Abstract
Levels of prostaglandin E2 and the prostaglandin-endoperoxide synthase-2 (PTGS2, or COX-2) increase in actively progressing periodontal lesions, but decrease in chronic disease. We hypothesized that chronic inflammation is associated with altered DNA methylation levels within the PTGS2 promoter, with effects on COX-2 mRNA expression. PTGS2 promoter methylation levels from periodontally inflamed gingival biopsies showed a 5.06-fold increase as compared with non-inflamed samples (p = 0.03), and the odds of methylation in a CpG site in the inflamed gingival group is 4.46 times higher than in the same site in the non-inflamed group (p = 0.016). The level of methylation at −458 bp was inversely associated with transcriptional levels of PTGS2 (RT-PCR) (p = 0.01). Analysis of the data suggests that, in chronically inflamed tissues, there is a hypermethylation pattern of the PTGS2 promoter in association with a lower level of PTGS2 transcription, consistent with a dampening of COX-2 expression in chronic periodontitis. These findings suggest that the chronic persistence of the biofilm and inflammation may be associated with epigenetic changes in local tissues at the biofilm-gingival interface.
Keywords: DNA methylation, PTGS2, CpG islands, periodontitis
INTRODUCTION
The production of prostaglandin E2 (PGE2) has been associated with periodontal inflammation (Goodson et al., 1974; Noguchi and Ishikawa, 2007). In periodontitis, increased levels of gingival crevicular fluid (GCF) PGE2 have been predictive of longitudinal periodontal attachment loss (Offenbacher et al., 1986), and associated with the clinical signs of bleeding on probing (Zhong et al., 2007), both of which would suggest that increased PGE2 expression is associated with progressive lesions (Champagne et al., 2003). The biosynthesis of PGE2 and other prostanoids is tightly coupled to the inducible expression of COX-2, and the transcriptional control of COX-2 levels appears to be the key regulatory gate for modulating tissue PGE2 levels (Noguchi et al., 2000). Although levels of PGE2 increase during certain stages of disease progression, little is known regarding the regulation of local PGE2 synthesis, in which some down-regulation must be needed to prevent a continued and ever-expansive loss of connective tissue. This homeostasis in chronic inflammation in the omnipresence of a microbial burden is probably due to the establishment of what has been referred as a “metastable” equilibrium (Feinberg and Tycko, 2004). This metastable equilibrium arises as the presence of a chronic inflammatory stimulus creates a new “set-point”, in which higher levels of inflammatory mediators are tolerated or down-regulated by some compensatory molecular mechanism(s) that prevent unrestricted tissue destruction and serve to dampen the uncontrolled inflammatory response.
Analysis of data in the literature provides some evidence of this down-regulatory mechanism. In a community study (Zhong et al., 2007), it was reported that the level of GCF PGE2 is negatively associated with attachment loss (as a marker of total history of disease activity), indicating that the greater the cumulative historical tissue damage, the lower the GCF-PGE2 level. Presumably, this is a result of a site-specific history of an episode of progression with increased attachment loss and elevations of local GCF PGE2 levels that eventually become dormant with lowered levels of PGE2.
Recently, we have found that certain periodontal bacteria can induce epigenetic alterations in host tissues, such as gene-specific methylation of CpG sequences (Bobetsis et al., 2007). In eukaryotes, DNA methylation occurs almost exclusively at the 5′ end of cytosine within the CpG dinucleotide context (Bird and Wolffe, 1999). It has been generally accepted that an increase of methylation in the gene promoter region is related to the decrease of gene expression, though exceptions have been identified (Kelavkar et al., 2007).
In this study, we sought to investigate the potential alteration in the DNA methylation pattern of the PTGS2 (COX2) gene promoter and its effect on the transcriptional control of COX-2. We also sought to identify potential feedback mechanisms that might lead to the suppression of PGE2 synthesis following periods of disease activity.
MATERIALS & METHODS
Participants and Tissue Specimens
In total, 16 participants, aged between 18 and 65 yrs, provided written informed consent and were enrolled into this study, which was approved by the Institutional Review Board of the University of North Carolina at Chapel Hill. Exclusion criteria included: (1) the use of either antibiotics or non-steroidal anti-inflammatory drugs within 1 mo prior to scheduled surgery; and (2) medical treatment for other diseases 3 mos prior to recruitment. Measurements included probing depths, clinical attachment level, and bleeding on probing at 6 sites per tooth. One interproximal gingival site was biopsied from each participant. Ten gingival biopsies were removed during routine periodontal flap surgeries from participants clinically diagnosed with chronic adult periodontitis. Those biopsied tissues were from sites with probing depths of 5 mm or more, bleeding on probing, and radiographic evidence of localized bone loss. These tissues are referred to as ‘inflamed’ in the data presentation. Non-inflamed tissues were collected from participants who were periodontally healthy or had localized mild gingivitis at non-study sites. Six non-inflamed gingival biopsies were removed from participants who were undergoing crown extension surgery at sites with probing depth measurements of 4 mm or less at all 4 inter-proximal probing sites and no bleeding on probing. Upon removal, gingival tissues were incubated with RNAlater ® (Applied Biosystems/Ambion, Austin, TX, USA) overnight at 4°C, decanted and transferred to a freezer at −80°C for storage.
DNA Preparation and Sodium Bisulfite Modification
Genomic DNA was isolated from gingival biopsies by the use of a DNeasy Tissue Kit (Qiagen Inc., Valencia, CA, USA). A 2-μg quantity of genomic DNA from each sample was treated with sodium bisulfite according to the method recommended (Grunau et al., 2001).
Bisulfite Specific PCR, Cloning, and Sequencing
The promoter sequence of PTGS2 (Appleby et al., 1994) was analyzed with MethPrimer software (Li and Dahiya, 2002). Two CpG islands encompassing -541 bp and -216 bp were identified. Sodium-bisulfite treated genomic DNA was amplified with primers that are specific to the CpG islands within the PTGS2 promoter. Primer sequences and PCR conditions are provided in the APPENDIX. A 334-bp PCR product including those 2 CpG islands was purified through electrophoresis, and the gel-purified PCR product was then cloned into a pGEM-T Easy vector (Promega, Madison, WI, USA). Colonies showing positive PCR fragment insertion were selected, and the insert was amplified with standard Sp6 and T7 primers (Promega) listed in the APPENDIX (including PCR conditions). PCR products from 4 to 7 clones for each individual gingival sample were sequenced at the UNC-CH genomic analysis facility.
RNA Isolation and Real-time PCR
Total RNA was isolated from RNAlater ®-treated gingival tissues with the use of an RNeasy Mini Kit (Qiagen). cDNA was then synthesized from 1 μg of total RNA with the Omniscript Kit (Qiagen) by random decamer primers (Applied Biosystems/Ambion). Real-time PCR was performed with 1 μL synthesized cDNA, TaqMan Universal PCR mix, and 20X on-demand primers (Applied Biosystems, Foster City, CA, USA) specific for the PTGS2 gene, in a 7000 Sequence Detection System apparatus (Applied Biosystems). Amplification of 18s rRNA from each gingival sample was included as internal control. The relative quantity of PTGS2 mRNA was calculated against 18S rRNA values (Livak and Schmittgen, 2001). Two samples, one from inflamed and one from non-inflamed gingival groups, did not provide enough RNA for analyses, and therefore were excluded from the quantitative RT-PCR study.
Statistical Analysis
Two-sample independent t tests were applied for the statistical analysis of clinical data and mRNA expression levels. We used the Mann-Whitney/Wilcoxon two-sample test (SAS v9.2) to compare overall percent methylation of each gingival sample between the 2 gingival groups. We used a generalized linear mixed model (GLMM) to estimate the odds ratio describing the relationship between methylation and inflamed vs. non-inflamed groups, conditional on CpG site and gingival tissue sample. Specifically, a three-level logistic regression model (Kim et al., 2006), with fixed effect for group and random effects for samples and sites within samples, was used. A 95% confidence interval for the odds ratio was constructed such that the observed difference between groups was assessed with respect to the variation between gingival tissue samples. We applied linear regression analysis to test for the significance of slope to evaluate the association between the percentage of methylation at a specific CpG dinucleotide (−458 bp) within the PTGS2 promoter region and the PTGS2 transcriptional level. We used chi-square approximation to test gender difference between participants in two groups. Alpha levels less than 0.05 were considered statistically significant.
RESULTS
Participants
There were no significant differences in age and gender between the two groups (Table). As expected, there were differences in mean probing depth, clinical attachment loss, and the presence of bone loss at inflamed sites as compared with sites in control individuals without inflammation.
Table.
Clinical Parameters of Study Participants
| Clinical Measurements | Inflamed (n = 10) | Non-inflamed (n = 6) |
|---|---|---|
| a Statistically significant difference by t test (p = 0.00001). Value reflects mean interproximal probing depth aggregated over 4 interproximal sites at the biopsy region. | ||
| b Statistically significant difference by t test (p = 0.01). Value reflects mean interproximal clinical attachment loss aggregated over 4 interproximal sites at the biopsy region. | ||
| Mean age (yrs) | 45.8 ± 7.4 | 44.2 ± 15.6 |
| Gender (Male/Female) | 5/5 | 1/5 |
| Mean probing depth (mm)a | 6.2 ± 0.6 | 2.5 ± 0.8 |
| Mean clinical attachment loss (mm)b | 3.8 ± 1.1 | 1.6 ± 1.1 |
| Alveolar bone loss | Yes | No |
Methylation Status of the CpG-rich PTGS2 Promoter Region
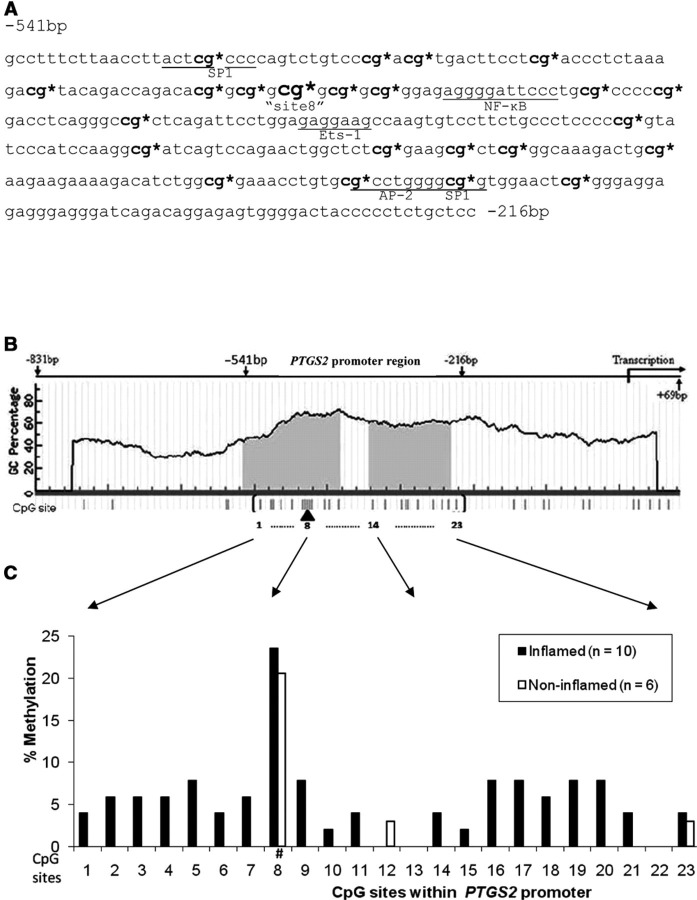
The overall methylation of the studied PTGS2 promoter region (−541 bp ~ −216 bp) in chronically inflamed gingival tissues was 5.06-fold higher than the methylation level exhibited in non-inflamed gingival tissues when the methylation levels of individual gingival tissue in both groups were compared [4.3% (1.8%, 8.5%) vs. 0.85% (0.7%, 1.3%), respectively, shown as median and interquartile range (IQR), p = 0.03] (Fig. 1). There are 23 CpG dinucleotides (Fig. 2A) present in 2 CpG islands (shaded area, Fig. 2B) within the upstream sequences of PTGS2 (−831 bp to +69 bp, Fig. 2B). Several transcription factor binding sequences (cis-elements), such as NF-κB, AP-2, and Sp-1 (Fig. 2A), were also identified within these CpG-rich regions. Therefore, we analyzed the methylation state of all 23 CpG sites within those regions to compare methylation levels between the two gingival sample groups. The individual clonal bisulfite sequencing map for the 16 participants appears in the APPENDIX Fig. It can be seen that the increased methylation shown in inflamed tissues extends over almost all of the 23 potential methylation sites analyzed (Fig. 2C). However, this diffuse methylation pattern was absent from the non-inflamed samples, in which methylation could be detected at only 3 CpG sites. In addition, the odds of methylation at a CpG site from a sample in the inflamed gingival group was 4.46 times higher (95% CI: 1.38, 14.35) than the odds of methylation at the same CpG site from a sample in the non-inflamed gingival group (p = 0.016), as estimated by the GLMM.
Figure 1.
PTGS2 promoter region (−541 bp ~ −216 bp) DNA methylation levels for both groups of gingival samples. The methylation level in chronic inflamed periodontitis samples [4.3% (1.8%, 8.5%)] was significantly higher than that in non-inflamed samples [0.85% (0.7%, 1.3%), *p = 0.03], as determined by the Mann-Whitney/Wilcoxon two-sample test. Box plot shows the median (center line in box), 25% quartile (bottom line in box), 75% quartile (top line in box), maximum (plus error bar), and minimum (minus error bar).
Figure 2.
Methylation level of individual CpG site within the promoter region of PTGS2. (A) Genomic sequence of PTGS2 promoter region ranging from −541 bp to −216 bp is presented. Sites for potential transcriptional factor binding are underlined. 23 CpG potential methylation sites are present in this region and marked with “*”. The 8th CpG (−458 bp) site is also indicated in a larger font size. (B) Diagrammatic representation of the PTGS2 gene promoter from −831 bp to +69 bp is shown. The shaded areas indicate CpG islands in this promoter region. The vertical bars underneath represent CpG potential methylation sites, with 23 sites within the CpG island in parentheses. (C) The methylation level of each individual CpG site within the identified CpG islands from inflamed periodontitis biopsies is compared with that in the non-inflamed samples. “Site 8” is marked with “#”.
Interestingly, the most heavily methylated site in both gingival tissue groups occurred at what we are designating as “site 8” (−458 bp, Fig. 2C), a CpG dinucleotide that is physically close to an NF-κB binding site (Fig. 2A). The methylation level of that particular CpG site was 23.5% in inflamed and 20.6% in non-inflamed gingival tissues, and higher than the methylation levels of other CpG dinucleotides in both groups (Fig. 2C).
CpG Methylation Status as Related to PTGS2 mRNA Expression Level
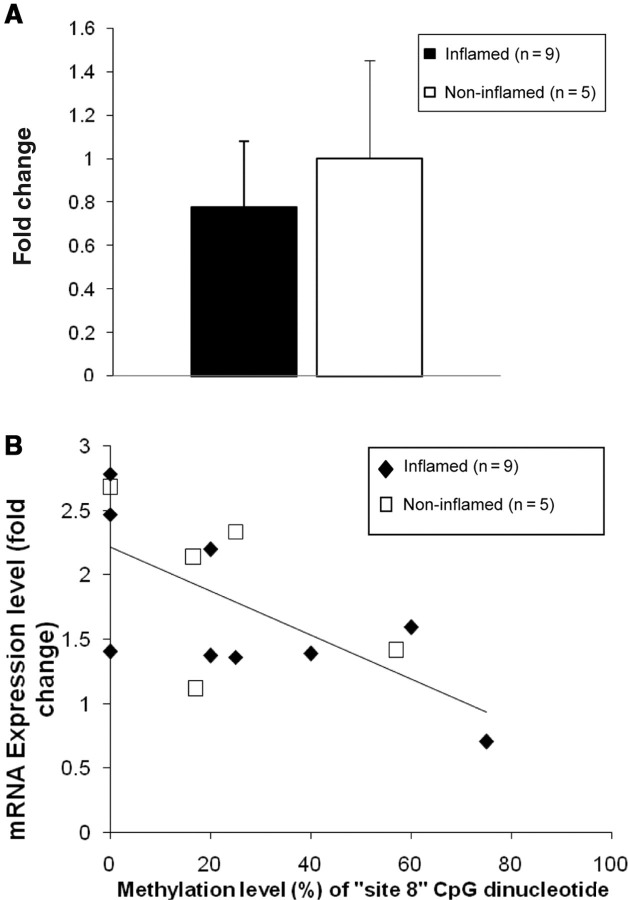
The mRNA level of PTGS2 in inflamed gingival samples, as determined by real-time PCR, was lower than the level in the non-inflamed group, although the difference was not significant (p = 0.36, Fig. 3A). We then plotted the percentage methylation level of “site 8” against the mRNA expression level of each individual sample. Samples from both gingival tissue groups are shown on this plot to demonstrate the association between methylation status and mRNA expression. The overall regression analysis pooling all clinical samples shows a statistically significant negative association between percentage of methylation of the CpG site at −458 bp and PTGS2 mRNA expression (p = 0.01, Fig. 3B).
Figure 3.
Transcriptional levels of PTGS2, determined by quantitative RT-PCR, from the inflamed and non-inflamed gingival samples. (A) mRNA expression of PTGS2 in the inflamed gingival group showed lower, yet non-statistically different (p = 0.36), levels as compared with non-inflamed gingival tissues. (B) The PTGS2 mRNA expression level of individual samples from both groups is inversely related to its methylation level of “site 8” CpG dinucleotides. Regression analysis indicates statistical significance (p = 0.01).
DISCUSSION
Although aging and environmental exposures can affect global genome methylation, new evidence indicates that some promoter methylation sites can be targeted by specific toxins, nutrient deficiencies, and infectious stimuli to modify methylation levels (Vanselow et al., 2006; Wasson et al., 2006). In this investigation, we report that chronically inflamed periodontal tissues exhibited an increased (5.06-fold) generalized methylation of the CpG-rich region of the PTGS2 promoter, as compared with non-inflamed periodontal tissues. In a previous study with a mouse model of Campylobacter rectus infection, we identified a hypermethylated Igf2 P0 promoter region, suggesting, for the first time, that infection with an oral pathogen can lead to epigenetic modifications (Bobetsis et al., 2007). The association between infection and alteration of DNA methylation is also supported by another study in an in vitro infection model (Yao et al., 2006), which reported that the promoter region of hMLH1 from a gastric cell line was hypermethylated by persistent Helicobacter pylori infection. Therefore, our findings are consistent with the concept that infections at mucosal surfaces can modify the epigenetic status of the tissues in a gene-specific manner. The role of inflammation in modulating methylation status, either by altering the density of cells at local sites of inflammatory infiltration, or by modifying the methylation status of existing cells, cannot be determined from these experiments. Furthermore, the intra-clonal differences of methylation status within the same gingival tissue sample (e.g., Appendix Fig.; participant 8 had 10 methylated sites in one clone, but none in 4 other clones) may reflect the epigenetic impact imposed by infection/inflammation on different cell types present in the biopsy samples.
The increase in methylation in chronic disease was associated with a metastable steady-state level of PTGS2 mRNA expression that was lower than that seen in non-inflamed participants with shallow sites. This finding is consistent with the report (Zhong et al., 2007) that shows lower PGE2 levels in deeper sites. It has been suggested (Beck et al., 1997) that most of the periodontal disease progression that occurs within participants likely involves shallow sites rather than deep sites. In this context, the chronic state of deep pockets reflects a historical episode of disease activity and a re-instatement of a new steady-state equilibrium, resulting in a metastable shift in COX-2 expression.
The site-specific methylation of the CpG dinucleotide at site −458 bp (“site 8”), which is located 12 bp upstream of an NF-κB binding site, was higher in both gingival groups. Since NF-κB activation enhances PTGS2 expression (Newton et al., 1997), the observed increase in methylation at this specific site may impair NF-κB activation. It has been reported (Kelavkar et al., 2007) that transcription can be critically affected by the methylation status of specific or “key” CpG sites within a regulatory region. In the present study, the methylated cytosine of this CpG dinucleotide, just upstream of the NF-κB binding site, may possibly either directly exclude the binding by this transcriptional activator or condense local chromatin structure by recruiting methyl CpG binding proteins (Bird and Wolffe, 1999; Vanselow et al., 2006).
One limitation of this report is the relatively small number of participants studied. Relationships between methylation status and clinical status are tenuous and should be confirmed in larger studies. However, this study provides the first evidence and proof-of-principle that epigenetic modifications of local tissues may occur in periodontal disease. Additional studies will be needed to understand whether epigenetic changes also occur in gingivitis or in other mucosal pathologies. Although we demonstrated that promoter methylation is linked to decreased mRNA expression, we do not have direct evidence that this is associated with altered levels of COX-2 protein expression or levels of PGE2 within the tissue. However, once the PTGS2 mRNA is translated into protein, the enzyme is unstable, having a short biological half-life (Perkins and Kniss, 1997). COX-2 does not exist in a zymogen form or as a pre-existing mRNA pool. Thus, the transcription of PTGS2 mRNA directly results in the synthesis of inducible COX-2 and appears to be the key regulatory gate for modulating tissue COX-2 activity.
Our working model of how epigenetic modification may affect periodontal status is based upon: (1) how changes in DNA methylation patterns alter gene expression profiles; (2) the fact that epigenetic changes are not readily reversible and are retained following cell division, creating a sustained change in gene expression and tissue phenotype that would persist, even following the reduction in the inflammatory infiltrate; (3) the fact that epigenetic alterations may induce tissue tolerance to the chronic stress imposed by the biofilm; and (4) the fact that epigenetic alterations may influence wound healing and the dynamics of biofilm emergence. This suggests that epigenetic modifications may result in long-lived alterations in the metastable state of the local periodontal tissues.
Supplementary Material
Acknowledgments
This work was supported by the University of North Carolina General Clinical Research Center (grant RR-00046, now UL1RR025747).
This study provides the first evidence and proof-of-principle that epigenetic modifications of local tissues may occur in periodontal disease.
Footnotes
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
REFERENCES
- Appleby SB, Ristimäki A, Neilson K, Narko K, Hla T -1994-. Structure of the human cyclo-oxygenase-2 gene. Biochem J 302-Pt 3-:723–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck JD, Sharp T, Koch GG, Offenbacher S -1997-. A study of attachment loss patterns in survivor teeth at 18 months, 36 months and 5 years in community-dwelling older adults. J Periodontal Res 32:497–505. [DOI] [PubMed] [Google Scholar]
- Bird AP, Wolffe AP -1999-. Methylation-induced repression—belts, braces, and chromatin. Cell 99:451–454. [DOI] [PubMed] [Google Scholar]
- Bobetsis YA, Barros SP, Lin DM, Weidman JR, Dolinoy DC, Jirtle RL, et al -2007-. Bacterial infection promotes DNA hypermethylation. J Dent Res 86:169–174. [DOI] [PubMed] [Google Scholar]
- Champagne CM, Buchanan W, Reddy MS, Preisser JS, Beck JD, Offenbacher S -2003-. Potential for gingival crevice fluid measures as predictors of risk for periodontal diseases. Periodontol 2000 31:167–180. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Tycko B -2004-. The history of cancer epigenetics. Nat Rev Cancer 4:143–153. [DOI] [PubMed] [Google Scholar]
- Goodson JM, Dewhirst FE, Brunetti A -1974-. Prostaglandin E2 levels and human periodontal disease. Prostaglandins 6:81–85. [DOI] [PubMed] [Google Scholar]
- Grunau C, Clark SJ, Rosenthal A -2001-. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res 29:E65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelavkar UP, Harya NS, Hutzley J, Bacich DJ, Monzon FA, Chandran U, et al -2007-. DNA methylation paradigm shift: 15-lipoxygenase-1 upregulation in prostatic intraepithelial neoplasia and prostate cancer by atypical promoter hypermethylation. Prostaglandins Other Lipid Mediat 82: 185–197. [DOI] [PubMed] [Google Scholar]
- Kim H, Preisser J, Rozier G, Valiyaparambil J -2006-. Multi-level analysis of group-randomized trials with binary outcomes. Community Dent Oral Epidemiol 34:241–251. [DOI] [PubMed] [Google Scholar]
- Li LC, Dahiya R -2002-. MethPrimer: designing primers for methylation PCRs. Bioinformatics 18:1427–1431. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD -2001-. Analysis of relative gene expression data using real-time quantitative PCR and the 2--Delta Delta C-T-- method. Methods 25:402–408. [DOI] [PubMed] [Google Scholar]
- Newton R, Kuitert LM, Bergmann M, Adcock IM, Barnes PJ -1997-. Evidence for involvement of NF-kappaB in the transcriptional control of COX-2 gene expression by IL-1beta. Biochem Biophys Res Commun 237:28–32. [DOI] [PubMed] [Google Scholar]
- Noguchi K, Ishikawa I -2007-. The roles of cyclooxygenase-2 and prostaglandin E2 in periodontal disease. Periodontol 2000 43:85–101. [DOI] [PubMed] [Google Scholar]
- Noguchi K, Yanai M, Shitashige M, Nishihara T, Ishikawa I -2000-. Cyclooxygenase-2-dependent prostaglandin production by peripheral blood monocytes stimulated with lipopolysaccharides isolated from periodontopathogenic bacteria. J Periodontol 71:1575–1582. [DOI] [PubMed] [Google Scholar]
- Offenbacher S, Odle BM, Van Dyke TE -1986-. The use of crevicular fluid prostaglandin E2 levels as a predictor of periodontal attachment loss. J Periodontal Res 21:101–112. [DOI] [PubMed] [Google Scholar]
- Perkins DJ, Kniss DA -1997-. Rapid and transient induction of cyclo-oxygenase 2 by epidermal growth factor in human amnion-derived WISH cells. Biochem J 321-Pt 3-:677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanselow J, Yang W, Herrmann J, Zerbe H, Schuberth HJ, Petzl W, et al -2006-. DNA-remethylation around a STAT5-binding enhancer in the alphaS1-casein promoter is associated with abrupt shutdown of alphaS1-casein synthesis during acute mastitis. J Mol Endocrinol 37:463–477. [DOI] [PubMed] [Google Scholar]
- Wasson GR, McGlynn AP, McNulty H, O’Reilly SL, McKelvey-Martin VJ, McKerr G, et al -2006-. Global DNA and p53 region-specific hypomethylation in human colonic cells is induced by folate depletion and reversed by folate supplementation. J Nutr 136: 2748–2753. [DOI] [PubMed] [Google Scholar]
- Yao Y, Tao H, Park DI, Sepulveda JL, Sepulveda AR -2006-. Demonstration and characterization of mutations induced by Helicobacter pylori organisms in gastric epithelial cells. Helicobacter 11:272–286. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Slade GD, Beck JD, Offenbacher S -2007-. Gingival crevicular fluid interleukin-1beta, prostaglandin E2 and periodontal status in a community population. J Clin Periodontol 34:285–293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.