Abstract
RNA processing is vital for the high fidelity and diversity of eukaryotic transcriptomes and the encoded proteomes. However, control of RNA processing is not fully established. Σ RNA is a class of conserved large noncoding RNAs (murine Hepcarcin; human MALAT-1) up-regulated in carcinomas. Using antisense technology, we identified that RNA post-transcriptional modification is the most significant global function of Σ RNA. Specifically, processing of the pre-mRNAs of genes including Tissue Factor and Endoglin was altered by hydrolysis of Σ RNA/MALAT-1. These results support the hypothesis that Σ RNA/MALAT-1 is a regulatory molecule exerting roles in RNA post-transcriptional modification.
Keywords: Hepcarcin, Σ RNA/MALAT-1, RNA processing, alternative splicing
1. Introduction
Transcription of eukaryotic genes generates pre-mRNA transcripts that are processed to mature mRNAs through various post-transcriptional modifications including capping, splicing, polyadenylation, degradation, and nuclear exportation. This is vital for the high fidelity and diversity of eukaryotic transcriptomes, where deregulation of the process is associated with diverse diseases such as cancer [1]. RNA-binding proteins including the serine/arginine rich (SR) proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs) play key roles in RNA splicing and degradation [2,3]. Interestingly, SR proteins and hnRNPs can be alternatively spliced themselves [4]. Despite these reasonably well-documented components and regulation of RNA processing by proteins, upstream control of RNA processing, particularly by RNA, has yet to be adequately elucidated.
Hepcarcin [5] and its human orthologue Alpha transcript (i.e., MALAT-1 or NEAT2) [6-8] are large noncoding RNAs (ncRNAs). In recognition that they are orthologous and the human ortholog is functional, as identified in this and other studies [9-10], we have proposed the term Σ RNA to categorize them as a novel functional group of RNA. Σ RNA/MALAT-1 is selectively expressed in tumor vascular endothelium [5], over-expressed in metastatic non-small cell lung carcinomas [6], up-regulated in six types of human carcinomas [5] and EpH4 cells transformed with the ErbB2 oncoprotein [11]. Interestingly, recent studies suggested that inhibition of MALAT-1 suppressed proliferation and invasion of CaSki cervical cancer cells [9] and impaired mobility of cancer cells [10], consistent with over-expression of MALAT-1 in cancer and its importance in cancer biology. In human Tig1 cells, Σ RNA/MALAT-1 associates with SC35 domains containing molecules including SR proteins that are involved in pre-mRNA modification [8]. These observations support the hypothesis that Σ RNA/MALAT-1 influences RNA processing.
Using antisense oligonucleotides (ASOs) as a tool to selectively mediate hydrolysis of Σ RNA/MALAT-1 in HeLa cells and HUVECs, this study supports the hypothesis that human Σ RNA/MALAT-1 may be a global regulator of RNA post-transcriptional modification. Processing of the pre-mRNAs of genes encoding Tissue Factor and Endoglin that play important roles in cancer biology is altered by hydrolysis of Σ RNA/MALAT-1. These observations indicate that Σ RNA/MALAT-1 is a large ncRNAs functioning as a regulator of RNA processing in higher eukaryotic cells.
2. Materials and Methods
2.1. Cells and reagents
Cells were originally obtained from the American Type Tissue Collection. HeLa cells were cultured in DMEM supplemented with 10% FCS and 2 mM glutamine in a humidified 5% CO2 incubator at 37°C. HUVECs were maintained in EGM medium and supplemented with 2% FCS (Lonza) in a humidified 5% CO2 incubator at 37°C.
2.2. ASO synthesis and transfection
Antisense phosphorothioate oligonucleotides modified with 2′-methoxyethyl were synthesized as described [12]. Unless otherwise indicated, transfection with ASOs were performed using Opti-MEM (Invitrogen) containing 75 nM of ASOs and 2.25 μg/ml Lipofectin (Invitrogen, or 1 μg/ml Lipofectamine 2000 for HUVECs) at 37°C for 4 hrs, at which time the medium was replaced with standard growth medium. Untreated control (UTC) or scrambled ASO (Con) at 75 nM were used as negative controls. The sequences of Σ RNA/MALAT-1 specific ASOs and their targeted regions are given in the supplementary Table S1.
2.3. RNA Isolation, Northern analyses, RT-PCR, and quantitative PCR
RNAs were isolated using RNeasy methodology (Qiagen). Northern blot analyses were performed as described [5]. Unless otherwise indicated, the probe for human Σ RNA/MALAT-1 recognizes nt 3581-4585 of EF177381. The probes for Endoglin recognize nt 881-1292 or nt 881-1070 of NM_000118.1. The probe for TF recognize nt 544-1125 of F3 (NM_001993), the gene encoding TF. cDNAs used for RT-PCR or quantitative PCR were generated with QuantiTect Reverse Transcription Kit (Qiagen). For RT-PCR analyses of TF transcripts, primers were designed according to the sequences of NM_001993 to amplify the region between exon 4 and 6 (nt 600-1183) of F3. The primers (IDT Technologies) were forward primer, 5′-ggacagccaacaattcagag-3′; reverse primer, 5′-tccagggtcttcatgctcc-3′. The RT-PCR products were resolved on 1.2% agarose gels. Quantitative PCR for GAPDH and Σ RNA/MALAT-1 were performed using Taqman universal PCR mix and Taqman gene expression assay mix that included FAM-dye labeled MGB probes (Applied Biosystems). Taqman gene expression assay mixes were Hs99999905_m1 for human GAPDH, and Hs00273907_s1 for human Σ RNA/MALAT-1, respectively (Applied Biosystems). The primers specifically detecting hTFalt were forward, exon 4-6 junction primer, 5′-caagttcaggaaagaaatattctac-3′; reverse primer, 5′-gaaacattcagtggggagttctcc-3′; and probe, 5′-[6-FAM]catccttgtcatcatcctggctatatc[TAMRA-6-FAM]. The qPCR primers for full length TF were forward primer, 5′-caggctggagtgcagtagca-3′; reverse primer, 5′-aggctgaggcagacaattgc-3′; and probe, 5′-[6-FAM]cttgcaccctccgtctctcggg[TAMRA-6-FAM].
2.4. Affymetrix exon array and Ingenuity pathway analysis (IPA)
Sixteen paired samples of total RNAs from cells transfected with control ASO (Con) or human Σ RNA/MALAT-1 specific ASOs (75 nM ASO254 for HeLa cells, or a mixture of 25 nM ASO254, ASO240, and ASO280 for HUVECs) were harvested 24 hrs after transfection. Total RNAs (2 μg) were depleted more than 95% of rRNAs using Ribominus (Invitrogen), and synthesized to double-stranded cDNA using random hexamers tagged with a T7 promoter sequence. Antisense cRNAs were generated from the cDNAs using T7 RNA polymerase, and used to generate single-stranded DNAs in the sense orientation using GeneChip Whole Transcript (WT) Sense Target Labeling Assay (Affymetrix). The single-stranded DNAs were fragmented and terminal-labeled with biotin, and hybridized to 16 GeneChip Human Exon 1.0 ST Arrays (Affymetrix). Gene chips were washed, stained using Genechip Fluidics Station 450 (Affymetrix) and Genechip Operating Software (GCOS), and scanned using Genechip Scanner 3000 7G (Affymetrix). The data were analyzed using XRAY (version 2.63) software, the Excel add-in from Biotique Systems Inc. The differentially expressed transcripts were uploaded to IPA (Ingenuity Systems, Redwood City, CA) for functional analysis.
2.5. Western blot analysis
HUVECs were grown to ~90% confluency. Total proteins were isolated by resuspending cells in 2X SDS reducing buffer followed by sonication to lyse the cells completely. Equal amounts of cell lysates were run on 8-16% gradient reducing gels (Invitrogen) and transferred by voltage gradient to positively charged nitrocellulose membranes in 20 minutes. The membranes were washed in 1X TBST (Tris-Buffered Saline Tween-20) buffer and incubated with 10% milk (blocking agent) at 4°C overnight. The membranes were incubated with polyclonal antibodies S2945 (Sigma), ab38813 (Abcam), and A2066 (Sigma) to detect SFRS12, SFRS1, and actin proteins, respectively, for 30 min with agitation and washed 3 times with 1X TBST. The membranes were then incubated with HRP-conjugated goat anti-rabbit antibody (Pierce) diluted 1:1000 with milk for 30 minutes with agitation and then washed 4 times with 1X TBST buffer. The membranes were developed with SuperSignal West Femto Chemiluminescent Substrate (Pierce); the signals were developed by exposure to HyBlot CL films (Denville) and quantified with Scion Image software.
3. Results and Discussion
3.1. Regulation of post-transcriptional modification by Σ RNA/MALAT-1
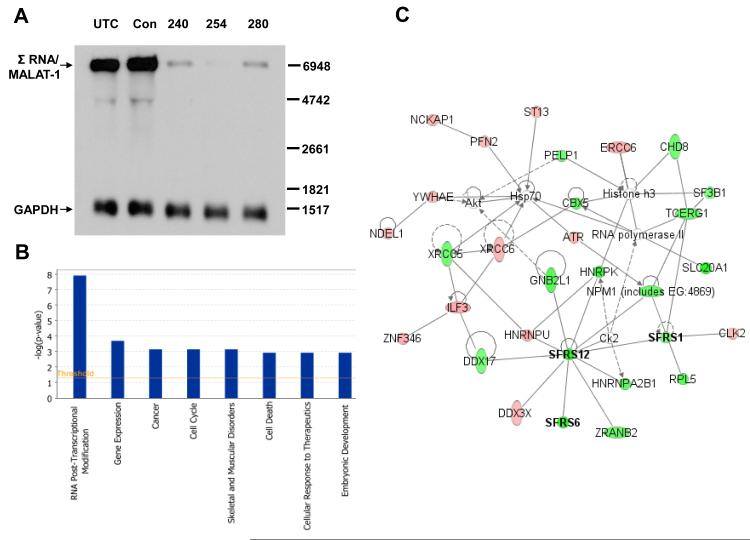
Antisense technology was applied to characterize the functional biology of Σ RNA/MALAT-1. Σ RNA/MALAT-1 was profoundly reduced 24 hrs after transfection with ASOs specifically targeted to the ncRNA in HeLa cells, with up to 100-fold target reduction by Northern and densitometric analyses using ASO254 and quantitative RT-PCR analysis (Fig. 1A and data not shown). Based on its predominant nuclear localization and association with SC35 splicing domains [5,8], we used Affymetrix exon arrays, which permit profiling of transcript architecture and evaluation of splicing, to explore transcripts that were differentially expressed and spliced after profound reduction of Σ RNA/MALAT-1. RNAs were harvested from HeLa cells 24 hrs after transfection with control ASO or ASO254, selected based on the highest specific target reduction (Fig. 1A), and processed for array analysis. Using P<0.001 as cut-off in the global analysis of the array data, 276 genes exhibited highly significant alternative splicing differences between ASO254 transfectants and the control (Table S2). Ingenuity Pathway Analysis (IPA) of the 276 genes indicated that RNA post-transcriptional modification was the function most significantly affected by reduction of Σ RNA/MALAT-1 by ASO254 (P<1.2×10−8, Fig. 1B). The molecules in this functional category and their interaction network are shown in Fig. 1C, many of which are SR proteins (SFRS1, SFRS6, and SFRS12) that play important roles in mRNA metabolism such as constitutive and alternative splicing, RNA stability, and mRNA quality control [2]. Also included in the network are hnRNP proteins (HNRNPU, HNRNPA2B1, and HNRPK) that participate in mRNA biogenesis. In cultured human umbilical vein endothelial cells (HUVECs), 668 genes exhibited alternative splicing differences (P<0.001, Table S3) 24 hrs after transfection with ASOs targeted to Σ RNA/MALAT-1. RNA post-transcriptional modification was also the most significant function (P<6.8×10−9, Fig. S1) affected by reduction of Σ RNA/MALAT-1, which included transcripts of SR proteins SFRS1, SFRS6, SFRS8, and SFRS21P (Table S3). These results are consistent with a prior report that all SR proteins analyzed were alternatively spliced [13]. Collectively, these data support that human Σ RNA/MALAT-1 influences the alternative splicing activity of a broad spectrum of molecules that are involved in RNA processing.
Fig. 1.
Evidence that Σ RNA/MALAT-1 regulates RNA post-transcriptional modification globally in HeLa cells. (A) Target reduction of Σ RNA/MALAT-1 by ASOs. RNAs from HeLa cell transfectants were used for Northern analyses. Position of Σ RNA/MALAT-1 is indicated. GAPDH, loading controls. UTC, untreated control; Con, scrambled ASO; 240, 254, and 280, Σ RNA/MALAT-1 specific ASOs. (B) IPA analyses of genes whose alternative splicing activity were significantly changed (P<0.001) by Σ RNA/MALAT-1 reduction in HeLa cells. (C) Components of the top class of differentially expressed genes and their network. Bold, transcripts of SR proteins. Red and green, up- and down- regulated transcripts, respectively, in the ASO254-transfected HeLa cells with significantly changed alternative splicing activity (P<0.001) due to Σ RNA/MALAT-1 reduction. Uncolored, genes in the network that were not identified in the exon array analysis.
3.2. Modulation of SR expression by perturbation of Σ RNA/MALAT-1
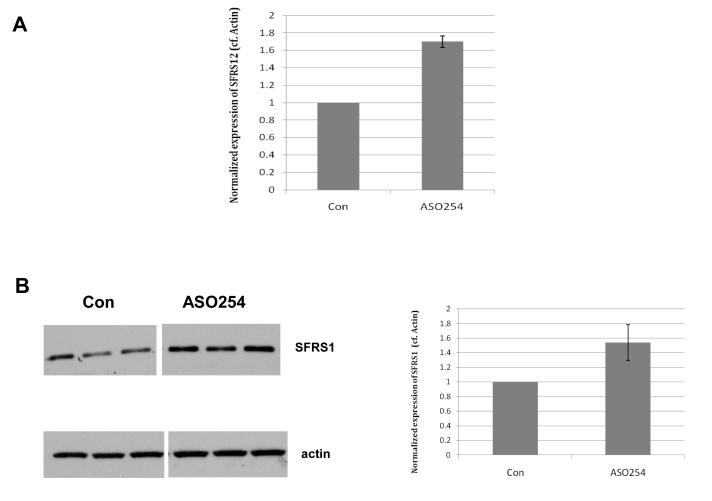
To confirm the regulation of SR proteins by Σ RNA/MALAT-1, we analyzed the expression of SFRS12 and SFRS1 in HUVECs upon suppression of Σ RNA/MALAT-1. Transfection of ASO254 resulted in a significant increase (P<0.05) of protein level of the major isoform (59.4 kDa) of SFRS12 (Fig. 2A), but not the minor isoform (71.6 kDa) of SFRS12 (data not shown), consistent with a hypothesis that perturbation of presence or levels of Σ RNA/MALAT-1 affects alternative splicing of SFRS12 mRNA and hence the encoded protein products. Additionally, transfection of ASO254 enhanced expression of SFRS1 protein in HUVECs (Fig. 2B). Collectively, the above data support the hypothesis that perturbation of Σ RNA/MALAT-1 with ASO254 changed expression of SR proteins SFSR12 and SFRS1, consistent with regulation by Σ RNA/MALAT-1 of RNA post-transcriptional modification.
Fig. 2.
Evidence that Σ RNA/MALAT-1 regulates SR proteins in HUVECs. Cells were transfected with Σ RNA/MALAT-1 ASO254 or control ASO. Proteins from 24 hrs transfectants were used for Western blot analysis. (A) Quantification of the expression levels of the 59.4 kDa major isoform of SFRS12 protein. (B) Images and quantification of the expression levels of the 27.7 kDa isoform of SFRS1. Actin immunoblotting was used as protein loading control.
3.3. Alteration of Endoglin transcripts by Σ RNA/MALAT-1
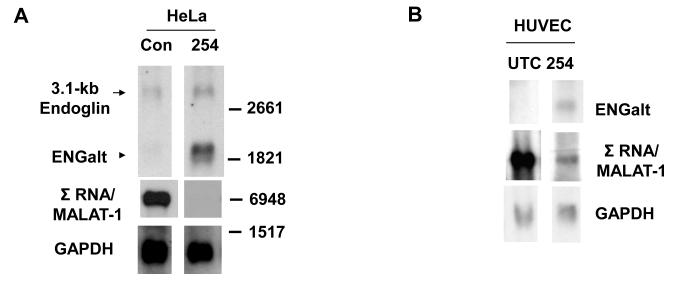
In addition to genes involved in RNA post-transcriptional modification, processing of other gene transcripts was also affected by Σ RNA/MALAT-1 suppression. One of the genes of interest was human Endoglin that encodes a homodimeric membrane glycoprotein [14] that is elevated in activated endothelial cells and solid tumors [15]. Numerous alternatively spliced Endoglin transcripts have been identified in health and disease [16]. Northern analyses with an antisense probe targeting nt 881-1292 of the 3.1-kb canonical Endoglin mRNA (GenBank accession # NM_000118.1) demonstrated expression in HeLa cells (Fig. 3A). Interestingly, an ~1.8-kb transcript was observed in HeLa cells and HUVECs following Σ RNA/MALAT-1 suppression by ASO254 (ENGalt, Fig. 3A and 3B), but barely detected in the control cells, consistent with the requirement of Σ RNA/MALAT-1 suppression for generating the ENGalt transcript.
Fig. 3.
Evidence that Σ RNA/MALAT-1 regulates processing of Endoglin transcript. Twenty-four hrs after transfection with 75nM ASOs, total RNAs were harvested from cells. Northern blot analyses of RNAs from HeLa (A) and HUVECs (B) are shown. UTC, untreated control; Con, scrambled ASO; 254, Σ RNA/MALAT-1 specific ASO. GAPDH, loading controls.
3.4. Regulation of Tissue Factor transcript repertoire by Σ RNA/MALAT-1
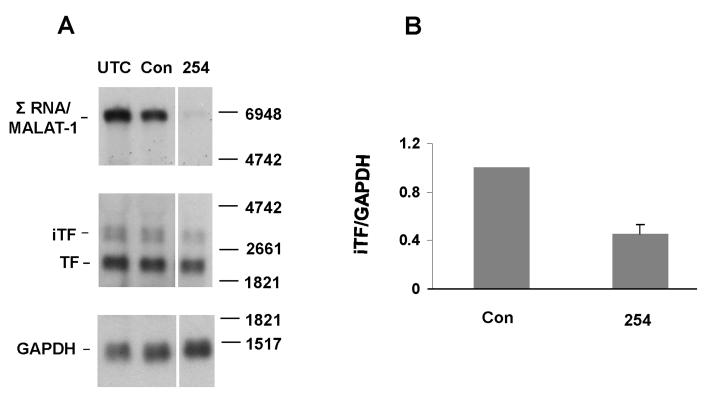
Tissue Factor (TF) was another transcript identified in our microarray analysis with altered RNA processing after knockdown of Σ RNA/MALAT-1. TF plays important roles in tumor growth, angiogenesis, and metastasis [17]. Two forms of alternative splicing events exist for human TF mRNA, i.e. intron 1 retained (e.g. AY940729, Fig. S2) and exon 5 skipped (e.g. AF497569). Twenty-four hrs following transfection, Σ RNA/MALAT-1 was markedly diminished in HeLa cells transfected with ASO254 (Fig. 4A). In addition to the major 2.2-kb form of mature TF, two minor ~3-kb transcripts, iTF, were observed, consistent with previous observations in other cells [18]. The iTF RNAs, but not the major 2.2-kb TF transcript, were identified with a probe for intron 1 of F3 (data not shown). The iTF transcripts were significantly decreased (P<0.005) in the ASO254 transfected cells compared with the control (Fig. 4A), with an ~2-fold difference (Fig. 4B). Interestingly, the mature TF transcript also appeared to be decreased in the ASO254 transfected cells (Fig. 4A). These results suggest that Σ RNA/MALAT-1 influences TF transcript repertoire.
Fig. 4.
Retention of intron 1 sequences in TF mRNA and the involvement of Σ RNA/MALAT-1. HeLa cells were transfected with 75nM ASO254 against Σ RNA/MALAT-1 or control ASO. Twenty-four hrs after transfection, total RNAs were harvested. (A) Northern analyses. UTC, untreated control. Top panels show knockdown of Σ RNA/MALAT-1 by ASO254. Middle panels show three distinguished bands of TF transcripts, a major ~2.2-kb (TF) and two minor ~3-kb forms (iTF). Bottom panels show GAPDH as loading controls. Molecular size markers are shown to the right. (B) Quantitative results of iTF (both forms) based on Northern analyses normalized with GAPDH levels, mean ± SD, from 3 independent experiments.
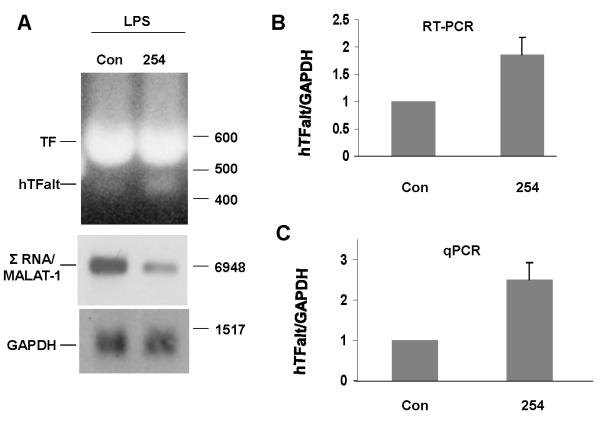
Both TF and TFalt are induced in endothelial cells by endotoxin [19] or inflammatory responses [20]. To address the role of Σ RNA/MALAT-1 in control of TF alternative splicing in endothelial cells, HUVEC were transfected with Σ RNA/MALAT-1 ASO254, then exposed to 10 μg/ml LPS for 4 hrs before harvesting RNA. Σ RNA/MALAT-1 was significantly reduced 24 hrs after transfection with ASO254 (Fig. 5A), with an average 8-fold target reduction. Semi-quantitative RT-PCRs detected both full-length TF and exon 5 skipped hTFalt in the LPS-treated cells. The amount of hTFalt transcript was significantly elevated in the Σ RNA/MALAT-1 suppressed cells (Fig. 5A and B). Quantitative PCR (qPCR) using primers specific to hTFalt indicated an ~2.5-fold increase of the exon 5 skipped hTFalt transcript by Σ RNA/MALAT-1 suppression (Fig. 5C). The specificity of the qPCR was confirmed by the appearance of a single band of qPCR product, and subsequent sequencing of this DNA band extracted from the agarose gel (data not shown). Collectively, these results indicate that Σ RNA/MALAT-1 influences the generation of alternatively spliced TF transcripts and thereby may influence the coagulation and cellular signaling pathways.
Fig. 5.
Involvement of Σ RNA/MALAT-1 in TF alternative splicing in LPS-treated HUVEC. HUVEC were transfected with 75nM ASO254 against Σ RNA/MALAT-1 or control ASO for 24 hrs, and treated with 10 μg/ml LPS for 4 hrs. (A) Northern analyses (middle for Σ RNA/MALAT-1, and bottom for GAPDH), or RT-PCR for exons 4-6 of F3 (top). Molecular size markers are shown to the right. (B) Quantitative results of the ratio of hTFalt/GAPDH transcripts, mean ± SD, based on the RT-PCR results. (C) Ratio of hTFalt/GAPDH transcripts based on qPCR results, mean ± SD.
In summary, the present study has identified that human Σ RNA/MALAT-1 modulates the transcript repertoire as a result of post-transcriptional modifications of primary transcripts. This follows from consideration of two scenarios. In a global view, RNA post-transcriptional modification is the most significant function influenced by reduction of Σ RNA/MALAT-1. With specific examples, we provide evidence for the controlling function of Σ RNA/MALAT-1 on primary transcript modification, i.e. Endoglin and Tissue Factor that both play roles in tumor biology [15,17]. Perturbation of Σ RNA/MALAT-1 altered splicing activity of genes that encode SR proteins and hnRNPs, which themselves participate in RNA processing. Collectively, these results are consistent with a hypothesis that Σ RNA/MALAT-1 is a master regulator of RNA post-transcriptional modification and perturbation of Σ RNA/MALAT-1 modifies processing of a wide variety of transcripts including the ones encoding SR proteins, phosphorylation of which is also shown to be modulated by the ncRNA [21], and TF and Endoglin that play important roles in tumor biology.
Supplementary Material
Acknowledgements
We wish to thank Dr. Wolfram Ruf for critical discussions, Elizabeth M. Boyd for illustrations, and Pablito Tejada for technical assistance. This is TSRI manuscript # 19612.
Abbreviations
- ASO
antisense oligo
- ENG
Endoglin
- ENGalt
alternatively spliced Endoglin
- hnRNP
heterogeneous nuclear ribonucleoprotein
- IPA
Ingenuity Pathway Analysis
- ncRNA
noncoding RNA
- nt
nucleotide
- SR protein
Serine/Arginine-rich protein.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- [2].Huang Y, Steitz JA. SRprises along a messenger’s journey. Mol. Cell. 2005;17:613–615. doi: 10.1016/j.molcel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- [3].Rossbach O, Hung LH, Schreiner S, et al. Auto- and crossregulation of the hnRNP L proteins by alternative splicing. Mol. Cell Biol. 2009;29:1442–1451. doi: 10.1128/MCB.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annual Review of Biochemistry. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- [5].Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- [6].van Asseldonk M, Schepens M, de Bruijn D, Janssen B, Merkx G, Geurts vK. Construction of a 350-kb sequence-ready 11q13 cosmid contig encompassing the markers D11S4933 and D11S546:mapping of 11 genes and 3 tumor-associated translocation breakpoints. Genomics. 2000;66:35–42. doi: 10.1006/geno.2000.6194. [DOI] [PubMed] [Google Scholar]
- [7].Ji P, Diederichs S, Wang W, Boing S, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- [8].Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Guo F, Li Y, Liu Y, Wang J, Li Y, Li G. Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta. Biochim. Biophys. Sin. 2010;42:224–229. doi: 10.1093/abbs/gmq008. [DOI] [PubMed] [Google Scholar]
- [10].Tano K, et al. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584:4575–4580. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- [11].Wilusz JE, Freier SM, Spector DL. 3′ End Processing of a Long Nuclear-Retained Noncoding RNA Yields a tRNA-like Cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Baker BF, et al. 2′-O-(2-Methoxy)ethyl-modified anti-intercellular adhesion molecule 1 (ICAM-1) oligonucleotides selectively increase the ICAM-1 mRNA level and inhibit formation of the ICAM-1 translation initiation complex in human umbilical vein endothelial cells. J. Biol. Chem. 1997;272:11994–12000. doi: 10.1074/jbc.272.18.11994. [DOI] [PubMed] [Google Scholar]
- [13].Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- [14].Fonsatti E, Sigalotti L, Arslan P, Altomonte M, Maio M. Emerging role of endoglin (CD105) as a marker of angiogenesis with clinical potential in human malignancies. Curr. Cancer Drug Targets. 2003;3:427–432. doi: 10.2174/1568009033481741. [DOI] [PubMed] [Google Scholar]
- [15].Dallas NA, et al. Endoglin (CD105): a marker of tumor vasculature and potential target for therapy. Clin. Cancer Res. 2008;14:1931–1937. doi: 10.1158/1078-0432.CCR-07-4478. [DOI] [PubMed] [Google Scholar]
- [16].Velasco S, Alvarez-Munoz P, Pericacho M, Dijke PT, Bernabeu C, Lopez-Novoa JM. L- and S-endoglin differentially modulate TGFbeta1 signaling mediated by ALK1 and ALK5 in L6E9 myoblasts. J. Cell Sci. 2008;121:913–919. doi: 10.1242/jcs.023283. [DOI] [PubMed] [Google Scholar]
- [17].Milsom C, Rak J. Tissue factor and cancer. Pathophysiol. Haemost. Thromb. 2008;36:160–176. doi: 10.1159/000175154. [DOI] [PubMed] [Google Scholar]
- [18].Gregory SA, Morrissey JH, Edgington TS. Regulation of tissue factor gene expression in the monocyte procoagulant response to endotoxin. Mol. Cell Biol. 1989;9:2752–2755. doi: 10.1128/mcb.9.6.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mackman N, Morrissey JH, Fowler B, Edgington TS. Complete sequence of the human tissue factor gene, a highly regulated cellular receptor that initiates the coagulation protease cascade. Biochemistry. 1989;28:1755–1762. doi: 10.1021/bi00430a050. [DOI] [PubMed] [Google Scholar]
- [20].Szotowski B, Antoniak S, Rauch U. Alternatively spliced tissue factor:a previously unknown piece in the puzzle of hemostasis. Trends Cardiovasc. Med. 2006;16:177–182. doi: 10.1016/j.tcm.2006.03.005. [DOI] [PubMed] [Google Scholar]
- [21].Tripathi V, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;24:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.