten mutations were associated with the clinical status of patients with chronic hepatitis B infection. The following mutations were observed: G1386M, C1485T, C1653T, T1753V, A1762T, and G1764A (Fig. 1).
Fig. 1.
The difference and frequencies of 5 mutational patterns (G1386M, C1485T, C1653T, T1753V, and A1762T/G1764A) according to the clinical status of each.
CH, chronic hepatitis; LC, liver cirrhosis; HCC, hepatocellular carcinoma.
The mutations A1762T and G1764A were frequently observed together. The frequency of the double mutation A1762T/G1764A was 148/194 (76.3%). The double mutation was observed in 148 out of 158 patients (93.7%) with either A1762T or G1764A mutations. The A1762T mutation alone (without G1764A) was observed in 2 patients, and single G1764A mutations were observed in 8 patients. The double mutation was considered to be a mutational pattern.
The frequency of 3 mutational patterns (G1386M, C1653T, and A1762T/G1764A) in patients with liver cirrhosis was significantly higher than that in patients with chronic hepatitis (35.4% vs 13.3%, p=0.004; 30.8% vs 13.3%, p=0.019; 81.5% vs 53.3 %, p=0.001; respectively).
The frequency of 5 mutational patterns (G1386M, C1485T, C1653T, T1753V, and A1762T/G1764A) in patients with HCC was significantly higher than that in patients with chronic hepatitis (37.7% vs 13.3%, p=0.002; 30.4% vs 15.0%, p=0.038; 41.2% vs 13.3%, p<0.001; 29.0% vs 5.0%, p<0.001; 91.5% vs 53.3%, p<0.001; respectively). Only the T1753V mutation was significantly more frequent in patients with HCC than in patients with liver cirrhosis (29.0% vs 12.3%, p=0.018) (Table 2).
Table 2.
Mutational Patterns of the HBV X Gene according to Clinical Status
Data are presented as number (%).
HBV, hepatitis B virus; CH, chronic hepatitis; LC, liver cirrhosis; HCC, hepatocelluar carcinoma.
*CH vs LC, p=0.004; CH vs HCC, p=0.002; LC vs HCC, p=0.783; †CH vs LC, p=0.125; CH vs HCC, p=0.038; LC vs HCC, p=0.583; ‡CH vs LC, p=0.019; CH vs HCC, p<0.001; LC vs HCC, p=0.212; §CH vs LC, p=0.150; CH vs HCC, p<0.001; LC vs HCC, p=0.018; ∥CH vs LC, p=0.001; CH vs HCC, p<0.001; LC vs HCC, p=0.098.
2. Significance of the mutations in the HBV X gene according to the clinical status
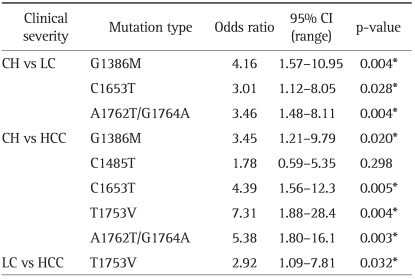
Although significant mutations were identified using univariate analysis, multivariate analysis was used to determine the association of the following mutations to the clinical status of HBV-infected patients: 1) Liver cirrhosis patients had more G1386M (odds ratio [OR], 4.16; 95% confidence interval [CI], 1.57-10.95; p=0.004], C1653T (OR, 3.01; 95% CI, 1.12-8.05; p=0.028), and A1762T/G1764A (OR, 3.46; 95% CI, 1.48- 8.11; p=0.004) mutations than chronic hepatitis patients; and 2) HCC patients had more G1386M (OR, 3.45; 95% CI, 1.21-9.79; p=0.020), C1653T (OR, 4.39; 95% CI, 1.56-12.3; p=0.005), T1753V (OR, 7.31; 95% CI, 1.88-28.4; p=0.004), and A1762T/G1764A (OR, 5.38; 95% CI, 1.80-16.10; p=0.003) mutations than the chronic hepatitis patients. In particular, T1753V mutation was the only base mutation showing difference in comparison between HCC and liver cirrhosis patients (29.0% vs 12.3%; OR, 2.92; 95% CI, 1.09-7.81; p=0.032; Table 3).
Table 3.
Multivariate Analysis of Mutations in the HBV X Gene according to Clinical Status
The significance of differences (p-value) in the indicated group is shown.
HBV, hepatitis B virus; CI, confidence interval; CH, chronic hepatitis; LC, liver cirrhosis; HCC, hepatocelluar carcinoma.
*p<0.05.
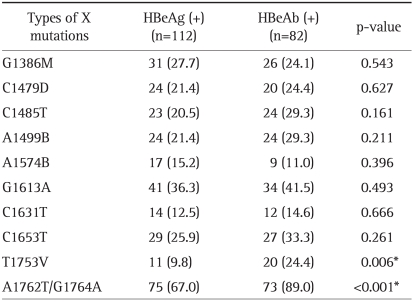
3. Different distribution of mutations in the HBV X gene according to the HBeAg serostatus
To evaluate the associations between the 10 mutations (>10% frequency) and HBeAg serostatus, the frequencies of various mutations were compared between HBeAg-positive and HBeAg-negative patients. The mutations T1753V (9.8% vs 24.4%, p=0.006) and A1762T/G1764A (67.0% vs 89.0%, p<0.001) were significantly more frequently found in HBeAg-negative than in HBeAg-positive patients (Table 4). Interestingly, T1753V mutation was not different between liver cirrhosis patients and HCC patients when the patients were HBeAg-positive, and its frequency was significantly higher in patients with HCC than in patients with liver cirrhosis only when the patients were HBeAg-negative (34.2% vs 12.5%, p=0.032).
Table 4.
Different Distributions of Mutations in the HBV X Gene according to HBeAg Serostatu
Data are presented as number (%). The significance of differences (p-value) in the indicated group is shown.
HBV, hepatitis B virus; HBeAg, hepatitis B e antigen; HBeAb, hepatitis B e antibody.
*p<0.05.
DISCUSSION
In contrast to several studies in which specific mutations in the X gene or enhancer II/core promoter regions of HBV were observed to be associated with the development of HCC, we focused our study on the association between the clinical status of the liver disease and mutations in the entire X gene region. We identified 6 specific mutations in the X gene that were associated with the clinical status of HBV-induced liver disease. These mutations, G1386M, C1485T, C1653T, T1753V, and A1762T/G1764A, result in the respective amino acid substitutions as follows: V5M/L, P38S, H94Y, I127T/N, and K130M/V131I. These amino acid substitutions induce changes in HBx, thereby altering its activities in transactivation and apoptosis.29,30
The most significant result in this study was the identification of the mutation G1386M (V5M/L) as a related factor in the progression from chronic hepatitis to liver cirrhosis or HCC. G1386M (V5M/L) mutations were significantly more prevalent in liver cirrhosis and HCC than in chronic hepatitis patients. This finding is consistent with a previous study that showed a significantly high prevalence of G1386M in patients with a severe liver disease such as chronic hepatitis B infection.24 However, in this study, the G1386M mutation was not significantly more prevalent in patients with HCC than in patients with liver cirrhosis, and there was no difference according to the HBeAg serostatus. In addition, in contrast to the previous study, the G1386M mutation was not associated with the double mutation A1762T/G1764A (not shown),24 thereby suggesting that this mutation may be a target of the host immune response against HBx. However, the epidemiologic data for this mutation and the study of its functional role are limited. Additional studies are required to determine the effects of this mutation.
In this study, the prevalence of the C1485T mutation (P38S) in HCC patients was significantly higher than that in chronic hepatitis patients. However, C1485T mutation as a risk factor in the progression from chronic hepatitis to HCC was eliminated after multivariate analysis. Also the prevalence of the C1485T mutation in liver cirrhosis was not significantly different in chronic hepatitis patients. Taken together, the frequency of the C1485T mutation was not significantly associated with the HBeAg serostatus; this finding is consistent with that of a previous study.24 These findings suggest that the C1485T mutation is not as important as other mutations in the progression from chronic hepatitis to HCC. This finding is also consistent with a previous experimental study in which a mutation in codon 38 did not affect the various signal transduction pathways.31
Our previous study had reported that B1499 mutation was significantly more frequent in the HCC group32 but in this study no difference was observed. The variation between the two studies comes from the difference between the two study groups. The previous study was limited to liver cirrhosis patients, and did not included chronic hepatitis patients. However this study included chronic hepatitis and liver cirrhosis patients, HCC patients in the previous study all had liver cirrhosis, but HCC patients in this study have either chronic hepatitis or liver cirrhosis. Therefore the lack of chronic hepatitis in the previous study group has proven to cause a variation in results between the two studies.
The frequencies of the mutations C1653T (H94Y) and A1762T/G1764A (K130M/V131I) in liver cirrhosis and HCC patients were higher than those in chronic hepatitis patients. The amino acid change in codon 94 caused by the C1653T mutation affects both the function and structure of HBx. C1653 in the box-alpha corresponds to the region associated with the target site for p53-mediated transcriptional repression and the binding site of the related factors. Nucleotide 1653 of HBV was placed at the center of the immunodominant antigenic domain.30 The C1653T mutation may enhance the binding affinity of related factors and enhancer II/core promoter activity,33-35 and cause activation of an indolent immune response, as well as abrogates both the antiproliferative and transactivation effects of wild-type HBx.36 These changes may be involved in inflammatory processes and/or hepatocarcinogenesis.
The double mutation A1762T/G1764A is well known to be associated with disease severity, such as HCC and fulminant hepatitis.9-11,13,14 In this study, the T1762 and A1764 mutations were almost in linkage disequilibrium, and their frequencies were as high as in previous studies.24,25,37 Although the frequency of the double mutation A1762T/G1764A was high (53.3%) in patients with chronic hepatitis, the mutation rate was significantly higher in patients with liver cirrhosis (81.5%) and HCC (91.5%). This finding is consistent with a previous study showing that the double mutation A1762T/G1764A is associated with more progressive forms of liver disease.24 The double mutation A1762T/G1764A has been associated with diminished HBeAg production, enhanced host immune response, and viral DNA replication.38-40 HBx including A1762T/G1764A mutant abrogates the antiproliferative effect compared to wild-type HBx.41 These effects may be involved in the pathogenesis of inflammatory processes or carcinogenesis.
In this study, the double mutation A1762T/G1764A was closely associated with liver cirrhosis and HCC rather than with chronic hepatitis. However, the prevalence of the pattern of T1762/A1764 mutations were not significantly different between cirrhosis and HCC patients. The 2 mutational patterns have been frequently found in patients with advanced liver disease such as cirrhosis and HCC.24,25 Recent studies have shown that these mutations would not be a predictive factor for HCC development in patients with cirrhosis,32,42 thereby suggesting that the significance of any mutation in the prediction of HCC development may be dependent on the differences in the frequency of cirrhosis in the HCC and non-HCC groups. The mutational patterns of C1653T and T1762/A1764 were associated with the development of HCC when the frequency of patients with cirrhosis in the HCC group was significantly higher than that in the non-HCC group.15,16
In our study, only the T1753V mutation (I127T/N) was increased as the HBV infection progressed from liver cirrhosis to HCC. T1753 mutation combined with T1762/A1764 showed enhanced viral replication in vitro,40 and HBV viral load is a factor associated with developing HCC. Also the change of amino acid by T1753V mutation may affect the function of HBx.12 Therefore the T1753V mutation may predict the development of HCC in patients with cirrhosis.37,42 As this tendency was observed only in HBeAg-negative patients but not in HBeAg-positive ones, T1753V mutation occurred more frequently in patients with HCC than in patients with liver cirrhosis only when the patients were HBeAg-negative. In the previous studies, it was demonstrated that T1753V mutations in HCC patients infected with C1/Cs were frequently found in cases of HBeAg-positive, whereas T1753V mutations in HCC patients infected with C2/Ce were frequently found in cases of HBeAg-negative.12 This is similar to our results because HBV genotype in Korea is C2/Ce. Therefore we suggested that the difference in the genotype is probably responsible for the different results, but further studies with more subjects are needed. In addition, this mutation may alter the binding activity of the ubiquitous transcription factor Sp1 and affect the core promoter activity.43 HBx including T1753V mutant induced a higher increase in the transactivational activity than wild-type HBx.41 These changes may contribute to the multistep process of hepatocarcinogenesis.
In conclusion, we believe that the mutations G1386M, C1485T, C1653T, T1753V, and A1762T/G1764A in the X gene are associated with the clinical status as chronic HBV infection progresses from chronic hepatitis to cirrhosis or HCC and from cirrhosis to HCC. Also our results showed a difference in the pattern of X gene mutations that was associated with each clinical status. These finding suggests that these mutations may be useful markers for predicting the clinical course of patients with chronic hepatitis B.
ACKNOWLEDGEMENTS
The work was supported by a grant from the Wonkwang University (2009).
References
- 1.Lee WM. Hepatitis B virus infection. N Engl J Med. 1997;337:1733–1745. doi: 10.1056/NEJM199712113372406. [DOI] [PubMed] [Google Scholar]
- 2.Ganem D, Varmus HE. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- 3.Schaller H, Fischer M. Transcriptional control of hepadnavirus gene expression. Curr Top Microbiol Immunol. 1991;168:21–39. doi: 10.1007/978-3-642-76015-0_2. [DOI] [PubMed] [Google Scholar]
- 4.Mayans MV, Calvet X, Bruix J, et al. Risk factors for hepatocellular carcinoma in Catalonia, Spain. Int J Cancer. 1990;46:378–381. doi: 10.1002/ijc.2910460307. [DOI] [PubMed] [Google Scholar]
- 5.London WT, Evans AA, McGlynn K, et al. Viral, host and environmental risk factors for hepatocellular carcinoma: a prospective study in Haimen City, China. Intervirology. 1995;38:155–161. doi: 10.1159/000150426. [DOI] [PubMed] [Google Scholar]
- 6.Omata M, Ehata T, Yokosuka O, Hosoda K, Ohto M. Mutations in the precore region of hepatitis B virus DNA in patients with fulminant and severe hepatitis. N Engl J Med. 1991;324:1699–1704. doi: 10.1056/NEJM199106133242404. [DOI] [PubMed] [Google Scholar]
- 7.Fattovich G, Brollo L, Alberti A, Pontisso P, Giustina G, Realdi G. Long-term follow-up of anti-HBe-positive chronic active hepatitis B. Hepatology. 1988;8:1651–1654. doi: 10.1002/hep.1840080630. [DOI] [PubMed] [Google Scholar]
- 8.Lindh M, Horal P, Dhillon AP, Norkrans G. Hepatitis B virus DNA levels, precore mutations, genotypes and histological activity in chronic hepatitis B. J Viral Hepat. 2000;7:258–267. doi: 10.1046/j.1365-2893.2000.00236.x. [DOI] [PubMed] [Google Scholar]
- 9.Thomas HC, Brown D, Routhier G, et al. Inducer and suppressor T-cells in hepatitis B virus-induced liver disease. Hepatology. 1982;2:202–204. doi: 10.1002/hep.1840020203. [DOI] [PubMed] [Google Scholar]
- 10.Eddleston AL, Mondelli M. Immunopathological mechanisms of liver cell injury in chronic hepatitis B virus infection. J Hepatol. 1986;3(Suppl 2):S17–S23. doi: 10.1016/s0168-8278(86)80096-4. [DOI] [PubMed] [Google Scholar]
- 11.Omata M, Yokosuka O, Imazeki F, et al. Correlation of hepatitis B virus DNA and antigens in the liver. A study in chronic liver disease. Gastroenterology. 1987;92:192–196. doi: 10.1016/0016-5085(87)90858-4. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka Y, Mukaide M, Orito E, et al. Specific mutations in enhancer II/core promoter of hepatitis B virus subgenotypes C1/C2 increase the risk of hepatocellular carcinoma. J Hepatol. 2006;45:646–653. doi: 10.1016/j.jhep.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 13.Kim JK, Chang HY, Lee JM, et al. Specific mutations in the enhancer II/core promoter/precore regions of hepatitis B virus subgenotype C2 in Korean patients with hepatocellular carcinoma. J Med Virol. 2009;81:1002–1008. doi: 10.1002/jmv.21501. [DOI] [PubMed] [Google Scholar]
- 14.Chou YC, Yu MW, Wu CF, et al. Temporal relationship between hepatitis B virus enhancer II/basal core promoter sequence variation and risk of hepatocellular carcinoma. Gut. 2008;57:91–97. doi: 10.1136/gut.2006.114066. [DOI] [PubMed] [Google Scholar]
- 15.Yuen MF, Tanaka Y, Shinkai N, et al. Risk for hepatocellular carcinoma with respect to hepatitis B virus genotypes B/C, specific mutations of enhancer II/core promoter/precore regions and HBV DNA levels. Gut. 2008;57:98–102. doi: 10.1136/gut.2007.119859. [DOI] [PubMed] [Google Scholar]
- 16.Shinkai N, Tanaka Y, Ito K, et al. Influence of hepatitis B virus X and core promoter mutations on hepatocellular carcinoma among patients infected with subgenotype C2. J Clin Microbiol. 2007;45:3191–3197. doi: 10.1128/JCM.00411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu JM, Lim SO, Oh SJ, Yoon SM, Seong JK, Jung G. HBx modulates iron regulatory protein 1-mediated iron metabolism via reactive oxygen species. Virus Res. 2008;133:167–177. doi: 10.1016/j.virusres.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 18.Shirakata Y, Koike K. Hepatitis B virus X protein induces cell death by causing loss of mitochondrial membrane potential. J Biol Chem. 2003;278:22071–22078. doi: 10.1074/jbc.M301606200. [DOI] [PubMed] [Google Scholar]
- 19.Kim S, Kim HY, Lee S, et al. Hepatitis B virus X protein induces perinuclear mitochondrial clustering in microtubule- and Dynein-dependent manners. J Virol. 2007;81:1714–1726. doi: 10.1128/JVI.01863-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee MO, Choi YH, Shin EC, et al. Hepatitis B virus X protein induced expression of interleukin 18 (IL-18): a potential mechanism for liver injury caused by hepatitis B virus (HBV) infection. J Hepatol. 2002;37:380–386. doi: 10.1016/s0168-8278(02)00181-2. [DOI] [PubMed] [Google Scholar]
- 21.Lara-Pezzi E, Majano PL, Gómez-Gonzalo M, et al. The hepatitis B virus X protein up-regulates tumor necrosis factor alpha gene expression in hepatocytes. Hepatology. 1998;28:1013–1021. doi: 10.1002/hep.510280416. [DOI] [PubMed] [Google Scholar]
- 22.Koike K. Hepatitis B virus X gene is implicated in liver carcinogenesis. Cancer Lett. 2009;286:60–68. doi: 10.1016/j.canlet.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 23.Bock CT, Toan NL, Koeberlein B, et al. Subcellular mislocalization of mutant hepatitis B X proteins contributes to modulation of STAT/SOCS signaling in hepatocellular carcinoma. Intervirology. 2008;51:432–443. doi: 10.1159/000209672. [DOI] [PubMed] [Google Scholar]
- 24.Kim HJ, Park JH, Jee Y, et al. Hepatitis B virus X mutations occurring naturally associated with clinical severity of liver disease among Korean patients with chronic genotype C infection. J Med Virol. 2008;80:1337–1343. doi: 10.1002/jmv.21219. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi K, Akahane Y, Hino K, Ohta Y, Mishiro S. Hepatitis B virus genomic sequence in the circulation of hepatocellular carcinoma patients: comparative analysis of 40 full-length isolates. Arch Virol. 1998;143:2313–2326. doi: 10.1007/s007050050463. [DOI] [PubMed] [Google Scholar]
- 26.Sung JJ, Tsui SK, Tse CH, et al. Genotype-specific genomic markers associated with primary hepatomas, based on complete genomic sequencing of hepatitis B virus. J Virol. 2008;82:3604–3611. doi: 10.1128/JVI.01197-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 29.Dong Q, Chan HL, Liu Z, et al. A1762T/G1764A mutations of hepatitis B virus, associated with the increased risk of hepatocellular carcinoma, reduce basal core promoter activities. Biochem Biophys Res Commun. 2008;374:773–776. doi: 10.1016/j.bbrc.2008.07.115. [DOI] [PubMed] [Google Scholar]
- 30.Stemler M, Weimer T, Tu ZX, et al. Mapping of B-cell epitopes of the human hepatitis B virus X protein. J Virol. 1990;64:2802–2809. doi: 10.1128/jvi.64.6.2802-2809.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muroyama R, Kato N, Yoshida H, et al. Nucleotide change of codon 38 in the X gene of hepatitis B virus genotype C is associated with an increased risk of hepatocellular carcinoma. J Hepatol. 2006;45:805–812. doi: 10.1016/j.jhep.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 32.Choi CS, Cho EY, Park R, Kim SJ, Cho JH, Kim HC. X gene mutations in hepatitis B patients with cirrhosis, with and without hepatocellular carcinoma. J Med Virol. 2009;81:1721–1725. doi: 10.1002/jmv.21591. [DOI] [PubMed] [Google Scholar]
- 33.Yuh CH, Chang YL, Ting LP. Transcriptional regulation of precore and pregenomic RNAs of hepatitis B virus. J Virol. 1992;66:4073–4084. doi: 10.1128/jvi.66.7.4073-4084.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.López-Cabrera M, Letovsky J, Hu KQ, Siddiqui A. Multiple liver-specific factors bind to the hepatitis B virus core/pregenomic promoter: trans-activation and repression by CCAAT/enhancer binding protein. Proc Natl Acad Sci U S A. 1990;87:5069–5073. doi: 10.1073/pnas.87.13.5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee H, Kim HT, Yun Y. Liver-specific enhancer II is the target for the p53-mediated inhibition of hepatitis B viral gene expression. J Biol Chem. 1998;273:19786–19791. doi: 10.1074/jbc.273.31.19786. [DOI] [PubMed] [Google Scholar]
- 36.Sirma H, Giannini C, Poussin K, Paterlini P, Kremsdorf D, Bréchot C. Hepatitis B virus X mutants, present in hepatocellular carcinoma tissue abrogate both the antiproliferative and transactivation effects of HBx. Oncogene. 1999;18:4848–4859. doi: 10.1038/sj.onc.1202867. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z, Tanaka Y, Huang Y, et al. Clinical and virological characteristics of hepatitis B virus subgenotypes Ba, C1, and C2 in China. J Clin Microbiol. 2007;45:1491–1496. doi: 10.1128/JCM.02157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyakawa Y, Okamoto H, Mayumi M. The molecular basis of hepatitis B e antigen (HBeAg)-negative infections. J Viral Hepat. 1997;4:1–8. doi: 10.1046/j.1365-2893.1997.00101.x. [DOI] [PubMed] [Google Scholar]
- 39.Kidd AH, Kidd-Ljunggren K. A revised secondary structure model for the 3'-end of hepatitis B virus pregenomic RNA. Nucleic Acids Res. 1996;24:3295–3301. doi: 10.1093/nar/24.17.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jammeh S, Tavner F, Watson R, Thomas HC, Karayiannis P. Effect of basal core promoter and pre-core mutations on hepatitis B virus replication. J Gen Virol. 2008;89(Pt 4):901–909. doi: 10.1099/vir.0.83468-0. [DOI] [PubMed] [Google Scholar]
- 41.Lin X, Xu X, Huang QL, et al. Biological impacts of "hot-spot" mutations of hepatitis B virus X proteins are genotype B and C differentiated. World J Gastroenterol. 2005;11:4703–4708. doi: 10.3748/wjg.v11.i30.4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu S, Zhang H, Gu C, et al. Associations between hepatitis B virus mutations and the risk of hepatocellular carcinoma: a meta-analysis. J Natl Cancer Inst. 2009;101:1066–1082. doi: 10.1093/jnci/djp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Günther S, Piwon N, Iwanska A, Schilling R, Meisel H, Will H. Type, prevalence, and significance of core promoter/enhancer II mutations in hepatitis B viruses from immunosuppressed patients with severe liver disease. J Virol. 1996;70:8318–8331. doi: 10.1128/jvi.70.12.8318-8331.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]