Abstract
The means by which various microevolutionary processes have acted in the past to produce patterns of cranial variation that characterize modern humans is not thoroughly understood. Applying a microevolutionary framework, within- and among-population variance/covariance (V/CV) structure was compared for several functional and developmental modules of the skull across a worldwide sample of modern humans. V/CV patterns in the basicranium, temporal bone, and face are proportional within and among groups, which is consistent with a hypothesis of neutral evolution; however, mandibular morphology deviated from this pattern. Degree of intergroup similarity in facial, temporal bone, and mandibular morphology is significantly correlated with geographic distance; however, much of the variance remains unexplained. These findings provide insight into the evolutionary history of modern human cranial variation by identifying signatures of genetic drift, gene flow, and migration and set the stage for inferences regarding selective pressures that early humans encountered since their initial migrations around the world.
1. Introduction
Recent studies have revealed that human cranial morphology, whether quantified using absolute linear dimensions or relative geometric morphometric techniques, largely reflects population history among humans [1–9]. Certain functional and developmental modules (FDMs) of the cranium have been identified as reflecting relationships based on neutral molecular markers [4–9]. In particular, patterns of variation in the 3D morphology of the temporal bone, upper face, basicranium, individual bones of the cranial vault [5–9], and to a lesser degree the mandible [10] have been demonstrated to reflect genetic distances among human populations. The assumption behind these findings is that the morphology of these FDMs is evolving largely neutrally, in a similar manner to neutral molecular loci, and can consequently be utilized in a similar manner to estimate genetic relationships among human specimens or samples in the absence of direct molecular data. The relationship between cranial morphology and genetic distances in humans indirectly suggests that human cranial morphology is evolving primarily neutrally; however, the influence of selection has not been explicitly tested and therefore cannot be definitively discounted. Alternative methods exist for directly testing the impact of microevolutionary processes on morphology, through the use of microevolutionary modeling.
The patterns of phenotypic diversity within a species are central to inferring its modes of evolutionary diversification. Lande's quantitative approach to evolutionary theory can be applied to assess the relative effects of genetic drift and selection in a sample [11]. Lande's model is based upon the formula: Bt = G(t/Ne), in which Bt is the between-population V/CV matrix during generation t, G is the additive V/CV matrix, and Ne is the effective population size [11]. For phenotypic data from contemporaneous groups, G is typically proportional to the within-group morphological V/CV matrix (W) [12–15]. Therefore, W can be substituted for G such that B ∝ W(t/Ne). Within a contemporaneous comparison, t and Ne will be constants, so the equation can be simplified further to B ∝ W. In other words, if populations have diversified primarily through neutral evolutionary processes, then their within-group variance/covariance (V/CV) matrices should be proportional to their between-group V/CV matrices [11, 16, 17]. Significant deviations from this pattern indicate that selection has impacted the morphology of a particular region, and the null hypothesis of neutrality is rejected [11, 16, 17]. This approach has been taken to evaluate the role of genetic drift in the evolution of cranial shape in New World Monkeys [18–21] and fossil hominins [22, 23]. Weaver and colleagues applied statistical tests based on Lande's model to human and Neandertal linear cranial dimensions and concluded that morphological divergence between the two species was consistent with a null hypothesis of drift [23]. However, to date, Lande's model has not been applied to the comparison of landmark-based cranial data among individual human populations with the purpose of assessing the impacts of selection and drift.
Several genetic models, including the Isolation by Distance (IBD) model [24–26] and the serial founder effect model [27, 28], predict that genetic divergence among demes increases with geographic distance between them. Therefore, one means of investigating the extent to which morphological diversity has been influenced by neutral or selective forces is to compare human craniometric variation to geographic distance. Human cranial morphology, as captured by linear dimensions, generally has an association with genetic distance on a global scale [2, 4]. However, Relethford [2] notes that the accordance of data to an IBD model does not definitively clarify the evolutionary history of the phenotype. Environmental conditions also grade geographically, and morphological clines can be adaptive. Thus, adherence of a phenotype to a cline does not conclusively indicate that neutral factors are dictating its variation.
In population genetics, adaptation is often identified by first accounting for variation that falls within the potential range of neutral evolution (e.g., [29–31]). This tenet derives from the Neutral Theory of Molecular Evolution, which argues that the majority of genetic variation is likely to be selectively neutral [29–31]. According to the Neutral Theory, the rate of divergence among groups will be determined by the mutation rate (μ), which is neutral [29]. Mutation generates new variation, and genetic drift acts upon it. There are two primary models in population genetics that describe neutral divergence patterns among groups [32–34]. According to the Constant Heritability (CH) model, the degree of heritability (h2) of a character dictates the rate of neutral divergence among groups [32, 33]. Under the Mutation-drift Equilibrium (MDE) model, the expected genetic variance is calculated as 2Neσm2, in which Ne is the effective population size and σm is the morphological variance. A population is considered to be in mutation-drift equilibrium when the rate of new mutations (causing new variation) is balanced by genetic drift (causing homozygosity). Therefore, morphological characters will reflect population history if populations approximate an equilibrium between mutation and genetic drift (MDE model), and/or new mutations impact the V/CV structure minimally (CH model). According to both models, the divergence rate among groups is determined by the strength of genetic drift, which is in turn dictated by Ne and h2.
Approaching the evolution of modern human cranial morphology from a population genetics framework, such as through the application of molecular-based models and microevolutionary modeling, provides a basis for understanding the patterns and variation that characterize humans today. Using an assumption of neutrality as the primary mode of microevolution unless demonstrated otherwise allows cranial form to be evaluated objectively, and deviations from the pattern expected under a neutral model can be investigated further and adaptive explanations sought out. This research expands upon previous studies investigating patterns of cranial morphology in modern humans, interpreting them in the context of geographic dispersion and migration, and revealing the microevolutionary processes which produced the variation in cranial shape observed among our species today.
2. Methods
2.1. Data Collection
Fifty-two landmarks capturing the shape of the basicranium and splanchnocranium were digitized in samples of sixteen modern human populations (Table 1). The three-dimensional coordinates of each landmark were recorded using a Microscribe G2 point digitizer (Immersion Corp.) and sent to a laptop computer. These landmarks are standard craniometric landmarks [35–37] and were selected for the current study following findings by Smith et al. [6, 7] that the morphology depicted by each set of landmarks overlying is significantly correlated with genetic distances in humans (Table 2). The landmarks were divided into subsets representing four different FDMs of the skull: the basicranium, upper face, mandible, and temporal bone. FDMs are portions of a morphological structure, in this case the skull, that share a common biomechanical function and/or developmental pathway.
Table 1.
Human population samples included in the present study, with sample sizes and museum locations. AMNH, American Museum of Natural History, NMNH, National Museum of Natural History, BNHM, British Natural History Museum. *Subset of populations included in the analysis of mandibular morphology.
| Population | Region | n | Museum locations |
|---|---|---|---|
| Cameroon* | Africa | 44 | AMNH |
| Khoisan* | Africa | 43 | AMNH, BNHM |
| Pare | Africa | 27 | AMNH |
| French | Europe | 50 | NMNH, BNHM |
| Russians | Europe | 36 | AMNH, NMNH |
| Han Chinese* | East Asia | 50 | AMNH, NMNH |
| Malay | East Asia | 55 | AMNH, NMNH |
| Japanese* | East Asia | 40 | BNHM, NMNH |
| Mongolian* | East Asia | 44 | AMNH |
| Siberian natives* | East Asia | 51 | AMNH, NMNH |
| Southern Indians* | South Asia | 50 | AMNH |
| Australian aborigines* | Oceania | 48 | AMNH, NMNH |
| Papua New Guineans* | Oceania | 34 | AMNH |
| Solomon Islanders | Oceania | 31 | AMNH, NMNH |
| Greenland Inuit* | North America | 43 | AMNH, NMNH |
| Mexican Indians | North America | 44 | NMNH |
Table 2.
Cranial landmarks included in each functional and developmental module.
| Landmark | Description |
|---|---|
| Basicranium | |
| Basion | Midline point on the anterior margin of the foramen magnum |
| Condylar foramen | The posterior point on the margin of the condylar foramen |
| Condyle anterior | Most anterior points on the occipital condyles |
| Condyle posterior | Most posterior points on the occipital condyles |
| Inferior nuchal | Midline point on the inferior nuchal line |
| Inion | Most posterior point on the external occipital protuberance |
| Mastoidale | Most inferior point on the mastoid process |
| Opisthion | Midline point at the posterior margin of the foramen magnum |
| Temporal Bone | |
| Anterior articular | Most anterior point on the articular surface of the articular eminence |
| Auriculare | A point on the lateral aspect of the root of the zygomatic process at the deepest incurvature |
| Entoglenoid | Most inferior point on the entoglenoid process |
| Jugular | Most lateral point of the jugular fossa |
| Lateral eminence | Point on the center of the lateral margin of the articular surface of the articular eminence |
| Lateral ovale | Most lateral point on the margin of the foramen ovale |
| Mandibular fossa | Deepest point within the mandibular fossa |
| Mastoidale | Center of the inferior tip of the mastoid process |
| Medial articular | Most inferior point on medial margin of articular surface of the articular eminence |
| Petrous apex | Apex of petrous part of the temporal bone |
| Porion | Most superior point of the external auditory meatus |
| Postglenoid | Most inferior point on the postglenoid process |
| Supraglenoid gutter |
Point of inflection, where the braincase curves laterally into the supraglenoid gutter, in the coronal plane of the mandibular fossa |
| Tympanic | Most inferolateral point on the tympanic element of the temporal |
| Zygion | Most lateral point on the zygomatic arch |
| Face | |
| Dacryon | Point on the medial orbit at which the frontal, lacrimal, and maxilla intersect |
| Ectoconchonion | The intersection of the most anterior surface of the lateral border of the orbit and a line bisecting the orbit along its long axis |
| Frontomalare Temporale |
Most laterally positioned point on the frontozygomatic suture |
| Glabella | Most anterior midline point on the frontal bone |
| Nasion | Point of intersection between the frontonasal suture and midsagittal plane |
| Orbitale | The lowest point on the margin of the orbit |
| Mandible | |
| Condylion laterale |
Most lateral point on the mandibular condyle |
| Coronoid process | Most superior point on the coronoid process of the mandible |
| Gnathion | Most inferior midline point on the mandible |
| Gonion | A point along the rounded posteroinferior corner of the mandible between the ramus and the body |
| Infradentale | Midline point at superior tip of the septum between the mandibular central incisors |
| M1-M2 contact | Projected (laterally) onto the alveolar margin |
| Mandibular notch | Most inferior point in the mandibular notch |
| Mesial P3 | Most mesial point on mandibular P3 alveolus, projected onto the alveolar margin |
| Pogonion | Most anterior point on the mental eminence |
The analyses of the basicranium, upper face, and temporal bone consisted of all 16 populations. However, due to the fragmentary nature of museum collections, many specimens were found to be missing mandibles. Consequently, the number of populations with adequate sample sizes to be included in the mandibular morphology analysis was reduced compared to the other analyses. Ten of the sixteen populations contained a sufficient number of mandibular specimens (Table 1); however, this reduced number is equivalent to those used in several other similar studies [4, 5, 7]. Sex determination was conducted by the author for each specimen, using traditional sexually dimorphic cranial traits [35], and approximately equal sex samples were then utilized for each population. Each population sample consisted exclusively of adult individuals, as indicated by the eruption of the third molars and/or fusion of the spheno-occipital synchondrosis. An approximation of the average geographic coordinates for each population was obtained using Google Earth.
2.2. Geometric Morphometric Analyses
Morphological coordinate data for each FDM were superimposed separately using Generalized Procrustes Analyses [38–41] in MorphoJ 1.02 [42]. During this step, all specimens were scaled, rotated, and translated, such that any remaining variation among them could be attributed directly to shape differences. Principal Components Analyses (PCA) were subsequently conducted, and Principal Component (PC) scores obtained. Mahalanobis D2 distances between pairs of populations were calculated based on the morphology of each FDM. In the context of geometric morphometric analyses of cranial morphology, D2 distances have been argued to be especially appropriate for estimating distances among groups due to their shared properties with the molecular distances with which they should covary if the morphology is evolving primarily neutrally (e.g., [43]). Patterns of morphological distances were also visualized using Multidimensional Scaling (MDS) plots, in which the relationships indicated by a distance matrix are forced onto three dimensions.
2.3. Geographic Comparisons
Geographic distances between each pair of populations were calculated from their approximate average geographic coordinates using great circle distances, a haversine formula in which the distance between two points (D) is D = 2R arctan(√havθ/√(1 − havθ)), in which θ is the central angle between the two points and R is the radius of the Earth. Additionally, following Ramachandran et al. [44], distance calculations included the consideration of migratory waypoints, geographic points through which early migrating humans would have likely passed in order to travel between continents. In accordance with Ramachandran et al. [44], the following waypoints were used: Anadyr, Russia (64N, 117E), Cairo, Egypt (30N, 31E), Istanbul, Turkey (41N, 28E), Phnom Penh, Cambodia (11N, 104E), and Prince Rupert, Canada (54N, 130W). This adjustment in geographic distance calculation for intercontinental distances generates a more realistic estimate of the actual amount of migratory distance between two human populations, taking potentially insurmountable geographic barriers such as large bodies of water into consideration.
In order to compare the statistical association between geographic distance and morphological distance, a Regression Analysis was conducted separately between the great circle distances for each pair of populations and the Mahalanobis distances based on the 3D morphology of each of the FDMs. The alpha level was set at α = 0.05 for significance, and the slope (β) was used to infer the degree of continuity between the variables. The R2 value was interpreted as the degree of morphological variation explained by geographic patterning. Geographic distances were also visualized using a Multidimensional Scaling (MDS) plot in order to compare the distribution of populations to those based on morphology.
2.4. Lande's Method
The likelihood that the patterns of cranial variation observed in humans today can be explained by genetic drift can be assessed by comparing among- and within-population V/CV matrices [16]. The V/CV matrices for each FDM were calculated in Statistica 9.1 using a Multiple Analysis of Variance (MANOVA) in which the morphological traits were treated as dependent variables and the populations as independent variables. Each V/CV matrix was subsequently simplified into its Principal Components for comparative purposes. Following Ackermann and Cheverud [19], PC scores were calculated for each population mean by multiplying population means by the standardized eigenvectors: Y = XE, in which Y is the PC score of a population mean, X is the matrix of population means, and E is the matrix of standardized eigenvectors. For each PC, the among-population variance was calculated as the variance among the population mean PC scores.
If the morphology of an FDM has diversified primarily through genetic drift, then Lande's model predicts that the between-group variation will be proportional to the within-group variation (B ∝ W). Here, this model is applied by comparing the within-population variances to the among-population variances. A Regression Analysis was conducted between the within- and among-population variances. For each FDM, a slope of β = 1.0 is consistent with the hypothesis of drift, while any significant deviations from 1.0 are suggestive of nonneutral patterns of microevolution.
As an additional test of whether the covariance structure was similar among and within populations, a Mantel test [45, 46] of these covariance matrices for each FDM was conducted in PopTools, an add-on for Microsoft Excel. FDMs, found to have significant correlations (P < .05), were interpreted as having similar covariances among and within populations. It should be noted, however, that this test considers only the lower diagonal of a matrix, and therefore the variances within each variable are not incorporated nor tested. As a result, it should not be considered a direct test of Lande's method but instead a means of further investigating covariance structure.
Alternate methods for investigating the proportionality of between- and among-group variance/covariance structure exist. Most notably, Flury [47] proposed a hierarchical method of testing for common principal components (CPCs) among matrices, which has been employed as a test for Lande's model [48, 49]. However, it has been noted that this approach may generate spurious results in analyses involving large samples, since even small deviations in biological variation can appear statistically significant and cause the null hypothesis of neutrality to be rejected [18, 20]. Therefore, that approach was not employed here.
3. Results
3.1. Morphological Variation among Populations and Geographic Distance Comparisons
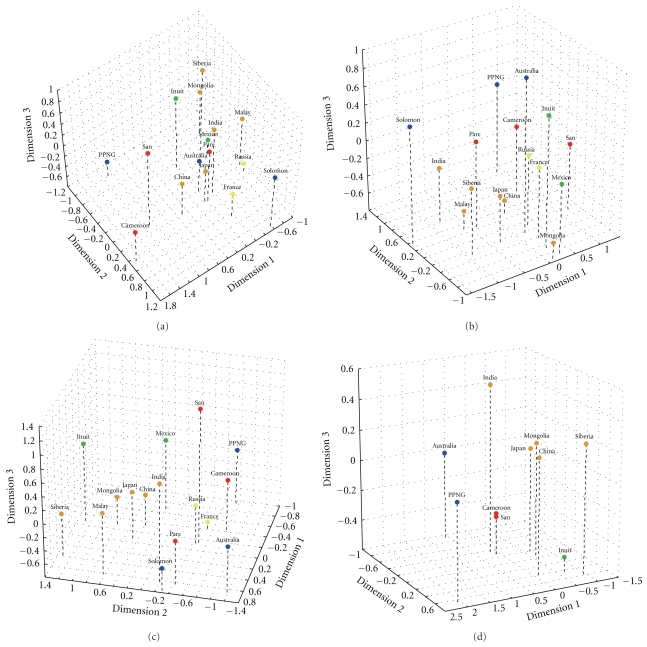
The matrices and multidimensional scaling plots of Mahalanobis D2 distances among populations indicated variation in the degree of similarity among groups in the morphology of the various FDMs (Figure 1). In general, most populations grouped roughly according to continent (Figure 1); however, the various continental groups exhibited differing degrees of dispersion and overlap with other groups.
Figure 1.
Multidimensional scaling (MDS) plot of Mahalanobis D2 distances among populations based on three-dimensional morphology of the: (a) basicranium, (b) temporal bone, (c) face, and (d) mandible. All plots use the following color scheme: Africa = red; Asia = orange; North America = green; Europe = yellow; Oceania = blue.
The three populations from Oceania (Australians, Papuans, and Solomon Islanders) were highly divergent in the morphology of the temporal bone, and basicranium as a whole. In fact, the D2 distances among the Oceanic populations were among the highest recovered for these FDMs. Despite some geographic patterning, the MDS plots revealed overlap between continental groups in basicranial and temporal bone morphology, and the African and Oceanic populations did not cluster together (Figures 1(a) and 1(b)).
The D2 distances based on facial morphology suggested some shared structure between the populations from Africa and those from Oceania (Figure 1(c)). The East Asian populations grouped together, as did the European samples. The New World groups were similar along two of the three dimensions, but the Inuit showed the greatest morphological affinity to the native Siberians.
With regard to mandibular morphology, the African populations clustered together, as did the East Asian populations (Figure 1(d)). However, the Oceanic populations were again widely divergent both from each other and from their Pacific neighbors in East Asia. The Inuit fell near the East Asian samples in dimensions 1 and 2 but exhibited a negative score along dimension 3.
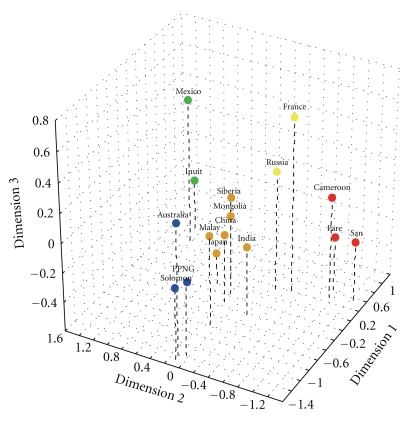
The great circle distances including waypoints among populations indicated the migratory distances required for each group to migrate to the center of the other's average geographic location (Table 3, Figure 2). The Regression Analyses between great circle distances and morphological distances revealed variation in the relationship between geography and shape for each FDM (Table 4). Morphological D2 distances based on the basicranium and mandible had moderate (R = 0.322 and 0.358, resp.) and significant (P = .0003 and .0156, resp.) correlations with great circle geographic distances. The morphological patterns of temporal bone shape were significant (P = .0367) but low (R = 0.191). Basicranial D2 distances, on the other hand, were nonsignificant (R = 0.110, P = .152). The R2 values of all comparisons were low (0.012–0.128).
Table 3.
Matrix of great circle distances using waypoints among human populations included in this study.
| Australia | Cameroon | China | France | India | Inuit | Japan | Malay | Mexico | Mongolia | Pare | PPNG | Russia | San | Siberia | Solomon | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Australia | 0 | 17337 | 8962 | 16434 | 9121 | 21859 | 13208 | 5276 | 20621 | 10226 | 17913 | 3384 | 13494 | 19758 | 12571 | 3644 |
| Cameroon | 17337 | 0 | 10026 | 6740 | 8462 | 20012 | 12767 | 12053 | 18774 | 9705 | 3110 | 16563 | 6367 | 3058 | 10732 | 17540 |
| China | 8962 | 10026 | 0 | 8611 | 3646 | 13233 | 3053 | 3678 | 11995 | 1264 | 10602 | 8188 | 5380 | 12447 | 3708 | 9165 |
| France | 16434 | 6740 | 8611 | 0 | 7871 | 17722 | 11034 | 11150 | 16484 | 8028 | 7316 | 15660 | 2691 | 9161 | 8609 | 16637 |
| India | 9121 | 8462 | 3646 | 7871 | 0 | 16703 | 6517 | 3837 | 15465 | 4496 | 9038 | 8347 | 5856 | 10883 | 7000 | 9324 |
| Inuit | 21859 | 20012 | 13233 | 17722 | 16703 | 0 | 11595 | 16575 | 6304 | 12211 | 20588 | 21085 | 13789 | 22433 | 9742 | 22062 |
| Japan | 13208 | 12767 | 3053 | 11034 | 6517 | 11595 | 0 | 7924 | 10357 | 3062 | 13343 | 12434 | 7355 | 15188 | 3279 | 13411 |
| Malay | 5276 | 12053 | 3678 | 11150 | 3837 | 16575 | 7924 | 0 | 15337 | 4942 | 12629 | 4662 | 8210 | 14474 | 7287 | 5650 |
| Mexico | 20621 | 18774 | 11995 | 16484 | 15465 | 6304 | 10357 | 15337 | 0 | 10973 | 19350 | 19847 | 12551 | 21195 | 8504 | 20824 |
| Mongolia | 10226 | 9705 | 1264 | 8028 | 4496 | 12211 | 3062 | 4942 | 10973 | 0 | 10281 | 9452 | 4519 | 12126 | 2554 | 10429 |
| Pare | 17913 | 3110 | 10602 | 7316 | 9038 | 20588 | 13343 | 12629 | 19350 | 10281 | 0 | 17139 | 6943 | 2699 | 11308 | 18116 |
| PPNG | 11702 | 16563 | 8188 | 15660 | 8347 | 21085 | 12434 | 4662 | 19847 | 9452 | 17139 | 0 | 12720 | 18984 | 11797 | 989 |
| Russia | 16061 | 6367 | 5380 | 2691 | 5856 | 13789 | 7355 | 8210 | 12551 | 4519 | 6943 | 12720 | 0 | 8788 | 4692 | 13697 |
| San | 19758 | 3058 | 12447 | 9161 | 10883 | 22433 | 15188 | 14474 | 21195 | 12126 | 2699 | 18984 | 15561 | 0 | 13153 | 19961 |
| Siberia | 12571 | 10732 | 3708 | 8609 | 7000 | 9742 | 3279 | 7287 | 8504 | 2554 | 11308 | 11797 | 4692 | 13153 | 0 | 12774 |
| Solomon | 3644 | 17540 | 9165 | 16626 | 9324 | 22062 | 13411 | 5650 | 20824 | 10345 | 18116 | 989 | 13697 | 20274 | 12774 | 0 |
Figure 2.
Multidimensional scaling (MDS) plot of geographic great circle distances among populations, incorporating waypoints. Geographic regions are depicted as follows: Africa = red; Asia = orange; North America = green; Europe = yellow; Oceania = blue.
Table 4.
Results of Regression Analysis of geographic great circle distances and Mahalanobis D2 distances based on the morphology of each functional and developmental module (FDM). Significant correlations are indicated in bold.
| FDM | R | R2 | P value |
|---|---|---|---|
| Basicranium versus geography | 0.110 | 0.012 | .1516 |
| Temporal bone versus geography | 0.191 | 0.037 | .0367 |
| Face versus geography | 0.322 | 0.103 | .0003 |
| Mandible versus geography | 0.358 | 0.128 | .0156 |
3.2. Lande's Model
The Regression Analysis of the PCs representing within- and among-population V/CVs indicated differences in the associations between these factors for the various FDMs (Table 5). For three of the four FDMs evaluated—the basicranium, face, and temporal bone—the slope was not significantly different than 1.0 (Table 5). The R2 values for these three FDMs indicated that a substantial proportion of variation was shared (86.5–95.6%) between the within- and among-population variances.
Table 5.
Results of Regression Analysis of within- and among-population variance/covariance (V/CV) matrices. Significant correlations are indicated in bold.
| FDM | β | 95% confidence | R2 | P value |
|---|---|---|---|---|
| Basicranium | 0.960 | 0.798–1.122 | 0.895 | .0096 |
| Face | 0.995 | 0.926–1.065 | 0.956 | .0048 |
| Mandible | 0.746 | 0.275–1.217 | 0.335 | .2539 |
| Temporal bone | 0.940 | 0.801–1.079 | 0.865 | <.0001 |
For mandibular morphology, however, a different pattern emerged. The slope of the regression equation was β = 0.746, although the correlation between the two variables was nonsignificant (P = .254) and the 95% confidence interval ranged widely from β = 0.275–1.217. The R2 value was 0.335 (Table 5).
The Mantel tests of within- and among-population covariances revealed highly significant correlations (P < .001) between the patterns for all four FDMs (Table 6).
Table 6.
Results of Mantel test comparing among- and within-population covariances. All correlations were significant.
| FDM | R | P value |
|---|---|---|
| Basicranium | 0.998 | <.001 |
| Face | 0.999 | <.001 |
| Mandible | 0.868 | <.001 |
| Temporal bone | 0.999 | <.001 |
4. Discussion
FDMs of the skull known to reflect population history in humans are generally assumed to be evolving primarily through neutral microevolutionary processes, such as genetic drift, gene flow, and mutation. As such, their patterns of variation behave very much like neutral molecular loci in that variation should accumulate at a relatively constant rate and grade geographically along clines. However, cranial morphology is affected by a number of complex and varied influences, including functional constraints and pressures of the masticatory apparatus, remodeling of osseous tissue, and indirectly through climate and diet, in ways that neutral genetic markers are not. As such, it is perhaps unreasonable to expect any functional aspect of morphology to behave according to a strict molecular model; however, the application of this framework can provide a starting point for identifying microevolutionary signatures that can be subsequently explored further.
The application of Lande's model [16] to subsets of cranial data indicated that the null hypothesis of predominantly neutral evolution cannot be rejected for the basicranium, temporal bone, and upper face, cranial regions which have been found previously to reflect population history [5–8]. The 3D shape of the temporal bone and face also track geography, with populations located in greatest proximity to each other sharing the most morphological affinities. However, despite the significance of these correlations, their R2 values are low, suggesting that much of the variance is unexplained by this factor alone.
Basicranial morphology was found to have a nonsignificant relationship with great circle distances. One possible explanation for this result is that, while this FDM is generally evolving primarily neutrally across humans as a species, a few populations may still be differentially affected by selection. In fact, this FDM contains some potentially adaptive aspects of morphology, in particular with respect to climate. Three samples in this study could be characterized as “cold-adapted,” the Inuit, Siberian natives, and, to a lesser degree, the Mongolians. These three groups cluster together on the MDS plot (Figure 1(a)), indicating affinities in basicranial morphology, as captured by the current set of landmarks. It has been noted that extremely cold climates may have a disproportionate effect on the basicranial morphology of populations exposed to them, causing them to differ from expected patterns of morphology [6, 46]. The functional pressures of a diet of tough and partially frozen items and paramasticatory behaviors may result in adaptive and plastic responses in the morphology of the masticatory apparatus, including the temporomandibular joint (TMJ), as these factors result in extensive stress on the masticatory apparatus [48–52]. Ethnographic literature has documented the Inuit practice of using the anterior dentition as a “third hand” [53], contributing to additional stresses to be accommodated by the skull. The skeletal structure of the Inuit skull has been modeled as a series of adaptations to generate and dissipate powerful occlusal forces, one of which involves a thickened tympanic plate [48, 49]. These factors have also been documented to affect some aspects of facial morphology; however, the facial landmarks utilized here specifically avoid the oral cavity and its associated alveolar bone. The extent of cold adaption in the basicranial morphology of the populations under consideration in the present study is difficult to tease apart definitively from geographic distance or population history, however, because the current cold-adapted populations are located comparatively closely to each other and share a fairly recent common ancestry [44]. Future studies incorporating cold-adapted samples from more distantly related groups, such as those from northern Europe, could shed additional light on this topic by separating out the effects of extreme climate in high latitudes from genetic relatedness.
One apparent deviation from geographic patterning in morphology is the widely divergent shape of the basicranium, temporal bone, and face among the three Oceanic populations. While these groups are located within a reasonably circumscribed geographic area (Australia, Papua New Guinea, and Solomon Islands), they are quite morphologically distinct. However, this is not surprising given that they are separated from each other by large bodies of water, which could have hindered the degree of possible gene flow among these groups by necessitating watercraft travel between islands. Additionally, several studies have suggested that Melanesia was likely colonized multiple times [54–58] and that speakers of the Papuan and Oceanic language groups have maintained a substantial degree of differentiation [54–56, 58]. Geographic distance is frequently used as a proxy for genetic distance, but, in cases of multiple colonization events of a region by genetically differentiated groups, this assumption may be unfounded. Friedlaender and colleagues found that molecular distances for populations within Melanesia are substantially higher than a worldwide average and show essentially no relationship with geographic distances [58]. Similarly, molecular studies have revealed substantial genetic delineation between Australian indigenous peoples and Papuans, which has been interpreted as separate colonization events from different ancestral stock [54–56]. In the case of basicranial and temporal bone microevolution, it is clear that simple geographic distances cannot explain the patterns of morphology among Oceanic groups, just as it does not explain their genetic relationships.
Genetic drift is rejected as the predominant mechanism influencing mandibular shape in Homo sapiens. This finding is consistent with previous studies that have attempted to approach this question in an alternate manner [7, 10]. Interpretations as to the utility of mandibular morphology for reconstructing population history have been varied and depend upon which populations are evaluated and the type of molecular data used for comparison. Smith [7] found that the shape of the mandible in nine human populations did not significantly correspond with their genetic distances based on short tandem repeat (STR) data; however, in a comparison to molecular distances based on Alu insertion polymorphisms and using a smaller number of populations, the association became statistically significant. Additionally, Nicholson and Harvati [10] found that mandibular shape tracks loosely with geography, and that functional demands such as diet and climate also play a role. These studies highlight the complexity of the patterns of mandibular morphology currently present in our species and the variety of microevolutionary processes that likely contributed to its current distribution.
Overall, the shape of the human skull, whether quantified using linear measurements or three-dimensional landmarks, reflects population history to a large degree [1–9]. Much of the microevolutionary history of this region, in particular the temporal bone and upper face, has involved largely neutral mechanisms and therefore reflects population history. However, simple geographic distance models [24–28] cannot explain the extent of the variation observed in individual FDMs, and specific functional aspects of the skull, such as the masticatory system, require selective explanations and interpretations. A population genetics framework provides a starting point for inferring the multicausal mechanisms of human cranial evolution, understanding these processes and their impacts, and retracing human evolution.
The subtext of many studies investigating human cranial evolution goes beyond understanding how the morphological variation has evolved. In the absence of molecular data in the paleoanthropological record, many researchers wish to identify phylogenetically informative aspects of morphology that can be used as a proxy for genetic data to address questions about hominin phylogenetic relationships and relative genetic distances among individual hominin specimens. Research into the relationship between cranial morphology and genetic relationships in humans is a crucial step in this process; however, the assumption that the patterns characterizing Homo sapiens will also apply to nonhuman species has not been tested. Future studies investigating similar factors in other catarhine primates can elucidate this question by using phylogenetic bracketing. Cranial FDMs found to be evolving predominantly neutrally and reflecting genetic relationships both within humans and among other Old World primates can be evaluated in the hominin fossil record to confidently infer phylogeny.
Acknowledgments
This project was supported by the National Science Foundation (BCS-0622570) and The Wenner-Gren Foundation (Grant no. 7499). The author would like to thank Chris Stojanowski for insightful discussions about microevolutionary modeling, Charles Roseman for guidance regarding the application of population genetics principles to the study of human cranial morphology, the late Charlie Lockwood for direction on geometric morphometrics and statistical methods, Verne Simons for help with the calculation of great circle distances, and Brent Adrian for assistance with references and discussions of inverse OWL matrices. She thanks Ian Tattersall, Gisselle Garcia, and Gary Sawyer of the AMNH, Dave Hunt of the NMNH, and Robert Kruszynski of the NHM for permission to measure physical anthropology collections in their care. An earlier version of this manuscript was improved by the comments of Bing Su and two anonymous reviewers.
References
- 1.Relethford JH. Global analysis of regional differences in craniometric diversity and population substructure. Human Biology. 2001;73(5):629–636. doi: 10.1353/hub.2001.0073. [DOI] [PubMed] [Google Scholar]
- 2.Relethford JH. Global patterns of isolation by distance based on genetic and morphological data. Human Biology. 2004;76(4):499–543. doi: 10.1353/hub.2004.0060. [DOI] [PubMed] [Google Scholar]
- 3.González-José R, Van Der Molen SV, González-Pérez E, Hernández M. Patterns of phenotypic covariation and correlation in modern humans as viewed from morphological integration. American Journal of Physical Anthropology. 2004;123(1):69–77. doi: 10.1002/ajpa.10302. [DOI] [PubMed] [Google Scholar]
- 4.Roseman CC. Detecting interregionally diversifying natural selection on modern human cranial form by using matched molecular and morphometric data. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(35):12824–12829. doi: 10.1073/pnas.0402637101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harvati K, Weaver TD. Human cranial anatomy and the differential preservation of population history and climate signatures. Anatomical Record A. 2006;288(12):1225–1233. doi: 10.1002/ar.a.20395. [DOI] [PubMed] [Google Scholar]
- 6.Smith HF, Terhune CE, Lockwood CA. Genetic, geographic, and environmental correlates of human temporal bone variation. American Journal of Physical Anthropology. 2007;134(3):312–322. doi: 10.1002/ajpa.20671. [DOI] [PubMed] [Google Scholar]
- 7.Smith HF. Which cranial regions reflect molecular distances reliably in humans? Evidence from three-dimensional morphology. American Journal of Human Biology. 2009;21(1):36–47. doi: 10.1002/ajhb.20805. [DOI] [PubMed] [Google Scholar]
- 8.Von Cramon-Taubadel N. Congruence of individual cranial bone morphology and neutral molecular affinity patterns in modern humans. American Journal of Physical Anthropology. 2009;140(2):205–215. doi: 10.1002/ajpa.21041. [DOI] [PubMed] [Google Scholar]
- 9.von Cramon-Taubadel N. Revisiting the homoiology hypothesis: the impact of phenotypic plasticity on the reconstruction of human population history from craniometric data. Journal of Human Evolution. 2009;57(2):179–190. doi: 10.1016/j.jhevol.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Nicholson E, Harvati K. Quantitative analysis of human mandibular shape using three-dimensional geometric morphometrics. American Journal of Physical Anthropology. 2006;131(3):368–383. doi: 10.1002/ajpa.20425. [DOI] [PubMed] [Google Scholar]
- 11.Lande R. Quantitative genetic analysis of multivariate evolution, applied to brain:body size allometry. Evolution. 1979;33:402–416. doi: 10.1111/j.1558-5646.1979.tb04694.x. [DOI] [PubMed] [Google Scholar]
- 12.Cheverud JM. A comparison of genetic and phenotypic correlations. Evolution. 1988;42(5):958–968. doi: 10.1111/j.1558-5646.1988.tb02514.x. [DOI] [PubMed] [Google Scholar]
- 13.Roff DA. The estimation of genetic correlations from phenotypic correlations: a test of Cheverud’s conjecture. Heredity. 1995;74(5):481–490. [Google Scholar]
- 14.Roff DA. The evolution of genetic correlations: an analysis of patterns. Evolution. 1996;50(4):1392–1403. doi: 10.1111/j.1558-5646.1996.tb03913.x. [DOI] [PubMed] [Google Scholar]
- 15.Roots KR, Gibson JP. Realized sampling variances of estimates of genetic parameters and the difference between genetic and phenotypic correlations. Genetics. 1996;143(3):1409–1416. doi: 10.1093/genetics/143.3.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lande R. Genetic variation and phenotypic evolution during allopatric speciation. American Naturalist. 1980;116:463–479. [Google Scholar]
- 17.Lofsvold D. Quantitative genetics of morphological differentiation in Peromyscus. II. Analysis of selection and drift. Evolution. 1988;42:54–67. doi: 10.1111/j.1558-5646.1988.tb04107.x. [DOI] [PubMed] [Google Scholar]
- 18.Ackermann RR, Cheverud JM. Phenotypic covariance structure in tamarins (genus Saguinus): a comparison of variation patterns using matrix correlation and common principal component analysis. American Journal of Physical Anthropology. 2000;111(4):489–501. doi: 10.1002/(SICI)1096-8644(200004)111:4<489::AID-AJPA5>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 19.Ackermann RR, Cheverud JM. Discerning evolutionary processes in patterns of tamarin (genus Saguinus) craniofacial variation. American Journal of Physical Anthropology. 2002;117(3):260–271. doi: 10.1002/ajpa.10038. [DOI] [PubMed] [Google Scholar]
- 20.Marroig G, Cheverud JM. A comparison of phenotypic variation and covariation patterns and the role of phylogeny, ecology, and ontogeny during cranial evolution of New World Monkeys. Evolution. 2001;55(12):2576–2600. doi: 10.1111/j.0014-3820.2001.tb00770.x. [DOI] [PubMed] [Google Scholar]
- 21.Marroig G, Cheverud JM. Did natural selection or genetic drift produce the cranial diversification of neotropical monkeys? American Naturalist. 2004;163(3):417–428. doi: 10.1086/381693. [DOI] [PubMed] [Google Scholar]
- 22.Ackermann RR, Cheverud JM. Detecting genetic drift versus selection in human evolution. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(52):17946–17951. doi: 10.1073/pnas.0405919102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weaver TD, Roseman CC, Stringer CB. Were neandertal and modern human cranial differences produced by natural selection or genetic drift? Journal of Human Evolution. 2007;53(2):135–145. doi: 10.1016/j.jhevol.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morton NE, Yee S, Harris DE, Lew R. Bioassay of kinship. Theoretical Population Biology. 1971;2(4):507–524. doi: 10.1016/0040-5809(71)90038-4. [DOI] [PubMed] [Google Scholar]
- 26.Cavalli-Sforza LL, Menozzi P, Piazza P. The History and Geography of Human Genes. Princeton, NJ, USA: Princeton University Press; 1994. [Google Scholar]
- 27.Von Cramon-Taubadel N, Lycett SJ. Brief communication: human cranial variation fits iterative founder effect model with African origin. American Journal of Physical Anthropology. 2008;136(1):108–113. doi: 10.1002/ajpa.20775. [DOI] [PubMed] [Google Scholar]
- 28.Betti L, Balloux F, Amos W, Hanihara T, Manica A. Distance from Africa, not climate, explains within-population phenotypic diversity in humans. Proceedings of the Royal Society B. 2009;276(1658):809–814. doi: 10.1098/rspb.2008.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217(5129):624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- 30.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, UK: Cambridge University Press; 1983. [Google Scholar]
- 31.King JL, Jukes TH. Non-Darwinian evolution. Science. 1969;167:788–798. doi: 10.1126/science.164.3881.788. [DOI] [PubMed] [Google Scholar]
- 32.Lande R. Natural selection and random genetic drift in phenotypic evolution. Evolution. 1976;30(2):314–334. doi: 10.1111/j.1558-5646.1976.tb00911.x. [DOI] [PubMed] [Google Scholar]
- 33.Lynch M, Hill WG. Phenotypic evolution by neutral mutation. Evolution. 1986;40:915–935. doi: 10.1111/j.1558-5646.1986.tb00561.x. [DOI] [PubMed] [Google Scholar]
- 34.Turelli M, Gillespie JH, Lande R. Rate tests for selection on quantitative characters during macroevolution and microevolution. Evolution. 1988;42:1085–1089. doi: 10.1111/j.1558-5646.1988.tb02526.x. [DOI] [PubMed] [Google Scholar]
- 35.Buikstra J, Ubelaker UH. Standards for Data Collection from Human Skeletal Remains. Fayetteville, Ark, USA: Arkansas Archaeological Survey; 1994. [Google Scholar]
- 36.Bass WM. Human Osteology: A Laboratory and Field Manual. 4th edition. Columbia, Mo, USA: Missouri Archaeological Society; 1995. [Google Scholar]
- 37.Lockwood CA, Kimbel WH, Lynch JM. Morphometrics and hominoid phylogeny: support for a chimpanzee-human clade and differentiation among great ape subspecies. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(13):4356–4360. doi: 10.1073/pnas.0306235101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gower JC. Generalized procrustes analysis. Psychometrika. 1975;40(1):33–51. [Google Scholar]
- 39.Rohlf FJ, Slice D. Extensions of the Procrustes method for the optimal superimposition of landmarks. Systematic Zoology. 1990;39:40–59. [Google Scholar]
- 40.Goodall CR. Procrustes methods and the statistical analysis of shape (with discussion) Journal of the Royal Statistical Society B. 1991;53:285–340. [Google Scholar]
- 41.Dryden IL, Mardia KV. Statistical Shape Analysis. Chichester, UK: John Wiley & Sons; 1998. [Google Scholar]
- 42.Klingenberg CP. MorphoJ. Manchester, UK: Faculty of Life Sciences, University of Manchester; 2008. [Google Scholar]
- 43.Lynch M. Methods for the analysis of comparative data in evolutionary biology. Evolution. 1991;45(5):1065–1080. doi: 10.1111/j.1558-5646.1991.tb04375.x. [DOI] [PubMed] [Google Scholar]
- 44.Ramachandran S, Deshpande O, Roseman CC, Rosenberg NA, Feldman MW, Cavalli-Sforza LL. Support from the relationship of genetic and geographic in human populations for a serial founder effect originating in Africa. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(44):15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Research. 1967;27(2):209–220. [PubMed] [Google Scholar]
- 46.Smouse PE, Long JC, Sokal RT. Multiple regression and correlation extensions of the Mantel test of matrix correspondence. Systematic Zoology. 1986;35:627–632. [Google Scholar]
- 47.Flury B. Common Principal Components and Related Multivariate Methods. New York, NY, USA: John Wiley & Sons; 1988. [Google Scholar]
- 48.Phillips PC, Arnold SJ. Hierarchical comparison of genetic variance-covariance matrices. I. Using the flury hierarchy. Evolution. 1999;53(5):1506–1515. doi: 10.1111/j.1558-5646.1999.tb05414.x. [DOI] [PubMed] [Google Scholar]
- 49.Hylander WL. The adaptive significance of Eskimo craniofacial morphology. In: Dahlberg AA, Graber T, editors. Orofacial Growth and Development. The Hague, The Netherlands: Mouton; 1977. pp. 129–169. [Google Scholar]
- 50.Balicki A. The Netsilik Eskimo. Garden City, NJ, USA: Natural History Press; 1970. [Google Scholar]
- 51.So JK. Human biological adaptation to arctic and subarctic zones. Annual Review of Anthropology. 1980;9:63–82. [Google Scholar]
- 52.Antón SC. Tendon-associated bone features of the masticatory system in Neandertals. Journal of Human Evolution. 1996;31(5):391–408. [Google Scholar]
- 53.De Poncins G. Kablooma. New York, NY, USA: Reynal; 1941. [Google Scholar]
- 54.Van Holst Pellekaan S, Frommer M, Sved J, Boettcher B. Mitochondrial D-loop diversity in Australian riverine and Australian desert Aborigines. Electrophoresis. 1997;18(9):1538–1543. doi: 10.1002/elps.1150180909. [DOI] [PubMed] [Google Scholar]
- 55.Van Holst Pellekaan SM, Frommer M, Sved JA, Boettcher B. Mitochondrial control-region sequence variation in aboriginal Australians. American Journal of Human Genetics. 1998;62(2):435–449. doi: 10.1086/301710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Redd AJ, Stoneking M. Peopling of Sahul: mtDNA variation in Aboriginal Australian and Papua New Guinean populations. American Journal of Human Genetics. 1999;65(3):808–828. doi: 10.1086/302533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Summerhayes GR. Island Melanesian pasts—a view from archaeology. In: Friedlaender JS, editor. Genes, Language, and Culture History in the Southwest Pacific. New York, NY, USA: Oxford University Press; 2007. pp. 10–35. [Google Scholar]
- 58.Friedlaender JS, Friedlaender FR, Reed FA, et al. The genetic structure of Pacific Islanders. PLoS Genetics. 2008;4(1, article e19) doi: 10.1371/journal.pgen.0040019. [DOI] [PMC free article] [PubMed] [Google Scholar]