Abstract
Gitelman's syndrome (GS) is a rare, autosomal recessive, salt-losing tubulopathy caused by mutations in the SLC12A3 gene, which encodes the thiazide-sensitive NaCl cotransporter (NCC). Because 18 to 40% of suspected GS patients carry only one SLC12A3 mutant allele, large genomic rearrangements may account for unidentified mutations. Here, we directly sequenced genomic DNA from a large cohort of 448 unrelated patients suspected of having GS. We found 172 distinct mutations, of which 100 were unreported previously. In 315 patients (70%), we identified two mutations; in 81 patients (18%), we identified one; and in 52 patients (12%), we did not detect a mutation. In 88 patients, we performed a search for large rearrangements by multiplex ligation-dependent probe amplification (MLPA) and found nine deletions and two duplications in 24 of the 51 heterozygous patients. A second technique confirmed each rearrangement. Based on the breakpoints of seven deletions, nonallelic homologous recombination by Alu sequences and nonhomologous end-joining probably favor these intragenic deletions. In summary, missense mutations account for approximately 59% of the mutations in Gitelman's syndrome, and there is a predisposition to large rearrangements (6% of our cases) caused by the presence of repeated sequences within the SLC12A3 gene.
Gitelman's syndrome (GS, MIM 263800) is a rare salt-losing tubulopathy1 characterized by hypokalemic metabolic alkalosis, hypomagnesaemia, and hypocalciuria. Loss of function mutations in the SLC12A3 gene encoding for the thiazide-sensitive NaCl cotransporter (NCC) are responsible for most of the cases.2,3 GS is inherited as an autosomal recessive trait, and homozygous and combined heterozygous mutations are expected.1 However, between 18 and 40% of patients with clinical GS are usually found to carry only one mutant allele after SLC12A3 screening.4–6 These incomplete genetic results raise the possibility of an excess of clinically suspected cases, genetic heterogeneity, or a failure in the mutation detection process. These issues have already been extensively discussed.7,8 However, genetic heterogeneity also exists, and a minority of patients with the GS phenotype harbor mutations at the CLCNKB gene.9,10
An important cause of apparently negative genetic testing could be large genomic rearrangements that are missed by direct sequencing and that usually account for 5 to 15% of the molecular defects responsible for autosomal recessive diseases.11,12 Several techniques exist for detecting these large deletions or insertions, such as the multiplex ligation-dependent probe amplification (MLPA) assay11,13 or the quantitative multiplex PCR of short fluorescent fragments (QMPSF),14 previously used in our laboratory.15 However, to date, a systematic screening for large genomic rearrangements at the SLC12A3 gene in GS is lacking, and only 116 of the 143 reported mutations in the Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/ac/index.php) is a large genomic rearrangement.
Here we report the molecular analysis of the SLC12A3 gene in a large cohort of 448 patients in whom GS was suspected. Direct sequencing showed homozygosity and combined heterozygosity in 70% of patients and only one mutated allele in 18% of patients, and a search for large genomic rearrangements was performed in 88 GS patients (51 heterozygous for point mutations 26 without mutation and 11 patients with homozygous mutations without consanguinity history). We show that large rearrangements may account for ≥6% of all mutations detected at the SLC12A3 gene in GS patients and therefore could improve the sensitivity of genetic testing to >80%.
RESULTS
Results of the First Screen by Direct Sequencing Analysis
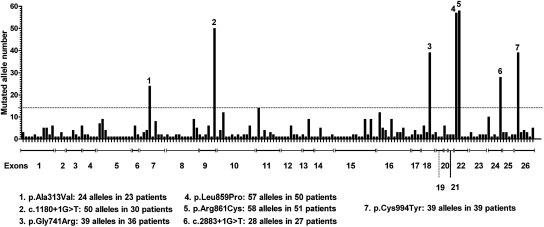
Of 448 index cases, two affected alleles were identified in 315 patients (70%): 79 of them were homozygous (25%) and 236 were compound heterozygous (74.9%). Only one mutant allele was detected in 81 patients (18%), and the wild-type genotype was detected in 52 patients (11.6%). The list of point mutations found is given in Supplementary Table 1: 172 different mutations were detected, spread throughout the gene. These included 64% missense, 14% frameshift, and 2% in-frame small deletions or insertions and 14% splice and 6% nonsense mutations. Figure 1 shows the distribution of the 711 mutated alleles along the 26 exons of the SLC12A3 gene. There were seven recurrent mutations, recurrence being defined arbitrarly by an allele frequency >2%, and included five missense amino acid changes found in 217 alleles (199 patients) and two splice mutations detected in 78 alleles (57 patients). They were mainly found in heterozygous compound subjects, except for c.1180 + 1G>T, which is highly prevalent in the Gypsy population.17 In the entire set of 172 point mutations, 100 have not been described before (53 missense, 19 frameshift, 16 splice, 6 nonsense, and 6 in-frame mutations; Supplementary Table 1, A–C). Supplementary Table 2 sums up the novel missense mutations and their in silico predictions.
Figure 1.
Frequency and distribution of the 172 detected mutations in 711 alleles. On the horizontal axis, each bar represents one mutation (there is no relation with the actual position in the exon). Dotted line corresponds to an allele frequency >2%.
Screening for Genomic Rearrangements in Simple Heterozygotes
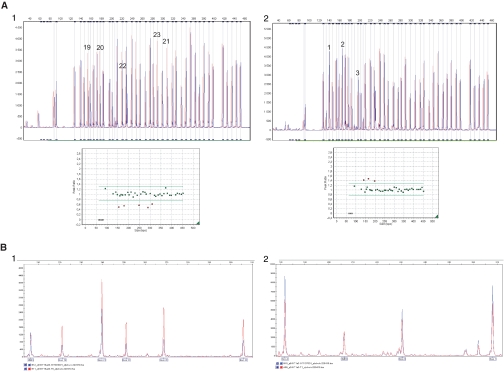
We decided to screen for large rearrangements at the SLC12A3 gene in patients with only one mutation detected by direct sequencing. Only 51 of the 81 samples were of sufficient DNA quality to be screened by MLPA or QMPSF. Rearrangements were found in 24 of them (47%, Table 1); single exon deletions were observed in 13 samples (E9del, n = 1; E14del, n = 2; E18del, n = 1; E26del, n = 9); deletions of two or more exons were detected in 9 samples (E1_E7del, n = 2; E2_E3del, n = 2; E4_E5del, n = 3; E19_E23del, n = 1; E24_E25del, n = 1); and duplications were detected in 2 samples (E1_E3dup and E1_E4dup). Two examples of these rearrangements are shown in Figure 2A.
Table 1.
Rearrangements detected by MLPA in 24 GS patients with one heterozygous mutation in the SLC12A3 gene detected by direct sequencing
| Patient | Nucleotide* | Protein | Exon/Intron | Reference | MLPA Heterozygous del or dup | Second Technique for Confirmation |
|---|---|---|---|---|---|---|
| BT038 | c.3077C>T | p.Thr1026Ile | 26 | 18 | E2_E3del | Long range PCR and breakpoint sequencing |
| BT213 | c.1046C>T | p.Pro349Leu | 8 | 1 | E1_E7del | SNPs analysis and oligo array comparative genomic hybridization |
| BT231 | c.1195C>T | p.Arg399Cys | 10 | 16 | E9del | Long range PCR |
| BT243 | c.1519C>T | p.Arg507Cys | 12 | This study | E4_E5del | Long range PCR and breakpoint sequencing + E6del by QMPSF |
| BT247 | c.938C>T | p.Ala313Val | 7 | 16 | E26del | QMPSF, Long range PCR and breakpoint sequencing |
| GT004 | c.1664C>T | p.Ser555Leu | 13 | 16 | E19_E23del | QMPSF, Long range PCR and breakpoint sequencing |
| GT034 | c.965–2_965–1dup | Splice defect | 7 | This study | E4_E5del | Long range PCR and breakpoint sequencing + E6del by QMPSF |
| GT059 | c.2891G>A | p.Arg964Gln | 25 | 1 | E26del | QMPSF, Long range PCR and breakpoint sequencing |
| GT121 | c.2981G>A | p.Cys994Tyr | 26 | 29 | E18del | QMPSF, Long range PCR and breakpoint sequencing |
| GT122 | c.2965G>A | p.Gly989Arg | 21 | 19 | E2_E3del | QMPSF, Long range PCR and breakpoint sequencing |
| GT137 | c.2576T>C | p.Leu859Pro | 22 | 1 | E1_E3dup | QMPSF |
| GT142 | c.2687G>A | p.Arg896Gln | 23 | 30 | E14del | QMPSF, Long range PCR and breakpoint sequencing |
| GT165 | c.2576T>C | p.Leu859Pro | 22 | 1 | E26del | QMPSF, Long range PCR and breakpoint sequencing |
| GT185 | c.1825 + 1del | Splice defect | 14 | This study | E1_E7del | QMPSF for exons 1 to 3 and 6 |
| GT187 | c.2576T>C | p.Leu859Pro | 22 | 1 | E26del | QMPSF, Long range PCR and breakpoint sequencing |
| GT196 | c.473G>A | p.Arg158Gln | 3 | 29 | E24_E25del | Long range PCR and breakpoint sequencing |
| GT243 | c.2981G>A | p.Cys994Tyr | 26 | 29 | E4_E5del | Long range PCR and breakpoint sequencing + E6del by QMPSF |
| GT278 | c.1387G>A | p.Gly463Arg | 11 | This study | E26del | QMPSF, Long range PCR and breakpoint sequencing |
| GT281 | c.626G>A | p.Arg209Gln | 5 | 16 | E26del | QMPSF, Long range PCR and breakpoint sequencing |
| GT285 | c.2929C>T | p.Arg977X | 25 | 1 | E14del | Long range PCR and breakpoint sequencing |
| GT291 | c.533C>T | p.Ser178Leu | 4 | 16 | E1_E4dup | QMPSF |
| B026 | c.1095 + 4A>G | Splice defect | 8 | This study | E26del | QMPSF, Long range PCR and breakpoint sequencing |
| B099 | c.2883 + 1G>T | Splice defect | 24 | 1 | E26del | QMPSF, Long range PCR and breakpoint sequencing |
| B104 | c.1883C>G | p.Ser628Trp | 15 | This study | E26del | QMPSF, Long range PCR and breakpoint sequencing |
Numbering is according to the cDNA sequence (GenBank : NM_000339.2). The A of the ATG of the initiator Methionine codon is denoted as nucleotide 1.
Figure 2.
MLPA and QMPSF electropherograms for SLC12A3 from two patients. For MLPA, each peak represents one exon of the SLC12A3 gene and 13 control probes. For QMPSF, each peak represents one analyzed exon and the HMBS internal control. Control samples are shown in red and patients' samples in blue. (A1) The MLPA half doses for exons 19 to 23 in patient GT004 and peak height ratio showing the deleted exons with ratio <0.7. (A2) MLPA duplication of exons 1 to 3 in patient GT137 and peak height ratio showing the duplicated exons with ratio >1.3. (B1) QMPSF half doses for exons 19 to 23 in patient GT004. (B2) QMPSF duplication of exons 1 to 3 in patient GT137.
As a second screening, we performed QMPSF analysis targeting the exon 6 of the SLC12A3 gene, not included in MLPA kit. This screening allowed the detection of E6del in only the three patients harboring the E4_E5del, thus extending the deletion to exon 6 (E4_E6del). QMPSF was also used to confirm E26del detected in nine patients, E19_E23del, and E1_E3dup (Figure 2B).
Clinical and biologic features of patients harboring one heterozygous rearrangement are shown in Table 2. All had a profound hypokalemia accompanied with metabolic alkalosis and usually hypomagnesaemia.
Table 2.
Clinical and biological characteristics of GS patients with one heterozygous mutation in the SLC12A3 gene and a large heterozygous rearrangements at the same gene
| Patient | Age at Diagnosis (years) | Gender | Clinical Presentation | Plasma Laboratory Findings |
Urinary Laboratory Findings |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Na (mmol/L) (reference range, 135 to 145) | K (mmol/L) (reference range, 3.5 to 4.5) | Cl (mmol/L) (reference range, 95 to 107) | HCO3 (mmol/L) (reference range, 22 to 28) | Mg (mmol/L) (reference range, 0.60 to 1.05) | Renin | Aldosterone | K (mmol/L) | Ca/creat (reference range, 0.04 to 0.37) | ||||
| BT038 | 26 | F | Fortuitous diagnosis | 138 | 2.7 | 99 | 27 | 0.40 | High | High | 90 | ND |
| BT213 | 16 | M | Growth retardation | 137 | 1.3 | 94 | 24 | 0.50 | High | Normal | 53 | ND |
| BT231 | 2 | M | Severe dehydration after diarrhea and growth retardation | 141 | 3.5 | 100 | 27 | ND | ND | High | 185 | ND |
| BT243 | 32 | M | Paresthesias and constipation | 138 | 2.6 | 102 | 26 | 0.84 | Normal | Normal | 35 | ND |
| BT247 | 31 | F | Growth retardation cramps, tetany, paresthesias | 139 | 2 | 98 | 27 | 0.44 | High | Normal | 51 | 0.03 |
| GT004 | 53 | F | Fortuitous diagnosis | ND | 2.5 | 97 | 31 | 0.40 | High | Normal | 68 | 0.15 |
| GT034 | 30 | F | Fortuitous diagnosis | ND | 2.8 | 98 | ND | 0.56 | ND | ND | 59 | 0.29 |
| GT059 | 28 | M | Fortuitous diagnosis | 137 | 2.4 | 97 | 30 | 0.55 | High | High | 59 | 0.28 |
| GT121 | 49 | M | Cramps, malaise with syncope | 142 | 2.9 | 100 | 33 | 0.62 | High | High | 62 | ND |
| GT122 | 13 | F | Cramps, polyuria | ND | 2.8 | 100 | ND | 0.70 | High | Normal | 23 | 0.01 |
| GT137 | 37 | M | Fortuitous diagnosis | 139 | 3.1 | 94 | 36 | 0.41 | ND | ND | 72 | ND |
| GT142 | 25 | F | Asthenia, chondrocalcinosis | 139 | 2.3 | ND | 26 | 0.34 | High | Normal | 96 | ND |
| GT165 | 37 | F | Fortuitous diagnosis | 139 | 2.5 | 96 | 29 | 0.50 | High | Normal | 32 | 0.01 |
| GT185 | 21 | M | Fatigue, cramps | 143 | 2.6 | ND | 31 | 0.53 | High | Normal | ND | 0.27 |
| GT187 | 23 | F | Palpitations, lypothymia, paresthesias | 139 | 2.7 | 102 | 32 | 0.57 | High | Normal | 34 | ND |
| GT196 | 12 | M | Growth retardation | 136 | 2.7 | 97 | 29 | 0.71 | ND | High | 47 | ND |
| GT243 | 5 | M | Growth retardation | ND | 2.9 | 102 | 26 | 0.67 | High | Normal | 134 | 0.03 |
| GT278 | 5 | M | Fortuitous diagnosis | 140 | 1.9 | 100 | 28 | 0.62 | Normal | Normal | 265 | ND |
| GT281 | 15 | M | Thoracic pain and palpitations | ND | 2.8 | 100 | 23 | 0.68 | High | High | 106 | 0.05 |
| GT285 | 32 | F | Chondrocalcinosis | 139 | 2.5 | 94 | 30 | 0.54 | ND | ND | 86 | 0.08 |
| GT291 | 47 | M | Right hemiparesis | 142 | 1.7 | 98 | 35 | 0.58 | Normal | Normal | 7 | ND |
| B026 | 48 | M | Familial hypokalemia | 142 | 3 | 99 | 21 | 0.64 | High | Normal | 97 | 0.09 |
| B099 | 2 | M | Abdominal pain | 138 | 2.6 | 97 | 27 | 0.60 | High | Normal | 52 | 0.3 |
| B104 | 6 | F | Abdominal pain | 133 | 2 | 91 | 33 | 0.83 | High | Normal | 94 | 0.28 |
F, female; M, male; ND, not determined.
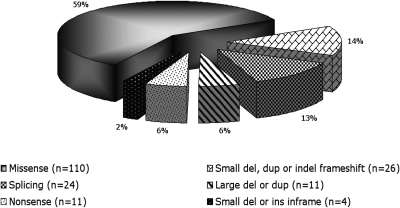
The updated spectrum of mutations detected in our cohort of GS patients, including genomic rearrangements, is shown in Figure 3. Whereas missense mutations were the majority (59%), small insertions or deletions detected by direct sequencing represent 14% and large rearrangements detected by MLPA and QMPSF accounted for 6% of the entire set of mutations.
Figure 3.
Pattern of mutations by type including genomic rearrangements at the SLC12A3 gene.
Further Genetic Screening in Subjects with No SLC12A3 Mutation
Because the phenotype caused by CLCNKB mutations may vary from the various types of Bartter syndrome to GS, we first analyzed the CLCNKB gene (sequencing and MLPA or QMPSF) in 49 of the 52 patients who tested negative for the SLC12A3 gene. Mutations were detected in 14 patients; Supplementary Table 4, A and B, summarizes the mutations detected and clinical data of these patients. We performed a search for genomic rearrangements in SLC12A3 gene by MLPA and QMPSF in 26 of those 38 remaining patients for whom the DNA quality was sufficient. No abnormality was detected, suggesting either another mutation at the SLC12A3 gene that was not detected by our screening or further genetic heterogeneity. Patients without mutations had similar features to the SLC12A3 mutated patients (Supplementary Table 3).
Further Genetic Screening in Homozygous Subjects without Notion of Consanguinity
In a subset of 28 patients belonging to the group of homozygous patients, the SNPs detected along the gene were homozygous, despite the absence of consanguinity. To exclude one heterozygous deletion, we were able to perform MLPA in 11 of them. Neither deletions nor duplications were detected in this group.
Characterization of the Deletion Breakpoints
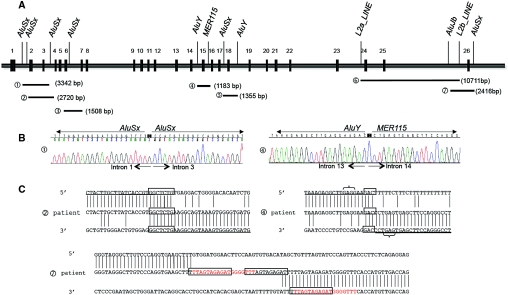
Long-range PCR completed by direct sequencing of the abnormal allele was first performed in nine patients (Supplementary Figure 1). Six different deletions (from 1.1 to 10.7 kb size) were found (Figure 4A): E2_E3del in two patients (c.282 + 667_c.506–205del and c.283–273_c.506–213del), the same E4_E6del (c.506–315_852 + 185del) in three patients, the same E14del (c.1169 + 773_c.1825 + 247del) in two other patients, a 1.3-kb E18del (c.2178 + 269 c.2285 + 685del) in one patient, and a 10.7-kb E24_E25del (c.2748–324_c.2952–505) in one patient.
Figure 4.
Mapping and characterization of breakpoints of SLC12A3 heterozygous deletions. (A) Schematic representation of the genomic organization of the 26 exons of the SLC12A3 gene and the location of breakpoints of 7 deletions: (A1) E2_E3del: 3342 bp deletion (c.282 + 667_c.506–205del). (A2) E2_E3del: 2720 bp deletion (c.283–273_c.506–213del). (A3) E4_E6del: 1508 bp deletion (c.506–315_852 + 185del). (A4) E14del: 1183 bp deletion (c.1169 + 773_c.1825 + 247del). (A5) E18del: 1355 bp deletion (c.2178 + 269_c.2285 + 685del). (A6) E24_E25del: 10711bp deletion (c.2748–324_c.2952–505). (A7) E26del: 2416bp deletion (c.2952–1593_*677delins25). The LCRs on or near the breakpoints are indicated. (B) Sequence analysis showing breakpoints of deletions 1 and 4. Breakpoints of deletion 1 are inside Alu repeats, suggesting a nonallelic homologous recombination. Breakpoints of deletion 4 are inside nonhomologous LCRs. (C) Sequence alignments at the breakpoints of two SLC12A3 heterozygous deletions probably originated from nonhomologous end-joining and one complex rearrangement. For deletions 2 and 4, boxes indicate the nucleotide microhomology. Brackets depict short motifs that might have facilitated the rearrangements. For deletion 7, boxes indicate the repetitive motifs; the read sequence is the 18-bp repetition that is present in inserted sequence and in the 3′UTR breakpoint.
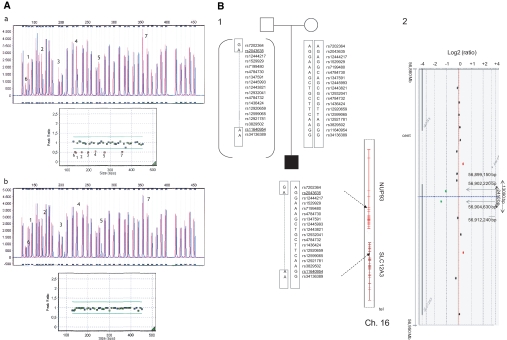
We also paid particular attention to a patient and his family with a large deletion containing the 5′ part of the gene up to exon 7 (Figure 5A). First, single nucleotide polymorphism (SNP) segregation analysis was performed by genotyping 16 SNPs at the SLC12A3 locus. The proband was homozygous for 14 of the16 SNPs, whereas his mother was heterozygous for 4 of the 7 SNPs lying in the intergenic region proximal to the SLC12A3 gene and for 7 of 9 SNPs in the NUP93 gene (Figure 5B1). Thus, the deletion mapped in an 89.9-kb region between rs2043635 (intron 5 of NUP93 gene; 56,818,987 Mb [hg19]) and rs11640954 (intron 8 SLC12A3 gene; 56,908,884 Mb [hg19]). To define this more precisely, we performed comparative genomic hybridization microarray analysis that allowed limiting the deletion size to 13,090 bp (Figure 5B2). On the centromeric side (q-arm), the closest probe that tested positive was mapped at position 56,899,150 Mb within the 5′UTR of SLC12A3. On the telomeric side, the closest probe that tested positive was located in exon 9 of SLC12A3 at position 56,912,240 Mb.
Figure 5.
Characterization of SLC12A3 E1_E7del for patient BT213. (A) MLPA electropherograms for the proband and his mother. Each peak represents one exon of the SLC12A3A gene and 13 control probes. (Aa) The electropherogram of the proband shows abnormal height peaks (in blue) in seven exons (numbered) compared with a normal control (in red). The peak ratio graph shows the 7 deleted exons for which the ratio was <0.7. (Ab) The electropherogram of the mother showing normal height peaks for all of the exons (blue) compared with a normal control (red). Normal peak ratios revealed an equivalent number of copies. (B1) Pedigree showing the constructed haplotypes resulting from biallelic SNP genotyping. The proband is shown in black. The SNPs covering the approximately 16-kb region upstream from the SLC12A3 gene are shown, including the two SNPs mapped to intron 8 of the SLC12A3 gene. The two deletion-flanking SNPs, covering a region of 89.9 kb, are underlined. Chromosome 16 showing both SLC12A3 and NUP93 (on the right). Cen, centromere; Tel, telomere. (B2) Results of molecular karyotyping. Array-comparative genomic hybridization log 2 ratio plot of chromosome 16 of the proband from the Agilent 244k, showing a 13-kb deletion from the probe lying within the 5′UTR of the SLC12A3 gene (56,899,150 Mb; hg19) to the probe that is specific to exon 9 (56,912,240 Mb; hg19). The deleted probes are in green, and unaltered probes (gain or loss) are in black.
High Number of Low Copy Repeats Sequences Favor Most Intragenic Deletions
Sequence alignment using RepeatMasker allowed identification of the presence of 41 Alu and 11 LINEs sequences within the SLC12A3 gene. Interestingly, the breakpoints of all of the genomic rearrangements identified in our patients contained low copy repeats (LCRs; Figure 4, A and B). For example, the 3342- and 2720-bp E2_E3del probably originated from the crossing over between two different AluSx repeats in intron 1 (312 and 237 bp) and the same AluSx repeat in intron 3 (256 bp). Similarly, the breakpoints for the 1508-bp deletion, which includes exons 4 to 6, were inside a sequence shared by the 256-bp AluSx repeat in intron 3 and the 309-bp AluSx repeat in intron 6. The breakpoints of the 1355-bp deletion were inside a sequence shared by the 297-bp AluSx repeat in intron 17 and the 308-bp AluY repeat in intron 18.
For two other deletions, the 1.1-kb E14del and the larger 10.7-kb E24_25del, the breakpoints were within nonhomologous LCRs: an AluY and a MER115 repeat in introns 13 and 14 and L2a_LINE and L2b_Line repeats in introns 23 and 25, respectively (Figure 4A). The sequence alignments at the breakpoint junctions for these two deletions showed six nucleotides with microhomology for the 10711-bp deletion and three nucleotides with microhomology and GAG rearrangement-promoting elements for the 1183-bp deletion (Figure 4C, deletions 2 and 4). These data strongly suggest nonhomologous end-joining as the causal mechanism for these deletions, facilitated by the presence of rearrangement-promoting elements inside the LCRs.
A Particular Rearrangement Was Observed for the Recurrent E26del
Taking into account the data obtained in the characterized deletions, we searched whether LCRs could also explain the recurrent E26del. Indeed, four AluSx and two AluJb are dispersed in intron 25 and one AluSx ∼500 bp after the stop codon. Long-range PCR amplification performed with a forward primer in exon 25 and a reverse primer distal to AluSx in the 3′ breakpoint resulted in the amplification of two bands in the nine patients with E26del detected by MLPA (Supplementary Figure 2). Direct sequencing of the short product showed the same complex rearrangement in the nine unrelated patients: a 2412-bp deletion with a 25-bp insertion (c.2952–1593_*677delins25). The 5′ breakpoint is 50 bp after an ALuJb repeat in intron 25, and the 3′ breakpoint is inside an AluSx repeat in the 3′UTR region (Figure 4A and 4C).
DISCUSSION
Based on a complete molecular investigation of the SLC12A3 gene on the largest cohort reported thus far, we showed the value of combining several techniques to finally achieve an 91% mutation detection rate in GS. Next to the identification of missense mutations (59% of the cases) and small insertions or deletions (14%) by direct sequencing, we used MLPA and QMPSF, which allowed the detection of large rearrangements in 24 of 51 GS patients (47%) known to be heterozygous for a point mutation. This suggests that almost one half of the patients suspected to have GS with only one mutated allele detected by direct sequencing (18% of our series and of the smaller series described by Ji et al.6) have a large genomic rearrangement on the other allele.
To date, >180 mutations in SLC12A3 have been reported, about 70% of which being missense mutations. In this study, direct sequencing allowed the detection of 110 different missense mutations in 290 subjects (64% of the mutations found), 51 of them being novel. Supplementary Table 2 sums up in silico predictions for these changes. None of these amino acid changes are known SNPs, and they were not detected in 200 control chromosomes. Furthermore, they were not detected either in 220 control chromosomes in another European study,5 and only one (p.Gly779Glu) was detected (allelic frequency of 0.02%) in 1985 unrelated subjects from the Framingham cohort.6 This missense change was predicted as functional and was associated with lower BP. Taken together, the novel 51 missense mutations described in this paper are rare variants very likely to be pathogenic.
Five previously described missense mutations were particularly frequent in our unrelated patients (Figure 1), raising the possibility of neutral polymorphisms. Two of them, p.Gly741Arg and p.Cys994Tyr detected in 36 and 38 patients, respectively, led to a total or partial loss of function when expressed in vitro into Xenopus laevis oocytes.18,19 Mutations p.Leu859Pro and p.Arg861Cys have not been expressed in vitro; however, three in silico methods predicted these amino acid changes to be pathogenic. Thus, there are strong arguments for p.Gly741Arg, p.Cys994Tyr, p.Leu859Pro, and p.Arg861Cys being hotspot mutations. In contrast, the previously reported p.Ala313Val mutation was predicted in silico as nondeleterious. This variant was detected in 19 GS patients: it was associated with a second pathogenic mutation in 18 patients and was homozygous in 1 patient, suggesting that it is deleterious.
All mutations described in this study, including large rearrangements and the 100 novel point mutations, will be available online at the European Network for the Study of Orphan Nephropathies website (http://www.eunefron.org/).
One of our major findings was the detection of large rearrangements in GS patients, representing about one half of the heterozygous patients tested. The breakpoints of these large-scale mutations were found to correspond to a high frequency of repetitive sequences within the SLC12A3 gene. More than a million of Alu sequences are dispersed throughout the genome, and regions with high Alu repeat content are prone to nonallelic homologous recombination, which may cause inherited diseases.20 The SLC12A3 gene contains 41 Alus and 11 LINEs dispersed in intronic regions corresponding to 22.5 and 6% of the gene sequence, respectively. Four of seven deletions were the consequence of Alu-mediated recombination: AluSx sequences involved in E2_E3del and in E4_E6del in two and three patients, respectively, and the AluSx and AluY sequences involved in E18del share >80% identity. The breakpoints of two other deletions were located in nonhomologous LCRs: AluY and MER115 for E14del and L2a and L2b_LINE for E24_E25del. However, we observed nucleotide microhomology and, in one case, the presence of the recombinant-promoting element GAG at breakpoint junctions. These data correspond to the characteristics observed in the nonhomologous end-joining mechanism.21 This mechanism, used by eukaryotic cells to repair double-strand DNA breaks, could be stimulated by genomic architecture (i.e., LCRs and sequence motifs).
The breakpoints analysis of the recurrent E26del detected in probands of nine unrelated families showed the presence of a complex indel mutation (2412-bp deletion and 25-bp insertion). The 5′ breakpoint was in close proximity to AluJb and the 3′ breakpoint was inside an AluSx. Repetitive motifs were detected in the insertion and in the 3′ breakpoint (Figure 4C, deletion 7). Similar indels have been described in the NF1 and CFTR genes.22,23 A multistep mechanism facilitated by the presence of LCRs is likely to favor them, as has been suggested for other complex rearrangements.24 The deletions and duplications detected have not been described as copy number variants in healthy controls (Database of Genomic Variants at http://projects.tcag.ca/variation/, Copy number variation project at the Children's Hospital of Philadelphia at http://cnv.chop.edu, and 1000 Genomes at http://browser.1000genomes.org/index.html).
Of note, large-scale mutations were not associated with a more severe phenotype when clinical and biochemical data from patients harboring a heterozygous deletion were compared with those from subjects homozygous or compound heterozygous for two missense mutations.
This was true for the age at diagnosis (17 [range, 2.25 to 63] versus 25.5 [range 2 to 53] years) or the importance of hypokalemia (2.62 [range, 1.8 to 3.4] versus 2.60 [range, 1.3 to 3.5] mmol/L). Also, the heterozygous E1_E7del was detected in two patients: one with a relatively severe phenotype and the other one with a mild phenotype. This deletion was identified previously in one homozygous and one heterozygous member of an Amish kindred,16 who were not mentioned as having a particularly severe phenotype compared with the 48 other GS patients. Thus, a null allele does not seem to be more detrimental than a punctual loss of function mutation, although the nature and position of SLC12A3 mutations have been thought to influence the severity of GS.5
Concerning the group of patients without mutations, most of them have a GS-like phenotype. Only three of them had an early presentation (BT100, B001, and B059 in Supplementary Table 2). In these patients, another cause of severe hypokalemic metabolic alkalosis such as congenital chloride diarrhea could be considered. Nevertheless, in these patients, there was no history of polyhydramnios, premature birth, or diarrhea. Furthermore, urinary electrolytes at diagnosis or on follow-up (5 to 10 years) showed sodium, potassium, and chloride wasting.
In conclusion, this molecular analysis of a large cohort of 448 GS patients, including the first search for large-scale mutations, showed that, despite the high efficiency of direct genomic sequencing in detecting the vast majority of the SLC12A3 mutations found in this disorder, a complementary technique is necessary to achieve a high mutation detection rate, especially for those patients in whom only one mutation had been detected. We confirmed that MLPA is an efficient technique for analyzing large genomic rearrangements, which account for ≥6% of mutations detected in our patients with GS. Moreover, we showed that nonallelic homologous recombination by Alu sequences and nonhomologous end-joining are most likely to be responsible for intragenic deletions. Finally, we detected CLCNKB mutations in 3% and excluded mutations and large rearrangements of the SLC12A3 gene in 8% (n = 36) of our GS patients, which questions the clinical diagnosis of GS and raises the possibility of genetic heterogeneity in this inherited tubulopathy.
CONCISE METHODS
Patients
Between January 2001 and August 2009, samples from 448 probands (219 males and 229 females) with a clinical diagnosis of GS were received at the Genetics Department at Hopital Européen Georges Pompidou, Paris. Most samples were sent from nephrology and endocrinology services thanks to the French Network for Tubulopathies and the European Network for the Study of Orphan Nephropathies (http://www.eunefron.org/). A few samples (n = 6) were also received from other countries (Austria, Canada, Luxemburg, and Portugal). Appropriate informed consent was obtained from all patients and their families. They were selected according to the classical criteria for GS: renal hypokalemia, metabolic alkalosis, hypomagnesaemia, hypocalciuria, and secondary hyperreninism and hyperaldosteronism. Nevertheless, because plasma renin and aldosterone were not measured in all cases at diagnosis and because the absence of hypomagnesaemia or hypocalciuria has been described in some genetically confirmed cases,25,26 we did not systematically require all of the criteria to be met before performing the genetic testing.
Detection of Point Mutations
Total DNA was extracted from blood peripheral leukocytes by standard procedures. Mutation analysis was performed by PCR amplification and direct sequencing of exons and flanking intronic sequences of the SLC12A3 gene, mainly as described previously4 (primers available upon request), on an ABI Prism 3730XL DNA Analyzer Sequencer (Perkin Elmer Applied Biosystems, Foster City, CA).
Two amino acid changes initially described as mutations and then as SNPs were considered as SNPs in this study: p.Arg913Gln (rs11643718) and p.Ala728Thr (rs61730207). In contrast, we considered the amino acid changes p.Arg209Trp (SNP rs28936388), p.Gly264Ala (SNP rs1529927), and p.Arg928Cys (SNP rs12708965) as loss of function mutations. Indeed, in vitro expression of p.Arg209Trp and p.Gly264Ala has been shown to produce a significant reduction in NCC activity,18,27 and the p.Arg928Cys change is predicted in silico to be deleterious and is also considered disease-causing.6 Eight novel missense changes detected in this study (p.Ala13Pro, p.Arg83Gln p.Val404Ile, p.Thr428Pro, p.Ser546Gly, pSer833Leu, p.Glu915Ala, and p.Gln1021Lys) were predicted in silico as nondeleterious (Supplementary Table 2) and were characterized as variants of unknown significance. The p.Ala322Val was also predicted as nondeleterious, but this change implicates the first nucleotide of exon 8 and may be considered as a splice mutation. Indeed, ESEfinder (at http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder) predicts a loss of two enhancer motifs (SRp55 and SF2/ASF).
MLPA Analysis
We used a commercially available kit, the SALSA MLPA P136 SLC12A3 Kit (MRC Holland, Amsterdam, The Netherlands) to detect large deletions or duplications in the SLC12A3 gene. The P136 kit contains 38 probes: 25 probes for SLC12A3 (one for each exon except exon 6) and 13 reference probes. The detailed procedure is described in Supplementary Materials.
QMPSF
We adapted the QMPSF method28 to detect large deletions or duplications at the SLC12A3 gene. Details of the procedure are given in Supplementary Materials and the corresponding primers in Supplementary Table 3. This method was used to amplify exon 6 of the SLC12A3 gene, which is not included in the MLPA SLC12A3 kit. It was also used to confirm deletions or duplications found by MLPA, especially when it was difficult to estimate the 3′ (exon 26) or 5′ breakpoints limits (duplication of exons 1 to 3 and 1 to 4) or if no amplification was obtained by long-range PCR (deletion of exons 19 to 23).
Mapping the Deletion Breakpoints by long-range PCR
Long-range PCR and sequencing analysis were performed in 10 patients to confirm the MLPA results and to determine the deletion breakpoints. Gene-specific primers located in proximal and distal nondeleted exons were designed for each type of deletion (Supplementary Table 4). Detailed of the procedure can be found in Supplementary Materials.
Characterization of Patient BT213's Deletion
Refinement of the deletion length by SNPs genotyping: a search for informative SNPs (with a minor allele frequency MAF >0.3) was performed on a region containing approximately 16 kb of sequence upstream from exon 1 of the SLC12A3 gene. Sixteen pairs of primers allowed the genotyping of 10 informative SNPs plus 6 additional SNPs with lower MAF (Supplementary Table 5). Two SNPs located in intron 8 (rs11640954 and rs34136389) were also genotyped in the patient and his mother to confirm heterozygosity in this region.
Further analysis by comparative genomic hybridization: to analyze the deletion breakpoints, we performed a whole genome array comparative genomic hybridization analysis using an Agilent 244k oligonucleotide array (Agilent, Santa Clara, CA). The complete procedure is given in Supplementary Materials.
Bioinformatic Analysis of Mutations
Mutation interpretation and amino acid conservation in orthologs were assessed using Alamut V.1.5 software (Interactive Biosoftware, Rouen, France; http://www.interactivebiosoftware.com/). For missense mutations, the Grantham chemical distance (Grantham R. 1974) between amino acids provided by Alamut software was used to test whether the changes between the residues were likely to affect physicochemical properties. Complementary analyses were performed with SIFT (Sorting Intolerant From Tolerant, http://www.Blocks.fhcrc.org/sift/SIFT.html), PolyPhen-2 (prediction of functional effects of human nsSNPs at http://genetics.bwh.harvard.edu/pph/), and Panther (evolutionary analysis of coding SNPs at http://www.pantherdb.org/tools/csnpScoreForm.jsp). LCRs in the SLC12A3 gene were found with RepeatMasker software at http://www.repeatmasker.org/.
DISCLOSURES
None.
Acknowledgments
The genetic department of the European Georges Pompidou Hospital is affiliated with the “Centre de Référence des Maladies Rénales Héréditaires de l′Enfant et de l'Adulte (MARHEA).” We thank Valérie Nau, Isabelle Roncelin, Valérie Boccio, Nelly Lepottier, Sylvie Cotigny, and Caroline Schmitt for technical assistance. We thank the nephrologists from the French tubulopathy network who referred the patients' DNA and gave access to their charts (especially Christophe Charasse, Bernard Charpentier, Jacques Dantal, Georges Deschênes, Philippe Eckart, Philippe Grimbert, Michèle Hall, Elisabeth Harvey, Bertrand Isidore, Jessica Leogite, Jacques Lombet, Férielle Louillet, Sebastien Maillez, Patrick Niaudet, Christine Pietrement and Sophie Taque) and Dr. Mounir Filali for his participation to this study. This study was supported by INSERM, Assistance Publique-Hôpitaux de Paris, and the European Community's 7th Framework Program (HEALTH-F2-200-201590, EUNEFRON program).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental information for this article is available online at http://www.jasn.org/.
REFERENCES
- 1. Simon DB, Nelson-Williams C, Bia MJ, Ellison D, Karet FE, Molina AM, Vaara I, Iwata F, Cushner HM, Koolen M, Gainza FJ, Gitleman HJ, Lifton RP: Gitelman's variant of Bartter's syndrome, inherited hypokalaemic alkalosis, is caused by mutations in the thiazide-sensitive Na-Cl cotransporter. Nat Genet 12: 24–30, 1996 [DOI] [PubMed] [Google Scholar]
- 2. Mastroianni N, De Fusco M, Zollo M, Arrigo G, Zuffardi O, Bettinelli A, Ballabio A, Casari G: Molecular cloning, expression pattern, and chromosomal localization of the human Na-Cl thiazide-sensitive cotransporter (SLC12A3). Genomics 35: 486–493, 1996 [DOI] [PubMed] [Google Scholar]
- 3. Melander O, Orho-Melander M, Bengtsson K, Lindblad U, Rastam L, Groop L, Hulthen UL: Genetic variants of thiazide-sensitive NaCl-cotransporter in Gitelman's syndrome and primary hypertension. Hypertension, 36: 389–394, 2000 [DOI] [PubMed] [Google Scholar]
- 4. Lemmink HH, Knoers NV, Karolyi L, van Dijk H, Niaudet P, Antignac C, Guay-Woodford LM, Goodyer PR, Carel JC, Hermes A, Seyberth HW, Monnens LA, van den Heuvel LP: Novel mutations in the thiazide-sensitive NaCl cotransporter gene in patients with Gitelman syndrome with predominant localization to the C-terminal domain. Kidney Int 54: 720–730, 1998 [DOI] [PubMed] [Google Scholar]
- 5. Riveira-Munoz E, Chang Q, Godefroid N, Hoenderop JG, Bindels RJ, Dahan K, Devuyst O: Transcriptional and functional analyses of SLC12A3 mutations: New clues for the pathogenesis of Gitelman syndrome. J Am Soc Nephrol 18: 1271–1283, 2007 [DOI] [PubMed] [Google Scholar]
- 6. Ji W, Foo JN, O'Roak BJ, Zhao H, Larson MG, Simon DB, Newton-Cheh C, State MW, Levy D, Lifton RP: Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet 40: 592–599, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gamba G: Molecular physiology and pathophysiology of electroneutral cation-chloride cotransporters. Physiol Rev 85: 423–493, 2005 [DOI] [PubMed] [Google Scholar]
- 8. Riveira-Munoz E, Devuyst O, Belge H, Jeck N, Strompf L, Vargas-Poussou R, Jeunemaitre X, Blanchard A, Knoers NV, Konrad M, Dahan K: Evaluating PVALB as a candidate gene for SLC12A3-negative cases of Gitelman's syndrome. Nephrol Dial Transplant 23: 3120–3125, 2008 [DOI] [PubMed] [Google Scholar]
- 9. Jeck N, Konrad M, Peters M, Weber S, Bonzel KE, Seyberth HW: Mutations in the chloride channel gene, CLCNKB, leading to a mixed Bartter-Gitelman phenotype. Pediatr Res 48: 754–758, 2000 [DOI] [PubMed] [Google Scholar]
- 10. Zelikovic I, Szargel R, Hawash A, Labay V, Hatib I, Cohen N, Nakhoul F: A novel mutation in the chloride channel gene, CLCNKB, as a cause of Gitelman and Bartter syndromes. Kidney Int 63: 24–32, 2003 [DOI] [PubMed] [Google Scholar]
- 11. Audrezet MP, Chen JM, Raguenes O, Chuzhanova N, Giteau K, Le Marechal C, Quere I, Cooper DN, Ferec C: Genomic rearrangements in the CFTR gene: Extensive allelic heterogeneity and diverse mutational mechanisms. Hum Mutat 23: 343–357, 2004 [DOI] [PubMed] [Google Scholar]
- 12. Bisceglia L, Fischetti L, Bonis PD, Palumbo O, Augello B, Stanziale P, Carella M, Zelante L: Large rearrangements detected by MLPA, point mutations, and survey of the frequency of mutations within the SLC3A1 and SLC7A9 genes in a cohort of 172 cystinuric Italian patients. Mol Genet Metab 99: 42–52, 2010 [DOI] [PubMed] [Google Scholar]
- 13. Desviat LR, Perez B, Ugarte M: Identification of exonic deletions in the PAH gene causing phenylketonuria by MLPA analysis. Clin Chim Acta 373: 164–167, 2006 [DOI] [PubMed] [Google Scholar]
- 14. Saugier-Veber P, Goldenberg A, Drouin-Garraud V, de La Rochebrochard C, Layet V, Drouot N, Le Meur N, Gilbert-Du-Ssardier B, Joly-Helas G, Moirot H, Rossi A, Tosi M, Frebourg T: Simple detection of genomic microdeletions and microduplications using QMPSF in patients with idiopathic mental retardation. Eur J Hum Genet 14: 1009–1017, 2006 [DOI] [PubMed] [Google Scholar]
- 15. Burnichon N, Rohmer V, Amar L, Herman P, Leboulleux S, Darrouzet V, Niccoli P, Gaillard D, Chabrier G, Chabolle F, Coupier I, Thieblot P, Lecomte P, Bertherat J, Wion-Barbot N, Murat A, Venisse A, Plouin PF, Jeunemaitre X, Gimenez-Roqueplo AP: The succinate dehydrogenase genetic testing in a large prospective series of patients with paragangliomas. J Clin Endocrinol Metab 94: 2817–2827, 2009 [DOI] [PubMed] [Google Scholar]
- 16. Cruz DN, Shaer AJ, Bia MJ, Lifton RP, Simon DB: Gitelman's syndrome revisited: An evaluation of symptoms and health-related quality of life. Kidney Int 59: 710–717, 2001 [DOI] [PubMed] [Google Scholar]
- 17. Coto E, Rodriguez J, Jeck N, Alvarez V, Stone R, Loris C, Rodriguez LM, Fischbach M, Seyberth HW, Santos F: A new mutation (intron 9 +1 G>T) in the SLC12A3 gene is linked to Gitelman syndrome in Gypsies. Kidney Int 65: 25–29, 2004 [DOI] [PubMed] [Google Scholar]
- 18. Kunchaparty S, Palcso M, Berkman J, Velazquez H, Desir GV, Bernstein P, Reilly RF, Ellison DH: Defective processing and expression of thiazide-sensitive Na-Cl cotransporter as a cause of Gitelman's syndrome. Am J Physiol 277: F643–F649, 1999 [DOI] [PubMed] [Google Scholar]
- 19. De Jong JC, Van Der Vliet WA, Van Den Heuvel LP, Willems PH, Knoers NV, Bindels RJ: Functional expression of mutations in the human NaCl cotransporter: Evidence for impaired routing mechanisms in Gitelman's syndrome. J Am Soc Nephrol 13: 1442–1448, 2002 [DOI] [PubMed] [Google Scholar]
- 20. Belancio VP, Hedges DJ, Deininger P: Mammalian non-LTR retrotransposons: For better or worse, in sickness and in health. Genome Res 18: 343–358, 2008 [DOI] [PubMed] [Google Scholar]
- 21. Gu W, Zhang F, Lupski JR: Mechanisms for human genomic rearrangements. Pathogenetics 1: 4, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lazaro C, Gaona A, Lynch M, Kruyer H, Ravella A, Estivill X: Molecular characterization of the breakpoints of a 12-kb deletion in the NF1 gene in a family showing germ-line mosaicism. Am J Hum Genet 57: 1044–1049, 1995 [PMC free article] [PubMed] [Google Scholar]
- 23. Ferec C, Casals T, Chuzhanova N, Macek M, Jr, Bienvenu T, Holubova A, King C, McDevitt T, Castellani C, Farrell PM, Sheridan M, Pantaleo SJ, Loumi O, Messaoud T, Cuppens H, Torricelli F, Cutting GR, Williamson R, Ramos MJ, Pignatti PF, Raguenes O, Cooper DN, Audrezet MP, Chen JM: Gross genomic rearrangements involving deletions in the CFTR gene: characterization of six new events from a large cohort of hitherto unidentified cystic fibrosis chromosomes and meta-analysis of the underlying mechanisms. Eur J Hum Genet 14: 567–576, 2006 [DOI] [PubMed] [Google Scholar]
- 24. Chuzhanova NA, Anassis EJ, Ball EV, Krawczak M, Cooper DN: Meta-analysis of indels causing human genetic disease: Mechanisms of mutagenesis and the role of local DNA sequence complexity. Hum Mutat 21: 28–44, 2003 [DOI] [PubMed] [Google Scholar]
- 25. Lin SH, Shiang JC, Huang CC, Yang SS, Hsu YJ, Cheng CJ: Phenotype and genotype analysis in Chinese patients with Gitelman's syndrome. J Clin Endocrinol Metab 90: 2500–2507, 2005 [DOI] [PubMed] [Google Scholar]
- 26. Tosi F, Bianda ND, Truttmann AC, Crosazzo L, Bianchetti MG, Bettinelli A, Ramelli GP: Normal plasma total magnesium in Gitelman syndrome. Am J Med 116: 573–574, 2004 [DOI] [PubMed] [Google Scholar]
- 27. Moreno E, Tovar-Palacio C, de los Heros P, Guzman B, Bobadilla NA, Vazquez N, Riccardi D, Poch E, Gamba G: A single nucleotide polymorphism alters the activity of the renal Na+:Cl- cotransporter and reveals a role for transmembrane segment 4 in chloride and thiazide affinity. J Biol Chem 279: 16553–16560, 2004 [DOI] [PubMed] [Google Scholar]
- 28. Houdayer C, Gauthier-Villars M, Lauge A, Pages-Berhouet S, Dehainault C, Caux-Moncoutier V, Karczynski P, Tosi M, Doz F, Desjardins L, Couturier J, Stoppa-Lyonnet D: Comprehensive screening for constitutional RB1 mutations by DHPLC and QMPSF. Hum Mutat 23: 193–202, 2004 [DOI] [PubMed] [Google Scholar]
- 29. Syren ML, Tedeschi S, Cesareo L, Bellantuono R, Colussi G, Procaccio M, Ali A, Domenici R, Malberti F, Sprocati M, Sacco M, Miglietti N, Edefonti A, Sereni F, Casari G, Coviello DA, Bettinelli A: Identification of fifteen novel mutations in the SLC12A3 gene encoding the Na-Cl Co-transporter in Italian patients with Gitelman syndrome. Hum Mutat 20: 78, 2002 [DOI] [PubMed] [Google Scholar]
- 30. Kurschat C, Heering P, Grabensee B: [Gitelman's syndrome: an important differential diagnosis of hypokalemia]. Dtsch Med Wochenschr 128: 1225–1228, 2003 [DOI] [PubMed] [Google Scholar]