Abstract
Population subgroups of the African malaria vector Anopheles gambiae have not been comprehensively characterized owing to the lack of unbiased sampling methods. In the arid savanna zone of West Africa, where potential oviposition sites are scarce, widespread collection from larval pools in the peridomestic human habitat yielded a comprehensive genetic survey of local A. gambiae population subgroups, independent of adult resting behavior and ecological preference. A previously unknown subgroup of exophilic A. gambiae is sympatric with the known endophilic A. gambiae in this region. The exophilic subgroup is abundant, lacks differentiation into M and S molecular forms, and is highly susceptible to infection with wild Plasmodium falciparum. These findings might have implications for the epidemiology of malaria transmission and control.
Much of the current genetic knowledge of African malaria vector populations is based on collection and analysis of indoor house-resting (endophilic) mosquitoes. Indoor capture of mosquitoes is often regarded as an unbiased collection method for epidemiologically important vectors of human malaria, including A. gambiae sensu stricto (ss), because they are thought to be ‘naturally endophilic’ (1). Outdoor-resting (exophilic) mosquitoes can contribute to malaria transmission but are underrepresented or absent from indoor collections, particularly if they also bite outdoors (2–7). Collection methods for exophilic mosquitoes are much less efficient than for indoor-resting mosquitoes, involving variants of i) artificial resting sites such as pits, boxes or pots, ii) manual aspiration from vegetation and holes, or iii) capture of mosquitoes landing on animal or human bait, now largely proscribed due to the risk of infection of human collectors (2). Further highlighting the challenge in sampling exophilic mosquitoes, resting sites thought to harbor large A. gambiae populations during the dry season have resisted detection for decades (8–10). Little is known about the population genetics of resting site choice, but the Garki malaria control project in Nigeria in the 1970s failed largely because pre-existing genetically-controlled exophilic behavior allowed persistent malaria transmission by outdoor-resting mosquitoes, despite widespread indoor residual insecticide spraying (3, 11).
Genetic division of A. gambiae populations into subgroups allows fine ecological partitioning by the species, mediating the expansion of malaria transmission spatially and temporally. Chromosome inversion polymorphisms define certain subgroups (10, 12). Frequency of the 2La inversion follows a geographic cline from the humid Central African forest, where the wild-type 2La+ allele is fixed, north to the arid West African savanna, where the inverted 2La allele is fixed (12). One likely phenotype of the 2La inversion is adaptation to aridity. Another example of niche expansion of A. gambiae by genetic subdivision is represented by two genetically diverged molecular forms, termed M and S (13). These molecular forms are detected by assays for fixed nucleotide differences on the X chromosome (Molecular Form Diagnostic SNPs, MFDS), and display other fixed SNPs in genomic ‘speciation islands’ (14). The M form dominates in marginal and disturbed habitats where S is less competitive (15).
We collected mosquito larvae from natural breeding pools in an arid ecological zone where availability of pools for oviposition is a limiting resource. Collection from pools offers unbiased sampling of population subgroups regardless of adult resting behavior, which is not currently possible by other methods. We made 68 collections at three village sites across a 400 km transect in the Sudan-Savanna ecological zone of Burkina Faso over two years, and supplementary collections over two additional years (Figures S1, S2 and (16)). All known classes of larval pools located ≤1km from human houses were sampled. Each collection consisted of ≤10 larvae per pool from ≥50 independent pools, to avoid oversampling siblings. Thus, larval mosquito populations occupying the peridomestic human habitat were deeply sampled. At matching sites and times, 12 collections were made of indoor-resting adult mosquitoes in village houses. Samples were genotyped for MFDS on the X chromosome, and for the 2La inversion.
Indoor-captured A. gambiae ss displayed the expected profile of characteristics for collections from the Sudan-Savanna zone (Figure 1): i) a low rate of MFDS hybrids (<1% (13, 14), and ii) near-fixation of the inverted allele 2La (96%) (12, 17). The expected genetic association was also observed between the MFDS and unlinked SNPs in a genomic ‘speciation island’ (>88% agreement between MFDS and chromosome 2L island (16)). However, mosquitoes raised from larval collections made simultaneously at the same transect sites had contrasting genetic composition to the indoor-captured collections. Larval collections included 35% MFDS hybrids and the 2La+ chromosome at a frequency of 58% (Figures 1, S3). We suspected the existence of a previously undescribed subgroup of A. gambiae ss in the larval pools, reproductively isolated from the population captured indoors at the same sites, because absence of M and S molecular form hybrids in the indoor-collected group could not occur if there were gene flow of hybrids from the sympatric larval-collected mosquitoes. To confirm and measure the extent of reproductive isolation, mosquitoes from larval pools and indoor-resting collections were genotyped at eight microsatellite loci on chromosome 3 (Chr3), and resulting genotypes were analyzed using STRUCTURE (18–20). We used microsatellite (short tandem repeat, STR) markers because individual STRs are 5–8 times more informative than single SNPs (21). Chr3 was chosen as a neutral genomic location because it does not carry features implicated in population substructure (16).
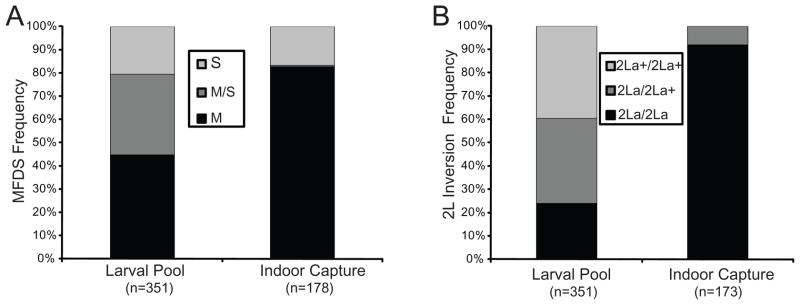
Figure 1. Comprehensive sampling of A. gambiae populations in the West African Sudan Savanna zone reveals unexpected genotypes.
Histograms compare populations of indoor-resting and larval pool collections of A. gambiae ss. Larval pools are a limiting resource in the arid zone, and larval pool sampling represents an unbiased survey of all population subgroups, while indoor house collections sample behaviorally endophilic mosquitoes. Frequencies of: A. molecular form M and S diagnostic SNP alleles (MFDS), and B. 2La inversion alleles are significantly different in larval pool and indoor-resting mosquito collections (for MFDS, χ2=89.669, p=3E-20; for 2La inversion, χ2=178.533, p=1.7E-39). The wild-type 2La+ chromosome allele and hybrids of the MFDS occur at unexpectedly high frequency in larval collections as compared to indoor-resting mosquito collections, suggesting population genetic substructure and behavioral partitioning. All samples are A. gambiae ss by molecular and cytogenetic species assays, collected ≤1km from human houses at 3 independent geographic sites across a 400 km transect in Burkina Faso (map, Figure S1) during 2007 and 2008 malaria transmission seasons.
Bayesian clustering indicates that the collected larvae did not form a panmictic group, but were comprised of two main subgroups (LnPD likelihood score optimal for k=2 clusters, Figure S4): i) individuals that clustered genetically with the indoor-collected samples, and ii) a separate, highly differentiated cluster (Wright’s fixation index Fst=0.1497, p<0.001, n=679, Figures 2, S4, S5). The novel cluster (ii) from larval pools is hereafter called the GOUNDRY subgroup of A. gambiae ss, named for a village within its range following convention (12). The indoor-collected cluster is hereafter called the ENDO subgroup of A. gambiae ss for comparative purposes, due to its consistent presence in collections of indoor-resting mosquitoes in this ecological zone. To control for the possibility that the pattern of GOUNDRY-ENDO differentiation might be an artifact of the markers used, we also used other STRs and SNPs to query the entire genome. All marker sets tested showed similar differentiation between ENDO and GOUNDRY, and consistent assignment of individual mosquitoes to either ENDO or GOUNDRY subgroups (16). Descriptions hereafter of ENDO and GOUNDRY mosquitoes refer to mosquitoes grown from larval collections, genotyped, and assigned to clusters as just described.
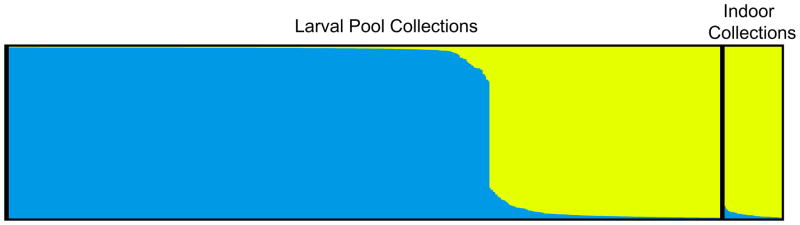
Figure 2. Bayesian cluster assignments divide A. gambiae ss Sudan-Savanna populations into two genetically and behaviorally differentiated subgroups.
Mosquitoes from indoor-resting and larval pool collections were clustered by multilocus genotypes using STRUCTURE. The indoor-resting mosquitoes consistently belonged to a single cluster, while the matched larval pools consistently contained two genetically differentiated subgroups: i) individuals that cluster robustly with the indoor-captured mosquitoes and thus are the source of the behaviorally endophilic subgroup (yellow), and ii) a distinct subgroup that are not captured among indoor-resting mosquitoes and thus are behaviorally exophilic (blue; LnPD statistic, Figure S4). Individuals are depicted as colored vertical bars (n=679). Samples to the right of the black line are indoor-captured mosquitoes, and to the left are larval pool collections.
The ENDO cluster of mosquitoes obtained from larval pool collections is genetically indistinguishable from indoor-resting adult mosquitoes (Figures 2, S4, S5). Consequently, the ENDO cluster collected from larval pools comprises a genetically and behaviorally defined subgroup that, among other properties, rests in human habitations after a bloodmeal, as evidenced by their morning capture in houses. In contrast, mosquitoes of the GOUNDRY cluster are not captured inside human dwellings in the morning by either exhaustive pyrethroid spray or manual aspirator catch, and thus are not indoor-resting by this empirical criterion (2). To rule out the possibility that GOUNDRY comprised a previously described taxon, a subset of samples (n=63) were typed to species by the original diagnostic assay, cytogenetic analysis of polytene chromosomes (22), and additional samples were tested using all available species molecular assays (four assays for the A. gambiae complex and one for A. nili (16)). All assays concordantly report mosquitoes belonging to both GOUNDRY and ENDO subgroups as A. gambiae ss. Both subgroups include chromosomal forms Mopti, Savanna, and hybrids, with no diagnostic or remarkable inversion differences (12, 16).
The GOUNDRY subgroup displays greater relative abundance in the local population (57% GOUNDRY, 43% ENDO, n=439; Figure 3A). GOUNDRY also consistently outnumbers ENDO within individual temporal collections, and thus the overall relative abundance is not an artifact of aberrant collections (GOUNDRY outnumbers ENDO in 15 of 20 temporal collections, Rank Sum Test p=0.003, Figure S7A). Despite their abundance we have not yet captured GOUNDRY mosquitoes as wild adults in pilot collections using CDC light traps or outdoor pit traps. Smaller numbers of samples from two additional years shows long-term presence and reproductive capacity of GOUNDRY mosquitoes in the population (Figure S7A). Locating GOUNDRY mosquito resting sites will require more extensive field sampling.
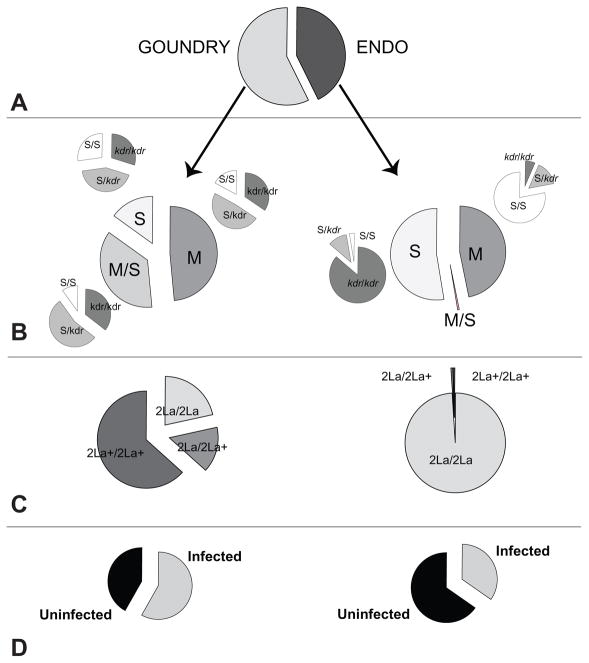
Figure 3. GOUNDRY and ENDO subgroups of A. gambiae ss display distinct genetic and phenotypic composition.
Larval site collections were genotyped to determine, A. ENDO and GOUNDRY relative population abundance, B. MFDS and the kdr-w pyrethroid resistance locus, C. the chromosome 2La inversion, and D. susceptibility to local P. falciparum parasites (n=439). Genetic characteristics of the ENDO subgroup are consistent with the canonical West African A. gambiaess ( <1% hybrid rate for MFDS, 2La inverted allele virtually fixed, kdr-w resistant allele strongly associated with molecular S form SNPs). In contrast, the GOUNDRY subgroup displays unexpected genotypes: the MFDS segregate as neutral molecular variation, the 2La+ inverted chromosome segregates at 68%, and the kdr-w alleles segregate independently of MFDS. Phenotypically, after feeding on blood with wild P. falciparum gametocytes, 58% of GOUNDRY and 35% of ENDO mosquitoes were permissive for parasite infection. For all features measured, frequencies are significantly different between ENDO and GOUNDRY subgroups (GOUNDRY greater relative abundance in 75% of collections, n=20, Rank Sum Test p=0.003; MFDS, χ2=113.8, p=1.9E-25; kdr associated with M-form SNPs, χ2=73.473, 1.1E-16; kdr in S-form SNPs, χ2=37.26, 1E-8; 2La inversion, χ2=257.045, p=1.5E-56; infection prevalence, χ2=23.1, p=1.5E-6).
The genome-wide genetic differentiation between GOUNDRY and ENDO population subgroups (Fst=0.1497, p<0.001, n=679) is an order of magnitude greater than the differentiation between M and S molecular forms within ENDO (Fst=0.015, p<0.001, n=190; Figure S6). The ENDO mosquitoes carrying distinct MFDS displayed the expected level of genetic differentiation, low MFDS hybrid rate, and genetic association between ChrX MFDS and Chr2L ‘speciation island’ SNPs (16). In contrast, the GOUNDRY subgroup displays i) no differentiation between mosquitoes carrying different MFDS genotypes (Fst=0 using Chr3 STRs, p=0.42, n=169), indicating that the M and S molecular forms do not exist within GOUNDRY, ii) a high rate of MFDS hybrids (36%), and frequencies of the three MFDS genotypes (M/M, M/S, S/S) in Hardy Weinberg Equilibrium (HWE) (χ2=2.449, p=0.29), iii) 39% association between MFDS and Chr2L ‘speciation island’ SNPs, which is not statistically different from random segregation (p=0.283) (16). Overall, these observations indicate that the molecular forms M and S and their incipient speciation are a feature only of the ENDO subgroup, and do not exist within GOUNDRY.
In agreement with previous studies (12), the ENDO subgroup, whether captured indoors as adults or from larval sites, is nearly fixed for the inverted 2La allele (98%). Unexpectedly for this zone, the wild-type 2La+ chromosome occurs at a frequency of 68% in the GOUNDRY subgroup, with all three resulting genotypes present at HWE frequencies (χ2=0.070, p=0.965, Figure 3C), but the genetic division between GOUNDRY and ENDO is not due to the presence of the 2La+ chromosome within GOUNDRY (Fig S5C).
Adult mosquitoes raised directly from wild larvae were fed on blood infected with wild P. falciparum. The susceptibility of GOUNDRY mosquitoes is significantly greater (58% infection prevalence) than ENDO mosquitoes (35% prevalence, χ2=22.181, p<0.001, n=439; Figure 3D). The difference in susceptibility between GOUNDRY and ENDO subgroups is robust to annual variation (Figure S7B, (16)), and is of the same magnitude as the difference between experimental and control mosquitoes in gene silencing studies of A. gambiae immune genes (23).
The ENDO subgroup is genetically more diverse than GOUNDRY (allelic richness 13.9 and 4.4, respectively; average heterozygosity 0.847 and 0.558 (16)). Given its lower diversity, the GOUNDRY subgroup may be evolutionarily more recent. At present, the geographic range of the GOUNDRY subgroup is unknown, though our current sampling covers at least 4×106 ha of the Sudan-Savanna zone, and our results suggest that the GOUNDRY/ENDO division may not exist in the ecologically distinct Guinea-Savanna zone to the SW (Figure S1). There are several reports from West Africa of rates of MFDS hybrids “higher than expected” (24, 25). Although no evidence of anthropophagy exists as yet for the GOUNDRY subgroup, any level of human feeding by mosquitoes displaying elevated parasite susceptibility would be of epidemiological concern (26–28). The GOUNDRY subgroup is abundant in its known range and would likely be selectively favored by indoor-based vector control measures such as insecticide spraying and insecticide treated bed nets. New tools including unbiased sampling methods should be developed to characterize and monitor this subgroup, as well as cryptic subgroups elsewhere.
Supplementary Material
Acknowledgments
We acknowledge support from NIH/NIAID to KDV, #AI073685 and AI042361.
REFERENCES AND NOTES
- 1.Pates H, Curtis C. Mosquito behavior and vector control. Annu Rev Entomol. 2005;50:53. doi: 10.1146/annurev.ento.50.071803.130439. [DOI] [PubMed] [Google Scholar]
- 2.Service MW. Mosquito ecology: Field sampling methods. 2. Chapman & Hall; London: 1993. [Google Scholar]
- 3.Coluzzi M, Sabatini A, Petrarca V, Di Deco MA. Behavioural divergences between mosquitoes with different inversion karyotypes in polymorphic populations of the Anopheles gambiae complex. Nature. 1977;266:832. doi: 10.1038/266832a0. [DOI] [PubMed] [Google Scholar]
- 4.Geissbuhler Y, et al. Interdependence of domestic malaria prevention measures and mosquito-human interactions in urban Dar es Salaam, Tanzania. Malar J. 2007;6:126. doi: 10.1186/1475-2875-6-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Odiere M, et al. Sampling outdoor, resting Anopheles gambiae and other mosquitoes (Diptera: Culicidae) in western Kenya with clay pots. J Med Entomol. 2007;44:14. doi: 10.1603/0022-2585(2007)44[14:soraga]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smits A, Roelants P, Van Bortel W, Coosemans M. Enzyme polymorphisms in the Anopheles gambiae (Diptera:Culicidae) complex related to feeding and resting behavior in the Imbo Valley, Burundi. J Med Entomol. 1996;33:545. doi: 10.1093/jmedent/33.4.545. [DOI] [PubMed] [Google Scholar]
- 7.Bockarie MJ, Service MW, Barnish G, Maude GH, Greenwood BM. Malaria in a rural area of Sierra Leone. III. Vector ecology and disease transmission. Ann Trop Med Parasitol. 1994;88:251. doi: 10.1080/00034983.1994.11812865. [DOI] [PubMed] [Google Scholar]
- 8.Charlwood JD, Vij R, Billingsley PF. Dry season refugia of malaria-transmitting mosquitoes in a dry savannah zone of east Africa. Am J Trop Med Hyg. 2000;62:726. doi: 10.4269/ajtmh.2000.62.726. [DOI] [PubMed] [Google Scholar]
- 9.Lehmann T, et al. Aestivation of the African malaria mosquito, Anopheles gambiae in the Sahel. Am J Trop Med Hyg. 2010;83:601. doi: 10.4269/ajtmh.2010.09-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toure YT, et al. The distribution and inversion polymorphism of chromosomally recognized taxa of the Anopheles gambiae complex in Mali, West Africa. Parassitologia. 1998;40:477. [PubMed] [Google Scholar]
- 11.Molineaux L, Shidrawi GR, Clarke JL, Boulzaguet JR, Ashkar TS. Assessment of insecticidal impact on the malaria mosquito’s vectorial capacity, from data on the man-biting rate and age-composition. Bull World Health Organ. 1979;57:265. [PMC free article] [PubMed] [Google Scholar]
- 12.Coluzzi M, Sabatini A, Petrarca V, Di Deco MA. Chromosomal differentiation and adaptation to human environments in the Anopheles gambiae complex. Trans R Soc Trop Med Hyg. 1979;73:483. doi: 10.1016/0035-9203(79)90036-1. [DOI] [PubMed] [Google Scholar]
- 13.della Torre A, et al. Molecular evidence of incipient speciation within Anopheles gambiae s.s. in West Africa. Insect Mol Biol. 2001;10:9. doi: 10.1046/j.1365-2583.2001.00235.x. [DOI] [PubMed] [Google Scholar]
- 14.White BJ, Cheng C, Simard F, Costantini C, Besansky NJ. Genetic association of physically unlinked islands of genomic divergence in incipient species of Anopheles gambiae. Mol Ecol. 2010 doi: 10.1111/j.1365-294X.2010.04531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costantini C, et al. Living at the edge: biogeographic patterns of habitat segregation conform to speciation by niche expansion in Anopheles gambiae. BMC Ecol. 2009;9:16. doi: 10.1186/1472-6785-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Supplementary Online Material including methods, figures, additional analysis, and primary data are available at Science Online.
- 17.Sharakhov IV, et al. Breakpoint structure reveals the unique origin of an interspecific chromosomal inversion (2La) in the Anopheles gambiae complex. Proc Natl Acad Sci U S A. 2006;103:6258. doi: 10.1073/pnas.0509683103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes. 2007;7:574. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenberg NA, Li LM, Ward R, Pritchard JK. Informativeness of genetic markers for inference of ancestry. Am J Hum Genet. 2003;73:1402. doi: 10.1086/380416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coluzzi M, Sabatini A. Cytogenetic observations on species A and B of the Anopheles gambiae complex. Parassitologia. 1967;9 [Google Scholar]
- 23.Mitri C, et al. Fine pathogen discrimination within the APL1 gene family protects Anopheles gambiae against human and rodent malaria species. PLoS Pathog. 2009;5:e1000576. doi: 10.1371/journal.ppat.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caputo B, et al. Anopheles gambiae complex along The Gambia river, with particular reference to the molecular forms of An. gambiae s.s. Malar J. 2008;7:182. doi: 10.1186/1475-2875-7-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliveira E, et al. High levels of hybridization between molecular forms of Anopheles gambiae from Guinea Bissau. J Med Entomol. 2008;45:1057. doi: 10.1603/0022-2585(2008)45[1057:hlohbm]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 26.Reisen WK, Boreham PF. Host selection patterns of some Pakistan mosquitoes. Am J Trop Med Hyg. 1979;28:408. doi: 10.4269/ajtmh.1979.28.408. [DOI] [PubMed] [Google Scholar]
- 27.Bruce-Chwatt LJ, Garrett-Jones C, Weitz B. Ten years’ study (1955–64) of host selection by anopheline mosquitos. Bull World Health Organ. 1966;35:405. [PMC free article] [PubMed] [Google Scholar]
- 28.White GB. Anopheles gambiae complex and disease transmission in Africa. Trans R Soc Trop Med Hyg. 1974;68:278. doi: 10.1016/0035-9203(74)90035-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.