Abstract
Defining the three body axes is a central event of vertebrate morphogenesis. Establishment of left-right (L-R) asymmetry in development follows the determination of dorsal-ventral (D-V) and anterior-posterior (A-P) body axes1,2, though the molecular mechanism underlying precise L-R symmetry breaking in reference to the other two axes is still poorly understood. Here by removing both Vangl1 and Vangl2, the two mouse homologues of a Drosophila core planar cell polarity (PCP) gene Van Gogh (Vang), we have uncovered a previously unappreciated function of PCP in initial breaking of lateral symmetry. The leftward nodal flow across the posterior notochord (PNC, also referred to as “the node”) has been identified as the earliest event in the de novo formation of L-R asymmetry3,4.We found that PCP is essential in interpreting the A-P patterning information and linking it to L-R asymmetry. In the absence of Vangl1 and Vangl2, cilia are positioned randomly around the center of the PNC cells and nodal flow is turbulent, which results in disrupted L-R asymmetry. Importantly, PCP in mouse, unlike what has been implicated in other vertebrate species, is not required for ciliogenesis, cilium motility, Sonic hedgehog (Shh) signaling or apical docking of basal bodies in ciliated tracheal epithelial cells. Our data suggest that PCP acts earlier than the unidirectional nodal flow during bilateral symmetry breaking in vertebrates and provide insight into the functional mechanism of PCP in organizing the vertebrate tissues in development.
Identifying the mechanism underlying the initial bilateral symmetry breaking has been an overriding goal in understanding how nature is able to produce organisms with consistent and distinct left and right anatomies. In mice, bilateral symmetry is broken by a leftward flow across a pit-like, teardrop-shaped PNC generated by posteriorly localized motile cilia3–6. However, an earlier asymmetric event that positions the cilia to the posterior sides of PNC cells in response to the A-P patterning signals remains to be identified. PCP, originally refers to asymmetry of epithelial cells along an axis orthogonal to the apical-basal axis, is an evolutionarily conserved mechanism by which positional information is transmitted and maintained among and within cells7. A group of core PCP proteins including the transmembrane protein Vang have been identified in Drosophila8. While mutants of core PCP genes have thus far failed to show L-R asymmetry defects, it is likely due to functional redundancy among members of a core PCP component. To overcome this and uncover important regulatory functions of PCP, we inactivated both Vangl1 and Vangl2, the two mouse homologues of the Drosophila Vang.
Vangl1 and Vangl2 were broadly expressed during L-R patterning (Supplementary Fig. 1). Importantly, prior to appearance of ciliated PNC cells, Vangl1 and Vangl2 protein levels were too low to be detected. However, after E7.75, both Vangl1 and Vangl2 protein levels were selectively and progressively upregulated in the PNC cells and asymmetrical localization of Vangl1 and Vangl2 protein was observed along the anterior-posterior (A-P) axis (Supplementary Fig. 2a–c), indicating that the PNC cells are indeed polarized and PCP may be required by the PNC cells to regulate early embryonic development. The Vangl1 and Vangl2 localization patterns are also consistent with the role of PCP in regulating convergent extension (CE) of neurulating vertebrate embryos9. In the sensory epithelia of the cochlea, Vangl1 and Vangl2 demonstrated similar asymmetrical localization pattern in both hair cells and supporting cells (Supplementary Fig. 2d, e). These data suggest that Vangl1 and Vangl2 play redundant roles in PCP, which should be severely disrupted by removing both Vangl genes.
A bona fide null allele of Vangl1 (Vangl1gt) was generated using a gene trap ES cell line (Supplementary Fig. 3). We also generated a floxed Vangl2 allele, Vangl2f, from which the null Vangl2Δ allele was derived (Supplementary Fig. 3). Vangl1 and Vangl2 regulated PCP in a gene dose-dependent manner (Fig. 1 and Supplementary Fig. 4). In the cochlea of the Vangl2Δ/Δ mutant, asymmetrical Vangl1 localization was still detected (Fig. 1a). The Vangl2Δ/Δ; Vangl1gt/gt embryos showed the most severe developmental defects compared to embryos with other genotypes, particularly when sensory hair cell polarity and CE were examined to assess PCP. Thus, Vangl1 and Vangl2 function of regulating PCP was removed in the Vangl2Δ/Δ; Vangl1gt/gt embryos.
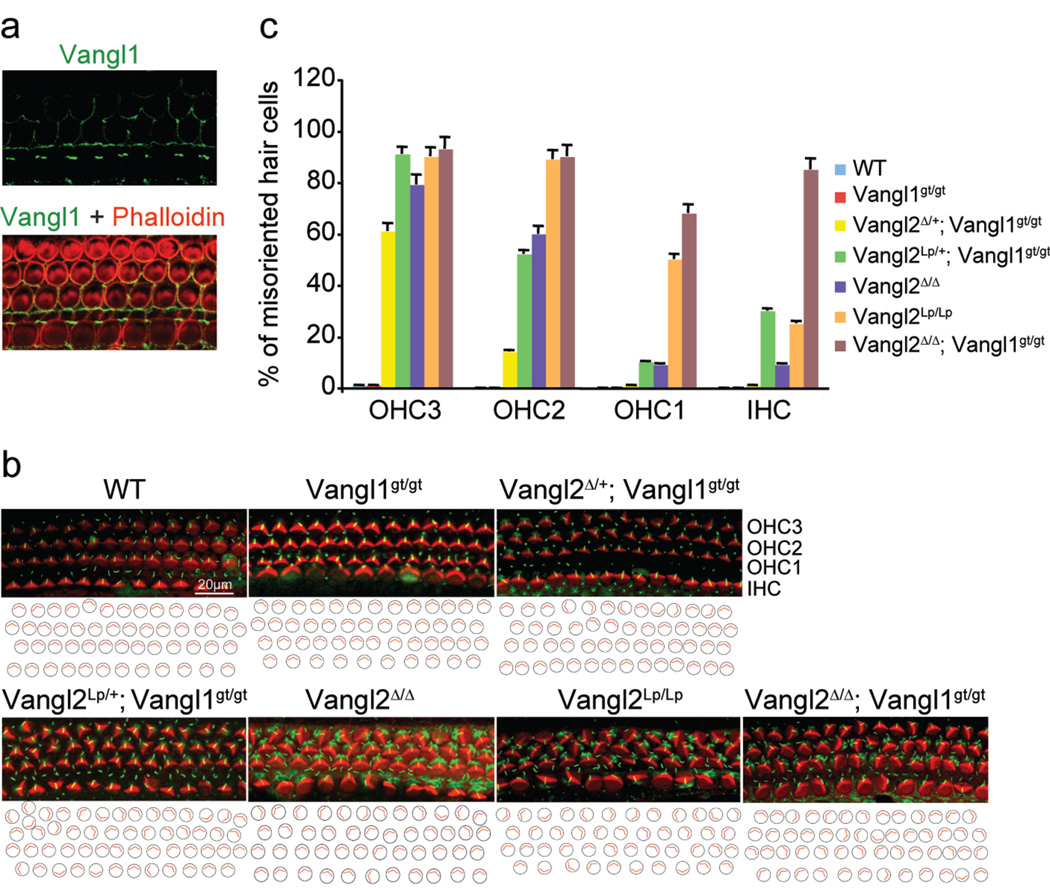
Figure 1. The Vangl2Δ/Δ; Vangl1gt/gt embryos exhibit the most severe polarity defects.
a, Asymmetrical localization of Vangl1 protein (green) in the sensory hair cells of the Vangl2Δ/Δ cochlea at E18.5. b, The sensory hair cell polarity at E18.5 is shown by the orientation of actin-based stereocilium bundles (red) and the microtubule-based kinocilia (green). c, Quantification of misoriented hair cells in embryos with different genotypes. Data are means ±SD from 3 samples. The orientation defects in the cochlea ranged from few misoriented cells (Vangl2Δ/+;Vangl1gt/gt) to several affected cells (Vangl2Δ/Δ, Vangl2Lp/+;Vangl1gt/gt and Vangl2 Lp̃Lp) to almost complete loss of polarity (Vangl2Δ/Δ; Vangl1gt/gt).
The Vangl2Δ/Δ; Vangl1gt/gt embryos exhibited multiple laterality defects, which were not detected in embryos with other genotypes (Fig. 2a). The Vangl2Δ/+; Vangl1gt/+ mice were fertile, morphologically normal and served as controls in our study. The Vangl2Δ/+; Vangl1gt/+ and Vangl2Δ/Δ; Vangl1gt/+ embryos always turned to the right side and their hearts correctly looped to the right (n>50), but in all of the Vangl2Δ/Δ; Vangl1gt/gt embryos assayed (n>10), both embryonic turning and heart looping failed. In addition, the normal mouse lung has five lobes, four on the right and one on the left. In the Vangl2Δ/Δ; Vangl1gt/gt embryos, however, mild left lung isomerism was observed with three lobes on the right side (4/9). Since PCP in visceral organs may also control later stage morphogenesis, to further assess the early L-R asymmetry defects in the Vangl2Δ/Δ; Vangl1gt/gt embryos, we examined the components in the Nodal signal cascade that are asymmetrically expressed and required for L-R patterning10. In control embryos, Nodal was first expressed symmetrically around the PNC (somite 0–1). Then between somite 2–7 stages, Nodal expression on the left side of the PNC became progressively stronger and its expression in the left lateral plate mesoderm (LPM) was also detected11,12. In the Vangl2Δ/Δ; Vangl1gt/gt embryos, however, Nodal expression around the PNC was reduced (6/11) (Fig. 2b, c). Strikingly, more robust Nodal expression on the right side of the PNC was also observed in the Vangl2Δ/Δ; Vangl1gt/gt embryos (5/11) (Fig. 2b, c). In the LPM of Vangl2Δ/Δ; Vangl1gt/gt embryos, reversed (1/8) or bilateral (3/8) Nodal expression sites were observed (Fig. 2c). Controlled by Nodal signaling, expression of Lefty1, Lefty2 and Pitx2 was only observed on the left side of the control embryos10,13,14,15. In the double mutant embryos, Lefty1 and 2 expression sites (1/2) were reversed (Fig. 2d) and bilateral Pitx2 expression (2/7) was found (Fig. 2e). These results demonstrate that PCP is required upstream of Nodal to pattern the L-R axis. The incomplete penetrance of the laterality defects in the Vangl1/2 mutant embryo is possibly due to other input(s) to the PCP pathway16.
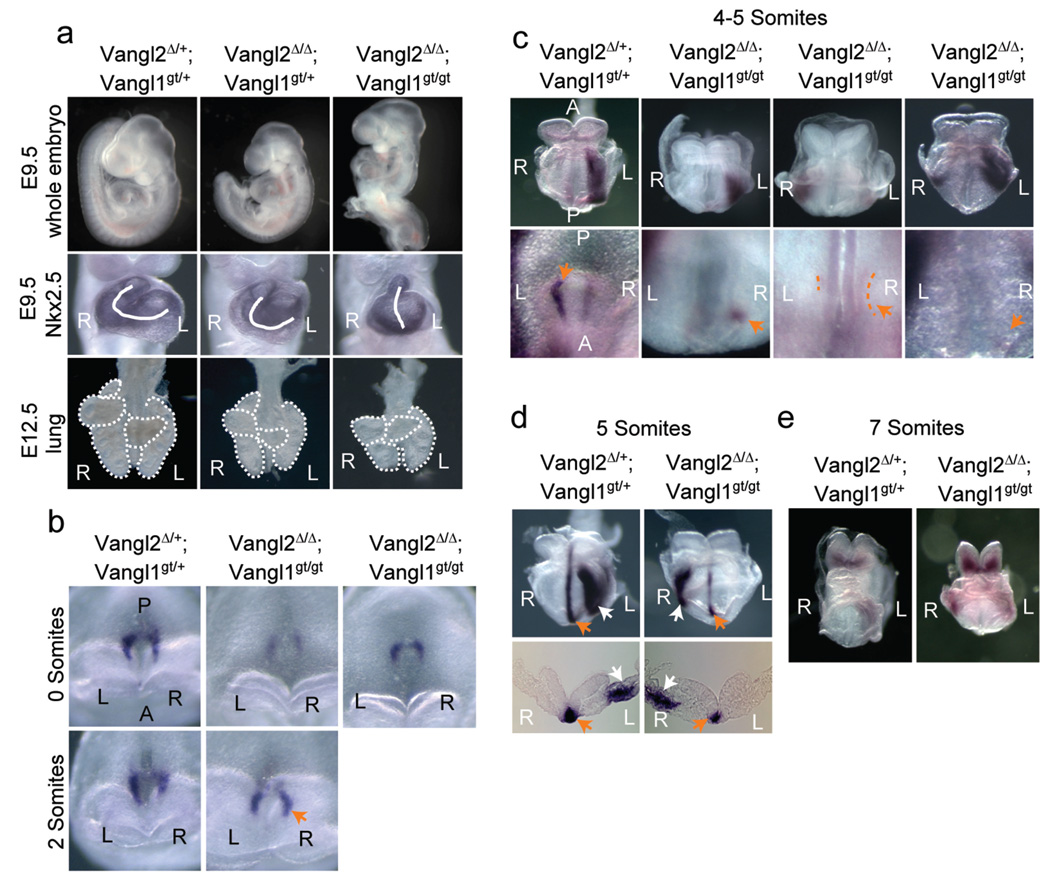
Figure 2. Laterality defects in the Vangl2Δ/Δ; Vangl1gt/gt embryos.
a, Failure of embryonic turning and heart (marked by Nkx2.5 expression) looping in the Vangl2Δ/Δ; Vangl1gt/gt embryos. The Vangl2Δ/Δ; Vangl1gt/gt lung has three lobes (outlined) instead of four on the right side. R: right; L: left. b, Perinodal Nodal expression is enhanced on the right side of the Vangl2Δ/Δ; Vangl1gt/gt embryo (arrow). c, Ventral views of Nodal expression in LPM (upper panel) and around the node (lower panel, arrows). Weak perinodal Nodal expression was outlined. d, Lefty1 expression (orange arrow) on the left side of prospective floor plate and Lefty2 expression (white arrow) in the LPM were shown in ventral views (upper panel) and transverse sections (lower panel). e, Ventral views of Pitx2 expression.
Since PCP has been implicated in ciliogenesis17, we examined the morphology and movement of cilia in PNC cells. Cilia are long, thin microtubule-based projections from basal bodies of most mammalian cells18. The PNC cell has a single motile cilium3, which is essential for establishing leftward fluid flow across the ventral surface of the PNC. When examined by scanning electron microscopy and immunofluorescent labeling, cilia appeared normal in the PNC of the Vangl2Δ/Δ; Vangl1gt/gt embryos (n=4 in each case) at E8.0 (Fig. 3a, b). Thus, the L-R asymmetry defects in the Vangl2Δ/Δ; Vangl1gt/gt embryos are not secondary to defects in ciliogenesis. The leftward nodal flow is enabled by the chirality of the cilium, as it rotates in a clockwise manner3, and the posterior ciliary localization in the PNC cell, which results in a posterior tilt of the cilium and its rotation axis5,6. Since it has been postulated that the posterior positioning of the basal body might be controlled by PCP6, we examined basal body localization in the PNC cells of the Vangl mutant embryos (Fig. 3c, d). The basal bodies were mostly localized posteriorly in both Vangl2Δ/+; Vangl1gt/+ and Vangl2Δ/Δ; Vangl1gt/+ embryos. However, in the Vangl2Δ/Δ; Vangl1gt/gt embryos, the PNC cell density was slightly decreased (Supplementary Fig. 5a) and the number of posteriorly localized basal bodies was significantly reduced. Most basal bodies remained around the center and anteriorly localized basal bodies were increased, resulting in an even distribution of basal bodies on the anterior and posterior sides of the PNC cells in the Vangl2Δ/Δ; Vangl1gt/gt embryos (Fig. 3d and supplementary Fig. 5b, c). Such loss of basal body posterior localization in the Vangl mutant is not a result of developmental delay (Supplemental Fig. 5c). Thus, posterior positioning of basal bodies in the PNC cells is indeed controlled by PCP, in the absence of which, basal bodies are mostly localized around the center. Cilia in the PNC emerge from the center of each cell at late streak (LS) stage (~E 7.25). By early head fold (EHF) stage (E7.75), most cilia are located in the posterior side of the PNC cells6,19. As positioning of the kinocilium in sensory hair cells is also an event downstream of PCP20, ciliary positioning may be a common readout of PCP.
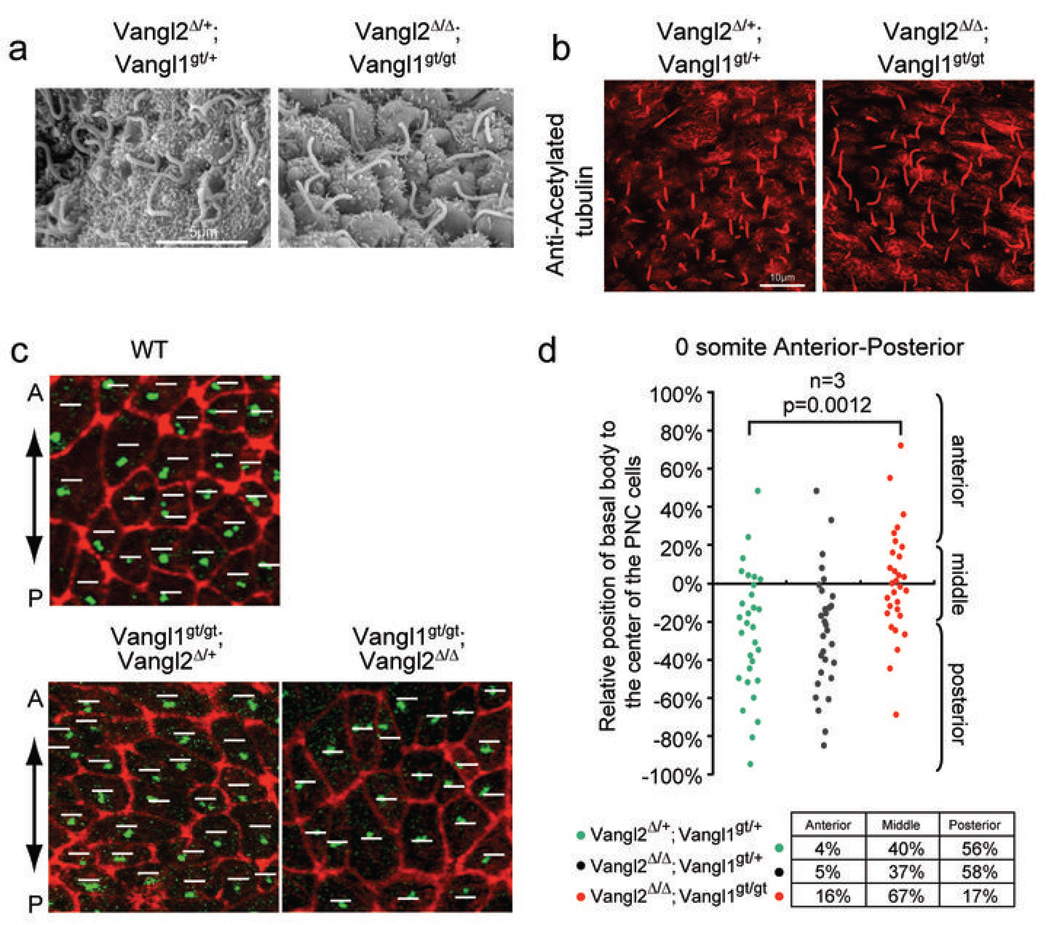
Figure 3. Randomized ciliary positioning in the Vangl2Δ/Δ; Vangl1gt/gt embryos.
a, Scanning electron microscopy of the PNC showing normal cilia at E8.0. b, Normal cilia (acetylated tubulin, red) in the PNC of the Vangl2Δ/Δ; Vangl1gt/gt embryo at E8.0. c, Basal 16 bodies (γ–tubulin, green) and cell boundaries (F-actin, red) of PNC cells at somite 0 stage. White lines indicate the middle position along the A-P axis of each PNC cell. A: anterior; P: posterior. d, Relative positions of basal bodies to the midline (along the A-P axis) of the PNC cells were plotted. Posteriorly biased basal body positioning was lost in the Vangl2Δ/Δ; Vangl1gt/gt embryos.
To test whether altered ciliary positioning in the PNC cells affect function, we examined nodal flow (Fig. 4a and Supplementary movie 1, 2). In control embryos, fast and smooth leftward bead movement across the PNC was observed. In the Vangl2Δ/Δ; Vangl1gt/gt embryos, bead movement was still biased leftward, though beads were often trapped in swirling vortices and requires more time to cross the PNC. While the nodal flow of control embryos generated straight and parallel bead paths, in the mutant, bead paths contained “knots”, abrupt turns and were often crossed with each other (2/2). When observed directly, the cilia in the PNC of the Vangl2Δ/Δ; Vangl1gt/gt embryo were beating at a speed and direction similar to that of the control (Fig. 4b and Supplementary movie 3, 4). The abnormal nodal flow in the Vangl2Δ/Δ; Vangl1gt/gt embryos also disrupted the left-biased perinodal rise of intracellular calcium in control embryos21 (Fig. 4c, d). Incidence of bilateral rise or reduction of intracellular calcium was increased in the Vangl2Δ/Δ; Vangl1gt/gt embryos. The ectopic calcium signal on the right side was weaker and the domain was narrower and shorter. Taken together, unidirectionality of nodal flow was significantly compromised in the Vangl2Δ/Δ; Vangl1gt/gt embryos. Thus, we have identified PCP as the first step in breaking L-R symmetry by transmitting the A-P positional information to the PNC cells to enable posterior ciliary positioning (Fig. 4e).
Figure 4. Vangl1 and Vangl2 are required for normal leftward nodal flow.
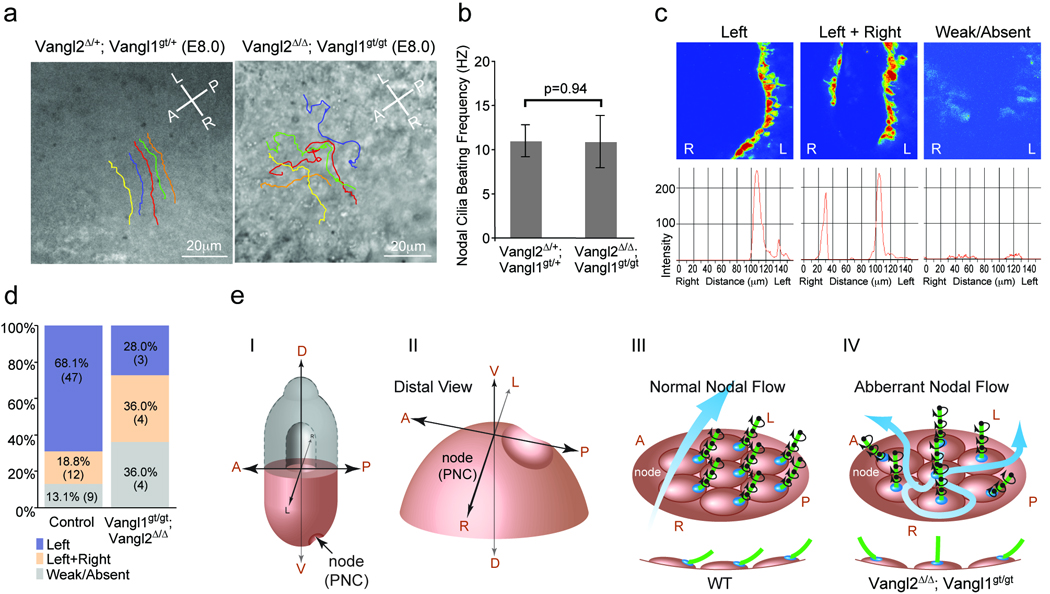
a, Nodal flow shown by tracing latex bead movement across the PNC with lines in different colors (Supplementary movie 1, 2). A: anterior; P: posterior; L: left; R: right. b, Ciliary beating was recorded by live imaging (Supplementary movie 3, 4) and the mean beating frequency was calculated from at least 10 nodal cilia of 3 embryos with indicated genotypes. Data are means ±SD. c, Intracellular calcium release around the node, intensity scales are shown in the lower panels. d, Distribution of embryos (percentage and number) according to calcium patterns. e, Schematic diagram showing the role of Vangl1 and Vangl2 in breaking the L-R symmetry. I. A mouse embryo at E8.0. (II) Enlarged distal view of (I). (III) Normal nodal flow generated by posteriorly positioned cilia. A side view of ciliated cells are shown in the lower panel. (IV) In the Vangl2Δ/Δ; Vangl1gt/gt embryo, randomly positioned cilia around the center of the PNC cells resulted in turbulent nodal flow.
Our data show that PCP is not required for ciliogenesis in mouse (Fig. 1b and supplementary Fig. 6c–f). In addition, Vangl1 and Vangl2 were not colocalized with cilia in any of the cells we had examined including the PNC cells, sensory hair cells, tracheal epithelial cells and floor plate cells (Supplementary Figs. 2a, d and 6a, b). In the Vangl2Δ/Δ; Vangl1gt/gt embryos at E18.5, there were no defects in ciliogenesis or docking of the basal bodies along the apical-basal axis of the multiciliated tracheal epithelial cells (Supplementary Fig. 6c, d). Normal Hedgehog signaling in the Vangl2Δ/Δ; Vangl1gt/gt embryos and cells provides further evidence that primary cilia are functionally normal (Supplementary Figs. 6g, h and 7).
To promote a robust break in symmetry, vertebrates have evolved mechanisms of early asymmetry initiation followed by asymmetry amplification and stabilization22. As PCP is a major conserved molecular mechanism by which directional cellular and tissue behaviors are controlled through reorganizing cytoskeleton, it could lead to both cilia-dependent and cilia-independent regulation of L-R asymmetry by controlling asymmetrical ciliary localization in ciliated PNC or cell movement around the node in chick which lacks ciliated PNC5,6,23. Involvement of PCP in regulating L-R asymmetry has been implicated before since mutations in Inversin (Inv), Bicaudal C and Seahorse lead to L-R asymmetry defects upstream of Nodal expression and these genes can interact with a core PCP gene Dishevelled (Dvl)24–26. Dvl has recently been found to regulate ciliary localization in the PNC and implicated to regulate L-R patterning27. However, it is unlikely that Inv acts mainly through PCP since Inv, but not Vangl1/2, is localized to PNC cilia and the phenotypes of the Inv and Vangl1/2 mutants are different in many aspects. As abnormal L-R patterning is also associated with multiple congenital diseases28, the availability of Vangl1 and Vangl2 conditional double mutants has opened a door to investigate the roles of PCP in many fundamental aspects of vertebrate development and diseases.
Note added in proof: While this work was in press, two independent studies29,30 were published, both also found that planar cell polarity mediated by Vangl2 is required for orienting and positioning motile cilia.
Methods Summary
Scanning electron microscopic analysis
Embryos were dissected at E7.75 and fixed in 2% glutaraldehyde, 4% paraformaldehyde, 0.1M sodium cacodylate, pH 7.4 at 4 °C overnight. Standard procedures were followed to prepare samples. Samples were examined by a Hitachi S-570 scanning electron microscope.
Analysis of calcium signaling
Embryos were dissected at E7.75–E8.0 and analyzed according to the established procedure21.
Video Microscopy
Embryos were dissected at E7.75–E8.0 in DMEM with 20% FBS. The node was placed facing down in a chamber with a cover slip on the bottom. Fluospheres (Invitrogen) were added into the medium to visualize the direction of nodal flow. Wide-field images were collected using a Personal DeltaVision system (Applied Precision Inc, Issaquah, WA, USA) mounted on an inverted Olympus IX71 microscope with an oil immersion PlanApo N 60×/1.42 objective lens. Images were acquired using a CoolSNAP ES2 camera with 2×2 binning and 128 pixels ×128 pixels imaging field.
Methods
Generation of and genotyping mouse lines
Generation of the floxed Vangl2 allele is described in Supplementary Fig. 1e. Vangl2Δ was generated by deleting exon 4 and 5 using the Sox2-Cre line31. Vangl1gt allele was generated using a Baygenomic gene trap ES cell line (XL802) (Supplementary Fig. 1a). Genotyping was done by PCR using genomic DNA prepared from yolk sac or tail. Vangl2P1 (5′-CCATCGATGCACATGTGGTATC-3′) and Vangl2P2 (5′-GACACTGTCCTCCATGTCCTTG-3′) were used to genotype the Vangl2f allele and Vangl2+ allele. Vangl2P1 and Vangl2P3 (5′-GCTATGACCAGCTACCTGAAGTC-3′) were used to genotype the Vangl2Δ allele. Vangl1P1 (5′-CCCTGGCTTTCTTGTTGGTC-3′) and Vangl1P3 (5′-GGGCTGGCTCTTGGAGTCAT-3′) were used to genotype the Vangl1+ allele. Vangl1P1 and Vangl1P2 (5′-CCTGGGGTTCGTGTCCTACA-3′) were used to genotype the Vangl1gt allele.
Whole mount in situ hybridization and LacZ staining
Whole mount in situ hybridization and LacZ staining were performed as described previously32. Embryos were cryosectioned at 20 µm.
Immunofluorescence and antibodies
Embryos were fixed in 4% paraformaldehyde for 30 min at 4 °C, washed in PBS and incubated overnight in 30% sucrose/PBS. Embryos were embedded in OCT compound (Tissue-Tek) and sectioned at 14 µm. Sections were permeabilized with 0.5% Triton X-100 in PBS and blocked in 3% bovine serum albumin, 0.1% Triton X-100 in PBS for 1 hour. Sections were incubated with primary antibodies at 4°C overnight. Secondary antibodies were applied at room temperature for 1 hour. Primary antibodies used in immunofluorescence include mouse anti-Pax6, Pax7, Nkx2.2, Lim1/2, Nkx6.1, Hnf3β and Islet-1 (Developmental Studies Hybridoma Bank, 1:50), rabbit anti-Olig2 (Abcam, 1:100), mouse anti-acetylated-tubulin (Sigma, 1:5,000), goat anti-γ-tubulin (Santa Cruz, 1:200), goat anti-Vangl2 (Santa Cruz, 1:100), rabbit anti-Vangl2 (gift of Dr. Mathew Kelley’s lab, 1:50) and goat anti-E-Cadherin (R&D, 1:500). Vangl1 antibodies were generated against Vangl1 (1-110 aa) polypeptides fused to glutathione S-transferase (GST) in rabbit. Anti-Vangl1 antibodies were purified using the immunizing peptide coupled to Affigel (Bio-Rad). Alexa Fluor 488 and 568 of donkey anti-mouse, rabbit and goat secondary antibodies were from Invitrogen. Confocal images were acquired using a Zeiss LSM 510 NLO microscope. Excitation wavelengths of 488, 561 and 740 nm were used for detection of secondary antibodies conjugated by Alexa Fluor 488, 568 and DAPI respectively.
Whole-mount Immunostaining of cochlea or early E6 – E8.5 embryos
Dissected cochlea or embryos were fixed in 4% PFA for 30 min at 4 °C. Cochlea or embryos were permeabilized with 0.5% Triton X-100 in PBS for 5 min and blocked in PBS with 3% BSA, 0.1% Triton X-100 at room temperature for 1 hour, followed by overnight incubation with primary antibodies at 4°C. Cochlea or embryos were washed in PBS with 0.1% Triton X-100 at room temperature for 1 h, followed by incubation with fluorescent secondary antibodies and Alexa Fluor-594-conjugated phalloidin (Invitrogen 1:200) at room temperature for 1 hour. The actin-based stereocilium bundles were shown by phalloidin staining whereas the microtubule-based kinocilia or PNC cilia were shown by staining using the mouse anti-acetylated-tubulin antibody (Sigma, 1:5,000). After washed with 0.1% Triton X-100 in PBS at room temperature for 1 hr, cochlea or the PNC region were flat mounted. Confocal images were acquired using a Zeiss LSM510 confocal microscope.
Immunolot Analysis
Embryonic tissues or cells were lysed in RIPA buffer. Cell lysates were analyzed by Immunoblot. Primary antibodies used include goat anti-Vangl2 (1:2000), rabbit anti-Vangl1(1:1000), rabbit anti-Gli3 (gift from Dr. Susan Mackem, 1:5000).
Analysis of calcium signaling
Embryos were dissected at E7.75–E8.0 and incubated with the fluorescent calcium indicator Fluo-4 (Invitrogen) at a concentration of 10 µM in DMEM with 20% FBS for 20 minutes at 37 °C. Embryos were then washed for 3 times in DMEM with 20% FBS and placed node facing up in a silicon chamber under a cover slip. Embryos were incubated at 37 °C for 20 minutes before confocal imaging. Confocal images were acquired using a Zeiss LSM510 confocal microscope. Excitation wavelength of 488 was used to detect Fluo-4. Embryos were genotyped after imaging. Data were analyzed with Image-Pro 6.3 software. High intensity was shown in red and low intensity in blue. Intensity scales of the calcium signal were plotted from left to right side of the node.
Video Microscopy
Embryos were dissected at E7.75–E8.0 in DMEM with 20% FBS. The node was placed facing down in a chamber with a cover slip bottom. For the nodal flow assay, Fluospheres (Invitrogen) were added into the medium to visualize the direction of nodal flow. Embryos were genotyped after imaging. Wide-field images were collected using a Personal DeltaVision system (Applied Precision Inc, Issaquah, WA, USA) mounted on an inverted Olympus IX71 microscope with an oil immersion PlanApo N 60×/1.42 objective lens. All images were acquired using a CoolSNAP ES2 camera with 2×2 binning and 128 pixels × 128 pixels imaging field. Videos were created using Applied Precision’s SoftWoRx software package version 4.0.0. Individual bead movement was traced and shown as a line with different color. The mean cilia beating frequency was calculated from at least 10 nodal cilia of embryos with indicated genotypes at E8.0. Data are means ±SD. The p value is calculated according to the student’s t-test.
Supplementary Material
Acknowledgement
We thank the Yang lab for stimulating discussion and Drs. Michael Kuehn, Amit Kumar and Jean Regard for critical reading of the manuscript. We also thank Julia Fekecs and Darryl Leja for preparing the graphics. We are grateful to Dr. Doris Wu for teaching us to dissect the mouse cochlea and Dr. Mathew Kelley for the Vangl2 antibodies. This study is supported by the intramural research program of National Human Genome Research Institute.
Footnotes
Author contributions
Hai Song generated most of the data in the manuscript. He designed and performed the experiments, interpreted and presented the data in figures. Jianxin Hu designed and performed some of the experiments, carried out statistical analysis, interpreted and put his data together in figures. Wen Chen analyzed and maintained the Vangl1 mutant mice. Gene Elliott generated the Vangl1 and Vangl2 mutant mice by performing blastocyst injection and teraploid aggregation. Philipp Andre and Bo Gao provided some critical technique support and help in writing the manuscript. Yingzi Yang designed and supervised the entire project, interpreted all data and wrote the manuscript.
References
- 1.Wilhelmi H. Experimentelle Untersuchungen ueber situs inversus viscerum. Archive der Entwicklungsmechanik. 1921;48:517–532. [Google Scholar]
- 2.Brown NA, Wolpert L. The development of handedness in left/right asymmetry. Development. 1990;109:1–9. doi: 10.1242/dev.109.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Nonaka S, et al. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- 4.Blum M, et al. Ciliation and gene expression distinguish between node and posterior notochord in the mammalian embryo. Differentiation. 2007;75:133–146. doi: 10.1111/j.1432-0436.2006.00124.x. [DOI] [PubMed] [Google Scholar]
- 5.Nonaka S, et al. De novo formation of left-right asymmetry by posterior tilt of nodal cilia. PLoS Biol. 2005;3:e268. doi: 10.1371/journal.pbio.0030268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okada Y, Takeda S, Tanaka Y, Belmonte JC, Hirokawa N. Mechanism of nodal flow: a conserved symmetry breaking event in left-right axis determination. Cell. 2005;121:633–644. doi: 10.1016/j.cell.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 7.Seifert JR, Mlodzik M. Frizzled/PCP signalling: a conserved mechanism regulating cell polarity and directed motility. Nat Rev Genet. 2007;8:126–138. doi: 10.1038/nrg2042. [DOI] [PubMed] [Google Scholar]
- 8.Axelrod JD, McNeill H. Coupling planar cell polarity signaling to morphogenesis. ScientificWorldJournal. 2002;2:434–454. doi: 10.1100/tsw.2002.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jessen JR, et al. Zebrafish trilobite identifies new roles for Strabismus in gastrulation and neuronal movements. Nat Cell Biol. 2002;4:610–615. doi: 10.1038/ncb828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamada H, Meno C, Watanabe D, Saijoh Y. Establishment of vertebrate left-right asymmetry. Nat Rev Genet. 2002;3:103–113. doi: 10.1038/nrg732. [DOI] [PubMed] [Google Scholar]
- 11.Lowe LA, et al. Conserved left-right asymmetry of nodal expression and alterations in murine situs inversus. Nature. 1996;381:158–161. doi: 10.1038/381158a0. [DOI] [PubMed] [Google Scholar]
- 12.Collignon J, Varlet I, Robertson EJ. Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature. 1996;381:155–158. doi: 10.1038/381155a0. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura T, et al. Generation of robust left-right asymmetry in the mouse embryo requires a self-enhancement and lateral-inhibition system. Dev Cell. 2006;11:495–504. doi: 10.1016/j.devcel.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Meno C, et al. lefty-1 is required for left-right determination as a regulator of lefty-2 and nodal. Cell. 1998;94:287–297. doi: 10.1016/s0092-8674(00)81472-5. [DOI] [PubMed] [Google Scholar]
- 15.Yoshioka H, et al. Pitx2, a bicoid-type homeobox gene, is involved in a lefty-signaling pathway in determination of left-right asymmetry. Cell. 1998;94:299–305. doi: 10.1016/s0092-8674(00)81473-7. [DOI] [PubMed] [Google Scholar]
- 16.Lawrence PA, Struhl G, Casal J. Planar cell polarity: one or two pathways? Nat Rev Genet. 2007;8:555–563. doi: 10.1038/nrg2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park TJ, Haigo SL, Wallingford JB. Ciliogenesis defects in embryos lacking inturned or fuzzy function are associated with failure of planar cell polarity and Hedgehog signaling. Nat Genet. 2006;38:303–311. doi: 10.1038/ng1753. [DOI] [PubMed] [Google Scholar]
- 18.Poole CA, Flint MH, Beaumont BW. Analysis of the morphology and function of primary cilia in connective tissues: a cellular cybernetic probe? Cell Motil. 1985;5:175–193. doi: 10.1002/cm.970050302. [DOI] [PubMed] [Google Scholar]
- 19.Lee JD, Anderson KV. Morphogenesis of the node and notochord: the cellular basis for the establishment and maintenance of left-right asymmetry in the mouse. Dev Dyn. 2008;237:3464–3476. doi: 10.1002/dvdy.21598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones C, et al. Ciliary proteins link basal body polarization to planar cell polarity regulation. Nat Genet. 2008;40:69–77. doi: 10.1038/ng.2007.54. [DOI] [PubMed] [Google Scholar]
- 21.McGrath J, Somlo S, Makova S, Tian X, Brueckner M. Two populations of node monocilia initiate left-right asymmetry in the mouse. Cell. 2003;114:61–73. doi: 10.1016/s0092-8674(03)00511-7. [DOI] [PubMed] [Google Scholar]
- 22.Tabin CJ. The key to left-right asymmetry. Cell. 2006;127:27–32. doi: 10.1016/j.cell.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 23.Gros J, Feistel K, Viebahn C, Blum M, Tabin CJ. Cell movements at Hensen's node establish left/right asymmetric gene expression in the chick. Science. 2009;324:941–944. doi: 10.1126/science.1172478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kishimoto N, Cao Y, Park A, Sun Z. Cystic kidney gene seahorse regulates cilia-mediated processes and Wnt pathways. Dev Cell. 2008;14:954–961. doi: 10.1016/j.devcel.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 25.Maisonneuve C, et al. Bicaudal C, a novel regulator of Dvl signaling abutting RNA-processing bodies, controls cilia orientation and leftward flow. Development. 2009;136:3019–3030. doi: 10.1242/dev.038174. [DOI] [PubMed] [Google Scholar]
- 26.Simons M, et al. Inversin, the gene product mutated in nephronophthisis type II, functions as a molecular switch between Wnt signaling pathways. Nat Genet. 2005;37:537–543. doi: 10.1038/ng1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hashimoto M, et al. Planar polarization of node cells determines the rotational axis of node cilia. Nat Cell Biol. 2010;12:170–176. doi: 10.1038/ncb2020. [DOI] [PubMed] [Google Scholar]
- 28.Sharma N, Berbari NF, Yoder BK. Ciliary dysfunction in developmental abnormalities and diseases. Curr Top Dev Biol. 2008;85:371–427. doi: 10.1016/S0070-2153(08)00813-2. [DOI] [PubMed] [Google Scholar]
- 29.Guirao B, et al. Coupling between hydrodynamic forces and planar cell polarity orients mammalian motile cilia. Nat Cell Biol. 2010;12:341–350. doi: 10.1038/ncb2040. [DOI] [PubMed] [Google Scholar]
- 30.Borovina A, Superina S, Voskas D, Ciruna B. Vangl2 directs the posterior tilting and asymmetric localization of motile primary cilia. Nat Cell Biol. 2010;12:407–412. doi: 10.1038/ncb2042. [DOI] [PubMed] [Google Scholar]
- 31.Hayashi S, Tenzen T, McMahon AP. Maternal inheritance of Cre activity in a Sox2Cre deleter strain. Genesis. 2003;37:51–53. doi: 10.1002/gene.10225. [DOI] [PubMed] [Google Scholar]
- 32.Topol L, et al. Wnt-5a inhibits the canonical Wnt pathway by promoting GSK-3-independent β-catenin degradation. J Cell Biol. 2003;162:899–908. doi: 10.1083/jcb.200303158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.