Abstract
Background:
KRAS mutation is a negative predictive factor for treatment with anti-epidermal growth factor receptor antibody in metastatic colorectal cancer (CRC). KRAS mutation analysis is usually performed on primary tumour tissue because metastatic tissue is often not available. However, controversial data are available on the concordance of test results between primary tumours and corresponding metastases. We assessed the concordance of KRAS mutation status in a study of 305 primary colorectal tumours and their corresponding liver metastases.
Methods:
Patients with histologically confirmed CRC who underwent surgical resection of the primary tumour and biopsy or surgical resection of the corresponding liver metastasis were included. KRAS mutation analysis was performed for codons 12 and 13.
Results:
KRAS mutation was detected in 108 out of 305 primary tumours (35.4%). In 11 cases (3.6%), we found a discordance between primary tumour and metastasis: 5 primary tumours had a KRAS mutation with a wild-type metastasis, 1 primary tumour was wild type with a KRAS mutation in the metastasis, and in 5 cases the primary tumour and the metastasis had a different KRAS mutation.
Conclusion:
We observed a high concordance of KRAS mutation status of 96.4% (95% CI 93.6–98.2%) between primary colorectal tumours and their corresponding liver metastases. In only six patients (2.0% 95% CI 0.7–4.2%), the discordance was clinically relevant. In this largest and most homogenous study to date, we conclude that both primary tumours and liver metastases can be used for KRAS mutation analysis.
Keywords: KRAS mutation, colorectal cancer, liver metastases, concordance
Recent advances in specific signalling pathways of cancer cells have introduced targeted therapy into treatment regimes for patients with metastatic colorectal cancer (CRC) (Tol and Punt, 2010). Cetuximab and panitumumab are monoclonal antibodies that bind to the extracellular domain of the epidermal growth factor receptor (EGFR). They inhibit ligand-induced stimulation of several intracellular signalling pathways, such as RAS/RAF/MAPK and phosphoinositide-3 pathway, which results in decreased stimulation of cell cycle progression, proliferation, angiogenesis, and stimulation of apoptosis (Scaltriti and Baselga, 2006). The KRAS oncogene is currently the most relevant molecular biomarker that predicts the response to EGFR-targeted therapy in CRC. An oncogenic mutation in KRAS leads to constitutive activation of the RAS/RAF signalling pathway independent from EGFR activation by binding of the ligand (Benvenuti et al, 2007). KRAS mutations occur in approximately 38% of colorectal tumours and involve codon 12 and 13 in >96% of cases (Oliveira et al, 2004). Metastatic CRC patients with tumours harbouring a KRAS mutation are resistant to treatment with anti-EGFR antibodies, showing lower response rates, decreased progression-free survival, and overall survival compared with patients with KRAS wild-type tumours (Karapetis et al, 2008; Tol et al, 2009; Van Cutsem et al, 2009). Therefore, the European Medicines Agency and the Food and Drug Administration have restricted the use of anti-EGFR antibodies in metastatic CRC to patients with KRAS wild-type tumours.
Cetuximab and panitumumab have shown efficacy both as monotherapy (Amado et al, 2008; Karapetis et al, 2008) and in combination with chemotherapy (Tol et al, 2009; Van Cutsem et al, 2009) in patients with KRAS wild-type metastatic CRC. Nevertheless, even among patients with KRAS wild-type tumours, the majority of patients do not respond to anti-EGFR therapy. Efficacy of anti-EGFR therapy was suggested to be further restricted to patients with BRAF wild-type tumours (Di Nicolantonio et al, 2008). An additional explanation for the suboptimal response rates to anti-EGFR antibodies in patients with KRAS wild-type tumours is discordance of KRAS mutation status between primary colorectal tumours and corresponding metastases. In the early dissemination model, tumour cells depart the primary lesion before the acquisition of a fully malignant phenotype to undergo new mutations and metastatic growth at a distant site (Klein, 2009). According to this model, a discordance in mutation status between primary tumours and metastases may occur, and as a consequence the mutation status of the primary tumour might not be adequate to predict the response of metastases to anti-EGFR treatment.
Current data on the concordance in KRAS mutation status between primary colorectal tumours and metastases are conflicting. Five studies showed a 100% concordance of KRAS mutation status in primary CRC and corresponding metastases (Losi et al, 1992; Suchy et al, 1992; Zauber et al, 2003; Weber et al, 2007; Etienne-Grimaldi et al, 2008). In contrast to these data, others have reported a discordance of KRAS mutation status in primary tumours and metastatic sites, with an overall discordance observed in 4–32% of the patients (Oudejans et al, 1991; Al-Mulla et al, 1998; Albanese et al, 2004; Oliveira et al, 2007; Artale et al, 2008; Santini et al, 2008; Cejas et al, 2009; Garm Spindler et al, 2009; Loupakis et al, 2009; Molinari et al, 2009; Perrone et al, 2009; Baldus et al, 2010; Italiano et al, 2010). These controversial results are probably due to the fact that these studies were underpowered with a small number of patients, and included a wide variety of metastatic sites. Therefore, it is still uncertain whether the evaluation of KRAS mutation status in the most commonly available primary tumour correctly reflects the KRAS mutation status of corresponding metastasis. This is highly relevant given the large number of CRC patients as well as the potential toxicity and costs of anti-EGFR therapy.
We assessed the concordance in KRAS mutation status in primary tumours and their corresponding liver metastases in an adequately powered study of 305 CRC patients.
Patients and methods
Patient selection
Patients with histologically confirmed CRC who underwent surgical resection of the primary tumour and biopsy or surgical resection of the corresponding liver metastasis were included in this analysis. Results were obtained from archived material of three large pathology laboratories and from material collected from the CAIRO2 study, a large multicentre trial of the Dutch Colorectal Cancer Group (Tol et al, 2009).
In patients with a discordance of KRAS mutation status between the primary tumour and metastasis, additional blocks of the primary tumour were obtained to exclude heterogeneity within the tumour. Lymph node metastases present at the time of diagnosis were also acquired in these patients.
Tumour DNA preparation
Formalin-fixed paraffin-embedded tissue blocks were cut at 4 μm thickness and stained with haematoxylin and eosin (HE). The presence of tumour tissue was marked by a pathologist. Subsequently the blocks were cut at 20–40 μm thickness and micro dissected for DNA extraction. Tumour tissue was dissolved in 200 μl lysis buffer (QIAamp DNA Micro Kit, Qiagen, Venlo, The Netherlands) and incubated with proteinase K overnight at 56 °C for two nights. DNA was extracted according to the manufacturer's protocol (QIAamp DNA Micro Kit, Qiagen), and DNA concentration was determined at 260 nm using the Nanodrop 26 ND-1000 spectrophotometer (Nanodrop Technologies Inc., Wilmington, NC, USA).
KRAS mutation analysis
For KRAS mutation analysis, exon 2 (codon 12 and 13) was amplified using a 50 μl reaction mixture containing 0.2 μm forward (5′-TGTAAAACGACGGCCAGTAGGCCTGCTGAAAATGACTG-3′) and reverse (5′-CAGGAAACAGCTATGACCTGGATCATATTCGTCCACAAAA-3′) primers (Invitrogen, Breda, The Netherlands); dATP, dCTP, dGTP and dTTP (GE Healthcare, Zeist, The Netherlands) at 0.2 mM each; 50 mM KCl; 10 mM Tris-HCl (pH 8.3); 2.5 mM MgCl2; 1 U AmpliTaq Gold polymerase (Applied Biosystems, Nieuwkerk a/d IJsel, The Netherlands) and 50 ng of template DNA. The PCR conditions were as follows: 94 °C for 10 min; 92 °C for 1 min, 60 °C for 1 min, 72 °C for 1 min (40 cycles); and 72 °C for 10 min.
All PCR products were purified with the MultiScreen HTS, 96 well Filtration System (Millipore, Carrigtwohill, Ireland). Subsequently, the purified products were sequenced using fluorescently labelled terminators (BigDye Terminators (v 1.1); Applied Biosystems, Foster City, CA, USA) with both M13-forward and M13-reverse sequencing primers. The sequencing products were analysed on an ABI 3730 DNA Analyser (Applied Biosystems) and the data analysis was performed using Sequencing Analysis Software Sequencing Analysis Software v5.3.1 with KBTM Basecaller. Sequence results were scored by visual inspection of the chromatograms (Applied Biosystems).
Statistical analysis
We considered a discordance level of 5% or more to be clinically relevant, that is, leading to substantial change in routine clinical practice. To exclude such level of discordance under the assumption that the true discordance was 2.5% or less, we set the sample size at 304 paired samples. With this sample size, the precision in the estimated percentage of discordance was 2.5% (i.e., s.e. 1.25, half-width of the 95% confidence interval equal to 2.5%).
The comparison of patient and primary tumour characteristics between patients with KRAS wild-type and KRAS mutant primary tumours was done using Wilcoxon's rank sum test or χ2 for numerical or categorical variables, respectively. Differences in KRAS mutation status between the primary tumour and corresponding metastasis were analysed by calculating the percentage of concordance, and (clinically relevant) discordance, together with the corresponding Clopper–Pearson 95% confidence intervals. Differences were considered to be statistically significant when the P-value was below 0.05. All statistical tests were two-sided.
Results
Patient characteristics
We analysed KRAS codon 12 and 13 mutations in 320 matched primary colorectal tumours and liver metastases. The tumour cell percentages in all primary tumours and metastases were above 30%. We failed to obtain a KRAS mutation status in 15 patients; therefore our further analyses were performed in 305 paired samples. Patient characteristics are shown in Table 1.
Table 1. Distribution of tumour characteristics according to KRAS status of the primary tumour.
| Overall, n=305 | KRAS mutation, n=108 | KRAS wild type, n=197 | P-value | |
|---|---|---|---|---|
| Age | 0.20 | |||
| Median (IQR) | 64 (57–70) | 65 (58–71) | 64 (57–70) | |
| Gender | 0.37 | |||
| Male | 191 (62.6%) | 64 (59.3%) | 127 (64.5%) | |
| Female | 114 (37.4%) | 44 (40.7%) | 70 (35.5%) | |
| Metastases presentation | 0.45 | |||
| Synchronous | 169 (55.4%) | 63 (58.3%) | 106 (53.8%) | |
| Metachronous | 136 (44.6%) | 45 (41.7%) | 91 (46.2%) | |
| Tumour location | 0.63 | |||
| Colon | 167 (54.8%) | 59 (54.6%) | 108 (54.8%) | |
| Rectum | 54 (17.7%) | 16 (14.8%) | 38 (19.3%) | |
| Rectosigmoid | 80 (26.2%) | 32 (29.6%) | 48 (24.4%) | |
| Unknown | 4 (1.3%) | 1 (0.9%) | 3 (1.5%) | |
| Histopathological subtype | 0.12 | |||
| Adenocarcinoma | 271 (88.9%) | 90 (83.3%) | 181 (91.9%) | |
| Adenocarcinoma with muc. component | 21 (6.9%) | 10 (9.3%) | 11 (5.6%) | |
| Mucinous adenocarcinoma | 8 (2.6%) | 5 (4.6%) | 3 (1.5%) | |
| Unknown | 5 (1.6%) | 3 (2.8%) | 2 (1.0%) | |
| Differentiation grade | 0.21 | |||
| Good | 33 (10.8%) | 13 (12.0%) | 20 (10.2%) | |
| Moderate | 196 (64.3%) | 65 (60.2%) | 131 (66.5%) | |
| Poor | 52 (17.0%) | 17 (15.7%) | 35 (17.8%) | |
| Unknown | 24 (7.9%) | 13 (12.0%) | 11 (5.6%) | |
| T stage | 0.62 | |||
| T1 | 4 (1.3%) | 2 (1.9%) | 2 (1.0%) | |
| T2 | 20 (6.6%) | 9 (8.3%) | 11 (5.6%) | |
| T3 | 231 (75.7%) | 81 (75.0%) | 150 (76.1%) | |
| T4 | 36 (11.8%) | 11 (10.2%) | 25 (12.7%) | |
| Unknown | 14 (4.6%) | 5 (4.6%) | 9 (4.6%) | |
| N stage | 0.10 | |||
| N0 | 114 (37.4%) | 46 (42.6%) | 68 (34.5%) | |
| N1 | 87 (28.5%) | 31 (28.7%) | 56 (28.4%) | |
| N2 | 86 (28.2%) | 26 (24.1%) | 60 (30.5%) | |
| Unknown | 18 (5.9%) | 5 (4.6%) | 13 (6.6%) | |
| Number of lymph nodes examined | 0.28 | |||
| Median (IQR) | 10 (6–15) | 10 (6–13) | 10 (6–16) | |
| Number of lymph node metastases | 0.15 | |||
| Median (IQR) | 1 (0–4) | 1 (0–3) | 1 (0–4) | |
Abbreviation: IQR=interquartile range.
KRAS mutation and histopathological parameters
A total of 108 patients (35.4%) had a KRAS mutation in the primary tumour; of which 37 patients had a Gly12Asp mutation, 28 patients a Gly12Val mutation, 14 patients a Gly13Asp mutation, 10 patients a Gly12Cys mutation, 7 patients a Gly12Ser mutation, 7 patients a Gly12Ala mutation, 3 patients a Gly12Arg mutation, 1 patient a Gly12Asp and Gly12Ala mutation and 1 patient a Gly12Phe mutation (Table 2). Histopathological characteristics of the primary tumour were comparable between patients with and without a KRAS mutation (Table 1).
Table 2. Distribution of KRAS mutation types.
| Codon 12/13 | Patients with KRAS mutation (n, %) |
|---|---|
| Gly12Asp | 37 (34%) |
| Gly12Val | 28 (26%) |
| Gly13Asp | 14 (13%) |
| Gly12Cys | 10 (9%) |
| Gly12Ser | 7 (6%) |
| Gly12Ala | 7 (6%) |
| Gly12Arg | 3 (3%) |
| Gly12Phe | 1 (1%) |
| Gly12Asp + Gly12Ala | 1 (1%) |
Concordance of KRAS status in primary tumours and corresponding liver metastases
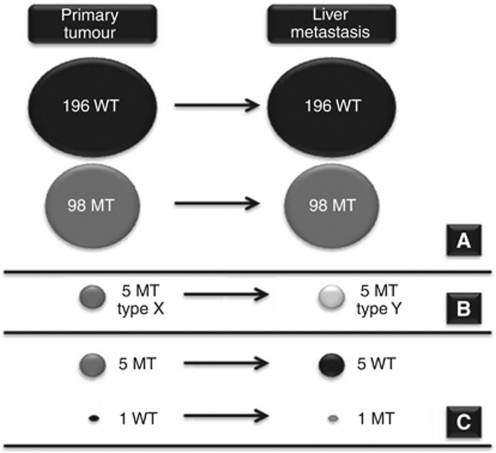
In 294 patients (96.4% 95% CI 93.6–98.2%), the same KRAS mutation status was obtained from the primary tumour and the corresponding liver metastasis. In 11 patients (3.6% 95% CI 1.8–6.4%), of which 7 had synchronous metastases at diagnosis and 4 developed metachronous metastases, we found a discordance between primary tumours and metastases. Five patients had a KRAS mutation in the primary tumour and not in the liver metastasis. Only one patient had a wild-type status of the primary tumour, while the metastasis showed a KRAS mutation. In five patients, the primary tumours had different KRAS mutations compared with the metastases. One of these patients had two primary tumours. Both primary tumours had the same KRAS mutation (Gly13Asp), while the liver metastasis had a different KRAS mutation (Gly12Ser). In another patient, the primary tumour had a double mutation (Gly12Asp/Gly12Val) and the metastasis had a Gly12Asp mutation (Figure 1, Table 3). Taken together, the observed discordance was clinically relevant in only six patients (2.0% 95% CI 0.7–4.2%).
Figure 1.
Overall concordance of the KRAS mutation status between primary tumour and liver metastasis (A), discordance without clinical impact (B), and discordance with clinical impact (C). Abbreviations: WT, wild type; MT, mutation.
Table 3. Patients with a discordant KRAS status between primary tumour and liver metastasis. Multiple blocks of primary tumour tissue and lymph node metastases were tested when available.
| KRAS status primary tumour | KRAS status 2nd tumour | KRAS status lymph node metastasis | KRAS status liver metastasis | |
|---|---|---|---|---|
| 1 | Gly12Ala | — | LN 1: Gly12Ala | WT |
| LN 2: Gly12Ala | ||||
| LN 3: Gly12Ala | ||||
| 2 | Gly12Asp | — | — | WT |
| Gly12Asp | ||||
| WT | ||||
| 3 | Gly12Cys | — | — | WT |
| 4 | Gly12Asp | — | LN 1: Gly12Asp | WT |
| Gly12Asp | LN 2: Gly12Asp | |||
| Gly12Asp | LN 3: Gly12Asp | |||
| Gly12Asp | LN 4: Gly12Asp | |||
| LN 5: WT | ||||
| 5 | Gly12Ser | — | — | WT |
| 6 | WT | — | — | Gly12Cys |
| 7 | Gly12Asp | — | LN 1: WT | Gly12Ala |
| LN 2: WT | ||||
| LN 3: WT | ||||
| 8 | Gly13Asp | Gly13Asp | LN 1: Gly13Asp | Gly12Ser |
| 9 | Gly12Ser | — | — | Gly12Ala |
| 10 | Gly12Cys | — | LN 1: Gly12Asp | Gly12Asp |
| Gly12Asp | LN 2: Gly12Asp | |||
| LN 3: Gly12Asp | ||||
| LN 4: Gly12Asp | ||||
| LN 5: Gly12Asp | ||||
| LN 6: WT | ||||
| 11 | Gly12Asp/Gly12Val | — | LN 1: Gly12Val | Gly12Asp |
| LN 2: Gly12Val | ||||
| LN 3: Gly12Val | ||||
| LN 4: Gly12Asp | ||||
| LN 5: Gly12Asp | ||||
| LN 6: Gly12Asp | ||||
| LN 7: Gly12Asp |
Abbreviation: WT=wild type.
Subsequent analyses in patients with a discordance of KRAS status
Several tests were performed to exclude bias of the test results. First, the HE coupes of all patients with a discordant KRAS mutation status between the primary tumour and liver metastasis were revised. The primary tumours and liver metastases had a mean tumour cell percentage of 65 and 60%, respectively. Subsequent independent reanalysis of the KRAS mutation status resulted in the same discordances.
Second, several mutation analyses were performed on different areas of the tumour and from different tumour blocks in order to establish possible tumour heterogeneity. Two patients showed heterogeneity of KRAS status within the primary tumour. One of these patients demonstrated two areas with a Gly12Asp mutation and one area with wild-type status, of which the latter resembled the liver metastasis. The other patient showed two different KRAS mutations within the same tumour, of which one is concordant with the liver metastasis (Table 3).
Third, 6 of the 11 patients with discordant results did have lymph nodes metastases at the time of diagnosis. KRAS mutation testing of all lymph nodes separately revealed overall concordant KRAS status between lymph node metastases and the primary tumour in three patients. The KRAS status of the lymph nodes in the other three patients showed heterogeneity, of which at least one lymph node metastases showed a different KRAS status compared with the primary tumour. However, this explains the discordance between the primary tumour and liver metastasis only in one patient (Table 3).
Discussion
This is the first adequately powered study in CRC that compares KRAS mutation status between primary tumours and their corresponding liver metastases. We showed that tissue from the primary tumour can reliably be used for KRAS mutation testing in order to select patients for anti-EGFR therapy.
We observed a concordant KRAS mutation status in 96.4% of 305 paired samples of colorectal tumours and liver metastases. However, the difference in KRAS status was not clinically relevant in 5 of the 11 patients with discordant results, because both primary tumour and metastasis had a different KRAS mutation. Given the high statistical power of our analysis, we were able to obtain a highly accurate estimate of the level of discordance that enabled us to conclude that the level of discordance was 2.0%. The high rate of concordance is in agreement with the notion that KRAS mutations are considered as early driving events in CRC progression, and associated with the growth of small adenoma to clinically significant size (Vogelstein et al, 1988). Therefore, KRAS mutation status is expected to be equal in both primary tumours and metastases (Klein, 2009).
The previously reported lower concordance levels between primary tumours and metastases are most likely due to bias caused by false-negative results in underpowered studies. We calculated that 304 paired cases were needed to reliably exclude a rate of discordance of >5%, while previous studies included only 10 to 110 patients (Table 4). Moreover, in these studies metastases of different sites were compared with the primary tumour. As the molecular patterns may differ between metastatic sites (Klein, 2009), more reliable results are obtained when KRAS mutation status is tested more rigorously for each metastatic site. The liver is the predominant site of metastases in the majority of metastatic CRC patients; therefore the results of our large series of 305 liver metastases provide a solid reference for clinical decision making as to anti-EGFR therapy. Another issue is the fact that KRAS testing is technically not as straightforward as is often assumed. Several quality assurance systems are now in place, and the first ‘round robin’ test indicates that at least 30% of the experienced pathology laboratories fail to pass the threshold level of the quality assurance programs (Bellon et al, 2011). Other important facts about KRAS testing are the correct evaluation of the amount of tumour tissue in the sample and the sensitivity of testing methods. In a previous study, we demonstrated in >500 samples that both sequencing and real-time PCR are reliable methods (Tol et al, 2010).
Table 4. Overview of studies providing data on KRAS status of primary tumour and related metastasis.
| Author study | Year | No. of pts | Analysed metastatic site | Method | KRAS mutation in PT (%) | KRAS mutation in PT, WT in M | KRAS WT in PT, mutation in M | Total percentage of discordance |
|---|---|---|---|---|---|---|---|---|
| Albanase | 2004 | 30 | Liver | SSCP analysis | 14 (47%) | 5/14 (36%) | 4/16 (25%) | 9/30 (30%) |
| Al-Mulla | 1998 | 26 | Liver | ASO/direct seq | 10 (38%) | 2/10 (20%) | 3/16 (19%) | 5/26 (19%) |
| 31 | Lymph node | ASO/direct seq | 10 (32%) | 1/10 (10%) | 5/21 (24%) | 6/31 (19%) | ||
| Artale | 2008 | 48 | Diverse, 81% liver | Direct seq | 11 (23%) | 1/11 (9%) | 2/37 (5%) | 3/48 (6%) |
| Baldus | 2010 | 20 | Visceral metastasis | Direct seq | 9 (45%) | 1/9 (11%) | 1/11 (9%) | 2/20 (10%) |
| 55 | Lymph node | Direct seq | 29 (53%) | 15/29 (52%) | 2/26 (8%) | 17/55 (31%) | ||
| Cejas | 2010 | 93 | Liver | Direct seq | 30 (32%) | 1/30 (3%) | 4/63 (6%) | 5/93 (5%) |
| 17 | Lung | Direct seq | 10 (59%) | 1/10 (10%) | 1/7 (14%) | 2/17 (12%) | ||
| Etienne-Grimaldi | 2008 | 48 | Liver biopsy | PCR-RFLP | 16 (33%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Italiano | 2009 | 59 | Not specified | Seq | 23 (39%) | 1/23 (4%) | 2/36 (6%) | 3/59 (5%) |
| Losi | 1992 | 19 | Local recurrence | Multiplex-ASPCR | 12 (63%) | 0 (0%) | 0 (0%) | 0 (0%) |
| 16 | Metastasis, 38% liver | Multiplex-ASPCR | 13 (81%) | 0 (0%) | 0 (0%) | 0 (0%) | ||
| Loupakis | 2009 | 43 | Liver | Seq | Not mentioned | 0 (0%) | 2/* | 2/43 (5%) |
| Molinari | 2009 | 37 | Diverse, 74% liver | Seq | 16 (43%) | 2/16 (13%) | 1/21 (5%) | 3/37 (8%) |
| 15 | Lymph node | Seq | 8 (53%) | 0 (0%) | 0 (0%) | 0 (0%) | ||
| Oliveira | 2006 | 28 | Lymph node | Not mentioned | 18 (64%) | 2/18 (11%) | 7/10 (70%) | 9/28 (32%) |
| Oudejans | 1991 | 31 | Liver and lung | Hybridization | 14 (45%) | 1/14 (7%) | 1/17 (6%) | 2/31 (6%) |
| Perrone | 2008 | 10 | Diverse, mainly liver | Direct seq | 2 (20%) | 1/2 (50%) | 1/8 (13%) | 2/10 (20%) |
| Santini | 2008 | 99 | Diverse, 80% liver | Seq | 38 (38%) | 3/38 (8%) | 1/61 (2%) | 4/99 (4%) |
| Garm Spindler | 2009 | 31 | Not specified | qPCR | 11 (35%) | 2/11 (18%) | 0/20 (0%) | 2/31 (6%) |
| Suchy | 1992 | 58 | Autopsy material, not specified | Dot-blot hybridization | 15 (26%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Weber | 2006 | 36 | Liver | Seq | 14 (39%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Zauber | 2003 | 42 | Diverse, 93% lymph node, 5% liver | SCCP analysis + seq | 22 (52%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Overall | 892 | All sites | All methods | 345/849 (41%) | 39/345 (11%) | 35/504 (7%) | 76/892 (9%) | |
| 276 | Liver | All methods | 84/233 (36%) | 8/84 (10%) | 11/149 (7%) | 21/276 (8%) | ||
| 129 | Lymph nodes | All methods | 65/129 (50%) | 18/65 (28%) | 14/64 (22%) | 32/129 (25%) |
Abbreviations: ASO=allele-specific oligonucleotide; ASPCR=allele-specific polymerase chain reaction; M=metastasis; pts=patients; PT=primary tumour; qPCR= quantitative PCR; RFLP=restriction fragment length polymorphism; SSCP=single strand conformational polymorphism; seq=sequencing. *Total number of cases not specified.
A discordant KRAS status between the primary tumour and metastasis was observed in a small number of patients (3.6%). In these cases, tumour cells may have departed the primary lesions before the acquisition of a fully malignant phenotype to undergo somatic mutations or deletions at a distant site (Klein, 2009). Another explanation for the discordant results may be heterogeneity of KRAS status within the primary tumour, although this was the case in only a small number of patients. Finally, a discordance may in theory be explained by metastases from a non-detected second primary.
Previously published data showed that a considerable fraction (25%, Table 4) of colorectal lymph node metastases does not resemble the primary tumour in terms of KRAS mutation status. In 5 of the 25 lymph node metastases that we tested the KRAS status was not concordant with the primary tumour, which is consistent with the literature (Table 4). Therefore, lymph node metastases do not seem suitable for determination of the KRAS mutation status of colorectal carcinomas. Discordance in KRAS mutation status might be due to clonal selection during the process of metastasis, however, heterogeneity in lymph node metastases could explain this discordance in only one patient.
Eight different KRAS mutation types were observed in our study, of which Gly12Asp showed the highest frequency. Five patients (1.6%) harboured different KRAS mutation types in the primary tumour compared with the metastases. This confirms the findings of Cejas et al (2009) and Albanese et al (2004), who reported a small number of patients (4 and 7%, respectively) with different mutation types between primary tumours and metastases. A different KRAS mutation type between primary lung adenocarcinomas and corresponding lymph node metastases was also observed in only 1% of the patients (Schmid et al, 2009). Currently, all patients with a KRAS mutation are excluded from treatment with anti-EGFR antibodies, independently of the mutation type. However, a recent paper indicated that codon 13 mutated tumours may be sensitive to cetuximab treatment (De Roock et al, 2010). As we observed a low frequency in KRAS mutation type discrepancies between primaries and metastases, this is not of clinical importance in selecting patients for anti-EGFR therapy.
In conclusion, we demonstrated a high level of concordance of 96.4% between primary tumours and liver metastases, which for clinical purposes to select CRC patients for anti-EGFR therapy was even higher with 98%. The implication of these results for general oncology practice is that both tissue of primary tumour or liver metastasis may be used for KRAS mutation testing. The results of our study are only valid for liver metastases and cannot be extrapolated to other metastatic locations. Furthermore, we demonstrated that discordance of test results between primary tumour and metastases cannot account for the failure rate of anti-EGFR therapy in patients with KRAS wild-type tumours. Therefore, novel predictive markers in addition to KRAS and BRAF mutation status are warranted.
Acknowledgments
This study was supported by a grant of the Dutch Colorectal Cancer Group (DCCG).
References
- Al-Mulla F, Going JJ, Sowden ET, Winter A, Pickford IR, Birnie GD (1998) Heterogeneity of mutant versus wild-type Ki-ras in primary and metastatic colorectal carcinomas, and association of codon-12 valine with early mortality. J Pathol 185: 130–138 [DOI] [PubMed] [Google Scholar]
- Albanese I, Scibetta AG, Migliavacca M, Russo A, Bazan V, Tomasino RM, Colomba P, Tagliavia M, La Farina M (2004) Heterogeneity within and between primary colorectal carcinomas and matched metastases as revealed by analysis of Ki-ras and p53 mutations. Biochem Biophys Res Commun 325: 784–791 [DOI] [PubMed] [Google Scholar]
- Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD (2008) Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 26: 1626–1634 [DOI] [PubMed] [Google Scholar]
- Artale S, Sartore-Bianchi A, Veronese SM, Gambi V, Sarnataro CS, Gambacorta M, Lauricella C, Siena S (2008) Mutations of KRAS and BRAF in primary and matched metastatic sites of colorectal cancer. J Clin Oncol 26: 4217–4219 [DOI] [PubMed] [Google Scholar]
- Baldus SE, Schaefer KL, Engers R, Hartleb D, Stoecklein NH, Gabbert HE (2010) Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res 16: 790–799 [DOI] [PubMed] [Google Scholar]
- Bellon E, Ligtenberg MJL, Tejpar S, Cox K, de Hertogh G, de Stricker K, Edsjö A, Gorgoulis V, Hoefler G, Jung A, Kotsinas A, Laurent-Puig P, López-Ríos F, Plato Hansen T, Rouleau E, Vandenberghe P, van Krieken JHJM, Dequeker E (2011) External quality assessment for KRAS testing is needed: set up of a European program and report of the first joined regional quality assessment rounds. Oncologist; doi:10.1634/theoncologist.2010-0429 [DOI] [PMC free article] [PubMed]
- Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, Zanon C, Moroni M, Veronese S, Siena S, Bardelli A (2007) Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res 67: 2643–2648 [DOI] [PubMed] [Google Scholar]
- Cejas P, Lopez-Gomez M, Aguayo C, Madero R, de Castro CJ, Belda-Iniesta C, Barriuso J, Moreno GV, Larrauri J, Lopez R, Casado E, Gonzalez-Baron M, Feliu J (2009) KRAS mutations in primary colorectal cancer tumors and related metastases: a potential role in prediction of lung metastasis. PLoS One 4: e8199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, Lamba S, Arena S, Frattini M, Piessevaux H, Van Cutsem E, O’Callaghan CJ, Khambata-Ford S, Zalcberg JR, Simes J, Karapetis CS, Bardelli A, Tejpar S (2010) Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA 304: 1812–1820 [DOI] [PubMed] [Google Scholar]
- Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A (2008) Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol 26: 5705–5712 [DOI] [PubMed] [Google Scholar]
- Etienne-Grimaldi MC, Formento JL, Francoual M, Francois E, Formento P, Renee N, Laurent-Puig P, Chazal M, Benchimol D, Delpero JR, Letoublon C, Pezet D, Seitz JF, Milano G (2008) K-Ras mutations and treatment outcome in colorectal cancer patients receiving exclusive fluoropyrimidine therapy. Clin Cancer Res 14: 4830–4835 [DOI] [PubMed] [Google Scholar]
- Garm Spindler KL, Pallisgaard N, Rasmussen AA, Lindebjerg J, Andersen RF, Cruger D, Jakobsen A (2009) The importance of KRAS mutations and EGF61A>G polymorphism to the effect of cetuximab and irinotecan in metastatic colorectal cancer. Ann Oncol 20: 879–884 [DOI] [PubMed] [Google Scholar]
- Italiano A, Hostein I, Soubeyran I, Fabas T, Benchimol D, Evrard S, Gugenheim J, Becouarn Y, Brunet R, Fonck M, Francois E, Saint-Paul MC, Pedeutour F (2010) KRAS and BRAF mutational status in primary colorectal tumors and related metastatic sites: biological and clinical implications. Ann Surg Oncol 17: 1429–1434 [DOI] [PubMed] [Google Scholar]
- Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au HJ, Langer C, Moore MJ, Zalcberg JR (2008) K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 359: 1757–1765 [DOI] [PubMed] [Google Scholar]
- Klein CA (2009) Parallel progression of primary tumours and metastases. Nat Rev Cancer 9: 302–312 [DOI] [PubMed] [Google Scholar]
- Losi L, Benhattar J, Costa J (1992) Stability of K-ras mutations throughout the natural history of human colorectal cancer. Eur J Cancer 28A: 1115–1120 [DOI] [PubMed] [Google Scholar]
- Loupakis F, Pollina L, Stasi I, Ruzzo A, Scartozzi M, Santini D, Masi G, Graziano F, Cremolini C, Rulli E, Canestrari E, Funel N, Schiavon G, Petrini I, Magnani M, Tonini G, Campani D, Floriani I, Cascinu S, Falcone A (2009) PTEN expression and KRAS mutations on primary tumors and metastases in the prediction of benefit from cetuximab plus irinotecan for patients with metastatic colorectal cancer. J Clin Oncol 27: 2622–2629 [DOI] [PubMed] [Google Scholar]
- Molinari F, Martin V, Saletti P, De Dosso S, Spitale A, Camponovo A, Bordoni A, Crippa S, Mazzucchelli L, Frattini M (2009) Differing deregulation of EGFR and downstream proteins in primary colorectal cancer and related metastatic sites may be clinically relevant. Br J Cancer 100: 1087–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira C, Velho S, Moutinho C, Ferreira A, Preto A, Domingo E, Capelinha AF, Duval A, Hamelin R, Machado JC, Schwartz Jr S, Carneiro F, Seruca R (2007) KRAS and BRAF oncogenic mutations in MSS colorectal carcinoma progression. Oncogene 26: 158–163 [DOI] [PubMed] [Google Scholar]
- Oliveira C, Westra JL, Arango D, Ollikainen M, Domingo E, Ferreira A, Velho S, Niessen R, Lagerstedt K, Alhopuro P, Laiho P, Veiga I, Teixeira MR, Ligtenberg M, Kleibeuker JH, Sijmons RH, Plukker JT, Imai K, Lage P, Hamelin R, Albuquerque C, Schwartz Jr S, Lindblom A, Peltomaki P, Yamamoto H, Aaltonen LA, Seruca R, Hofstra RM (2004) Distinct patterns of KRAS mutations in colorectal carcinomas according to germline mismatch repair defects and hMLH1 methylation status. Hum Mol Genet 13: 2303–2311 [DOI] [PubMed] [Google Scholar]
- Oudejans JJ, Slebos RJ, Zoetmulder FA, Mooi WJ, Rodenhuis S (1991) Differential activation of ras genes by point mutation in human colon cancer with metastases to either lung or liver. Int J Cancer 49: 875–879 [DOI] [PubMed] [Google Scholar]
- Perrone F, Lampis A, Orsenigo M, Di Bartolomeo M, Gevorgyan A, Losa M, Frattini M, Riva C, Andreola S, Bajetta E, Bertario L, Leo E, Pierotti MA, Pilotti S (2009) PI3KCA/PTEN deregulation contributes to impaired responses to cetuximab in metastatic colorectal cancer patients. Ann Oncol 20: 84–90 [DOI] [PubMed] [Google Scholar]
- Santini D, Loupakis F, Vincenzi B, Floriani I, Stasi I, Canestrari E, Rulli E, Maltese PE, Andreoni F, Masi G, Graziano F, Baldi GG, Salvatore L, Russo A, Perrone G, Tommasino MR, Magnani M, Falcone A, Tonini G, Ruzzo A (2008) High concordance of KRAS status between primary colorectal tumors and related metastatic sites: implications for clinical practice. Oncologist 13: 1270–1275 [DOI] [PubMed] [Google Scholar]
- Scaltriti M, Baselga J (2006) The epidermal growth factor receptor pathway: a model for targeted therapy. Clin Cancer Res 12: 5268–5272 [DOI] [PubMed] [Google Scholar]
- Schmid K, Oehl N, Wrba F, Pirker R, Pirker C, Filipits M (2009) EGFR/KRAS/BRAF mutations in primary lung adenocarcinomas and corresponding locoregional lymph node metastases. Clin Cancer Res 15: 4554–4560 [DOI] [PubMed] [Google Scholar]
- Suchy B, Zietz C, Rabes HM (1992) K-ras point mutations in human colorectal carcinomas: relation to aneuploidy and metastasis. Int J Cancer 52: 30–33 [DOI] [PubMed] [Google Scholar]
- Tol J, Dijkstra JR, Vink-Borger ME, Nagtegaal ID, Punt CJ, van Krieken JH, Ligtenberg MJ (2010) High sensitivity of both sequencing and real-time PCR analysis of KRAS mutations in colorectal cancer tissue. J Cell Mol Med 14: 2122–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tol J, Koopman M, Cats A, Rodenburg CJ, Creemers GJ, Schrama JG, Erdkamp FL, Vos AH, van Groeningen CJ, Sinnige HA, Richel DJ, Voest EE, Dijkstra JR, Vink-Borger ME, Antonini NF, Mol L, van Krieken JH, Dalesio O, Punt CJ (2009) Chemotherapy, bevacizumab, and cetuximab in metastatic colorectal cancer. N Engl J Med 360: 563–572 [DOI] [PubMed] [Google Scholar]
- Tol J, Punt CJ (2010) Monoclonal antibodies in the treatment of metastatic colorectal cancer: a review. Clin Ther 32: 437–453 [DOI] [PubMed] [Google Scholar]
- Van Cutsem E, Kohne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D’Haens G, Pinter T, Lim R, Bodoky G, Roh JK, Folprecht G, Ruff P, Stroh C, Tejpar S, Schlichting M, Nippgen J, Rougier P (2009) Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med 360: 1408–1417 [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL (1988) Genetic alterations during colorectal-tumor development. N Engl J Med 319: 525–532 [DOI] [PubMed] [Google Scholar]
- Weber JC, Meyer N, Pencreach E, Schneider A, Guerin E, Neuville A, Stemmer C, Brigand C, Bachellier P, Rohr S, Kedinger M, Meyer C, Guenot D, Oudet P, Jaeck D, Gaub MP (2007) Allelotyping analyses of synchronous primary and metastasis CIN colon cancers identified different subtypes. Int J Cancer 120: 524–532 [DOI] [PubMed] [Google Scholar]
- Zauber P, Sabbath-Solitare M, Marotta SP, Bishop DT (2003) Molecular changes in the Ki-ras and APC genes in primary colorectal carcinoma and synchronous metastases compared with the findings in accompanying adenomas. Mol Pathol 56: 137–140 [DOI] [PMC free article] [PubMed] [Google Scholar]