Abstract
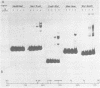
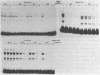
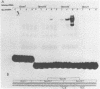
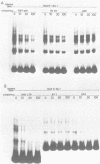
We have documented that the Rous sarcoma virus (RSV) internal enhancer functions in the nontransformed Baby Hamster Kidney (BHK) cell line. The sequences within this region were assayed for their ability to bind to specific factors present in BHK nuclear extracts using the gel retardation assay and DNAse I footprinting. At least two sequences within the internal enhancer which can specifically bind nuclear factors in vitro have been identified. These regions are located between nucleotides 813-850 and 856-877. These sites map within the overall region of the internal enhancer which has been shown to be essential for enhancer activity and within the specific region which can function as an orientation independent enhancer. Using the DNase I footprinting and binding data to design an oligonucleotide, we have demonstrated that an oligonucleotide extending from nucleotides 804-877 will substitute efficiently as an enhancer. We also demonstrate that the SV40 enhancer does not compete for the factors which bind to the RSV internal enhancer, whereas an oligonucleotide to the binding site for EFII in the LTR can compete for factor binding to the internal enhancer.
Full text
PDF


















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arrigo S., Yun M., Beemon K. cis-acting regulatory elements within gag genes of avian retroviruses. Mol Cell Biol. 1987 Jan;7(1):388–397. doi: 10.1128/mcb.7.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Carthew R. W., Chodosh L. A., Sharp P. A. An RNA polymerase II transcription factor binds to an upstream element in the adenovirus major late promoter. Cell. 1985 Dec;43(2 Pt 1):439–448. doi: 10.1016/0092-8674(85)90174-6. [DOI] [PubMed] [Google Scholar]
- Chiswell D. J., Gillespie D. A., Wyke J. A. The changes in proviral chromatin that accompany morphological variation in avian sarcoma virus-infected rat cells. Nucleic Acids Res. 1982 Jul 10;10(13):3967–3980. doi: 10.1093/nar/10.13.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B. R., Raymond K., Ju G. Transcriptional activity of avian retroviral long terminal repeats directly correlates with enhancer activity. J Virol. 1985 Feb;53(2):515–521. doi: 10.1128/jvi.53.2.515-521.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson B. M., Felsenfeld G. Specific factor conferring nuclease hypersensitivity at the 5' end of the chicken adult beta-globin gene. Proc Natl Acad Sci U S A. 1984 Jan;81(1):95–99. doi: 10.1073/pnas.81.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson B. M., Lewis C. D., Felsenfeld G. Interaction of specific nuclear factors with the nuclease-hypersensitive region of the chicken adult beta-globin gene: nature of the binding domain. Cell. 1985 May;41(1):21–30. doi: 10.1016/0092-8674(85)90057-1. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura F. K. Nuclear activity from F9 embryonal carcinoma cells binding specifically to the enhancers of wild-type polyoma virus and PyEC mutant DNAs. Nucleic Acids Res. 1986 Apr 11;14(7):2845–2861. doi: 10.1093/nar/14.7.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Merlino G. T., Willingham M. C., Pastan I., Howard B. H. The Rous sarcoma virus long terminal repeat is a strong promoter when introduced into a variety of eukaryotic cells by DNA-mediated transfection. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6777–6781. doi: 10.1073/pnas.79.22.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Groudine M., Conkin K. F. Chromatin structure and de novo methylation of sperm DNA: implications for activation of the paternal genome. Science. 1985 May 31;228(4703):1061–1068. doi: 10.1126/science.2986289. [DOI] [PubMed] [Google Scholar]
- Hearing P., Shenk T. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell. 1983 Jul;33(3):695–703. doi: 10.1016/0092-8674(83)90012-0. [DOI] [PubMed] [Google Scholar]
- Herbomel P., Saragosti S., Blangy D., Yaniv M. Fine structure of the origin-proximal DNAase I-hypersensitive region in wild-type and EC mutant polyoma. Cell. 1981 Sep;25(3):651–658. doi: 10.1016/0092-8674(81)90172-0. [DOI] [PubMed] [Google Scholar]
- Herr W., Clarke J. The SV40 enhancer is composed of multiple functional elements that can compensate for one another. Cell. 1986 May 9;45(3):461–470. doi: 10.1016/0092-8674(86)90332-6. [DOI] [PubMed] [Google Scholar]
- Herr W., Gluzman Y. Duplications of a mutated simian virus 40 enhancer restore its activity. Nature. 1985 Feb 21;313(6004):711–714. doi: 10.1038/313711a0. [DOI] [PubMed] [Google Scholar]
- Jones K. A., Kadonaga J. T., Rosenfeld P. J., Kelly T. J., Tjian R. A cellular DNA-binding protein that activates eukaryotic transcription and DNA replication. Cell. 1987 Jan 16;48(1):79–89. doi: 10.1016/0092-8674(87)90358-8. [DOI] [PubMed] [Google Scholar]
- Khoury G., Gruss P. Enhancer elements. Cell. 1983 Jun;33(2):313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- Laimins L. A., Tsichlis P., Khoury G. Multiple enhancer domains in the 3' terminus of the Prague strain of Rous sarcoma virus. Nucleic Acids Res. 1984 Aug 24;12(16):6427–6442. doi: 10.1093/nar/12.16.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciw P. A., Bishop J. M., Varmus H. E., Capecchi M. R. Location and function of retroviral and SV40 sequences that enhance biochemical transformation after microinjection of DNA. Cell. 1983 Jul;33(3):705–716. doi: 10.1016/0092-8674(83)90013-2. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Nordheim A., Rich A. Negatively supercoiled simian virus 40 DNA contains Z-DNA segments within transcriptional enhancer sequences. Nature. 1983 Jun 23;303(5919):674–679. doi: 10.1038/303674a0. [DOI] [PubMed] [Google Scholar]
- Ohlsson H., Edlund T. Sequence-specific interactions of nuclear factors with the insulin gene enhancer. Cell. 1986 Apr 11;45(1):35–44. doi: 10.1016/0092-8674(86)90535-0. [DOI] [PubMed] [Google Scholar]
- Peterson C. L., Orth K., Calame K. L. Binding in vitro of multiple cellular proteins to immunoglobulin heavy-chain enhancer DNA. Mol Cell Biol. 1986 Dec;6(12):4168–4178. doi: 10.1128/mcb.6.12.4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piette J., Kryszke M. H., Yaniv M. Specific interaction of cellular factors with the B enhancer of polyoma virus. EMBO J. 1985 Oct;4(10):2675–2685. doi: 10.1002/j.1460-2075.1985.tb03987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintrell N., Hughes S. H., Varmus H. E., Bishop J. M. Structure of viral DNA and RNA in mammalian cells infected with avian sarcoma virus. J Mol Biol. 1980 Nov 15;143(4):363–393. doi: 10.1016/0022-2836(80)90218-1. [DOI] [PubMed] [Google Scholar]
- Robinson H. L., Blais B. M., Tsichlis P. N., Coffin J. M. At least two regions of the viral genome determine the oncogenic potential of avian leukosis viruses. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1225–1229. doi: 10.1073/pnas.79.4.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson H. L., Jensen L., Coffin J. M. Sequences outside of the long terminal repeat determine the lymphomogenic potential of Rous-associated virus type 1. J Virol. 1985 Sep;55(3):752–759. doi: 10.1128/jvi.55.3.752-759.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saragosti S., Moyne G., Yaniv M. Absence of nucleosomes in a fraction of SV40 chromatin between the origin of replication and the region coding for the late leader RNA. Cell. 1980 May;20(1):65–73. doi: 10.1016/0092-8674(80)90235-4. [DOI] [PubMed] [Google Scholar]
- Sealey L., Chalkley R. At least two nuclear proteins bind specifically to the Rous sarcoma virus long terminal repeat enhancer. Mol Cell Biol. 1987 Feb;7(2):787–798. doi: 10.1128/mcb.7.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow N. D., Wilkie N. M. An improved technique for obtaining enhanced infectivity with herpes simplex virus type 1 DNA. J Gen Virol. 1976 Dec;33(3):447–458. doi: 10.1099/0022-1317-33-3-447. [DOI] [PubMed] [Google Scholar]
- Swanstrom R., DeLorbe W. J., Bishop J. M., Varmus H. E. Nucleotide sequence of cloned unintegrated avian sarcoma virus DNA: viral DNA contains direct and inverted repeats similar to those in transposable elements. Proc Natl Acad Sci U S A. 1981 Jan;78(1):124–128. doi: 10.1073/pnas.78.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenke M., Grundström T., Matthes H., Wintzerith M., Schatz C., Wildeman A., Chambon P. Multiple sequence motifs are involved in SV40 enhancer function. EMBO J. 1986 Feb;5(2):387–397. doi: 10.1002/j.1460-2075.1986.tb04224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]