Abstract
Signaling events controlled by calcineurin promote cardiac hypertrophy, but the degree to which such pathways are required to transduce the effects of various hypertrophic stimuli remains uncertain. In particular, the administration of immunosuppressive drugs that inhibit calcineurin has inconsistent effects in blocking cardiac hypertrophy in various animal models. As an alternative approach to inhibiting calcineurin in the hearts of intact animals, transgenic mice were engineered to overexpress a human cDNA encoding the calcineurin-binding protein, myocyte-enriched calcineurin-interacting protein-1 (hMCIP1) under control of the cardiac-specific, α-myosin heavy chain promoter (α-MHC). In unstressed mice, forced expression of hMCIP1 resulted in a 5–10% decline in cardiac mass relative to wild-type littermates, but otherwise produced no apparent structural or functional abnormalities. However, cardiac-specific expression of hMCIP1 inhibited cardiac hypertrophy, reinduction of fetal gene expression, and progression to dilated cardiomyopathy that otherwise result from expression of a constitutively active form of calcineurin. Expression of the hMCIP1 transgene also inhibited hypertrophic responses to β-adrenergic receptor stimulation or exercise training. These results demonstrate that levels of hMCIP1 producing no apparent deleterious effects in cells of the normal heart are sufficient to inhibit several forms of cardiac hypertrophy, and suggest an important role for calcineurin signaling in diverse forms of cardiac hypertrophy. The future development of measures to increase expression or activity of MCIP proteins selectively within the heart may have clinical value for prevention of heart failure.
The protein phosphatase calcineurin plays a critical role in the processes by which several types of cells respond to extracellular signals or environmental stresses through changes in gene expression. Calcineurin-dependent signal transduction pathways have been characterized extensively in T lymphocytes responding to antigen stimulation (1), and the current success of organ transplantation in humans was made possible by the discovery of calcineurin antagonist drugs with potent immunosuppressive effects. In skeletal myocytes, calcineurin influences myogenic differentiation (2), transduces effects of motor nerve stimulation to alter specialized properties of different myofiber subtypes (3, 4), and mediates hypertrophic responses to insulin-like growth factor-1 (IGF-1) (5, 6). In the heart, an activated calcineurin transgene drives hypertrophic growth that progresses to dilated cardiomyopathy and heart failure, in a manner that recapitulates the natural history of several widely prevalent forms of human heart disease (7).
Numerous recent studies have sought to determine whether calcineurin serves an important signaling function in forms of cardiac hypertrophy that are relevant to human disease. The calcineurin antagonist drugs cyclosporin and FK506 have been observed by several laboratories to block hypertrophic responses of the heart in animal models of pressure overload or genetic cardiomyopathy (8–13), but other groups by using ostensibly similar models have failed to observe such effects (14, 15). Different experimental approaches are necessary to resolve this controversy.
Here, we address this goal by generating transgenic mice that overexpress the calcineurin inhibitory protein myocyte-enriched calcineurin-interacting protein (MCIP) 1 selectively in the heart. In cultured skeletal myocytes, MCIP1 blocks calcineurin signaling by binding directly to the catalytic subunit (CnA) of the calcineurin holoenzyme and inhibiting its activating effects on nuclear factor of activated T cells (NFAT) and myocyte enhancer factor-2 (MEF2) proteins that transduce calcineurin-generated signals to target genes (16). In mice and humans, MCIP1 is expressed primarily in cardiac and skeletal muscles (16), and transcription of the MCIP1 gene is potently stimulated by activated calcineurin (17), thereby establishing a negative feedback mechanism that presumably serves to protect cells from otherwise deleterious consequences of unrestrained calcineurin activity. Before its function was known, the gene encoding MCIP1 was annotated as DSCR1 (18), based on its location within the critical region of human chromosome 21, trisomy of which leads to Down syndrome. Two other genes encoding closely related proteins that we term MCIP2 and MCIP3 were annotated originally as ZAKI-4/DSCR1L1 and DSCR1L2, respectively (19). Proteins encoded by these genes have also been termed calcipressins (20).
Several other proteins have been found to bind and inhibit calcineurin. These include cabin/cain and AKAP79 (21–23), as well as the immunosuppressive drug targets cyclophilin and FK506 binding protein (FKBP) (24), but MCIP1 is distinctive among this set of proteins in several respects. Unlike cyclophilin and FKBP, no exogenous chemicals are required for calcineurin inhibition by MCIP1. Unlike cabin/cain and AKAP79, MCIP1 is preferentially expressed in striated myocytes. Finally, and perhaps most importantly, only the expression of MCIP1 has been shown to be induced by calcineurin, thus providing a mechanism for negative feedback regulation of calcineurin activity (17).
Forced overexpression of MCIP1 in hearts of transgenic mice was found to attenuate hypertrophic responses to each of several different stimuli, and to prevent progression to dilated cardiomyopathy that otherwise results from chronic activation of calcineurin in the myocardium. Notably, concentrations of MCIP1 sufficient to inhibit cardiac hypertrophy produced no apparent deleterious effects in unstressed animals. These data support a role for calcineurin-dependent signaling in diverse forms of cardiac hypertrophy, and suggest that measures to augment expression of MCIP1 or related proteins within the heart could have clinical value.
Materials and Methods
Plasmid Constructs and Generation of Transgenic Mice.
A full-length human MCIP1 cDNA encoding the exon 4 splice variant of hMCIP1 with an HA epitope tag from the human influenza hemagglutinin protein (hMCIP1-HA) was cloned 3′ to a 5.5-kb segment of the α-myosin heavy chain promoter (α−MHC) and 5′ to a 0.6-kb polyadenylation signal from the human GH gene (Fig. 1A), carried in the pBluescriptII SK+ vector (Stratagene). The transgene was linearized and separated from prokaryotic sequences after digestion with NotI, and microinjected into fertilized oocytes from C57BL/6 mice, which were introduced into pseudopregnant females to generate lines of transgenic mice, by using standard techniques. Animals were genotyped by Southern blot analysis of tail genomic DNA digested with EcoRI and probed with the hMCIP1 transgene (Fig. 1B). Animals carrying the α−MHC-hMCIP1 transgene were crossed with transgenic mice expressing a constitutively activated form of calcineurin, also under the control of the α−MHC promoter (25) to produce doubly transgenic mice (α−MHC-hMCIP1 × α−MHC-CnA*). MCIP expression plasmids were constructed in the pTARGET vector (Promega) under the control of the cytomegalovirus (CMV) promoter. pCMV-hMCIP1 encodes an HA-tagged full-length hMCIP1 (amino acids 1–197). pCMV-ΔhMCIP1 encodes an HA-tagged truncated hMCIP1 (amino acids 81 to 197). Other expression vectors and reporter genes have been previously described (16).
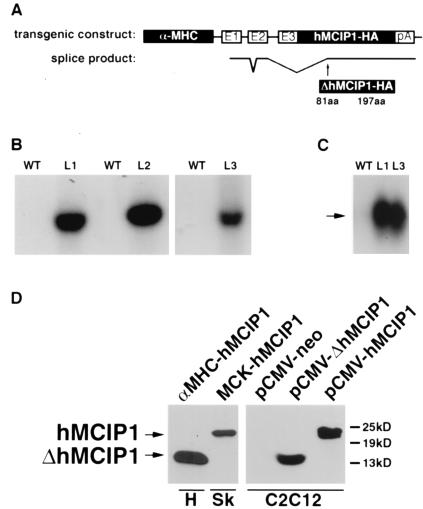
Figure 1.
Design and expression of the α−MHC-hMCIP1 transgene. (A) Schematic illustration of components of the transgene, including a 5.5-kb α−MHC promoter fragment with three noncoding exons (E1, E2, and E3) and intervening nontranscribed segments of the α−MHC gene, followed by a full-length human MCIP1 cDNA with a carboxyl terminal epitope tag (HA) and a polyadenylation (pA) signal from the human GH gene. The lower line illustrates the unexpected pattern of mRNA splicing observed in vivo, resulting in translation of a truncated protein (ΔhMICP1) initiated at amino acid 81 relative to the wild-type (WT) protein. (B) Southern blot of genomic DNA from WT mice and each of three lines (L1, L2, and L3) of α−MHC-hMCIP1 transgenic mice by using a probe specific to the human MCIP1 gene sequence. (C) Northern blot of heart mRNA from WT and two of the α−MHC-hMCIP1 transgenic mice lines (L1 and L3) by using a probe specific to human MCIP1 mRNA. Transgene expression was higher in the L2 line (data not shown). The arrow indicates the anomalously spliced 1.2-kb α−MHC-hMCIP1 transgene transcript (ΔhMCIP). Expression of the transgene was at least an order of magnitude higher than the endogenous 2.2-kb mMCIP1 transcript (data not shown). (D) Western blots probed to detect the hMCIP-HA protein in extracts from an L1 α−MHC-hMCIP1 transgenic heart (H) showing the 14-kDa truncated ΔhMCIP1 protein product. A positive control for comparison to a correctly spliced full length 24-kDa hMCIP1 protein was drawn from a skeletal muscle extract (Sk) from a line of transgenic mice expressing hMCIP1 under the control of the muscle creatine kinase promoter (MCK-hMCIP1). C2C12 cells were transfected with plasmids expressing the indicated products for size comparisons [pCMV-neo, pCMV-ΔhMCIP1 (amino acids 81 to 197) and pCMV-hMCIP1 (amino acids 1 to 197)].
Cell Culture Studies.
C2C12 myoblasts were maintained and transfected as previously described (16). Luciferase assays of whole cell extracts were performed as previously described (16). All results were corrected for variations in transfection efficiency by normalization to expression of a cotransfected pCMV-LacZ plasmid.
Intact Animal Studies.
In addition to the genetic cross to α−MHC-CnA* animals, α−MHC-hMCIP1 transgenic mice and wild-type littermates were subjected to two independent stimuli known to provoke cardiac hypertrophy. Twelve-week-old and 28-week-old male mice were surgically implanted with a s.c. miniosmotic pump [Alzet (Palo Alto, CA) model 2001] that released isoproterenol in 0.9% NaCl at a rate of 28 μg per hour per 25 kg of body weight over a 7-day period. Control pumps delivered a 0.9% NaCl solution. Hearts were harvested on day eight. The heart body weight ratios (htw/bw) of isoproterenol-treated animals were compared with the mean htw/bw ratios of age-matched saline-treated animals of the same genotype. For exercise-induced hypertrophy, 28-week-old male animals were placed in individual cages where they had free access to a running wheel. The number of revolutions was monitored continuously for 28 days. The animals ran an average of 3 to 5 km a day. The htw/bw ratios of the exercised animals were compared with the mean htw/bw ratios of age-matched sedentary animals of the same genotype. A Wilcoxon rank sum test was used to determine statistically significant differences among treatment groups (P < 0.05). All animal protocols were reviewed and approved by the appropriate institutional review boards.
RNA Isolation and Analysis.
Total RNA was prepared from mouse heart and skeletal muscle by using RNAzol (Life Technologies, Rockville, MD) following the manufacturer's protocol. Northern blot analysis was performed with 20 μg of total RNA in each lane and probed in Ultrahyb (Ambion, Austin, TX) with a DNA fragment from the coding region of the human MCIP1 transgene. RNA dot blots prepared with 2 μg of total RNA were probed with end-labeled oligonucleotides specific for mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH), α−MHC, beta-myosin heavy chain (β−MHC), α-skeletal actin or atrial natriuretic factor (ANF). Bound probes were detected on a Storm PhosphorImager (Molecular Dynamics) and quantified by using imagequant (version 1.2).
Western Blot Analysis.
Protein extracts were made by homogenizing frozen mouse heart and skeletal muscle in PBS plus 20% glycerol, 1% Triton X-100 and 1 mM DTT by using a polytron homogenizer (Kinematica, Lucerne, Switzerland). Samples were boiled in protein loading buffer containing SDS, then passed over glass wool to remove DNA. Extracts were prepared from tissue culture cells by lysis directly in protein loading buffer. Proteins were separated by SDS/PAGE. Epitope-tagged, transgene-derived proteins were detected by using a primary rat anti-HA monoclonal antibody (Roche, Nutley, NJ) and a horse radish peroxidase-tagged goat anti-rat secondary antibody (Bio-Rad).
Gross and Histological Analysis of Hearts.
Hearts were perfusion fixed, embedded in paraffin, and sectioned at 5 μm for histologic evaluation. Sections were stained with hematoxylin/eosin and Masson's trichrome and photomicrographed by using standard brightfield optics.
Transthoracic Echocardiography.
A 12-MHz probe (Hewlett–Packard) was applied to the shaved chest wall of mice sedated with Avertin (0.1–0.15 cc/mg body weight) and maneuvered to obtain a parasternal short axis view of the left ventricular cavity. Two-dimensional and M-mode echocardiography images were digitized and stored on an optical disk. Data were collected by using a sweep speed of 150. Fractional shortening (FS) was calculated from the left ventricular end diastolic dimension (LVEDD) minus the left ventricular end systolic dimension (LVESD) divided by the LVEDD.
Results
Expression and Function of the α−MHC-hMCIP1 Transgene Product.
Three independent lines of α−MHC-hMCIP1 transgenic mice were established (Fig. 1B). In all three of these lines, both mRNA and protein products of the transgene were expressed in hearts of animals in the F1 generation (Fig. 1 C and D). However, the protein produced by the transgene was 14 kDa, much smaller than the predicted 24-kDa full-length hMCIP1 protein. Reverse transcriptase was used to generate a cDNA copy of the transgene product. The sequence of the cDNA demonstrated anomalous RNA splicing of the transgene from a cryptic splice donor site within the second noncoding exon of the α−MHC gene to an acceptor site within the hMCIP1 cDNA (illustrated in Fig. 1A), thereby removing the authentic translational initiation codon. Fortuitously, a stable but truncated protein product is initiated from an internal, in-frame methionine codon at amino acid 81. This truncated MCIP1 protein accumulates to measurable levels within the heart (Fig. 1D).
To test whether the 14-kDa transgene product inhibits calcineurin activity, the C2C12 myoblast cell line was transfected with a luciferase reporter gene driven by a calcineurin-responsive IL-2 promoter (IL-2-Luc). Cotransfection of a constitutively active calcineurin expression vector (pCMV-CnA*) with either an empty control vector (pCMV-neo) or a plasmid expressing a full-length hMCIP1 (pCMV-hMCIP1) demonstrated the ability of hMCIP1 to almost completely inhibit calcineurin induction of the IL-2-Luc reporter gene (Fig. 2). A plasmid expressing the truncated cDNA from the transgene (pCMV-ΔhMCIP1) was as effective an inhibitor of calcineurin as the wild-type, full-length MCIP1 protein (Fig. 2), verifying that the truncated 14-kDa transgene product found in the hearts of the α-MHC-hMCIP1 mice is a powerful inhibitor of calcineurin activity. Western analysis comparing cardiac ΔhMCIP1 protein levels in the three founder lines showed comparable transgenic protein levels in L1 and L3, whereas in the L2 line, ΔhMCIP1 protein levels were two to three times higher (data not shown).
Figure 2.
Functional activity of the truncated ΔhMCIP1 protein product compared with full-length hMCIP1, assessed by inhibition of calcineurin-dependent activation in C2C12 myoblasts. C2C12 cells were transfected with a luciferase reporter gene driven by a calcineurin-responsive IL-2 promoter (IL-2-Luc). Cells were cotransfected with a constitutively active calcineurin expression plasmid (pCMV-CnA*) and either an empty control vector (pCMV-neo) or the indicated MCIP-encoding plasmid (pCMV-hMCIP1 or pCMV-ΔhMCIP1).
Strong cardiac-specific expression is initiated around the time of birth under the control of the α-MHC promoter (25). The α-MHC-hMCIP1 transgene was inherited in a normal 1 to 1 Mendelian ratio in L1 and L3 with no evidence of deleterious effects in adult mice. Transmission of the L2 transgene was slightly reduced, suggesting partial embryonic lethality, possibly because of embryonic expression of the transgene in this line. After birth, however, L2 mice demonstrated normal growth and life spans. All subsequent experiments were done by using animals from the L1 and L3 founder lines, which performed comparably.
MCIP1 Inhibits Cardiac Hypertrophy in a Genetic Model of Cardiomyopathy.
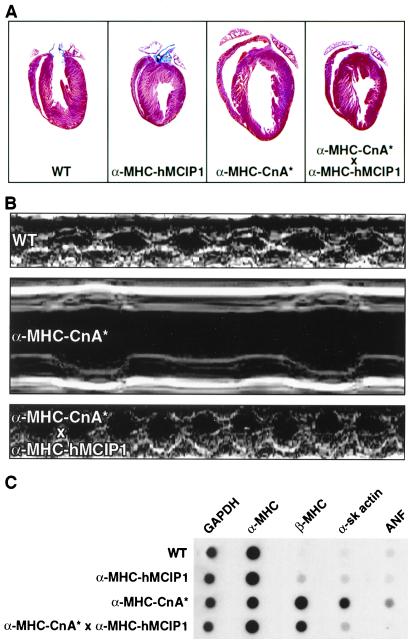
As reported previously (7), we observed massive cardiac hypertrophy in α−MHC-CnA* transgenic mice. The extent of hypertrophic growth induced by the α−MHC-CnA* transgenic was attenuated, however, by concomitant expression of the α−MHC-hMCIP1 transgene (Fig. 3A). Animals carrying the α−MHC-hMCIP1 transgene were viable and fertile, and exhibited no obvious cardiovascular abnormalities, with the exception of a mild (5–10%) reduction in cardiac mass compared with wild-type animals (Table 1). In mice bearing both the α−MHC-CnA* and α−MHC-hMCIP1 transgenes, the extent of cardiac hypertrophy was reduced to approximately 28% of the response noted with the α−MHC-CnA* transgene alone (Table 1 and Fig. 3A). By using a Wilcoxon rank sum test for two groups of independent samples of observations, the α-MHC-CnA * × α-MHC-hMCIP1 double transgenic mice developed significantly less cardiac hypertrophy than the α-MHC-CnA* transgenic mice (P < 0.01). Expression of the hMCIP1 transgene also prevented dilated cardiomyopathy induced by the CnA* transgene, as assessed by transthoractic echocardiography of a matched set of four male littermates (Fig. 3B and Table 2). Fractional shortening (FS) as an indication of the ejection volume of the heart was restored to wild-type levels in the α-MHC-hMCIP1 × α−MHC-CnA* double transgenic (FS = 0.63) compared with the animal carrying the α−MHC-CnA* transgene alone, in which cardiac contractility was greatly impaired (FS = 0.30). Whereas the α−MHC-CnA* transgenic mice frequently suffer sudden death (7), no such early mortality was apparent in the α-MHC-hMCIP1 × α−MHC-CnA* double transgenic mice. In addition to reducing the morphological and functional consequences of the α−MHC-CnA* transgene, hMCIP1 overexpression reduced the reinduction of the fetal program of gene expression that is characteristic of this and other models of cardiac hypertrophy. The levels of β-MHC, α-skeletal actin and ANF transcripts were elevated in the α−MHC-CnA* transgenic hearts compared with wild-type hearts. In the α−MHC-hMCIP1 × α−MHC-CnA* double transgenic, both α-skeletal actin and ANF transcript levels were comparable to those found in wild-type hearts. Similarly, β-MHC expression, while greater than wild-type levels, was reduced relative to the levels detected in the hearts of α−MHC-CnA* mice (Fig. 3C).
Figure 3.
Effects of hMCIP1 on cardiac hypertrophy induced by activated calcineurin. (A) Gross morphology of hearts from wild-type, α−MHC-hMCIP1 transgenic, α−MHC-CnA* transgenic, and doubly transgenic (α−MHC-hMCIP1 × α−MHC-CnA*) mice aged 20 weeks. (B) M-mode echocardiographic analysis from male littermates of the indicated genotypes at an age of 20 weeks. Heart rate was markedly slowed in the α-MHC-CnA* animals relative to mice of other genotypes. Echocardiographic measurements of these animals are found in Table 2. (C) RNA dot blot analysis of gene expression of GAPDH, α-MHC, β-MHC, α-skeletal actin, and ANF in hearts of mice of the indicated genotypes.
Table 1.
Heart weight/body weight ratios of transgenic mice
| Wild type | α-MHC-hMCIP1 | α-MHC-CnA* | α-MHC-CnA* × α-MHC-hMCIP1 |
|---|---|---|---|
| 4.33 ± 0.09 | 4.10 ± 0.23 | 9.92 ± 1.06 | 5.69 ± 0.35 |
| (n = 16) | (n = 26) | (n = 5) | (n = 7) |
Data are calculated as heart weight/body weight (mg/g; mean ± SEM). The numbers of animals of each genotype that were tested are shown in parentheses. Wild-type and α-MHC-hMCIP1 mice were 12–15 weeks of age. Median age of α-MHC-CnA* and α-MHC-hMCIP1 × α-MHC-CnA* mice was 12 weeks.
Table 2.
Echocardiographic assessment of the left ventricular chamber
| Genotype | LVEDD, cm | LVESD, cm | FS |
|---|---|---|---|
| Wild type | 0.237 | 0.081 | 0.66 |
| α-MHC-hMCIP1 | 0.253 | 0.098 | 0.62 |
| α-MHC-CnA* | 0.479 | 0.339 | 0.30 |
| α-MHC-CnA* × α-MHC-hMCIP1 | 0.298 | 0.113 | 0.63 |
Data collected from 20-week-old male littermates. LVEDD, left ventricular end-diastolic dimension; LVESD, left ventricular end-systolic dimension; FS, fractional shortening; FS = (LVEDD − LVESD)/LVEDD.
MCIP1 Limits Hypertrophic Responses to Other Stimuli in Intact Animals.
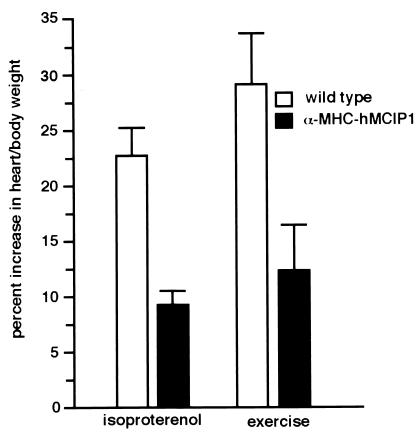
Calcineurin activation has been implicated in the progression of both pathological and adaptive cardiac hypertrophy (26). Chronic administration, over a 7-day period, of the β-adrenergic receptor agonist isoproterenol by using implantable osmotic minipumps produced a modest degree of cardiac hypertrophy in male wild-type animals, as described previously (27) (22.8 ± 1.95% increase in htw/bw ratio). This hypertrophic response was diminished, although not abolished, by transgenic overexpression of hMCIP1 (9.4 ± 0.87% increase; Fig. 4). Similarly, physiological adaptive hypertrophy resulting from 28 days of nocturnal wheel running in wild-type male mice was reduced in animals carrying the α−MHC-hMCIP1 transgene (29.2 ± 4.23% increase in wild-type versus 12.3 ± 4.15% increase in the transgenic animals; Fig. 4). By using a Wilcoxon rank sum test for two groups of independent samples of observations, the α−MHC-hMCIP1 mice developed significantly less cardiac hypertrophy in response to either isoproterenol or exercise (P < 0.05).
Figure 4.
Effects of hMCIP1 on cardiac hypertrophy in response to β-adrenergic receptor stimulation or exercise in intact animals. The graph indicates the percentage increase in htw/bw of male wild-type (n = 7) and α−MHC-hMCIP1 transgenic (n = 9) mice subjected to chronic infusion of isoproterenol over 7 days or the percentage increase in htw/bw of male wild-type (n = 4) and α−MHC-hMCIP1 transgenic (n = 4) mice after 28 days of voluntary wheel running exercise. The α−MHC-hMCIP1 mice developed significantly less cardiac hypertrophy in response to either isoproterenol or exercise (P < 0.05).
Discussion
Substantial progress has been made in defining signaling pathways that control hypertrophic growth of cardiomyocytes. A great diversity of different signaling molecules, including cell surface receptors, G proteins, protein kinases, and transcription factors have the capacity to stimulate this process when activated in cultured cardiomyocytes or in hearts of transgenic mice. However, a central issue in this field is whether the different hypertrophic signaling pathways revealed by such experiments are parallel and redundant, or whether certain signaling molecules constitute nodal points that are required for responses to many different hypertrophic stimuli. From the perspective of translational research, the latter possibility presents greater opportunities to intervene in the process in a manner that ultimately would benefit patients at risk for sudden death or heart failure as a consequence of cardiac hypertrophy.
The recent discovery that calcineurin, a calcium-regulated protein phosphatase, participates in hypertrophic signaling events (7) was greeted with special interest because of its potential therapeutic implications. Drugs capable of inhibiting calcineurin were already in clinical use, and initial observations in intact animal models suggested that pharmacological inhibition of calcineurin-dependent signaling could inhibit cardiac hypertrophy arising from a diversity of primary stimuli (7, 9). However, controversy over the mechanistic importance of calcineurin-dependent signaling events in the development and progression of cardiac hypertrophy has arisen because of seemingly contradictory reports from different laboratories as to the effects of calcineurin antagonist drugs in various animal models (8–12, 14, 15). As we (and others) have argued (28), however, responses to immunosuppressive drugs, or the lack thereof, should not be interpreted as the sine qua non for defining the participation of calcineurin in hypertrophic signaling pathways. Toxic consequences of these drugs based on inhibition of calcineurin in other cell types, or based on nonspecific drug effects, may preclude the achievement of effective inhibitory concentrations within the myocardium or may confound the interpretation of results. The relatively high level of calcineurin expression in the heart compared with lymphocytes (29) adds further difficulty to achieving complete suppression of calcineurin activity by systemic delivery of currently available pharmacologic inhibitors. The ability to inhibit calcineurin selectively within the myocardium by mechanisms distinct from the action of immunosuppressive drugs, as reported here, should help to resolve this controversy.
The present data support several new conclusions. First, hMCIP1 is capable of antagonizing calcineurin-dependent signaling events in tissues of intact animals, demonstrating that previous descriptions of calcineurin inhibition by MCIP proteins (16, 20, 30) are not an artifact of the high concentrations that can be generated transiently within the cell culture environment. Second, concentrations of hMCIP1 sufficient to inhibit calcineurin signaling in the heart have no apparent deleterious effects in unstressed animals, other than a slight reduction in cardiac mass. Third, hMCIP1 is capable of reducing hypertrophic responses to a variety of genetic (CnA*), physiologic (exercise training), or pharmacologic (isoproterenol) stimuli. The latter finding strongly implicates calcineurin activation as an obligate downstream step in the intracellular pathways that couple activity and β-adrenergic stimulation to cardiac growth.
Our conclusions based on cardiac-specific expression of hMCIP1 are corroborated by independent studies conducted contemporaneously by DeWindt et al. (31), who generated transgenic mice that overexpress truncated forms of two other proteins that have been shown to inhibit calcineurin activity, cabin/cain and AKAP79. Like hMCIP1, these other endogenous inhibitors of calcineurin also blocked hypertrophic responses of the heart induced by β-adrenergic receptor stimulation. In addition, transgene-mediated calcineurin inhibition by cabin/cain or AKAP79 reduced cardiac hypertrophy in response to pressure overload (aortic banding), without compromising aortic pressure or precipitating circulatory failure. Whereas cardiac-specific overexpression of any one of the three calcineurin inhibitory proteins studied in these two reports is capable of inhibiting cardiac hypertrophy, only MCIP1 is likely to function as an endogenous regulator of hypertrophic signaling, because it is expressed abundantly within the myocardium and its transcription is up-regulated by calcineurin activity (17).
Collectively, these two reports support the viewpoint that calcineurin plays an important role in the hypertrophic response of the myocardium triggered by different initiating stresses through multiple pathways. Therefore, downstream events controlled by calcineurin are likely to be shared among medically relevant forms of cardiac hypertrophy. Preclinical studies that seek to develop means to inhibit calcineurin by drug or gene therapy are a rational step toward the development of novel strategies to prevent heart failure in humans. Additional research also is needed to determine whether the MCIP family of proteins has other functions, distinct from calcineurin inhibition, that may be pertinent to its impact on cardiac hypertrophy. The extent to which principles established in rodent models of cardiac hypertrophy can be extrapolated to human cardiomyopathies remains conjectural. Nevertheless, further studies of endogenous calcineurin inhibitory proteins such as MCIP are warranted, and could lead to the development of clinically effective anti-hypertrophic countermeasures.
Acknowledgments
We thank B. Mercer and A. Hawkins for technical assistance and Dr. D. Srivastava for comments on the manuscript. This work was supported by the National Institutes of Health (AR40849, HL07360, and HL61544) and by the D. W. Reynolds Center for Clinical Cardiovascular Research. R.B.V. is supported by a National Institutes of Health Training Grant in Cardiovascular Research (National Heart, Lung, and Blood Institute). T.A.M. is a Pfizer Fellow of the Life Sciences Research Foundation. R.L.N. was supported by a postdoctoral fellowship from the National Institutes of Health.
Abbreviations
- CMV
cytomegalovirus promoter
- CnA*
constitutively active catalytic subunit of calcineurin
- HA
epitope tag from the human influenza hemagglutinin protein
- htw/bw
heart weight to body weight ratio
- MCIP
myocyte-enriched calcineurin-interacting protein
- hMCIP
human MCIP
- α-MHC
alpha-myosin heavy chain
- ANF
atrial natriuretic factor
Footnotes
See commentary on page 2947.
References
- 1.Clipstone N A, Crabtree G R. Nature (London) 1992;357:695–697. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- 2.Friday B B, Horsley V, Pavlath G K. J Cell Biol. 2000;149:657–666. doi: 10.1083/jcb.149.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chin E R, Olson E N, Richardson J A, Yang Q, Humphries C, Shelton J M, Wu H, Zhu W, Bassel-Duby R, Williams R S. Genes Dev. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu H, Naya F J, McKinsey T A, Mercer B, Shelton J M, Chin E R, Simard A R, Michel R N, Bassel-Duby R, Olson E N, Williams R S. EMBO J. 2000;19:1963–1973. doi: 10.1093/emboj/19.9.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Musaro A, McCullagh K J, Naya F J, Olson E N, Rosenthal N. Nature (London) 1999;400:581–585. doi: 10.1038/23060. [DOI] [PubMed] [Google Scholar]
- 6.Semsarian C, Wu M J, Ju Y K, Marciniec T, Yeoh T, Allen D G, Harvey R P, Graham R M. Nature (London) 1999;400:576–581. doi: 10.1038/23054. [DOI] [PubMed] [Google Scholar]
- 7.Molkentin J D, Lu J R, Antos C L, Markham B, Richardson J, Robbins J, Grant S R, Olson E N. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim H W, De Windt L J, Mante J, Kimball T R, Witt S A, Sussman M A, Molkentin J D. J Mol Cell Cardiol. 2000;32:697–709. doi: 10.1006/jmcc.2000.1113. [DOI] [PubMed] [Google Scholar]
- 9.Sussman M A, Lim H W, Gude N, Taigen T, Olson E N, Robbins J, Colbert M C, Gualberto A, Wieczorek D F, Molkentin J D. Science. 1998;281:1690–1693. doi: 10.1126/science.281.5383.1690. [DOI] [PubMed] [Google Scholar]
- 10.Murat A, Pellieux C, Brunner H R, Pedrazzini T. J. Biol. Chem. 2000. [DOI] [PubMed] [Google Scholar]
- 11.Meguro T, Hong C, Asai K, Takagi G, McKinsey T A, Olson E N, Vatner S F. Circ Res. 1999;84:735–740. doi: 10.1161/01.res.84.6.735. [DOI] [PubMed] [Google Scholar]
- 12.Shimoyama M, Hayashi D, Takimoto E, Zou Y, Oka T, Uozumi H, Kudoh S, Shibasaki F, Yazaki Y, Nagai R, Komuro I. Circulation. 1999;100:2449–2454. doi: 10.1161/01.cir.100.24.2449. [DOI] [PubMed] [Google Scholar]
- 13.Hill J A, Karimi M, Kutschke W, Davisson R L, Zimmerman K, Wang Z, Kerber R E, Weiss R M. Circulation. 2000;101:2863–2869. doi: 10.1161/01.cir.101.24.2863. [DOI] [PubMed] [Google Scholar]
- 14.Ding B, Price R L, Borg T K, Weinberg E O, Halloran P F, Lorell B H. Circ Res. 1999;84:729–734. doi: 10.1161/01.res.84.6.729. [DOI] [PubMed] [Google Scholar]
- 15.Zhang W, Kowal R C, Rusnak F, Sikkink R A, Olson E N, Victor R G. Circ Res. 1999;84:722–728. doi: 10.1161/01.res.84.6.722. [DOI] [PubMed] [Google Scholar]
- 16.Rothermel B, Vega R B, Yang J, Wu H, Bassel-Duby R, Williams R S. J Biol Chem. 2000;275:8719–8725. doi: 10.1074/jbc.275.12.8719. [DOI] [PubMed] [Google Scholar]
- 17.Yang J, Rothermel B, Vega R B, Frey N, McKinsey T A, Olson E N, Bassel-Duby R, Williams R S. Circ Res. 2000;87:61e–68e. doi: 10.1161/01.res.87.12.e61. [DOI] [PubMed] [Google Scholar]
- 18.Fuentes J J, Pritchard M A, Estivill X. Genomics. 1997;44:358–361. doi: 10.1006/geno.1997.4866. [DOI] [PubMed] [Google Scholar]
- 19.Strippoli P, Lenzi L, Petrini M, Carinci P, Zannotti M. Genomics. 2000;64:252–263. doi: 10.1006/geno.2000.6127. [DOI] [PubMed] [Google Scholar]
- 20.Kingsbury T J, Cunningham K W. Genes Dev. 2000;14:1595–1604. [PMC free article] [PubMed] [Google Scholar]
- 21.Lai M M, Burnett P E, Wolosker H, Blackshaw S, Snyder S H. J Biol Chem. 1998;273:18325–18331. doi: 10.1074/jbc.273.29.18325. [DOI] [PubMed] [Google Scholar]
- 22.Kashishian A, Howard M, Loh C, Gallatin W M, Hoekstra M F, Lai Y. J Biol Chem. 1998;273:27412–27419. doi: 10.1074/jbc.273.42.27412. [DOI] [PubMed] [Google Scholar]
- 23.Sun L, Youn H D, Loh C, Stolow M, He W, Liu J O. Immunity. 1998;8:703–711. doi: 10.1016/s1074-7613(00)80575-0. [DOI] [PubMed] [Google Scholar]
- 24.Rusnak F, Mertz P. Physiol Rev. 2000;80:1483–1521. doi: 10.1152/physrev.2000.80.4.1483. [DOI] [PubMed] [Google Scholar]
- 25.Gulick J, Subramaniam A, Neumann J, Robbins J. J Biol Chem. 1991;266:9180–9185. [PubMed] [Google Scholar]
- 26.Eto Y, Yonekura K, Sonoda M, Arai N, Sata M, Sugiura S, Takenaka K, Gualberto A, Hixon M L, Wagner M W, Aoyagi T. Circulation. 2000;101:2134–2137. doi: 10.1161/01.cir.101.18.2134. [DOI] [PubMed] [Google Scholar]
- 27.Friddle C J, Koga T, Rubin E M, Bristow J. Proc Natl Acad Sci USA. 2000;97:6745–6750. doi: 10.1073/pnas.100127897. . (First Published May 30, 2000; 10.1073/pnas.100127897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olson E N, Williams R S. Cell. 2000;101:689–692. doi: 10.1016/s0092-8674(00)80880-6. [DOI] [PubMed] [Google Scholar]
- 29.Crabtree G R. Cell. 1999;96:611–614. doi: 10.1016/s0092-8674(00)80571-1. [DOI] [PubMed] [Google Scholar]
- 30.Fuentes J J, Genesca L, Kingsbury T J, Cunningham K W, Perez-Riba M, Estivill X, Luna S. Hum Mol Genet. 2000;9:1681–1690. doi: 10.1093/hmg/9.11.1681. [DOI] [PubMed] [Google Scholar]
- 31.De Windt L J, Lim H W, Bueno O F, Liang Q, Delling U, Braz J C, Glascock B J, Kimball T F, del Monte F, Hajjar R J, Molkentin J D. Proc Natl Acad Sci USA. 2001;98:3322–3327. doi: 10.1073/pnas.031371998. [DOI] [PMC free article] [PubMed] [Google Scholar]