Abstract
Background & Aims
Genetic studies of the serum expression of antibodies to microbial antigens may yield important clues to the pathogenesis of Crohn’s disease. Our aim was to conduct a linkage study using expression of anti-CBir1, anti-I2, anti-OmpC, and ASCA as quantitative traits.
Methods
Expression of antibodies to microbial antigens was measured by ELISA and a standard ~10cM whole genome microsatellite study was conducted. SNP genotyping was performed using either Illumina or TaqMan MGB technology. NFKB1 activation in cells from EBV-transformed cell lines was assessed using EMSA and protein was measured using ELISA and Western blotting.
Results
Evidence for linkage to anti-CBir1 expression was detected on human chromosome 4 (LOD 1.82 at 91cM). We therefore directly proceeded to test the association of haplotypes in NFKB1, a candidate gene. One haplotype, H1, was associated with anti-CBir1 (P = 0.003) and another, H3, was associated with ASCA (P = 0.023). Using cell lines from CD patients with either H1 or H3, NF-κB activation and NFKB1 p105 and p50 production were significantly lower for patients with H1 compared to patients with H3.
Conclusions
These results suggest that NFKB1 haplotypes induce dysregulation of innate immune responses by altering NFKB1 expression. The results also show the use of EBV-transformed lymphoblastoid cell lines to conduct phenotypic studies of genetic variation.
Keywords: NFKB1, anti-CBir1 antibody, innate immunity, Crohn’s Disease, genome-wide association studies
Introduction
Chronic relapsing, remitting intestinal inflammation is a characteristic common to all patients with Crohn’s disease (CD), however, CD demonstrates significant heterogeneity as patients can exhibit a broad spectrum of disease phenotypes and expression of serum antibodies to intestinal microbes.[1, 2, 3] CD has been associated with the presence of antibodies to Clostridium-related flagellin (anti-CBir1),[4, 5] a Pseudomonas fluorescens-related peptide (anti-I2),[6, 7] Escherichia coli outer membrane protein C (anti-OmpC),[2, 8] and Saccharomyces cerevisiae oligomannan (ASCA).[9, 10] We have observed that expression of antibodies to more than one of these antigens, as well as the magnitude of that expression, is associated with a more aggressive CD phenotype of stricturing, internal perforating disease and a greater requirement for small bowel surgery.[11] In addition, antibody to flagellin (anti-CBir1) alone identifies a CD subgroup with a more complicated CD phenotype [5] and a more aggressive CD behavior in pediatric patients.[12] We and others have also observed that ASCA and anti-OmpC are familial traits,[13, 14, 15] suggesting that genetic variation may lead to alterations in the expression of antibodies to microbial antigens. Taken together, these observations support the overall paradigm that genetic variation may alter responses to commensal microbes by either the innate, adaptive or both immune systems and that such alterations may lead to variations in CD phenotype. Therefore, genetic studies of these various responses may yield important clues to the alterations and pathways related to CD pathogenesis.
This concept is further supported by evidence from experimental mouse models in which alteration of one or more host genes results in colitis when mice are raised under normal conditions, but not when raised in microbe-free conditions.[16] Pertinent to this study are the findings that targeted disruption of the IL10 gene in C3H/HeJBir mice causes a severe colitis but this does not occur in C57BL/6J mice.[17, 18] Using this differential susceptibility/resistance, a major modifying locus for this colitis was identified on mouse chromosome 3, the Cytokine deficiency-induced colitis susceptibility (Cdcs) 1 locus.[19] Subsequently, Beckwith and co-workers refined the location of Cdcs1 to a 7 Mb interval spanning the nfkb1 gene.[20] Using a series of congenics, these authors demonstrated that: a) all aspects of the colitis phenotype were confined to this interval; b) macrophages from the C3H/HeJBir strain expressed higher levels of nuclear factor (NF)-kappa (κ)-B p50; c) C3H/HeJBir showed reduced innate responsiveness both in vivo and in vitro to bacterial ligands, including flagellin; and d) C3H/HeJBir showed an increased CD4 T cell response compared to the C57BL/6J strain.
This study is part of our ongoing research on the genetics of sub-phenotypes of IBD, in particular, the genetics of serum expression of anti-CBir1 antibodies. Beginning with a whole genome linkage study of antibody expression as a quantitative trait, we report (1) the evidence for linkage of anti-CBir1 expression to a human region syntenic to the mouse locus Cdcs1, (2) the association of NFKB1 haplotypes to anti-CBir1 expression, and (3) functional differences between NFKB1 haplotypes.
Materials and Methods
Three studies were conducted: a linkage study of the expression of each of four serum antibodies (anti-CBir1, anti-I2, anti-OmpC, and anti-Saccharomyces cerevisiae, ASCA), a case-control study of the association of the NFKB1 gene, and a genotype-phenotype study of NFKB1 function using lymphoblastoid cell lines from CD subjects with various NFKB1 haplotypes.
Subjects
Recruitment of subjects at the Cedars-Sinai IBD Center was conducted under the approval of the local Institutional Review Board. Disease phenotype was assigned using a combination of standard endoscopic, histological, and radiographic features as previously described.[11] The characteristics of the subjects studied are found in Table 1: a) The “Linkage Subjects” were members of 80 “CD-only” families, families with at least two members affected with CD and no known members affected with UC, and 57 “Mixed” families, families with at least two members affected with either CD or UC: (b) The “Case Control Subjects” consisted of 763 CD patients and 254 controls, mainly spouses, matched on ethnicity (Ashkenazi Jewish and non-Jewish); c) “Genotype-Phenotype Subjects” were case-control subjects selected based on NFKB1 haplotype.
Table 1.
Characteristics of Subjects
| a. Linkage Subjects | ||
|---|---|---|
| "CD Only" Families | "Mixed" Families | |
| Number of Families | 80 | 57 |
| Number of subjects with CD | 202 | 84 |
| Number of subjects with UC | 71 | |
| Number of subjects unaffected | 285 | 238 |
| Total number of subjects genotyped & with complete serum measurements for all 4 antibodies | 358 | 292 |
| b. Case-Control Subjects | ||
| CD Cases | Controls | |
| Jewish | 306 | 51 |
| Non-Jewish | 441 | 201 |
| c. Genotype-Phenotype Subjects-zEBV-transformed Cell Lines | ||
| NFKB1 H1/H1 | 11 | |
| NFKB1 H1/X | 3 | |
| NFKB1 H3/H3 | 8 | |
| NFKB1 H3/X | 3 | |
| NFKB1 X/X | 13 | |
Genotyping
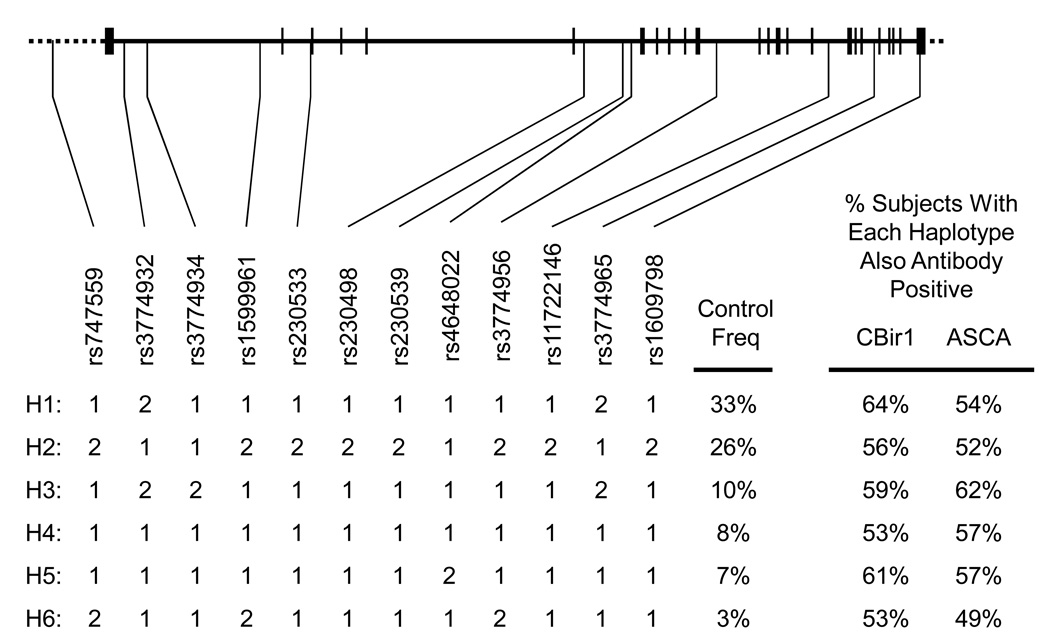
(a) Linkage Study. “Linkage Subjects” (Table 1a) were genotyped for 381 autosomal microsatellite markers from Weber set 9, average spacing ~10 cM. The length of dye-labeled polymerase chain reaction products were measured by computer-aided electrophoresis (Applied Biosystems, Foster City, CA).[21] (b)Case-Control Study. Single nucleotide polymorphisms (SNPs) markers were genotyped using either Illumina Golden Gate technology (Illumina, Foster City, CA) [22]or TaqMan MGB technology (Applied Biosystems, Foster City, CA). [23, 24] The SNPs studied are shown in Figure 3.
Figure 3. NFKB1 SNPs and haplotypes.
This figure showing the location of NFKB1 SNPs studied was created with the aid of the UCSC Genome Browser (http://genome.ucsc.edu). Diagram of the NFKB1 gene. Vertical lines show the position of exons. [31] List of SNPs studied. Accession numbers (“rs numbers”) in the dbSNP of the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov) are listed along with arrows showing their position along the NFKB1. The most common NFKB1 haplotypes as observed in this study. The SNP combinations that make up the six major haplotypes are shown, along with the frequency in the controls of this study are listed. "2" refers to the minor allele observed in the controls. Except for rs3774934 and rs1609798 the pairwise r2 between all SNPs shown is >85%. The r2 for rs3774934 ranged from >90% with rs747559 to rs1599961 and 76% with rs230533 to rs3774968. The r2 for rs1609798 was >88% except with rs3774934 (66%) and rs4648022 (53%).
Enzyme-Linked Immunosorbent Assay (ELISA) of Anti-CBir1, anti-I2, anti-OmpC, and ASCA antibody
Sera from all subjects in both the Linkage Study and the Case-Control Study were analyzed for anti-CBir1, anti-I2, anti-OmpC, and ASCA expression in a blinded fashion at the Cedars-Sinai Medical Center with a fixed ELISA as described previously.[2, 5] Anti-CBir1, anti-I2, and anti-OmpC positive was defined as the mean ± 2 SD of the healthy controls. Sera showing ASCA reactivity (IgG, IgA, or both) exceeding the reference range were termed ASCA positive (ASCA+).
Nuclear and Cytosolic Cell Extracts
EBV-transformed lymphoblastoid cell lines were selected based on NFKB1 haplotype in CD patients (Genotype-Phenotype Subjects, Table 1c) [25]. Nuclear and cytosolic proteins were extracted from these cell lines from subjects with specific haplotypes. Briefly, cells were harvested and resuspended in lysing buffer for 5 min. The supernatant (cytosolic fraction) was stored at − 80 °C until use. The pelletted nuclei were rinsed twice with 1ml of Nonidet P-40-free lysing buffer. After centrifugation, the pelletted nuclei were resuspended in 60 µl of nuclear suspension buffer. Nuclear suspensions were centrifuged and the supernatant was collected for nuclear protein. Protein concentrations were determined by the Bradford method.
Electrophoretic Mobility Shift Assay (EMSA)
DNA probe containing a consensus κB enhancer element was used (sc-2505, Santa Cruz Biotechnology, Santa Cruz, CA). The probe was 5’-endlabeled with γ-32P ATP using T4 polynucleotide kinase (Promega Corporation, Madison, WI). Nuclear protein extracts (5 µg) were incubated with a 5’-end-labeled probe. Reaction products were separated on 6% polyacrylamide gels. The gels were dried and visualized by autoradiography. For analysis of NF-κB activation, the digitized quantification of the specific NF-κB bands in one H1/H1 cell line was set as 1.0, and the intensities of the bands in other samples were expressed in relation to this reference value.
NF-κB Activity Assays
NF-κB p50 activities from 5 µg nuclear protein were measured using the Active Motif TransAM NF-κB ELISA, with bound oligonucleotide containing the NF-κB consensus binding site, as recommended by the manufacturer (Active Motif, Carlsbad, CA).
Western Blot Analysis
10 µg nuclear and cytosolic proteins were separated by 10% SDS-polyacrylamide gel electrophoresis and transferred onto PDVF membranes (Invitrogen, Carlsbad, CA). NF-κB subunit p65, p50 and p105 were stained with a rabbit anti-human NF-κB/p65 antibody (Santa Cruz Biotechnology) and p50 and its precursor p105 with a rabbit anti-human NF-κB/p50 and p105 antibody (Santa Cruz Biotechnology). HRP-conjugated goat anti-rabbit IgG was used for secondary antibody.
Statistical Analysis
The antibody serum levels were log transformed to enable analyses based on the normal distribution. These levels were used as a quantitative trait in the Linkage Study and the Case-Control Study. Variance components linkage analysis as implemented in the SOLAR computer program was used to test for linkage of the microsatellite markers.[26] The LOD score was used, not to establish linkage, but as a method of ranking the genetic regions in which to look for candidate genes. For the Case-Control Study, haplotypes were assigned from individual SNP data using the computer program PHASE v2.[27] Association of NFKB1 haplotypes was tested using chi-square and the significance of results was assessed by applying a permutation test to the data in order to correct for the multiple testing of haplotypes in Haploview.[28],[29] Wilcoxon test was used to analyze the results in ELISA and densitometric data of EMSA and Western blotting. Differences were considered significant at P < 0.05.
Results
Linkage Study: Linkage of anti-CBir1 expression to human chromosome 4
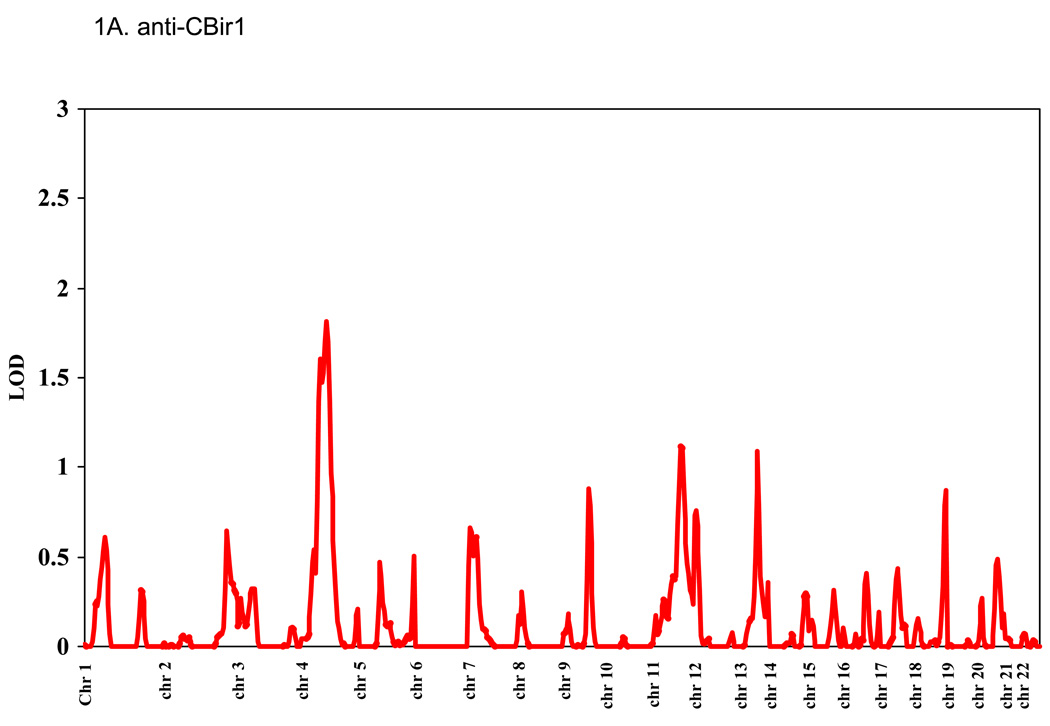
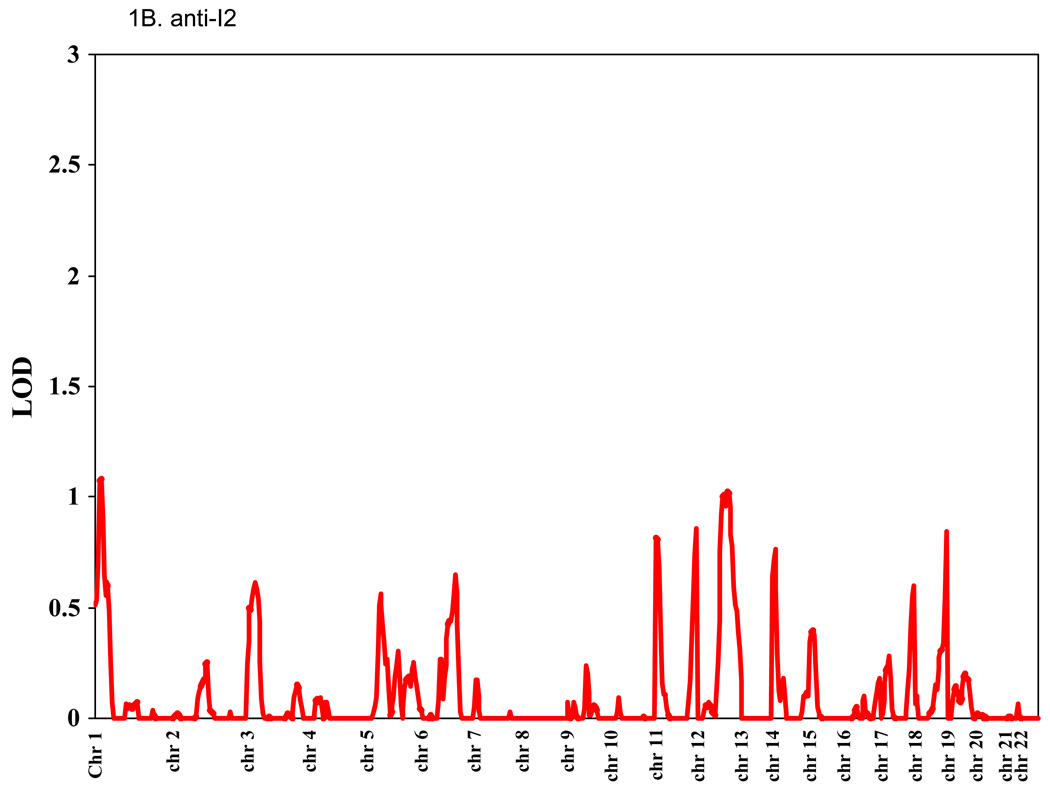
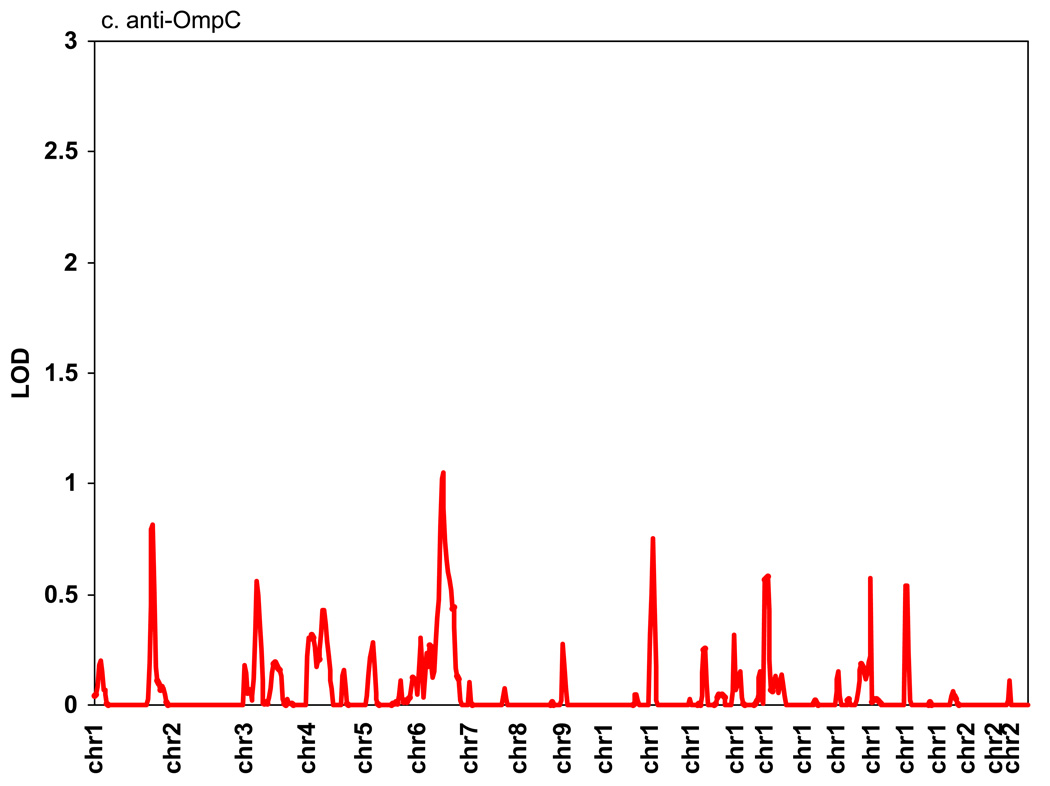
Genomic regions showing support for linkage to anti-CBir1, anti-I2, anti-OmpC, and ASCA are shown in Figure 1. The highest LOD score for anti-CBir1 was 1.82 on Chr4, for anti-I2 was 1.14 on Chr1, for anti-OmpC was 1.1 on Chr 6, and for ASCA was 1.8 on Chr 5. The heritability for anti-CBir1 level was 0.19 (p = 0.0005). Since we have previously observed that CD severity is associated with the number of antibodies expressed in a given subject, bivariate linkage of two antibodies was calculated pairwise. No new peaks with LOD > 1 were observed, and only one peak showed a very modest increase (data not shown except for anti-CBir1 and ASCA expression and Chr 4, see below).
Figure 1. Linkage study of antibody expression.
The levels of serum expression of anti-CBir1, anti-I2, anti-OmpC, and ASCA were treated as quantitative traits and LOD scores were calculated using the variance components method as implemented in the SOLAR computer program.[26] Average spacing of microsatellite markers was ~10 cM. (A) anti-CBir1, (B) anti-I2, (C) anti-OmpC, and (D) ASCA.
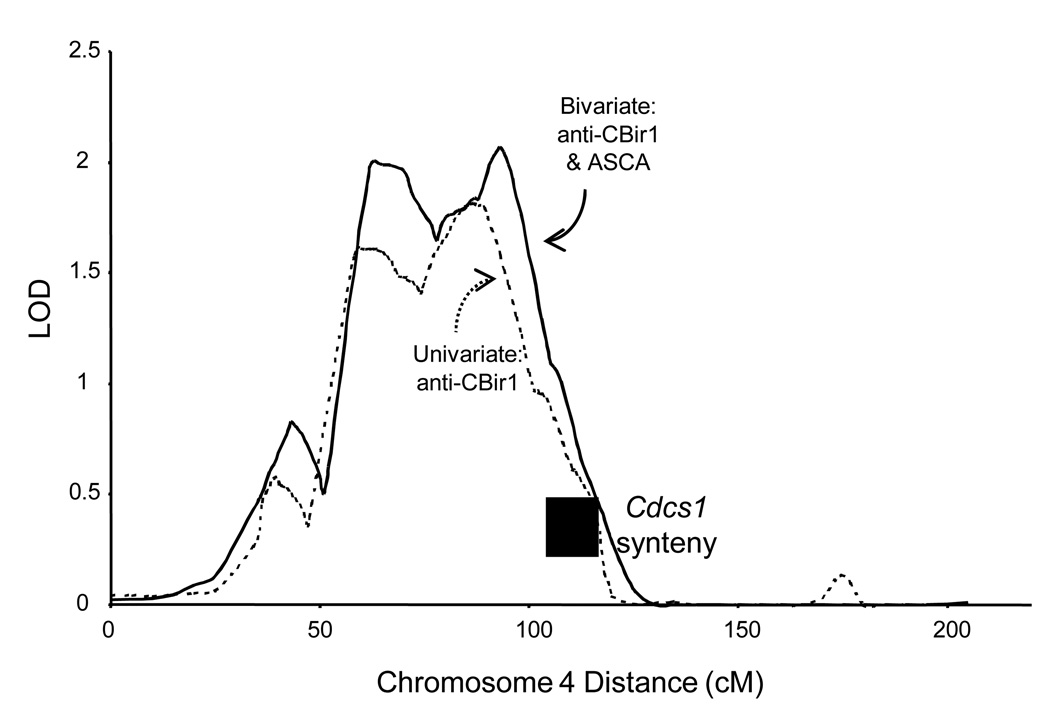
With bivariate linkage analysis of anti-CBir1 and ASCA together, the anti-CBir1 peak on Chr 4 increased from 1.82 to 1.99 (Figure 2). We observed that the −1 LOD region of this peak included both the human NFKB1 gene, located as shown in Fig 1B, and the region syntenic to the mouse Cdcs1 locus, approximately located between human markers MFD324 and AFM248ZG9 (~105–113cM).[30] Since aspects of the mouse Cdcs1 phenotype are similar to the human expression of anti-CBir, we directly tested NFKB1 as a candidate gene for anti-CBir1 and ASCA expression.
Figure 2. Linkage of anti-CBir1 expression to human chr 4.
Detail for the highest LOD score on chromosome 4 (LOD = 1.82 at 91 cM). The dotted line represents anti-CBir1 expression alone, and the solid line is for a bivariate analysis of anti-CBir1 and ASCA together. The position of the human region syntenic to the mouse sequence for the Cdcs1 locus is also shown.
Case-Control Study: Association of anti-CBir1 and ASCA with NFKB1
We observed 6 NFKB1 haplotypes with frequency greater than 1% (Figure 3). Haplotype 1 (H1) was associated with the presence of anti-CBir1 expression (Figure 4A) and H3 was associated with the presence of ASCA expression (Figure 4B). We did not observe an association between CD or UC and these NFKB1 haplotypes, nor with a previously reported NFKB1 insertion/deletion polymorphism (data not shown; [31]). When the families with NFKB1 H1 were removed, there was a modest decrease in the observed LOD, from 1.8 to 1.6.
Figure 4. Association of anti-CBir1 and ASCA with NFKB1 haplotypes.
The proportion of subjects positive for either anti-CBir1(a) or ASCA (b) is shown in relation to NFKB1 haplotype. Haplotypes of NFKB1 were tested for association with the presence or absence of antibody expression by chi-square test. P-values are empirical P-values by permutation test in order to correct for multiple comparisons (n=6 haplotypes).
Genotype-Phenotype Study: The anti-CBir1-related NFKB1 haplotype (H1) is associated with the down-regulation of NF-κB
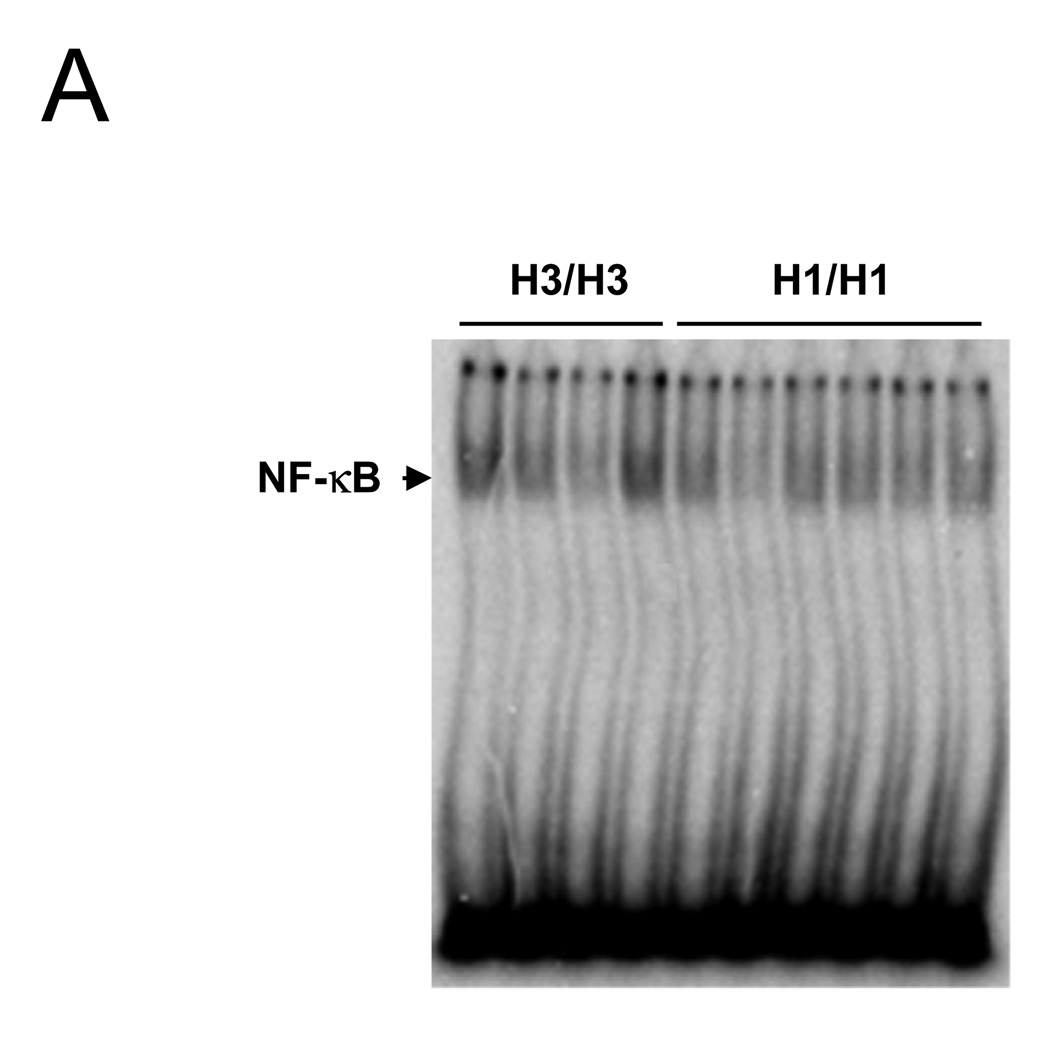
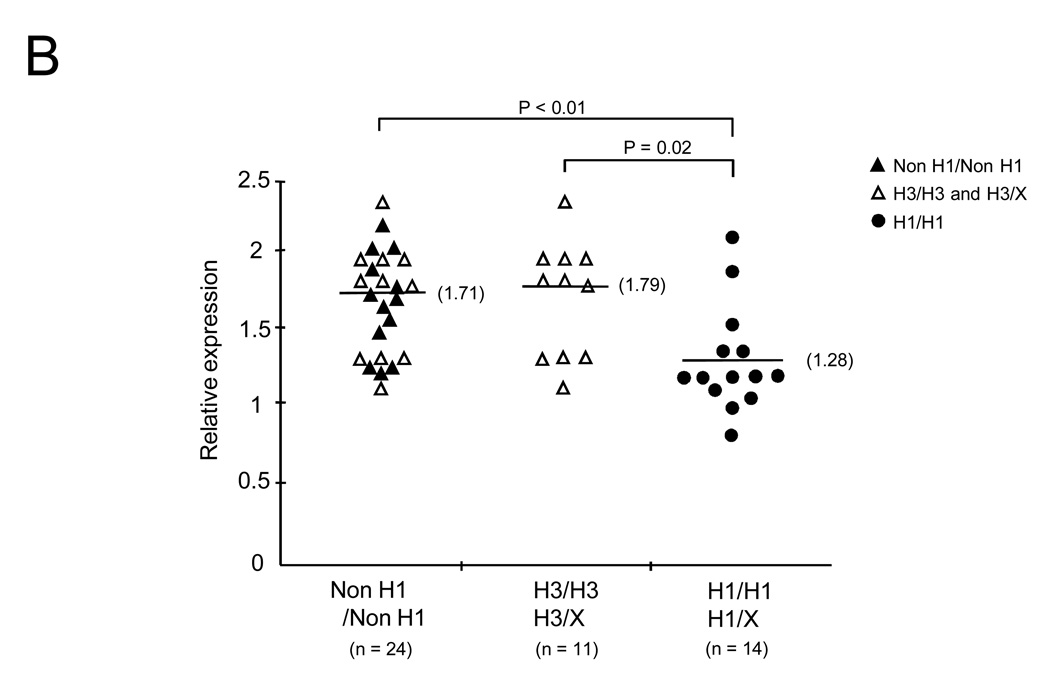
To investigate whether anti-CBir1 related NFKB1 haplotype (H1) affects NF-κB activation, EBV transformed B cell lines were generated from peripheral blood cells of CD patients known to be homozygous for specific haplotypes. We prepared 14 cell lines from H1 positive (+) patients and 24 cell lines from H1 negative (−) patients including 11 from ASCA associate haplotype H3. First, nuclear proteins were extracted from these cell lines and EMSA was performed to measure NF-κB activation (Figure 5A). NF-κB activation was quantitated by densitometric analysis. The median NF-κB activation was significantly lower for patients with H1 (+) compared with patients with cells without H1 (−)(Figure 5B open and close triangle) (median NF-κB relative ratio for H1(+) was 1.28 compared to H1 (−) 1.71, P < 0.01) as well as H1(+) compared to H3(+)(Figure 5B open triangle)(median NFKB relative ratio for H1 1.28 compared to H3 1.79 P = 0.02) There were too few H1(+) heterozygotes (n = 3) to establish any gene dose effect.
Figure 5. Down-regulation of NF-κB activation in B cell lines from Crohn’s Disease patients with anti-CBir1 related NFKB1 haplotype.
Haplotype specific B cell lines were prepared from peripheral blood of CD patients. EMSA analysis was performed in non H1/non H1 (n = 24) including H3/H3 and H3/X (non H1) (n = 11) and H1/H1 and H1/X (non H3) (n = 14). (A) NF-κB-specific band from H3/H3 and H1/H1 was shown. (B) Densitometric readings were obtained. The median NF-κB relative expression was lower for H1/H1 and H1/X (closed circle) (1.28) compared to non H1 (closed and opened triangle). The median NF-κB relative expression was lower for H1/H1 and H1/X (1.28) compared to H3/H3 and H3/X (opened triangle). All P-values are generated by Wilcoxon test.
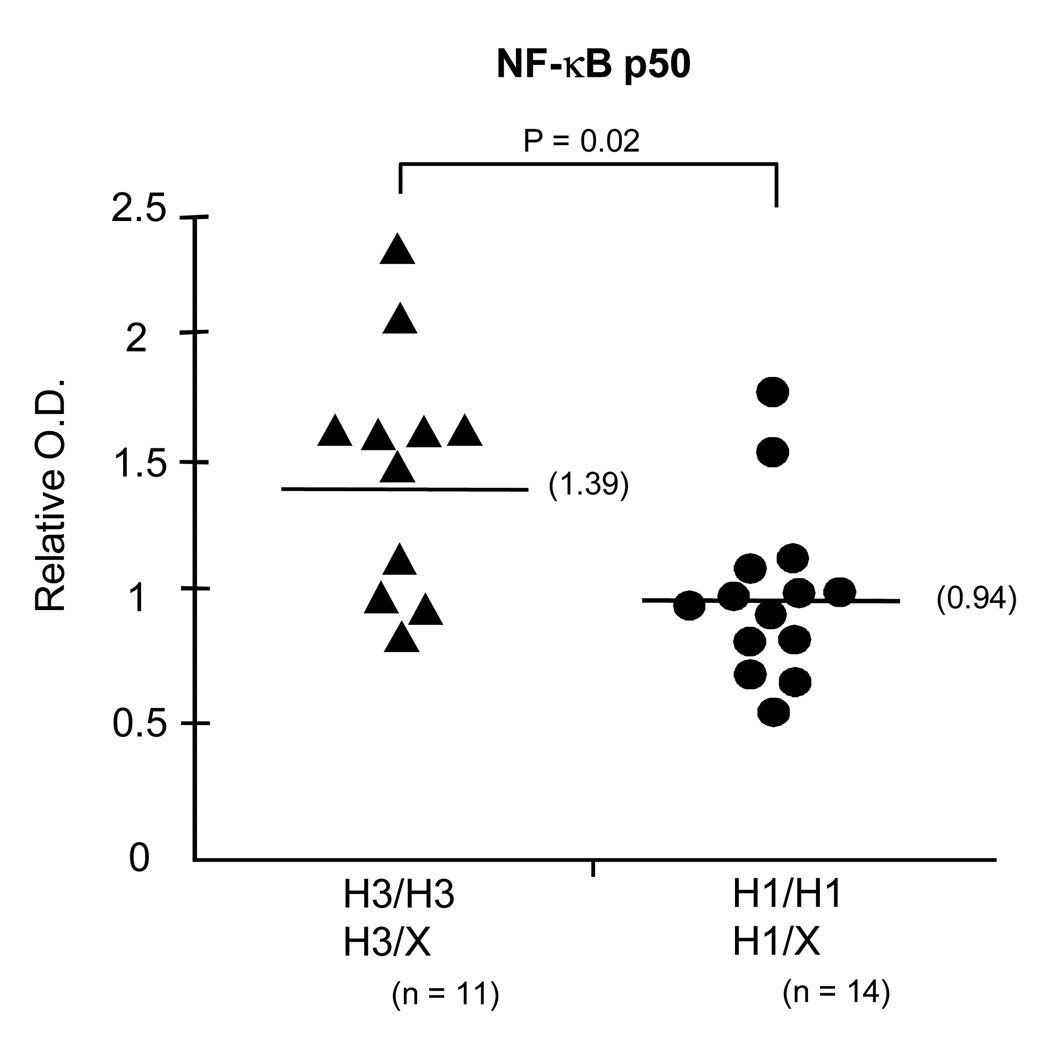
NF-κB is a heterodimer consisting of NF-κB p50 from NFKB1 and p65 from ReIA. To further assess NFKB1/p50 expression, we performed ELISA to measure the quantitation of NF-κB p50 for nuclear protein-DNA binding to consensus NF-κB motif by using NF-κB p50 antibodies. These data show that NF-κB p50 from H1 (+) cells significantly bound the consensus NF-κB motif less than p50 from H3 (+) cells (median NF-κB relative ratio for H1 was 0.94 compared to H3, 1.39, P=0.02) (Figure 6).
Figure 6. Association of NF-κB p50 with NFKB1 haplotypes.
ELISA analysis of NF-κB p50 from nuclear protein in H3/H3 and H3/X (non H1) (n = 11) and H1/H1 and H1/X (non H3) (n = 14) subjects. The median NF-kB p50 expression was lower for H1 compared to H3 patients.
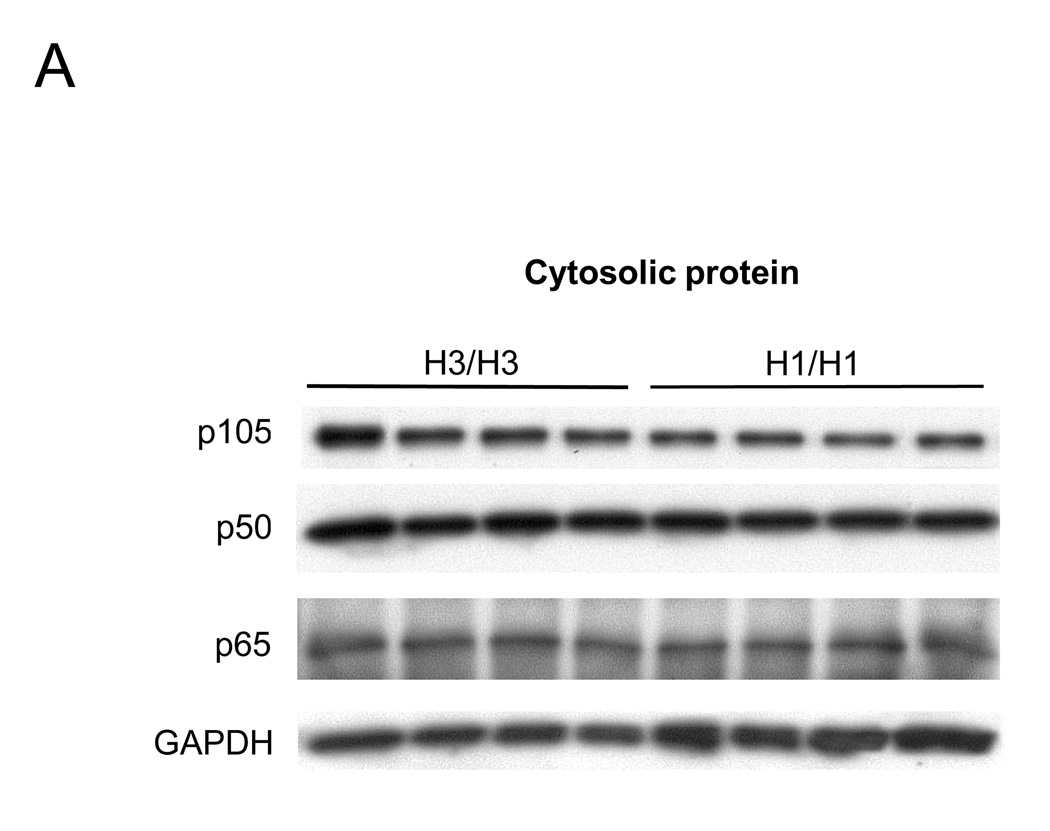
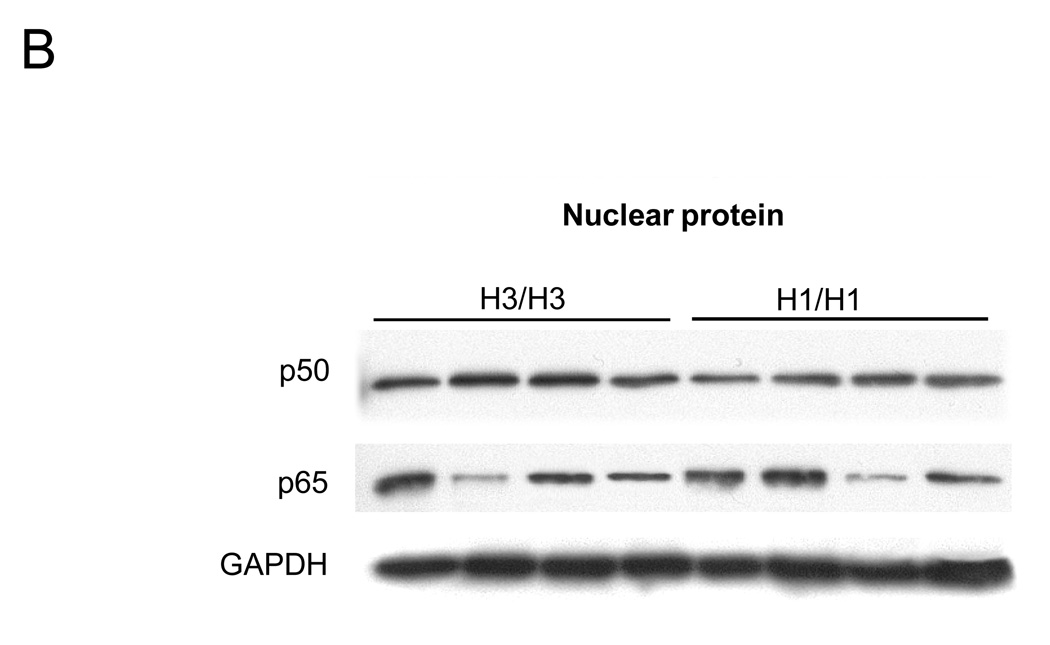
Because NF-κB1/p105 is the inactive precursor of p50 and is localized to the cytoplasm, with p50 being translocated into the nucleus upon activation, we next measured the production of the NF-κB1 proteins (both p105 and p50) in cytosolic and nuclear proteins by Western blot to investigate whether the different NFKB1 haplotypes influence the production of NF-κB1 proteins such as p105 and p50. In cytosolic protein, levels of both p105 and p50 proteins from H1 homozygotes were lower than the levels seen in H3 patients by densitometrical analysis (median p105 relative ratio for H1 was 0.85 compared to H3, 1.00, P = 0.02, and median p50 relative ratio for H1 was 0.93 compared to H3 1.00, P = 0.035) (Figure 7A). There was no significant difference in NF-κB p65 cytosolic or nuclear protein production between H1 and H3. In nuclear protein, p50 from H1 group was lower than from H3 B cells (median p50 relative ratio for H1 was 0.84 compared to H3 1.00, P = 0.02) (Figure 7B). Taken together, these functional studies demonstrated the anti-CBir1-related NFKB1 haplotype is associated with decreased p105 and p50 expression and function, leading to down-regulated NF-κB activation.
Figure 7. Productions of NF-κB p105, p50 and p65 with NFKB1 haplotypes in cytosolic and nuclear protein.
NF-κB p105, p50 and p65 expression from cytosolic (A) and p50 and p65 expressions from nuclear protein (B) in H3/H3 and H1/H1 subjects were analyzed by Western blot analysis. (A)
Discussion
A genome wide linkage study of antibody expression in Crohn’s disease patients has led, by way of human synteny to a mouse locus for a related phenotype, to the identification of NFKB1 haplotypes associated with anti-CBir1 and ASCA expression. Furthermore, an anti-CBir1-related haplotype, NFKB1 H1, results in lowered expression of NFKB1 in human H1/H1 EBV-transformed lymphoblastoid cell lines. These observations support the concepts that the NFKB1 gene is involved in human antibody expression and that changes in NF-κB1 expression are involved in anti-CBir1-expression in human CD. Since we have observed the association of antibody expression and severity of CD, perhaps alterations in NF-κB expression may be involved in disease progression or severity as well. The observations reported here afford a great opportunity to study further details of the relationships between NFKB1 variation, NF-κB expression, immune dysregulation, and serum expression of antibodies to microbial antigens in CD patients.
Our linkage evidence supports a region of chromosome 4 as a quantitative trait locus for the serum expression of anti-CBir1 and ASCA in CD subjects. However, we observed only a modest reduction of the variance components LOD score when subjects with NFKB1 H1 were omitted from analysis. There are several possible reasons for this finding: first, expression of anti-CBir1 and ASCA probably have a large environmental component that complicates the analysis of this complex trait. Second, the number of NFKB1 mutations in CD subjects may be very large, so that a family-based study will detect transmission to affected subjects, but an association study based on major haplotypes may not detect mutations on a varied haplotypic background. Third, there may be other genetic variation at this locus that determines response to microbial antigens. While the human region syntenic to the mouse Cdcs1 locus is relatively narrow, the linkage region is not. The minus 1 LOD interval, roughly between GATA72G09N and GATA62A12, spans other candidate genes, most notably IGJ, CXCL1–6, 9–11, 13, and IL8. This is not unusual with a complex trait. There is evidence that the three common CD associated variants within NOD2 do not explain the entire CD linkage signal at the IBD1 locus on chromosome 16 [32] and that TCF7L2 does not explain the majority of the chromosome 10 linkage signal in type 2 diabetes[33]. Interestingly, despite the fact that TCF7L2 is the most-widely replicated loci in type 2 diabetes, the original linkage signal in Caucasians implicating this region revealed a multipoint LOD score of less than 2.0[34].
The only other locus with a LOD score of greater than 1.5 was linkage of ASCA to chromosome 5. The most interesting candidate gene located under this peak is PTGER4. A Franco-Belgian GWAS identified a number of variants in a gene desert on chromosome 5p13 strongly associated with CD[35]. Using a study of global expression these investigators identified that these variants were associated with altered expression of the prostaglandin receptor PTGER4 (EP4)[36] and this locus in a cohort of ASCA serotyped CD cases is worthy of further research. These results support the experimental approach of using expression of antibodies to CD associated microbial antigens and population wide genetic techniques to search and find genetic variants otherwise not found in the population at large.
Linkage disequilibrium mapping of the entire minus 1 LOD interval on chromosome 4 is currently underway in order to find other genetic determinants of expression of anti-CBir1 antibody and therefore determinants of the response to bacterial flagellin. It is also possible that these haplotypes could be in LD with variants in neighboring genes. The data from the HapMap [37] project suggest that in Caucasians there is a recombination hot spot centromeric to NFKB1 with little if any LD in this direction. In the telomeric direction, LD extends beyond NFKB1 into the gene encoding lysosomal mannosidase beta (MANBA) which contains genetic variants associated with the autosomal recessive lysosomal storage disease beta-mannosidosis (mental retardation, cutaneous manifestations and an absence of any reported gastrointestinal manifestations).
The NF-κB family consists of RelA (p65), NF-κB1 (p50/p105), NF-κB2 (p52/p100), c-Rel, and RelB. In many cell types, the most abundant form of NF-κB is the p50/p65 heterodimer, which regulates the expression of genes involved in the immune and acute-phase responses. Aberrant activation of NF-κB controls the expression of many genes of inflammatory cytokines involved in the pathogenesis of IBD.[38] The three CD associated mutations in the NOD2/CARD15 gene on chromosome 16 all have a defect in their ability to activate NF-κB.[39] Recent evidence suggests that this may result in a defect in the innate immune system’s ability to protect the gut against invasive bacteria.[40] Furthermore, an insertion/deletion variant (−94ins/del ATTG) in the promoter of NFKB1 has previously been associated with ulcerative colitis in humans from North America, with homozygosity for the deletion conferring a moderate increase in ulcerative colitis (UC) risk (OR 1.57, 95% CI 1.14–2.16).[31] Therefore, the association between NF-κB and the dysregulation of innate immune responses may be involved in the pathogenesis of IBD.
EBV-transformed B cell lines have been used recently to demonstrate heritable variation in human gene expression,[41] and specific germ-line determinants that regulate gene expression have been identified.[42] Here, we demonstrate that immortalized B cell lines can reflect the functional consequences associated with different haplotypes of a specific gene. Furthermore, the effects of individual polymorphisms that impact NF-κB activation were detected in this study. Given that this assay has detected herein significant effects of SNPs in NFKB1, it is likely that it can be used to detect novel polymorphisms with similar phenotypes and that it provides a tool to perform ex vivo human genetic studies. In this study, we showed that B cell lines with the anti-CBir1-associated haplotype (H1) had lower NFKB1 activity than non-H1+ B cell lines. Furthermore, B cell lines with the anti-CBir1-associated haplotype (H1) also had lower protein production of NFKB1 than the ASCA associated haplotype (H3), which was likely the reason for the reduced NF-κB activation in H1 cell lines. The difference in p105/p50 expression using Western blots were not dramatic, nonetheless, significant, where as p65 was not. Importantly, however, these data are completely in accordance with the magnitude of differences seen with two other assays (EMSA & ELISA).
Taken together, we demonstrated that the genetic variation identified by haplotypes affects NFKB1 differentially. Previous animal studies provide evidence in vivo and in vitro that the C3Bir Cdcs1 susceptibility allele confers a reduced innate immune response to bacterial antigens signaling through Toll-like receptors (TLRs), including TLR2, TLR5 and TLR9, and NOD2. Our results in humans are consistent with the reduced induction of NFKB1 driven inflammatory genes that is associated with this colitogenic Cdcs1 locus in the mouse. Our present findings indicate that NF-κB is less activated in anti-CBir1 positive CD patients and suggests that NF-κB may abrogate the innate immune response to bacteria in this subpopulation of CD patients. This relative defect could be permissive to an upregulated adaptive immune response leading to intestinal inflammation.
Acknowledgments
Grant Support: Supported by USPHS Grants PO1DK071176 and PO1DK046763, the Feintech Chair in Inflammatory Bowel Disease (SRT) and the Cedars-Sinai Board of Governor’s Chair in Medical Genetics (JIR). Genotyping was supported in part by M01-RR00425 to the Cedars-Sinai GCRC genotyping core (KDT).
Abbreviations
- Ab
Antibody
- ASCA
anti-Saccharomyces cerevisiae antibody
- Cdcs
Cytokine deficiency-induced colitis susceptibility
- EBV
Epstein-Barr virus
- ELISA
enzyme-Linked Immunosorbent Assay
- EMSA
electro-mobility shift assay
- I2
a Pseudomonas fluorescens-related peptide
- NF-κB
nuclear factor-kappa-B
- OmpC
a Escherichia coli outer membrane protein C
- SNPs
single nucleotide polymorphisms
- TLR
Toll-like receptor
Footnotes
"The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or non exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd and its Licensees to permit this article (if accepted) to be published in Gut editions and any other BMJPGL products to exploit all subsidiary rights, as set out in our licence (http://gut.bmjjournals.com/ifora/licence.dtl)."
Competing Interest: None to declare.
References
- 1.Yang H, Taylor KD, Rotter JI. Inflammatory bowel disease. I. Genetic epidemiology. Mol Genet Metab. 2001;74:1–21. doi: 10.1006/mgme.2001.3199. [DOI] [PubMed] [Google Scholar]
- 2.Landers CJ, Cohavy O, Misra R, et al. Selected loss of tolerance evidenced by Crohn's disease-associated immune responses to auto- and microbial antigens. Gastroenterology. 2002;123:689–699. doi: 10.1053/gast.2002.35379. [DOI] [PubMed] [Google Scholar]
- 3.Vermeire S, Rutgeerts P. Antibody responses in Crohn's disease. Gastroenterology. 2004;126:601–604. doi: 10.1053/j.gastro.2003.12.020. [DOI] [PubMed] [Google Scholar]
- 4.Lodes MJ, Cong Y, Elson CO, et al. Bacterial flagellin is a dominant antigen in Crohn disease. J Clin Invest. 2004;113:1296–1306. doi: 10.1172/JCI20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Targan SR, Landers CJ, Yang H, et al. Antibodies to CBir1 flagellin define a unique response that is associated independently with complicated Crohn's disease. Gastroenterology. 2005;128:2020–2028. doi: 10.1053/j.gastro.2005.03.046. [DOI] [PubMed] [Google Scholar]
- 6.Wei B, Huang T, Dalwadi H, et al. Pseudomonas fluorescens encodes the Crohn's disease-associated I2 sequence and T-cell superantigen. Infect Immun. 2002;70:6567–6575. doi: 10.1128/IAI.70.12.6567-6575.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iltanen S, Tervo L, Halttunen T, et al. Elevated serum anti-I2 and anti-OmpW antibody levels in children with IBD. Inflamm Bowel Dis. 2006;12:389–394. doi: 10.1097/01.MIB.0000218765.84087.42. [DOI] [PubMed] [Google Scholar]
- 8.Arnott ID, Landers CJ, Nimmo EJ, et al. Sero-reactivity to microbial components in Crohn's disease is associated with disease severity and progression, but not NOD2/CARD15 genotype. Am J Gastroenterol. 2004;99:2376–2384. doi: 10.1111/j.1572-0241.2004.40417.x. [DOI] [PubMed] [Google Scholar]
- 9.Lindberg E, Magnusson KE, Tysk C, et al. Antibody (IgG, IgA, and IgM) to baker's yeast (Saccharomyces cerevisiae), yeast mannan, gliadin, ovalbumin and betalactoglobulin in monozygotic twins with inflammatory bowel disease. Gut. 1992;33:909–913. doi: 10.1136/gut.33.7.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quinton JF, Sendid B, Reumaux D, et al. Anti-Saccharomyces cerevisiae mannan antibodies combined with antineutrophil cytoplasmic autoantibodies in inflammatory bowel disease: prevalence and diagnostic role. Gut. 1998;42:788–791. doi: 10.1136/gut.42.6.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mow WS, Vasiliauskas EA, Lin YC, et al. Association of antibody responses to microbial antigens and complications of small bowel Crohn's disease. Gastroenterology. 2004;126:414–424. doi: 10.1053/j.gastro.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 12.Dubinsky MC, Lin YC, Dutridge D, et al. Serum immune responses predict rapid disease progression among children with Crohn's disease: immune responses predict disease progression. Am J Gastroenterol. 2006;101:360–367. doi: 10.1111/j.1572-0241.2006.00456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vermeire S, Peeters M, Vlietinck R, et al. Anti-Saccharomyces cerevisiae antibodies (ASCA), phenotypes of IBD, and intestinal permeability: a study in IBD families. Inflamm Bowel Dis. 2001;7:8–15. doi: 10.1097/00054725-200102000-00002. [DOI] [PubMed] [Google Scholar]
- 14.Sutton CL, Yang H, Li Z, et al. Familial expression of anti-Saccharomyces cerevisiae mannan antibodies in affected and unaffected relatives of patients with Crohn's disease. Gut. 2000;46:58–63. doi: 10.1136/gut.46.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mei L, Targan SR, Landers CJ, et al. Familial expression of anti-Escherichia coli outer membrane porin C in relatives of patients with Crohn's disease. Gastroenterology. 2006;130:1078–1085. doi: 10.1053/j.gastro.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 16.Elson CO, Cong Y, McCracken VJ, et al. Experimental models of inflammatory bowel disease reveal innate, adaptive, and regulatory mechanisms of host dialogue with the microbiota. Immunol Rev. 2005;206:260–276. doi: 10.1111/j.0105-2896.2005.00291.x. [DOI] [PubMed] [Google Scholar]
- 17.Berg DJ, Davidson N, Kuhn R, et al. Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4(+) TH1-like responses. J Clin Invest. 1996;98:1010–1020. doi: 10.1172/JCI118861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahler M, Leiter EH. Genetic and environmental context determines the course of colitis developing in IL-10-deficient mice. Inflamm Bowel Dis. 2002;8:347–355. doi: 10.1097/00054725-200209000-00006. [DOI] [PubMed] [Google Scholar]
- 19.Farmer MA, Sundberg JP, Bristol IJ, et al. A major quantitative trait locus on chromosome 3 controls colitis severity in IL-10-deficient mice. Proc Natl Acad Sci U S A. 2001;98:13820–13825. doi: 10.1073/pnas.241258698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beckwith J, Cong Y, Sundberg JP, et al. Cdcs1, a major colitogenic locus in mice, regulates innate and adaptive immune response to enteric bacterial antigens. Gastroenterology. 2005;129:1473–1484. doi: 10.1053/j.gastro.2005.07.057. [DOI] [PubMed] [Google Scholar]
- 21.Birren B, Green E, Klapholz S, et al. Analyzing DNA Volume 1.1 ed. Cold Spring Harbor Laboratory. 1997:675. [Google Scholar]
- 22.Shen R, Fan JB, Campbell D, et al. High-throughput SNP genotyping on universal bead arrays. Mutat Res. 2005;573:70–82. doi: 10.1016/j.mrfmmm.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ. Allelic discrimination using fluorogenic probes and the 5' nuclease assay. Genet Anal. 1999;14:143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 24.Kutyavin IV, Afonina IA, Mills A, et al. 3'-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 2000;28:655–661. doi: 10.1093/nar/28.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pressman S, Rotter JI. Epstein-Barr virus transformation of cryopreserved lymphocytes: prolonged experience with technique. Am J Hum Genet. 1991;49:467. [PMC free article] [PubMed] [Google Scholar]
- 26.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 29.SAS/STAT. SAS/STAT User's Guide. 4th ed. North Carolina: SAS Institute Inc; 1990. [Google Scholar]
- 30.Kuhn RM, Karolchik D, Zweig AS, et al. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karban AS, Okazaki T, Panhuysen CI, et al. Functional annotation of a novel NFKB1 promoter polymorphism that increases risk for ulcerative colitis. Hum Mol Genet. 2004;13:35–45. doi: 10.1093/hmg/ddh008. [DOI] [PubMed] [Google Scholar]
- 32.Hampe J, Frenzel H, Mirza MM, et al. Evidence for a NOD2-independent susceptibility locus for inflammatory bowel disease on chromosome 16p. Proc Natl Acad Sci U S A. 2002;99:321–326. doi: 10.1073/pnas.261567999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grant SF, Thorleifsson G, Reynisdottir I, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 34.Reynisdottir I, Thorleifsson G, Benediktsson R, et al. Localization of a susceptibility gene for type 2 diabetes to chromosome 5q34-q35.2. Am J Hum Genet. 2003;73:323–335. doi: 10.1086/377139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Libioulle C, Louis E, Hansoul S, et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dixon AL, Liang L, Moffatt MF, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 37.The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 38.Rogler G, Brand K, Vogl D, et al. Nuclear factor kappaB is activated in macrophages and epithelial cells of inflamed intestinal mucosa. Gastroenterology. 1998;115:357–369. doi: 10.1016/s0016-5085(98)70202-1. [DOI] [PubMed] [Google Scholar]
- 39.Bonen DK, Ogura Y, Nicolae DL, et al. Crohn's disease-associated NOD2 variants share a signaling defect in response to lipopolysaccharide and peptidoglycan. Gastroenterology. 2003;124:140–146. doi: 10.1053/gast.2003.50019. [DOI] [PubMed] [Google Scholar]
- 40.Hisamatsu T, Suzuki M, Reinecker HC, et al. CARD15/NOD2 functions as an antibacterial factor in human intestinal epithelial cells. Gastroenterology. 2003;124:993–1000. doi: 10.1053/gast.2003.50153. [DOI] [PubMed] [Google Scholar]
- 41.Cheung VG, Conlin LK, Weber TM, et al. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 42.Morley M, Molony CM, Weber TM, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]