Abstract
Stable isotope labeling (SIL) methods coupled with nanoscale liquid chromatography and high resolution tandem mass spectrometry are increasingly useful for elucidation of the proteome-wide differences between multiple biological samples. Development of more effective programs for the sensitive identification of peptide pairs and accurate measurement of the relative peptide/protein abundance are essential for quantitative proteomic analysis. We developed and evaluated the performance of a new program, termed UNiquant, for analyzing quantitative proteomics data using stable isotope labeling. UNiquant was compared with two other programs, MaxQuant and Mascot Distiller, using SILAC-labeled complex proteome mixtures having either known or unknown heavy/light ratios. For the SILAC-labeled Jeko-1 cell proteome digests with known heavy/light ratios (H/L = 1:1, 1:5, and 1:10), UNiquant quantified a similar number of peptide pairs as MaxQuant for the H/L = 1:1 and 1:5 mixtures. In addition, UNiquant quantified significantly more peptides than MaxQuant and Mascot Distiller in the H/L = 1:10 mixtures. UNiquant accurately measured relative peptide/protein abundance without the need for post-measurement normalization of peptide ratios, which is required by the other programs.
Keywords: Quantitative proteomics, Stable isotope labeling, LC-MS/MS, Software Development
Introduction
In quantitative shotgun proteomics, proteolysis-derived peptides are commonly measured with liquid chromatography-tandem mass spectrometry (LC-MS/MS) and used as surrogates of their parent proteins for relative quantification.1, 2 In a label-free approach, the proteomes under comparison are analyzed separately in standardized LC-MS/MS runs. Peptide intensities, spectra counts and extracted ion chromatography (XIC) are used to measure protein abundances.3–5 Alternatively, by employing stable isotope labeling (SIL), the proteomes under comparison are combined and analyzed together in one LC-MS/MS run. Comparison of the signal intensities of the same peptides and their stable isotope labeled analogues yields an estimate of protein abundances.6, 7 In general, SIL methods minimize the variability during sample processing steps and LC-MS/MS analyses and provide results with less systematic errors and higher reproducibility comparing to the label-free approach.8
Recently, a variety of SIL approaches have been developed including the labeling of elements (15N labeling), specific amino acids (ICAT and SILAC), and terminals (iTRAQ and 18O labeling) in peptides.9–14 A number of academically developed software tools, such as ASAPRatio,15 Census,16 MSQuant,17 MaxQuant,18 Vista,19 and WaveletQuant,20 and some commercial software, such as Mascot Distiller (Matrix Science), have been produced to analyze SIL-based quantitative proteomics datasets21. Moreover, many efforts have been made to improve proteomic data analysis; these include enhancing the dynamic range22 and mass accuracy,23 combining database searches,24 and estimating the false discovery rate (FDR).25 However, efficient and accurate measurement of MS intensity data is still a very challenging aspect of quantitative proteomics analysis.
Previously, we developed a series of algorithms and software for processing high-resolution tandem mass spectrometry data,26 for eliminating mass error,27, 28 for quantifying 18O-based peptides14 and for obtaining confident peptide identifications29 in bottom-up proteomics measurements, using hybrid Fourier transform (FT) mass spectrometry instruments.30 Building upon these algorithms and software, we report a new program, named UNiquant, for SIL-based quantitative proteomic data analysis. In UNiquant, we aim to establish a user-friendly software solution for accurate measurement of relative peptide/protein abundance that supports a broad spectrum of SIL methods and database search engines for quantitative proteomics applications.
Materials and Methods
Sample preparation
The human lymphoma cell lines Jeko-1 and DHL16 were cultured in RPMI 1640 medium with 10% fetal bovine serum. Cells were used in 2 types of proteomics experiments employing either SILAC or pulse SILAC (pSILAC) labeling:31
Doublet SILAC labeling was used to prepare standard proteome mixtures. Jeko-1 cells were grown in either SILAC “light” (l-arginine and l-lysine) or “heavy” (l-13C6-argnine and l-13C6-lysine, doubly labeled) medium for two weeks (more than 5 cell cycles). Cells were harvested in the lysis buffer (7 M urea, 2 M thiourea, 50 mM ammonium bicarbonate, pH 7.4). Protein concentrations of the clarified lysates were determined with the Coomassie protein assay. Lysates from the two growth conditions were mixed to create standard proteome mixtures in three heavy/light (H/L) ratios: 1:1, 1:5, and 1:10.
Triplet pSILAC labeling was used to prepare proteome mixtures of unknown protein ratios. Two flasks of DHL16 cells were grown in SILAC light (l-arginine and l-lysine) media for two weeks. One flask was transiently transfected with microRNA-155 vector and the other one was transfected with GFP vector. At the time of transfection, the medium for the microRNA-155 transfection was replaced with SILAC medium-heavy (l-13C6-argnine and l-13C6-lysine) medium, while the medium for the GFP transfection was replaced with SILAC heavy (l-13C6 15N4-argnine and l-13C6 15N2-lysine) medium. Cells from the medium-heavy and heavy labeling were harvested 4 hrs and 48 hrs respectively after treatment. Protein concentrations of clarified lysates were determined with the Coomassie protein assay. The two samples were mixed equally based on their total protein abundances. The experiments were repeated twice.
Mixed cell lysates were desalted using centrifugal filter devices with a low mass cutoff of 3000 Da (Millipore, Billerica, MA). The desalted lysates were reduced with 10 mM dithiothreitol at 95°C for 10 min then alkylated with 55 mM iodoacetamide in the dark at room temperature for 1 hr. Then the samples were diluted 8-fold using 50 mM ammonium bicarbonate and digested with sequencing grade trypsin solution [1µg/µL at a trypsin/protein ratio of 1:50 (wt/wt)] over night at 37°C. The digests were desalted with Sep-Pak Cartridges C18 Plus (Waters, Milford, MA) and dried in a vacuum centrifuge. The peptides were re-suspended in 25 mM ammonium bicarbonate. Peptides concentrations were determined using BCA assay (Pierce) and dried again in a vacuum centrifuge.
Strong Cation Exchange Fractionation
The dried peptide sample was re-suspended in 150 µL of 10 mM ammonium formate, 25% acetonitrile, pH 3.0. Strong cation exchange (SCX) liquid chromatography was performed on an Ultimate 3000 HPLC system, Dionex (Sunnyvale, CA) utilizing a PolySulfoethyl A 1.0×150 mm (5-µm, 300-Å) column (Poly LC, Columbia, MD) at a flow rate of 50 µL/min. The mobile phases consisted of 10 mM ammonium formate (pH 3.0)/25% acetonitrile (solvent A), and 500 mM ammonium formate (pH 6.8)/25% acetonitrile (solvent B). After loading the sample onto the column, elution was maintained at 100% solvent A for 10 min. Peptides were then separated by using a gradient from 0–60% solvent B over 79 min, followed by a gradient of 60–100% solvent B over 10 min. The gradient was then held at 100% solvent B for 10 min. 109 vials were collected (1 min/fraction) from each run. The vials were combined into 5 fractions for each doublet SILAC sample and 20 fractions for each triplet SILAC sample. The final peptide concentration in each fractions was 0.1~1 µg/µL.
LC-MS/MS Analysis
LC-MS/MS analysis was performed on each pool using a NanoLC-2D LC system (Eksigent, Dublin, CA) and an LTQ-Orbitrap XL mass spectrometer equipped with a nanospray ionization source (Thermo Scientific, San Jose, CA). Digested peptides were loaded at 20 µL/min onto a 5 mm × 300 µm reverse phase C18 trap column (5 µm, 100Ǻ, Dionex, Sunnyvale, CA) and washed with 0.1% formic acid for 20 min. Peptides were moved from the trap column onto a 75 µm inner diameter PicoFrit®C18 analytical column with an integrated 15 µm emitter (New Objective, Woburn, MA) at 300 nL/min. The analytical column was eluted with a gradient of: 0–5% B for the first 5 min, 5–60% B for 65 min, 60–100% B for 20 min, 100-0% B for 10 min, and 0% B for the last 20 min (solvent A = 0.1% (vol/vol) formic acid in water, solvent B = 90% acetonitrile in 0.1% formic acid in water). The mass spectrometer was operated in the data-dependent mode to automatically switch between MS and MS/MS acquisition. Survey full scan MS spectra (from m/z 375 to 1,575) were acquired in the Orbitrap with resolution R=100,000. The most intense ions (up to five, depending on signal intensity) were sequentially isolated for fragmentation in the linear ion trap using collision induced dissociation (CID) with normalized collision energy of 30% and a target value of 5000. Former target ions selected for MS/MS were dynamically excluded for 75 s.
Peptide Identification
DeconMSn26 (http://omics.pnl.gov/software/) was used to determine and refine the monoisotopic mass and charge state of parent ions from the LTQ-Orbitrap raw data, and to create a peak list of these ions in .mgf format. The peak list contained the fragment information such as the MS/MS spectra, refined precursor ion and charge state. DtaRefinery28 (http://omics.pnl.gov/software/) was used to improve mass measurement errors for parent ions of tandem MS/MS data by modeling systematic errors based on putative peptide identifications using the algorithm as described. A script written in Python (programming language) was used to automate the process of generating .mgf files from raw data using DeconMSn and DtaRefinery. The resulting .mgf file was submitted to Mascot (version 2.2, Matrix Science, London, UK) database searching against i) a concatenated database containing 73,928 proteins from international protein index (IPI) database (version 3.52), ii) the commonly observed 262 contaminants (forward database), and iii) the reversed sequences of all proteins (reverse database). Carbamidomethylation was set as the fixed modification and oxidation of methionine was set as the variable modification. The initial mass deviation tolerance of precursor ion was set to 10 ppm and fragment ion tolerance was set to 0.5 Da. A maximum of 2 missed cleavages were allowed in peptide identification.
Peptide identification score given by the search engines and mass accuracy are two important parameters for calculating the FDR.14 We found that when the Mascot peptide identification score (a minimum of 10) is divided by the square root of the precursor ion mass error, these two parameters contribute equally to the final FDR. We have defined this new factor as the Quality of Peptide Identification (QPI). Here the Mascot peptide score representing a measure of confidence that the peptide identification assigned to the observed MS/MS spectrum of the candidate peptide. The total QPI of a peptide was taken as the sum of the QPI values for all MS/MS spectra that were matched to this peptide sequence. Identified peptides were sorted by a descending order of QPI values and a cutoff was applied to ensure a total FDR < 0.01.
Peptide and Protein Quantitation
Precursor ion intensity, measured in the high resolution full MS, was extracted by UNiquant and used as an abundance measurement for each identified peptide. The input files for quantitation were the MS raw data and the Mascot output .dat file. UNiquant can also utilize the search results from other database search engines, but the identified peptides have to be converted into tab delimited text files containing the filtered peptide sequence, identification score, scan number, observed m/z, and charge state information. The quantitation algorithm in UNiquant was first developed for hybrid-FT-MS instruments.14 Briefly, the two types of information generated in each LC-MS duty cycle, MS/MS spectra (fragment ions) and LC-FT MS high resolution full MS spectra (precursor ions), were combined via the accurate mass information. Theoretical molecular mass for a peptide (labeled or unlabeled) is calculated, and the search is performed within a range of the high resolution MS spectra. A histogram of mass differences between calculated and measured molecular masses is used to determine a useful mass accuracy matching range (< 20 ppm). The histogram maximization approach is applied to correct the mass calibration, which improves mass accuracy and confidence of identifications.27 Intensities of both unlabeled and labeled precursor ions are measured on the full MS spectrum for each identified peptide. The accuracy of the abundance measurement was reported to be highly correlated with the signal-to-noise (S/N) ratios.19 In order to ensure the quality of our quantitation result, a default S/N ratio cut-off for all peptide peaks was set at 2 and this value can be adjusted. The output file of UNiquant includes a list of peptides with refined m/z, mass error, S/N ratios, and intensities of light and heavy species.
Frequently, a given peptide appears more than once in the LC-MS/MS output. The spectra count is the number of times that a peptide identified by database search. The relative abundance of each identified peptide was calculated as the sum (based on spectra counts) of peak intensities (PI) for the heavy species of the peptide divided by the sum of intensities for the light species of the peptide:
| (Equation 1) |
where n is the spectra count for a specific peptide, PIH is the peak intensity of the heavy species and PIL is the peak intensity of the light species. Similarly, the relative abundance of each identified protein was calculated by dividing the sum of the intensities of all peptide heavy species for the protein by the sum of the intensities of all peptide light species.
Quantitation by MaxQuant and Distiller
The same raw data were processed by MaxQuant (version 1.0.13.13) and Distiller (version 2.2.1.2) using the same proteome database and the same searching parameters submitted to the Mascot engine. The identification procedure of MaxQuant was set without filtering for labeled amino acids, a peptide and protein FDR of 0.01, a threshold peptide posterior error probability (PEP) of 1, a minimal peptide length of 6, and a minimal ratio count for protein quantitation of 1. All the quantified ratios obtained from MaxQuant are the reported ratios before normalization.
For Distiller, the default processing options were applied for MS data processing. The peaks were un-centroided, re-gridded with 20 points per Da, and aggregated using the sum scan group method. The profile type of spectrum format was used with a minimum of one peak per scan with a maximum charge state of 3 MS/MS processing: peaks were un-centroided, re-gridded with 20 points per Da, formatted in profile form, and aggregated using the time domain method. A minimum of 10 peaks were required per spectrum with a maximum charge state of plus 2 and the precursor mass tolerance was 3 Da. Precursor charge state was determined directly from the parent ion scan, or from the raw data file information. If neither of these methods was successful the data were processed with both a plus 2 and a plus 3 charge state. Time domain: the minimum precursor mass was 700 Mr and the maximum 16,000 Mr with a tolerance of 1 m/z, a maximum intermediate time of 30 seconds and a maximum intermediate scan count of 5. Minimum number of scans was 1. MS peak picking: the filtering correlation threshold was 0.7 with a minimum S/N of 10, a minimum peak m/z of 50 and a maximum peak m/z of 100,000. The peak profile used a minimum peak width of 0.02 Da, an expected peak width of 0.2 Da, and a maximum peak width of 2 Da. The isotope distribution fit method was used with maximum peak iteration per scan of 500. MS/MS processing parameters were the same as MS peak picking. The quantified ratios are not normalized.
Results
Software Development
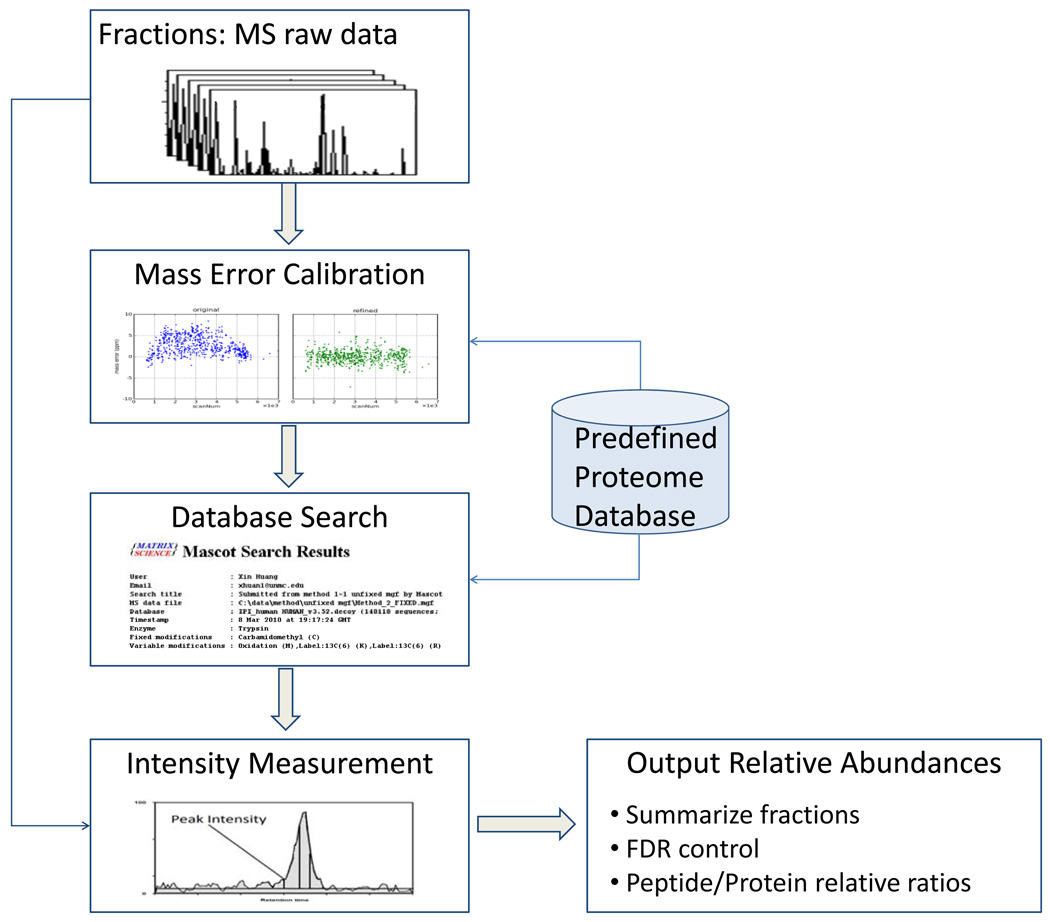
A schematic diagram of data analysis procedures in UNiquant is shown in Figure 1. The five major components in this data processing pipeline are the raw data deisotoping, mass error calibration, database search, peptide intensity measurement, and summary of quantitation results. Deisotoping of MS spectra, precursor ion mass correction and calibration were accomplished by DeconMSn and DtaRefinery, respectively. After a database search, the confident peptide identification results were filtered based on a specific FDR cutoff (Supplemental Figure 1) and the peptide intensities were measured and exported. The UNiquant software was developed on the platform of Microsoft .net framework. The programming language was Microsoft VB.net and C#. UNiquant was installed on an Intel dual-core PC with 3G RAM. For each LC-MS/MS dataset, the generation of mgf file with corrected precursor masses by DeconMSn and DTARefinery lasted about 1 hour. The mgf files were submitted to Mascot for peptide identification. After database searching, the extraction of peptide intensities and subsequent quantification procedures by UNiquant took about 10 minutes.
Figure 1.
Components and data flow in UNiquant. Five major components are involved in this data processing pipeline: deisotoping of raw data, mass error calibration, database search, peptide intensity measurement, and summary of quantitation results. The programs used to accomplish these tasks are DeconMSn, DtaRefinery, Mascot and UNiquant, respectively.
Identification of Peptide Pairs from Proteome Mixtures with Known Ratios
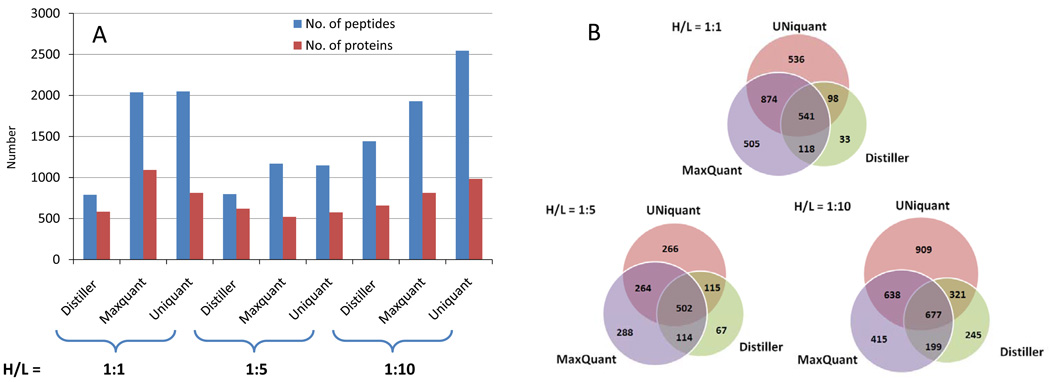
The performance of UNiquant was compared to MaxQuant and Distiller using the SILAC labeled proteome digests of Jeko-1 cells with known heavy/light (H/L) ratios (H/L = 1:1, 1:5, and 1:10). For identification of the peptides/proteins, we used the same database search engine (Mascot), with identical searching parameters, and searched the data against the same proteome database (IPI version 3.52). Using the same FDR cutoff of 0.01 (based on the decoy database approach), the number of peptide pairs and proteins being identified by each program is shown in Figure 2A. UNiquant and MaxQuant identified nearly equal numbers of peptide pairs in the H/L = 1:1 and 1:5 standard proteome mixtures, while Distiller identified only approximately 39% and 69% as many peptide pairs at the same mixture conditions, respectively. For the H/L = 1:10 standard proteome mixture, UNiquant identified 34% more peptide pairs and proteins than MaxQuant and 76% more than Distiller.
Figure 2.
(A) The number of peptides and proteins identified and quantified from complex proteome mixtures with known heavy/light (H/L) ratios (H/L = 1:1, 1:5, and 1:10) by Distiller, MaxQuant, and UNiquant, respectively. The H/L ratios of proteome mixtures are indicated at the bottom of the figure. (B) Venn diagram compares the number of peptides identified and quantified by Distiller, MaxQuant, and UNiquant, respectively. Three programs shows different coverage of standard proteome mixture in different mixtures (H/L = 1:1, 1:5, and 1:10).
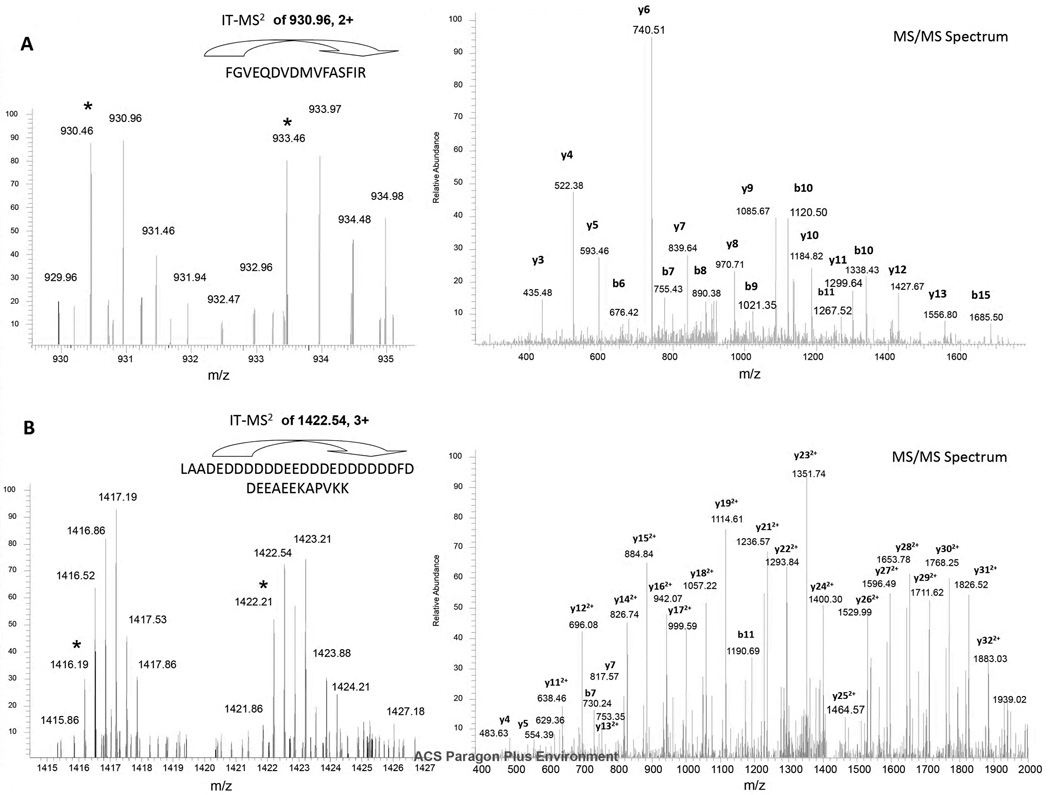
The three programs provided different proteome coverage of Jeko-1 cells (Figure 2B). In the H/L = 1:1 standard mixture, 541 peptide pairs were identified by MaxQuant, UNiquant, and Distiller. Three programs identified 2705 peptide pairs in total. There were only 33 peptide pairs exclusively identified by Distiller. However, 536 and 505 peptide pairs were identified by either UNiquant or MaxQuant, respectively. In the H/L = 1: 5 and 1:10 standard mixtures, 502 and 677 peptide pairs were identified by all three programs, respectively. Among the 536 peptide pairs exclusively identified by UNiquant from the H/L = 1:1 standard proteome mixture, two peptide pairs were illustrated. In Figure 3A, the abundance of the peptide pair in the full MS spectrum was relatively low (S/N ratio = 3.5), however, it was successfully identified and quantified by UNiquant pipeline as “FGVEQDVDMVFASFIR” with a good quality of the MS/MS spectrum. The SILAC doublet appeared at least 8 times in the full MS scans with low S/N ratios. Four MS/MS spectra derived from two full MS scans were collected for the precursor ions of the SILAC doublet. The H/L ratio was accurately quantified as 0.96 by UNiquant from the two MS full scans. Figure 3B indicates a large molecular weight peptide with two missed cleavage sites that was detected only by UNiquant. More than 8 isotopic peaks were seen in either light or heavy labeled peptide. The mass-shift was 18 Da (3 labeled amino acids) for this peptide pair. In this case, a non-lightest isotopic peak (m/z = 1422.54, charge = 3+) of the heavy precursors ions was selected as the precursor ions and successfully identified.
Figure 3.
Two examples of peptide pairs identified and quantified by UNiquant. (A) Identification of peptide “FGVEQDVDMVFASFIR”, with S/N = 3.5 and good quality of MS/MS spectrum. (B) Identification of peptide “LAADEDDDDDDEEDDDEDDDDDDFDDEEAEEKAPVKK”. This peptide contains two missed cleavage sites. The symbol * labels the monoisotopic peak for the light and heavy form of SILAC labeled peptide.
Quantification of Peptide Pairs from Proteome Mixtures with Known Ratios
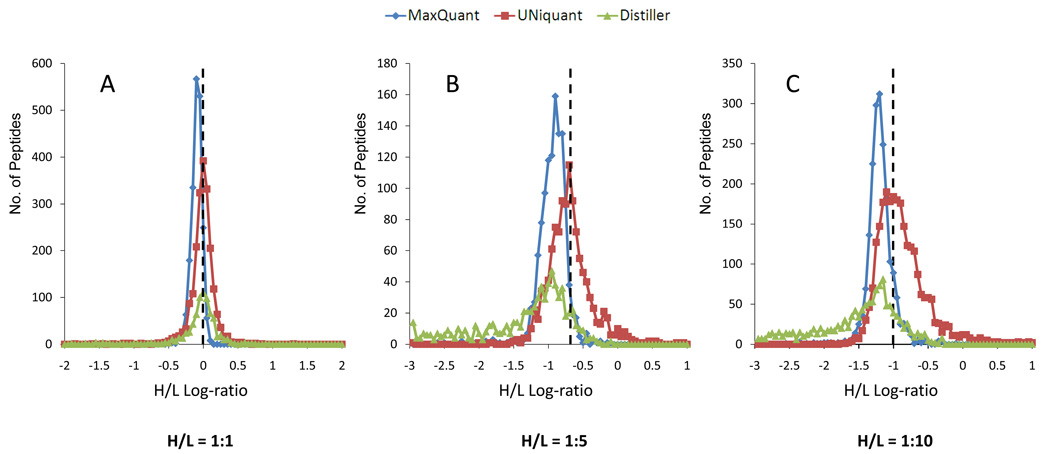
The distributions of peptides H/L log-ratio (log10 based) obtained by Distiller, Maxquant and UNiquant for three proteome mixtures with known ratios (H/L = 1:1, 1:5, and 1:10) without post-quantitation normalization were plotted. The true log-ratio of each proteome mixture with known H/L ratio is indicated as a dashed line in each panel of Figure 4. As shown, the median log-ratio of peptides quantified by UNiquant was generally equal to the true value of the log-ratio in each proteome mixtures. In the H/L = 1:1 proteome mixture, the median log-ratio of peptides quantified by UNiquant was −0.029, which is closer to the true log-ratio zero, compared to −0.042 and −0.113 obtained by Distiller and MaxQuant, respectively (Figure 4A). In the H/L = 1:5 and 1:10 proteome mixtures, the peptide log-ratio quantified by UNiquant and MaxQuant are generally Gaussian distributed, while the distributions quantified by Distiller were negatively skewed. Similarly, the median of peptides quantified by UNiquant was much closer to the true value than that obtained by the other two programs (Figure 4B and 4C).
Figure 4.
Distributions of the log-ratios (heavy/light, log10 based) for the peptides identified and quantified from complex proteome mixtures with known heavy/light (H/L) ratios (H/L = 1:1, 1:5, and 1:10) by Distiller (green), MaxQuant (blue), and UNiquant (red), respectively. The dashed vertical line in panels A, B and C indicates the true/expected log-ratios for the peptides identified and quantified from complex proteome mixtures with known heavy/light (H/L) ratios (H/L = 1:1, 1:5, and 1:10), respectively.
Application of UNiquant to Proteome Mixtures with Unknown Ratios
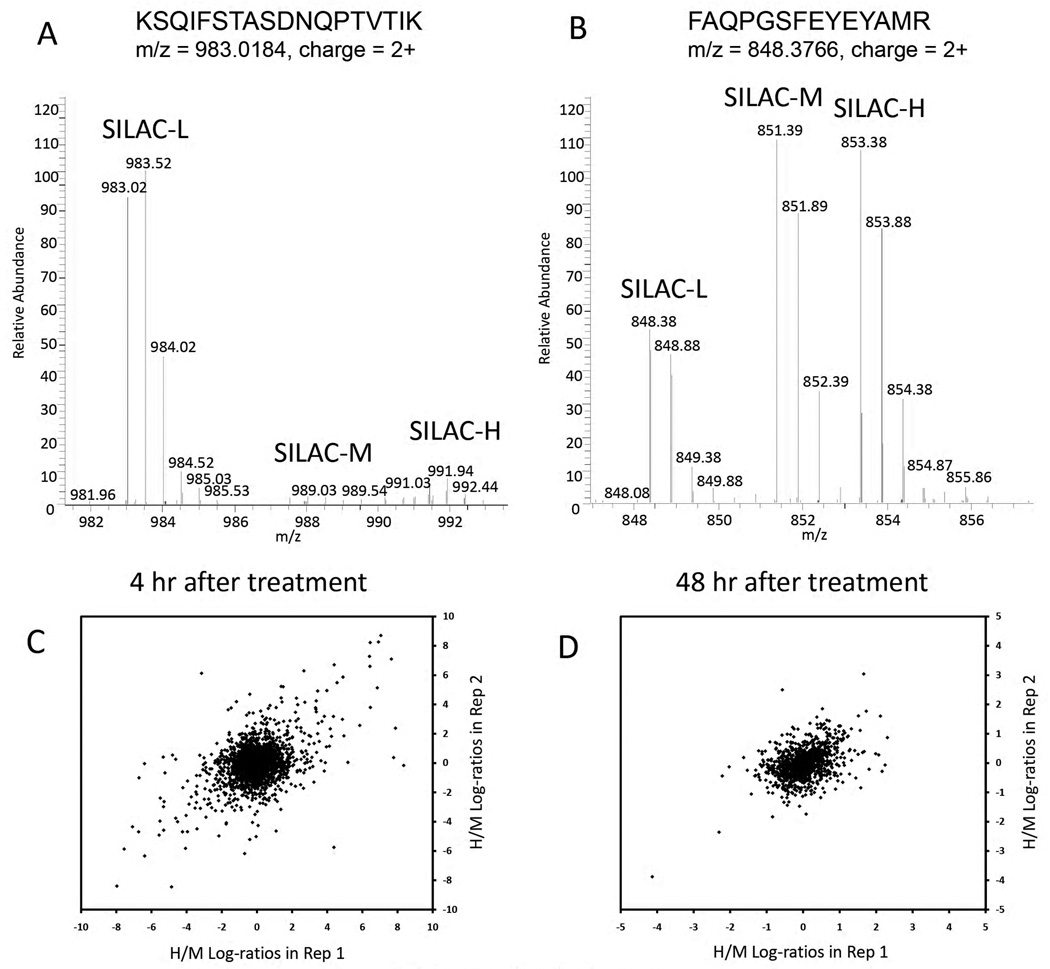
UNiquant was used to analyze newly synthesized proteins affected by microRNAs using the pSILAC technique31. Table 1 tabulates the number of peptides/proteins identified and quantified by UNiquant and MaxQuant from DHL16 cells after transfection of microRNA-155. After 4 hr of transfection, UNiquant quantified 1637 and 1602 proteins in the 2 replicates, respectively, while MaxQuant quantified 581 and 478 proteins. In this dataset, UNiquant quantified more than twice the number of proteins compared to MaxQuant. Two typical peptide triplets in the MS full scan of 4 hr and 48 hr datasets after microRNA treatment are displayed (Figure 5A and 5B), respectively. In the 48 hr dataset, peptide “FAQPGSFEYEYAMR” was quantified by both UNiquant and MaxQuant. However, the peptide “KSQIFSTASDNQPTVTIK” was only detected by UNiquant in the 4 hr dataset. As shown, there were fewer newly synthesized peptides (medium-heavy and heavy) 4 hr after microRNA transfection while 48 hr after transfection, there were more newly synthesized peptides than those remaining light species before treatment.
Table 1.
Number of peptides and proteins quantified by MaxQuant and UNiquant, respectively, in the triple SILAC dataset.
| MaxQuant | UNiquant | |||
|---|---|---|---|---|
| No. of peptides | No. of proteins | No. of peptides | No. of proteins | |
| 4 hr Rep1 | 1131 | 581 | 4799 | 1637 |
| 4 hr Rep2 | 854 | 478 | 4396 | 1602 |
| In common | 436 | 319 | 2421 | 1225 |
| 48 hr Rep1 | 1177 | 567 | 2422 | 954 |
| 48 hr Rep2 | 1543 | 730 | 2523 | 909 |
| In common | 537 | 386 | 1237 | 671 |
Figure 5.
(A–B) Typical full MS spectra of newly synthesized peptides and original peptides after (A) 4 hr and (B) 48 hr transfection of microRNA. The triplet peptide sequence, m/z, and charge status were indicated accordingly. The SILAC-L, SILAC-M, and SILAC-H peptides indicate the original peptides (light), newly synthesized peptides after microRNA transfection (medium-heavy), and newly synthesized peptides after GFP transfection (heavy). (C–D) Correlation of peptides H/M log-ratios (heavy/medium-heavy, log2 based) quantified by UNiquant for the 2 replicates in the (C) 4 hr and (D) 48 hr triple pSILAC dataset.
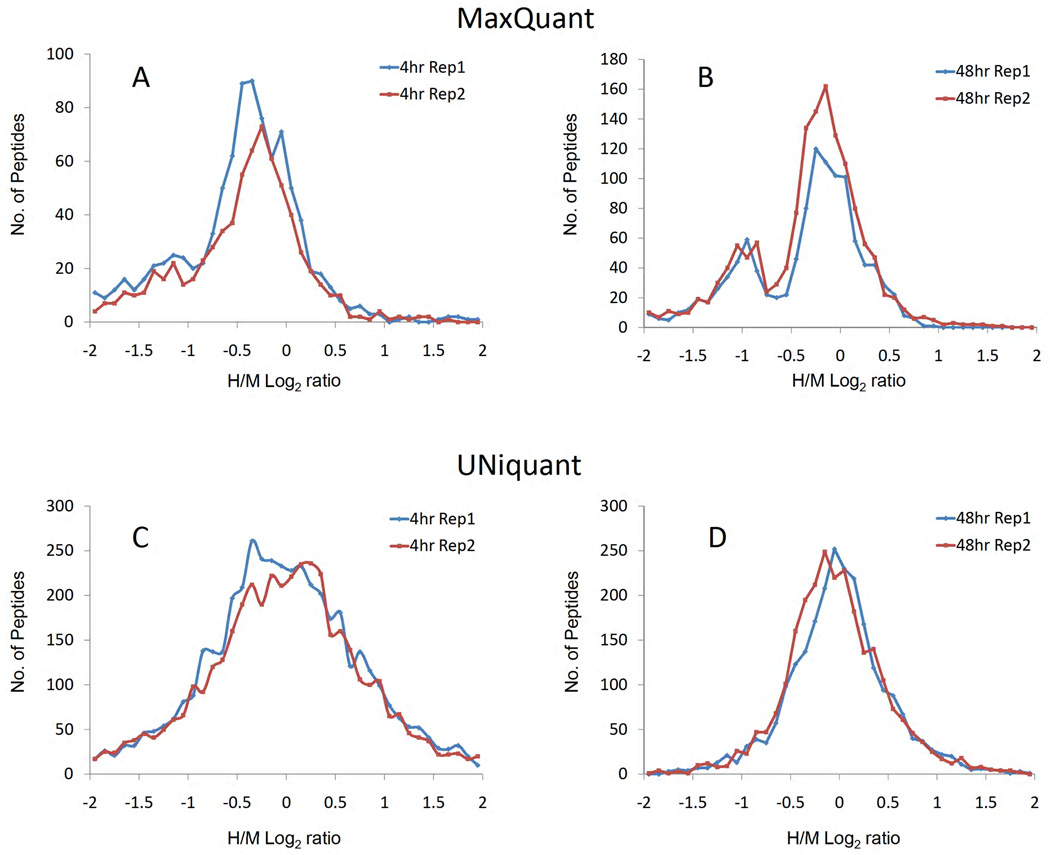
A scatter plot of the first versus second replicates was prepared for the heavy/medium-heavy (H/M) log-ratios (log2 based) of quantified peptides, and for both the 4 hr and 48 hr dataset, respectively. The log dynamic range of the 4 hr treatment dataset was less than 8.0, and the correlation between the two replicates was relatively high (Figure 5C, r = 0.455, p < 0.001). The log dynamic range of the 48 hr treatment was less than 2.0 and the two replicates were significantly correlated (Figure 5D, r = 0.435, p < 0.001). Moreover, the distributions of peptide log-ratios obtained by UNiquant and MaxQuant were compared. The log-ratio distributions quantified by MaxQuant were negatively skewed in both replicates in the 4 hr dataset (Figure 6A), while the distributions quantified by MaxQuant showed double-peak profiles in the 48 hr dataset (Figure 6B). All the log-ratios quantified by UNiquant displayed general Gaussian distributions (Figure 6C and 6D). The medians of peptide log-ratios quantified by MaxQuant were −0.499 and −0.472 for the 2 replicates in the 4 hr dataset, and −0.249 and −0.271 for the 2 replicates in the 48 hr datasets, respectively. The medians of peptide log-ratios quantified by UNiquant were −0.055 and −0.013 for the 2 replicates in the 4 hr dataset, and −0.015 and −0.061 in the 48 hr datasets, respectively.
Figure 6.
(A–B) are the distributions of the log-ratios (heavy/medium, log10 based) obtained by MaxQuant for the newly synthesized peptides after 4 hr (A) and 48 hr (B) transfection, respectively. (C–D) are the distributions of the log-ratios (heavy/medium, log10 based) obtained by UNiquant for the newly synthesized peptides after 4 hr (C) and 48 hr (D) transfection, respectively. The newly synthesized peptides after microRNA transfection and mock GFP transfection are labeled with SILAC-M (medium), and SILAC-H (heavy), respectively.
Discussion
Strategy for Peptide Pair Detection
In SIL-based quantification, information on peptide abundance is derived either from the intensity of the peptide precursor ion signal (ICAT, SILAC, 18O labeling), or from the intensity of report ion after MS/MS fragmentation (iTRAQ). UNiquant was designed to quantify the relative abundance based on the full MS spectrum. However, unlike isobaric labeling of peptides in iTRAQ (quantitation was based on MS/MS spectrum), the pair of precursor ions illustrates many more isotopic peaks in the full MS scans.7 In a data-dependent acquisition mode, tandem FT-MS instruments such as LTQ-FT or LTQ-Orbitrap capture a number of the most abundant precursor ions to perform MS/MS fragmentation. Therefore, the selected isotopic peaks for MS/MS fragmentation can be derived from either unlabeled (light species) or labeled (heavy species) peptides. UNiquant does not detect SIL-peptide pairs before identification. After database search, theoretical masses for both unlabeled and labeled species for each identified peptide were determined and intensities were calculated based on the confident identifications. This strategy was also applied by other programs such as Vista19. In contrast, MaxQuant uses an alternative strategy for peak pair detection, one which identifies pairs of light and heavy peptides from the MS data prior to peptide identification.18 Advantages of this strategy are that the resulting peak-list is much cleaner than the peak-list from the original raw data, the peaks have a high S/N ratio, and co-eluting peptides can be readily identified. However, this strategy may result in some loss of pairs, especially in the case of peptide pairs with low intensity or high noise background as seen in Figure 3 and Figure 5A. On hybrid high resolution mass spectrometry, the precursor scan is performed in an Orbitrap/FT mass analyzer and the MS/MS fragmentation is usually accomplished in the linear ion trap mass analyzer. Consequently, low signal levels in MS scans do not necessarily predict the quality of the fragmentation spectra and subsequent peptide identification. It is not surprising that signals with high S/N ratios and high intensity can be identified and quantified by all quantitative proteomics programs; however, low intensity MS signals does present a substantial challenge for the use of full MS spectra to quantify low-abundant proteins by various programs.19 Based on our observation, the intensity cutoff for peptide peak detection is different for the 3 programs we compared in this study. Maxquant has a higher intensity cutoff of quantified peptides (log10 intensity range: about 3~7) than UNiquant (log10 intensity range: 3~9) and Mascot Distiller (log10 intensity range: 4~9). In addition, we noticed that MaxQuant applied a filter of selecting 6 most intense fragment ions per 100 Da in the MS/MS spectra, whereas the DeconMSn module in UNiquant pipeline submitted the entire spectra of deisotoped fragment ions for database search. Finally, the way for calculating FDR is slightly different for three programs although all 3 programs use a decoy database approach. UNiquant used QPI, while Maxquant used a posterior error probability based on the peptide P-score distribution to set the cutoff of FDR.18, 32 All of these factors contribute by various degrees to the sensitivity of peptide pair detection and explain why the proteome coverage is different for the 3 programs (Figure 2) and why UNiquant quantifies more proteins and peptides in a sample where newly synthesized proteins are being analyzed in cells after miRNA treatment (Figure 6). Overall, we showed that UNiquant is designed to better meet the challenge of detecting and quantifying low abundant proteins.
Strategy for Protein Quantification
In UNiquant, once a peptide is identified, from one or multiple MS/MS spectra, the corresponding parent MS spectrum is found. The SIL-peptide pair is then quantified by the intensity of a pair of precursor ions which have passed a strict tolerance filter in the full MS spectrum. This filter confirms the identity of detected pair-wise peptides using three parameters: co-elution (same parent MS spectrum), high accuracy of predefined mass-shift (< 20ppm) and S/N threshold (>= 2). For the quantitation of a SIL-peptide pair with more than one MS/MS spectrum, intensities of heavy and light peaks are summed up separately from each corresponding full MS spectrum (Equation 1). This algorithm is equivalent to calculating the peak area from an extra ion chromatogram for the co-eluted pair of SIL-peptides, which was adopted in Census.16 However, our MS/MS-directed quantitation strategy avoids chromatogram alignment (which is highly dependent on the chromatography performance and the time-span of every full MS scan) and defining the co-elution window from a complex background. Carrillo et al compared several algorithms for combining peptide intensities to estimate relative protein abundance, and concluded that the sum of intensities method provided one of the most accurate estimates.33 In UNiquant, the sum of intensities method was used to calculate the peptide relative abundance from multiple identified MS/MS spectra, and protein relative abundance from multiple quantified peptides.
In quantitative proteomics data analysis, a normalization approach can be applied based on the assumption that the amounts of most proteins in the sample will be unchanged by the variable being tested. Thus, the averaged relative abundances of the proteins can be adjusted to one. In MaxQuant, the relative peptide/protein abundances before and after normalization are both provided.18 To accomplish this adjustment, a correction factor is then applied to all the proteins to obtain the modified log-ratios. However, in some cases, this normalization approach may not be applied. For instance, the use of phosphatase inhibitors will affect a broad range of cellular phosphorylation events34; thus the assumption of normalization is not valid if only the phosphoproteome was investigated. Furthermore, the assumption that the majority of proteins are unchanged might be incorrect when the specific treatment could affect a board range of protein concentration, such as transcription factor, microRNA, etc. Therefore, a quantitative proteomic solution for accurate relative protein abundance measurement is necessary. We looked at newly synthesized proteins affected by microRNA-155 using the pSILAC technique. Since all the proteins in the sample are unlabeled before microRNA transfection, the true ratio for the majority of the SILAC labeled proteins can’t equal one. In this study, quantitation by UNiquant suppresses the systematic errors in measurement and reduces the need for data normalization (Figure 4, Figure 6C and 6D).
Compatibility with existing programs and labeling methods
Another important issue for the quantitative proteomics software design is the compatibility with the existing programs. However, most of the currently used quantitation programs can only work with a specific or a limited number of MS instruments, database engines, and isotope labeling methods.21 In our pipeline, the UNiquant module reads a list of identified peptides containing the filtered peptide sequence, identification score, scan number, observed m/z, and charge state information, then searches back to the MS raw data to retrieve the intensity information. Finally, it outputs the relative abundance of identified peptides. Therefore, it can support the various outputs of database engines for peptide identification. Here we have demonstrated the performance of UNiquant based on the Mascot output, and UNiquant was applied to analyze the SEQUEST output as well. The next goal of UNiquant is to establish a platform-free application by supporting the extensible markup language (XML) formatted MS raw data (mzXML) and peptide identification data (pepXML). Furthermore, by defining the SIL context, UNiquant is capable of any precursor ion-based quantitation of relative protein abundance. In this paper, we have demonstrated two examples of UNiquant for the SILAC quantitative applications. Furthermore, we have applied UNiquant for quantitative proteomic analysis using trypsin-catalyzed 16O/18O labeling approach (data not shown).
Conclusion
In summary, we have developed a new software pipeline, named UNiquant, for SILAC-based quantitative proteomic data analysis. The MS/MS directed quantitation strategy employed by UNiquant allows for sensitive peptide pair identification and accurate quantitation without the need for post-measurement normalization of peptide ratios and chromatogram alignment. UNiquant is potentially compatible with a broad spectrum of SIL methods and database search engines.
Supplementary Material
Acknowledgement
We thank Dr. Lawrence Schopfer for the editing of this manuscript. Mass spectrometry data were obtained in the Mass Spectrometry & Proteomics Core Facility at the University of Nebraska Medical Center which is supported by the Nebraska Research Initiative. This work was financially supported by NIH U01 CA114778-03 (W.C.C), NEHHS LB606 (S.J.D); and X.H was supported by a scholarship from China Scholarship Council.
Abbreviations
- SIL
stable isotope labeling
- SILAC
stable isotope labeling of amino acids in cell culture
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- XIC
extracted ion chromatography
- ICAT
isotope-coded affinity tags
- iTRAQ
isobaric tag for relative and absolute quantitation
- FDR
false discovery rate
- FT
Fourier transform
- CID
collision induced dissociation
- QPI
quality of peptide identification
- H/L
heavy/light
- H/M
heavy/medium-heavy
- S/N
signal-to-noise
- PI
peak intensities
- PEP
posterior error probability
- IPI
international protein index
- XML
extensible markup language
References
- 1.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol. 2005;1(5):252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 2.Mann M, Kelleher NL. Precision proteomics: the case for high resolution and high mass accuracy. Proc Natl Acad Sci U S A. 2008;105(47):18132–18138. doi: 10.1073/pnas.0800788105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fang R, Elias DA, Monroe ME, Shen Y, McIntosh M, Wang P, Goddard CD, Callister SJ, Moore RJ, Gorby YA, Adkins JN, Fredrickson JK, Lipton MS, Smith RD. Differential label-free quantitative proteomic analysis of Shewanella oneidensis cultured under aerobic and suboxic conditions by accurate mass and time tag approach. Mol Cell Proteomics. 2006;5(4):714–725. doi: 10.1074/mcp.M500301-MCP200. [DOI] [PubMed] [Google Scholar]
- 4.Finney GL, Blackler AR, Hoopmann MR, Canterbury JD, Wu CC, MacCoss MJ. Label-free comparative analysis of proteomics mixtures using chromatographic alignment of high-resolution muLC-MS data. Anal Chem. 2008;80(4):961–971. doi: 10.1021/ac701649e. [DOI] [PubMed] [Google Scholar]
- 5.Gao BB, Stuart L, Feener EP. Label-free quantitative analysis of one-dimensional PAGE LC/MS/MS proteome: application on angiotensin II-stimulated smooth muscle cells secretome. Mol Cell Proteomics. 2008;7(12):2399–2409. doi: 10.1074/mcp.M800104-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geiger T, Cox J, Ostasiewicz P, Wisniewski JR, Mann M. Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat Methods. 2010;7(5):383–385. doi: 10.1038/nmeth.1446. [DOI] [PubMed] [Google Scholar]
- 7.Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7(12):952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 8.Qian WJ, Petritis BO, Kaushal A, Finnerty CC, Jeschke MG, Monroe ME, Moore RJ, Schepmoes AA, Xiao W, Moldawer LL, Davis RW, Tompkins RG, Herndon DN, Camp DG, Smith RD. Plasma Proteome Response to Severe Burn Injury Revealed by (18)O-Labeled "Universal" Reference-Based Quantitative Proteomics. J Proteome Res. 2010;9(9):4779–4789. doi: 10.1021/pr1005026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Washburn MP, Ulaszek R, Deciu C, Schieltz DM, Yates JR., 3rd Analysis of quantitative proteomic data generated via multidimensional protein identification technology. Anal Chem. 2002;74(7):1650–1657. doi: 10.1021/ac015704l. [DOI] [PubMed] [Google Scholar]
- 10.Fu C, Hu J, Liu T, Ago T, Sadoshima J, Li H. Quantitative analysis of redox-sensitive proteome with DIGE and ICAT. J Proteome Res. 2008;7(9):3789–3802. doi: 10.1021/pr800233r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pflieger D, Junger MA, Muller M, Rinner O, Lee H, Gehrig PM, Gstaiger M, Aebersold R. Quantitative proteomic analysis of protein complexes: concurrent identification of interactors and their state of phosphorylation. Mol Cell Proteomics. 2008;7(2):326–346. doi: 10.1074/mcp.M700282-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.de Hoog CL, Foster LJ, Mann M. RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell. 2004;117(5):649–662. doi: 10.1016/s0092-8674(04)00456-8. [DOI] [PubMed] [Google Scholar]
- 13.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131(1):58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 14.Ding SJ, Wang Y, Jacobs JM, Qian WJ, Yang F, Tolmachev AV, Du X, Wang W, Moore RJ, Monroe ME, Purvine SO, Waters K, Heibeck TH, Adkins JN, Camp DG, 2nd, Klemke RL, Smith RD. Quantitative phosphoproteome analysis of lysophosphatidic acid induced chemotaxis applying dual-step (18)O labeling coupled with immobilized metal-ion affinity chromatography. J Proteome Res. 2008;7(10):4215–4224. doi: 10.1021/pr7007785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li XJ, Zhang H, Ranish JA, Aebersold R. Automated statistical analysis of protein abundance ratios from data generated by stable-isotope dilution and tandem mass spectrometry. Anal Chem. 2003;75(23):6648–6657. doi: 10.1021/ac034633i. [DOI] [PubMed] [Google Scholar]
- 16.Park SK, Venable JD, Xu T, Yates JR., 3rd A quantitative analysis software tool for mass spectrometry-based proteomics. Nat Methods. 2008;5(4):319–322. doi: 10.1038/nmeth.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mortensen P, Gouw JW, Olsen JV, Ong SE, Rigbolt KT, Bunkenborg J, Cox J, Foster LJ, Heck AJ, Blagoev B, Andersen JS, Mann M. MSQuant, an open source platform for mass spectrometry-based quantitative proteomics. J Proteome Res. 2010;9(1):393–403. doi: 10.1021/pr900721e. [DOI] [PubMed] [Google Scholar]
- 18.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 19.Bakalarski CE, Elias JE, Villen J, Haas W, Gerber SA, Everley PA, Gygi SP. The impact of peptide abundance and dynamic range on stable-isotope-based quantitative proteomic analyses. J Proteome Res. 2008;7(11):4756–4765. doi: 10.1021/pr800333e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mo F, Mo Q, Chen Y, Goodlett DR, Hood L, Omenn GS, Li S, Lin B. WaveletQuant, an improved quantification software based on wavelet signal threshold denoising for labeled quantitative proteomic analysis. BMC Bioinformatics. 2010;11:219. doi: 10.1186/1471-2105-11-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mueller LN, Brusniak MY, Mani DR, Aebersold R. An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J Proteome Res. 2008;7(1):51–61. doi: 10.1021/pr700758r. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt A, Kellermann J, Lottspeich F. A novel strategy for quantitative proteomics using isotope-coded protein labels. Proteomics. 2005;5(1):4–15. doi: 10.1002/pmic.200400873. [DOI] [PubMed] [Google Scholar]
- 23.Haas W, Faherty BK, Gerber SA, Elias JE, Beausoleil SA, Bakalarski CE, Li X, Villen J, Gygi SP. Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol Cell Proteomics. 2006;5(7):1326–1337. doi: 10.1074/mcp.M500339-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Alves G, Wu WW, Wang G, Shen RF, Yu YK. Enhancing peptide identification confidence by combining search methods. J Proteome Res. 2008;7(8):3102–3113. doi: 10.1021/pr700798h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kall L, Storey JD, MacCoss MJ, Noble WS. Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J Proteome Res. 2008;7(1):29–34. doi: 10.1021/pr700600n. [DOI] [PubMed] [Google Scholar]
- 26.Mayampurath AM, Jaitly N, Purvine SO, Monroe ME, Auberry KJ, Adkins JN, Smith RD. DeconMSn: a software tool for accurate parent ion monoisotopic mass determination for tandem mass spectra. Bioinformatics. 2008;24(7):1021–1023. doi: 10.1093/bioinformatics/btn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tolmachev AV, Monroe ME, Jaitly N, Petyuk VA, Adkins JN, Smith RD. Mass measurement accuracy in analyses of highly complex mixtures based upon multidimensional recalibration. Anal Chem. 2006;78(24):8374–8385. doi: 10.1021/ac0606251. [DOI] [PubMed] [Google Scholar]
- 28.Petyuk VA, Jaitly N, Moore RJ, Ding J, Metz TO, Tang K, Monroe ME, Tolmachev AV, Adkins JN, Belov ME, Dabney AR, Qian WJ, Camp DG, 2nd, Smith RD. Elimination of systematic mass measurement errors in liquid chromatography-mass spectrometry based proteomics using regression models and a priori partial knowledge of the sample content. Anal Chem. 2008;80(3):693–706. doi: 10.1021/ac701863d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tolmachev AV, Monroe ME, Purvine SO, Moore RJ, Jaitly N, Adkins JN, Anderson GA, Smith RD. Characterization of strategies for obtaining confident identifications in bottom-up proteomics measurements using hybrid FTMS instruments. Anal Chem. 2008;80(22):8514–8525. doi: 10.1021/ac801376g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu T, Belov ME, Jaitly N, Qian WJ, Smith RD. Accurate mass measurements in proteomics. Chem Rev. 2007;107(8):3621–3653. doi: 10.1021/cr068288j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 32.Olsen JV, Mann M. Improved peptide identification in proteomics by two consecutive stages of mass spectrometric fragmentation. Proc Natl Acad Sci U S A. 2004;101(37):13417–13422. doi: 10.1073/pnas.0405549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carrillo B, Yanofsky C, Laboissiere S, Nadon R, Kearney RE. Methods for combining peptide intensities to estimate relative protein abundance. Bioinformatics. 2010;26(1):98–103. doi: 10.1093/bioinformatics/btp610. [DOI] [PubMed] [Google Scholar]
- 34.Pan C, Gnad F, Olsen JV, Mann M. Quantitative phosphoproteome analysis of a mouse liver cell line reveals specificity of phosphatase inhibitors. Proteomics. 2008;8(21):4534–4546. doi: 10.1002/pmic.200800105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.