Abstract
Orexin-producing neurons are clearly essential for the regulation of wakefulness and sleep because loss of these cells produces narcolepsy. However, little is understood about how these neurons dynamically interact with other wake- and sleep-regulatory nuclei to control behavioral states. Using survival analysis of wake bouts in wild-type and orexin knockout mice, we found that orexins are necessary for the maintenance of long bouts of wakefulness, but orexin deficiency has little impact on wake bouts <1 min. Since orexin neurons often begin firing several seconds before the onset of waking, this suggests a surprisingly delayed onset (>1 min) of functional effects. This delay has important implications for understanding the control of wakefulness and sleep because increasing evidence suggests that different mechanisms are involved in the production of brief and sustained wake bouts. We incorporated these findings into a mathematical model of the mouse sleep/wake network. Orexins excite monoaminergic neurons and we hypothesize that orexins increase the monoaminergic inhibition of sleep-promoting neurons in the ventrolateral preoptic nucleus. We modeled orexin effects as a time-dependent increase in the strength of inhibition from wake- to sleep-promoting populations and the resulting simulated behavior accurately reflects the fragmented sleep/wake behavior of narcolepsy and leads to several predictions. By integrating neurophysiology of the sleep/wake network with emergent properties of behavioral data, this model provides a novel framework for investigating network dynamics and mechanisms associated with normal and pathologic sleep/wake behavior.
INTRODUCTION
Orexin-producing neurons play an essential role in the regulation of wakefulness and sleep. Loss of the orexin neurons or the orexin neuropeptides (orexin-A and -B, also known as hypocretin-1 and -2) causes narcolepsy, a common sleep disorder characterized by excessive daytime sleepiness, rapid eye movement (REM) sleep soon after sleep onset, disturbed nocturnal sleep, and cataplexy (Dauvilliers et al. 2007; Scammell 2003). Mice, rats, and dogs with disrupted orexin signaling all have sleepiness and cataplexy strikingly similar to that seen in people with narcolepsy (Beuckmann et al. 2004; Chemelli et al. 1999; Hungs and Mignot 2001).
These features of narcolepsy highlight the necessity of orexins, but little is understood about how the orexin neurons dynamically interact with other wake- and sleep-regulatory nuclei to modulate sleep/wake behavior. The orexin neurons are strictly wake-active (Lee et al. 2005; Mileykovskiy et al. 2005) and send excitatory projections to many state-regulatory nuclei (Peyron et al. 1998). Although orexin knockout (OXKO) mice have normal hourly amounts of wakefulness and sleep, their wake bouts are much shorter than normal (Mochizuki et al. 2004), suggesting that orexins mainly stabilize sleep/wake behavior. Several groups have proposed that long bouts of wakefulness may be generated by different neural mechanisms than those that produce brief wake bouts (Halasz et al. 2004; Lo et al. 2004). Thus we hypothesize that orexins selectively influence the production of long wake bouts.
To examine these network effects of orexins, we first compared survival distributions of wakefulness, non-REM (NREM) sleep, and REM sleep bout durations in OXKO and wild-type (WT) mice. We then integrated these findings into a neurobiologically based mathematical model of the sleep/wake network that simulates realistic mouse sleep/wake behavior and links the activity of specific neuronal populations to the expression of wakefulness, NREM sleep, and REM sleep (Diniz Behn et al. 2007). This combination of survival analysis and mathematical modeling provides new insights into the effects of orexins on sleep/wake behavior and identifies mechanisms through which the absence of orexins destabilizes network dynamics.
METHODS
Animals
Founder OXKO mice were on a C57BL/6J-129/SvEV background and their offspring were backcrossed with C57BL/6J mice for eight generations. We recorded sleep/wake behavior in eight male OXKO mice and seven WT littermates, all 5–6 mo old and weighing 30–35 g. All experiments were approved by the Institutional Animal Care and Use Committees of Beth Israel Deaconess Medical Center and Harvard Medical School.
Surgery and electroencephalogram-electromyogram recordings
Mice were anesthetized with ketamine-xylazine (100 and 10 mg/kg, administered intraperitoneally) and implanted with electroencephalogram (EEG) and electromyogram (EMG) electrodes as described previously (Mochizuki et al. 2004). EEG signals were recorded using two ipsilateral stainless steel screws (1.5 mm to the right of the sagittal suture, 1 mm anterior to bregma, and 1 mm anterior to lambda). EMG signals were acquired by a pair of multistranded stainless steel wires inserted into the neck extensor muscles. Nine days after surgery, mice were transferred to individual recording cages in a sound-attenuated chamber with a 12 h/12 h light/dark (LD) cycle (30 lux; lights on at 7:00 am and off at 7:00 pm) and a constant temperature of about 23°C. They had unrestricted access to food and water and acclimated to the recording cables for another 5 days.
Two weeks after surgery, we recorded spontaneous sleep/wake behavior. EEG/EMG signals were amplified (Model 12, Grass Technologies, West Warwick, RI) and digitized at 128 Hz using a sleep-scoring system (Sleep Sign, Kissei Comtec, Matsumoto, Japan). The signals were digitally filtered (EEG: 0.3–30 Hz; EMG: 2–100 Hz) and semiautomatically scored in 10-s epochs as wakefulness, NREM sleep, or REM sleep. This preliminary scoring was visually inspected and corrected when appropriate. We scored epochs as cataplexy based on the following criteria: wakefulness preceding cataplexy onset had to last ≥40 s (Fujiki et al. 2006); cataplexy onset was marked by an abrupt transition from wakefulness to periods of high EEG theta activity (4 –9 Hz) and atonia in the nuchal muscles. Simultaneous video recordings showed that during cataplexy the mouse was often prone or lying on its side in a posture atypical of sleep and the cataplexy almost always occurred outside of the usual nest (Mochizuki et al. 2004). These episodes were always followed by a direct transition back to wakefulness. Cataplexy never occurred in WT mice.
Survival analysis of sleep/wake bouts
In humans, mice, and other species, Lo and colleagues (2004) reported that wake-bout durations follow a power-law distribution (proportional to t−∞) and sleep-bout durations follow an exponential distribution. In mice, the power-law distribution holds for brief awakenings lasting <1–2 min, but longer wake bouts appear to be governed by different dynamic principles. Because orexins help consolidate sleep/wake behavior, we first determined whether these distributions were altered in OXKO mice.
We calculated Kaplan–Meier survival distributions for wake-, NREM sleep-, and REM sleep-bout durations of individual mice and for data pooled by genotype (SAS, Cary, NC). We also analyzed cataplexy-bout durations in OXKO mice. This analysis measures the probability that a given bout will “survive” long enough to reach a given duration. Twenty-four– hour data, light-period data, and dark-period data were analyzed separately. Due to the properties of the wake-bout survival curves, we also calculated separate survival distributions for short (<60 s), intermediate (60 –1,000 s), and long (>1,000 s) wake bouts. The analysis by Lo and colleagues (2004) suggested that there is an emergent threshold in the range of 1–2 min that distinguishes short wake bouts; using this threshold as a guideline, we increased the upper bound for short durations until the differences in survival approached significance. This resulted in the 60-s threshold for short wake bouts. We chose the threshold of 1,000 s to separate the plateau and tail regions of the survival curves, but the results for intermediate- and long-bout durations were independent of this choice. This partitioning removed the possible biasing effect of long durations from the analysis of shorter bouts.
Statistical analysis of survival curves
To determine the statistical distributions of each bout type (wake, NREM sleep, REM sleep, and cataplexy), we performed survival analyses in which we plotted data in log–log and semilog coordinates and used linear regression to fit the data (Blumberg et al. 2005). Goodness-of-fit to power-law and exponential distributions, respectively, were evaluated with r2 values. We considered the data to be consistent with a given distribution when r2 values were >0.95.
In additional comparisons between genotypes, we tested the null hypothesis of similar risk of bout termination using both log-rank and Wilcoxon statistical tests. The Wilcoxon is a weighted version of the log-rank test that is less sensitive to differences in long bouts (Allison 1995). These tests were performed on 24-h group data, separate light-and dark-period group data, and light-period group data for wake bouts separated into short, intermediate, and long durations.
Mathematical modeling of orexinergic effects
In a prior paper, we described the full derivation of a mathematical model of the mouse sleep/wake network, based on neurophysiologic properties of the relevant neuronal populations (Diniz Behn et al. 2007). Briefly, three relaxation oscillators were used to model activity in wake-, sleep-, and REM sleep-promoting neuronal populations; excitatory/inhibitory coupling between populations was based on the known anatomy and physiology of sleep/wake regulatory populations (Fig. 1A). High levels of activity in the wake-, sleep-, and REM sleep-promoting populations correspond to wakefulness, NREM sleep, and REM sleep, respectively. Therefore model output can be summarized in a hypnogram of simulated sleep/wake behavior (Fig. 1B).
FIG. 1.
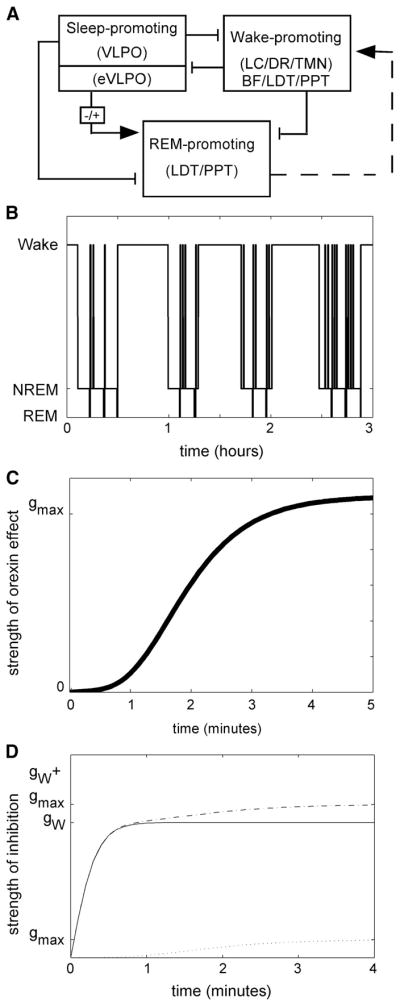
The network model of the sleep/wake network simulates sleep/wake behavior. A: the physiologically based model includes wake-, sleep-, and REM sleep-promoting neuronal populations. The wake-promoting population also includes basal forebrain (BF) and pontine cholinergic populations. Inhibitory (lines) and excitatory (arrows) couplings are based on reported anatomy and physiology for these populations. The dashed line represents indirect excitation from laterodorsal tegmental nucleus/pedunculopontine tegmental nucleus (LDT/PPT) to monoaminergic populations. B: hypnogram summarizes 3 h of sleep/wake behavior generated by state-dependent activity in each of these populations. C: the sigmoidal activation function describing the onset of orexin signaling minimizes orexin effects during the first minute of wakefulness. The time course and shape of the activation function for orexin effects explicitly reflect the properties predicted by the survivor plot data. D: orexin effects time-dependently increase the strength of inhibition from wake- to sleep-promoting populations. The profiles of this strength of inhibition are shown in the absence (solid line) and the presence (dashed line) of orexin signaling. Because orexin effects are small during the first minute of wakefulness, the strength of inhibition is similar initially; however, as the wake bout continues, the presence of orexin signaling is associated with higher levels of inhibition (gW + gmax vs. gW only).
Circadian influences on sleep/wake behavior, including such phenomena as diurnal variations in amounts of wake and sleep, lengths of bouts, and gating of REM sleep, are not included in the model. Although the anatomic substrate for circadian modulation of sleep/wake behavior is becoming more clear (Saper et al. 2005), the physiologic and dynamic mechanisms that achieve this modulation are not well understood (Aston-Jones et al. 2001; Lu et al. 2000; Mistlberger 2005; Saint-Mleux et al. 2004, 2007). Therefore we analyzed and modeled average sleep/wake behavior over 24 h to investigate baseline interactions of the sleep/wake network. In modeling the role of orexins in sleep/wake behavior, we focused on the abnormally short wake bouts of OXKO mice, which are present at all circadian times (Mochizuki et al. 2004).
The flip-flop mechanism between wake-promoting monoaminergic populations [locus coeruleus (LC), dorsal raphe (DR), and tube-romammillary nucleus (TMN)] and sleep-promoting populations [ventrolateral preoptic nucleus (VLPO) and other preoptic nuclei] forms the basis for fast transitions between wakefulness and sleep (Diniz Behn et al. 2007; Saper et al. 2001). In addition, both projections from VLPO to basal forebrain, laterodorsal tegmental nucleus (LDT), and pedunculopontine tegmental nucleus (PPT) (Lu et al. 2002; Saper et al. 2001) and physiology (Datta and Siwek 2002; Gallopin et al. 2000; Saint-Mleux et al. 2004; Strecker et al. 2000; Szymusiak et al. 2000) support the inclusion of these nuclei in the modeled wake-promoting population despite the limited anatomic evidence for inhibition of VLPO by these regions (Chou et al. 2002). Additional LDT/PPT neurons that are mainly active during REM sleep are associated with the modeled REM sleep-promoting population. Coupling strengths between populations were modulated by variables representing homeostatic NREM and REM sleep drives and this modulation initiated transitions between states. In addition, network dynamics permitted the expression of intrinsic oscillatory properties of the modeled wake-promoting population, resulting in the production of brief awakenings with durations independent of homeostatic NREM sleep drive. Therefore although the model assumed a single wake-promoting population, it included different mechanisms for brief and sustained wake bouts. The original model did not address the role of orexins or the disrupted sleep/wake dynamics in orexin-deficient animals (Diniz Behn et al. 2007).
In the current expansion of this model, we considered orexin neurons separately from other modeled wake-promoting neuronal populations because of their many physiologic and anatomic differences. Because loss of orexins does not substantially affect the amount of time spent in sleep/wake states, we theorized that orexins are necessary for the consolidation but not the production of wakefulness. Therefore we included orexin activity as a modulatory element in the model rather than separately modeling a population of orexin neurons. Our survival analysis of wake-bout durations in WT and OXKO mice suggested that brief wake bouts are not influenced by orexins, but that orexins are necessary for the consolidation of long wake bouts (see RESULTS). If different wake-promoting populations were responsible for brief and sustained wake bouts, then these results would indicate that orexins affect only populations involved in sustained wake bouts. However, in our modeling framework, a single wake-promoting population under different dynamic configurations drives both brief and sustained wake bouts. Therefore we hypothesize that orexins mainly affect long wake bouts because of the time course of orexin signaling, rather than targeted sites of action.
To evaluate the time course of orexin signaling, we modeled the effects of orexins with a saturating sigmoidal function of time awake. This function, gOX(tW), is given by
where the parameter gmax scales the strength of the effect, the parameter ρ controls the rate of onset of the effect, and tW tracks the time elapsed since the onset of the wake bout. The exponents helped produce a more gradual growth of gOX(tW) between initial minimal values and saturating values. This function worked well because the sigmoidal shape of gOX(tW) minimized orexin effects for small values of tW, reflected activation of orexin signaling with a time course determined by ρ, and eventually saturated in a manner consistent with biological signaling. However, other functional forms for gOX(tW) with these properties, such as hyperbolic tangent functions with a delay, could also be used (see RESULTS).
We examined the time course of orexin onset by varying ρ in our model. Based on the results of our survival analysis, the parameter ρ was chosen to satisfy the following conditions: 1) gOX(tW) is near zero for tW < 60 s and 2) gOX(tW) reaches half of its maximal activity at tW = 120 s. This time course results in a delayed increase in the strength of inhibition from the wake-promoting population to the sleep-promoting population (Fig. 1C). The parameter values used in the simulations are given in the APPENDIX.
Modeling orexin’s site of action
Orexins may act through several mechanisms to stabilize wakefulness and sleep. The orexin neurons project to many wake-promoting regions including the LC, DR, TMN, basal forebrain, and pontine cholinergic neurons, and orexins have direct excitatory effects on these targets (Burlet et al. 2002; Eggermann et al. 2001; Eriksson et al. 2001; España et al. 2001; Fadel and Frederick-Duus 2008; Hagan et al. 1999; Horvath et al. 1999; Huang et al. 2001; Liu et al. 2002; Peyron et al. 1998). Orexin fibers also innervate sleep-promoting regions such as the VLPO (Chou et al. 2002; Lu et al. 2000) and, although orexins may not directly influence VLPO neurons (Eggermann et al. 2001), they may act on nearby monoaminergic or cholinergic nerve terminals. Based on these findings, we examined three potential sites of orexin action. These sites were specified by constants (OXW, OXS, OXh) that multiply the function gOX(tW), and thus modulate the system in a time-dependent manner (mathematical details in the APPENDIX).
Circadian variations in cerebrospinal fluid orexin-A levels in squirrel monkeys suggest that wake-promoting orexin signaling may oppose accumulating sleep drive during the day (Zeitzer et al. 2003). Although circadian drives are not included in our current model, we explored the effect of time-dependently modulating the accumulating sleep drive through the parameter OXh. This parameter affects the rate at which homeostatic sleep drive is accumulated. Low values of OXh reflect a more rapid accumulation of homeostatic sleep drive. In the model, low OXh shortened the duration of long wake bouts, but it did not alter the duration or frequency of brief bouts of wakefulness (data not shown). We did not pursue this implementation because the increase in brief wake bouts is an important feature of the fragmented behavior observed in OXKO mice.
Orexins may directly excite wake-promoting neurons (Burlet et al. 2002; Eriksson et al. 2001; Hagan et al. 1999; Huang et al. 2001; Liu et al. 2002). Therefore we chose the parameter OXW to represent an increase in tonic excitatory drive to wake-promoting populations. Due to the form of coupling used in the model, minor changes to the activity level of the wake-promoting populations do not affect the strength of inhibition exerted by these populations. Thus decreasing OXW does not increase the strength of inhibition to sleep-promoting populations. This effect is considered separately (see following text). Instead, decreasing OXW altered state-transition mechanisms that involve the activity of the wake-promoting populations and resulted in fewer brief wake bouts (data not shown; mechanism explained in Diniz Behn et al. 2007). This behavior was in direct contrast to the behavior observed in OXKO mice, so this mechanism was not implemented.
Orexins may indirectly inhibit sleep-promoting neurons. Wake-promoting neurotransmitters including norepinephrine (NE), serotonin (5-HT), and acetylcholine (ACh) inhibit the VLPO (Gallopin et al. 2000; Osaka and Matsumura 1995). Orexins have no direct inhibitory effect on VLPO neurons (Eggermann et al. 2001), but orexins may inhibit VLPO and other sleep-active neurons by increasing monoaminergic and cholinergic signaling. We used the parameter OXS to reflect these orexin-related changes in the strength of inhibition from wake- to sleep-promoting populations. When we decreased OXS, the resulting simulated behavior closely mimicked the state fragmentation of OXKO mice.
Using different initial conditions and noise conditions, we generated eight 24-h segments of simulated WT mouse sleep/wake behavior in which orexins enhance the inhibition from wake- to sleep-promoting populations and eight 24-h segments of simulated OXKO mouse sleep/wake behavior with no orexin signaling (OXS = 0). We then compared these simulated results with experimental results obtained from seven WT mice and eight OXKO littermates.
RESULTS
Survival analysis of wake bouts
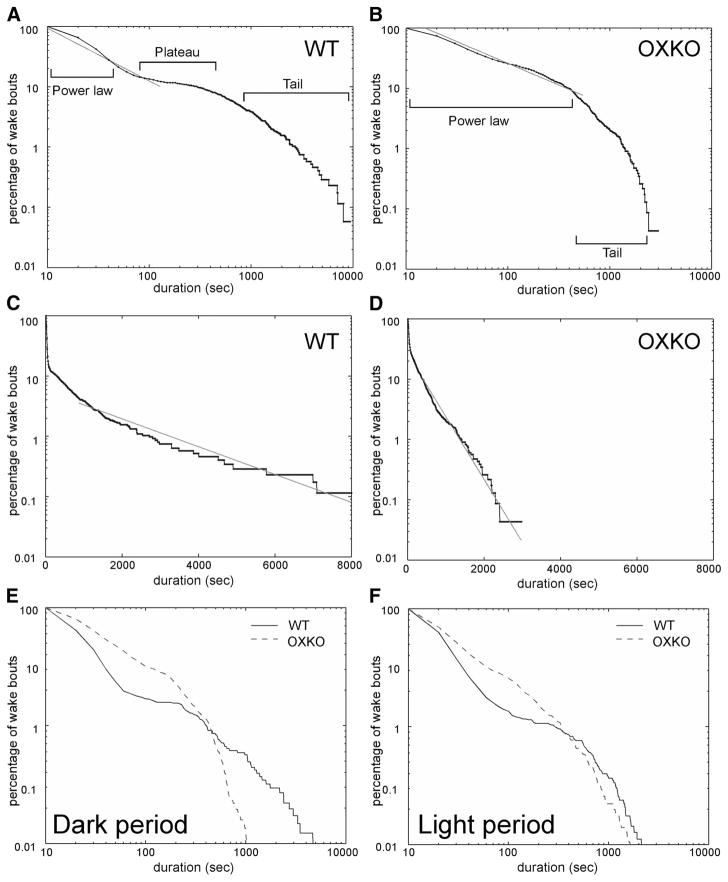
We examined the effects of orexins on the maintenance of wakefulness using survival analysis of wake-bout durations. We calculated survival of bouts for each individual and for data pooled by genotype using 24-h EEG/EMG recordings of WT (n = 7) and OXKO mice (n = 8). WT and OXKO mice had an average of 189 ± 19 and 290 ± 19 wake bouts, respectively. In WT mice, wake bouts lasting <60 s followed a power-law distribution as revealed by linear regression of log–log data (r2 = 0.98) (Fig. 2). The power-law distribution accounted for almost 90% of all wake bouts and is consistent with prior reports (Blumberg et al. 2007; Lo et al. 2004). As in WT mice, 90% of wake bouts in OXKO mice followed a power-law distribution (r2 = 0.96); however, in OXKO mice this power-law distribution extended across the first 600 s of wake.
FIG. 2.
Survival of wake bouts is disrupted in orexin knockout (OXKO) mice (n = 8) compared with wild-type (WT) mice (n = 7). A: in WT mice, the wake-bout survival curve shows 3 distinct regions: power law, plateau, and tail. Wake bouts <60 s obey a power-law distribution as shown by linear regression on log–log WT data (r2 = 0.98). B: in OXKO mice, the survival curve lacks the plateau region, the power-law behavior holds for wake bouts ≤600 s (r2 = 0.96), and the tail is much shorter. C: in WT mice, wake bouts >1,000 s follow an exponential distribution as shown by linear regression on semilog data (r2 = 0.97). D: in OXKO mice, wake bouts >600 s show this exponential behavior (r2 = 0.97). E and F: these differences in survival of wake bouts persist in separate analyses of the dark and light periods, although the difference in the right-hand tails is attenuated during the light period.
In WT mice, the power-law region was followed by a prominent plateau in which few wake bouts terminated between 100 and 1,000 s. Because the appearance of the plateau may be influenced by differences in maximal bout durations between genotypes, we separately examined survival curves of wake bouts during the light and dark periods. The plateau region persisted in both periods and maximal wake-bout durations for WT and OXKO mice were similar during the light period. This suggests that the plateau is a robust physiologic feature, rather than an artifact of a few very long wake bouts in WT mice. In contrast, OXKO mice lacked this plateau during both the light and dark periods (P < 0.05 by log-rank and Wilcoxon statistical tests). Instead, the persistence of the power-law distribution for wake bouts lasting as long as 600 s in these animals represents a significant fall-off of intermediate-length bouts and indicates a substantial dysfunction of the mechanisms that maintain wakefulness.
In both WT and OXKO mice, the right-hand tails of the wake-bout survival curves fall off sharply. Linear regression of semilog data (Fig. 2, C and D) showed that wake bouts >1,000 s in WT mice and >600 s in OXKO mice follow an exponential distribution (r2 = 0.97 for both WT and OXKO mice). This difference was very apparent in the dark period, but absent in the light period when WT mice do not produce very long wake bouts. For 24-h data, the characteristic timescale τ was smaller in OXKO mice, reflecting the steeper slope associated with the tail of the OXKO mouse data; however, in the light period, τ values were similar between the two groups, suggesting a circadian element to the differences between genotypes.
To eliminate the possibility that survival curves could be biased by differences in maximal bout durations and to better understand the differences between the two curves, we separately analyzed survival distributions for short (<60 s), intermediate (60 –1,000 s), and long (>1,000 s) bouts of wakefulness (Fig. 3). By separately considering the survival of these subsets, we could identify differences in fall-offs between genotypes; however, since this analysis was performed on subsets of the full data set, it does not maintain the power-law structure of the overall survival curve. Wake bouts <60 s did not differ between groups, but intermediate and long wake bouts were more likely to terminate early in OXKO mice (log-rank and Wilcoxon statistical tests, P < 0.05). These findings suggest that, although orexins do not significantly affect survival of wake bouts lasting <60 s, they are important for sustaining longer periods of wakefulness.
FIG. 3.
OXKO mice fail to maintain intermediate and long wake bouts even during the light period. A: WT and OXKO mice have similar survival of short wake bouts (<60 s). B: intermediate wake bouts (60 –1,000 s) terminate early in OXKO mice due to an absence of the plateau region (log-rank and Wilcoxon tests, P < 0.05). C: although the difference in the right-hand tail region is less striking in the light period, long wake bouts (>1,000 s) terminate earlier in OXKO mice, maintaining the significant difference between genotypes (log-rank and Wilcoxon tests, P < 0.05). This analysis was performed on light-period wake bouts to avoid possible biasing effects of the very long wake bouts in WT mice during the dark period. These plots show the survival of subsets of all wake bouts; the power-law behavior in survival of wake-bout durations is apparent only when all wake bouts are considered in the analysis due to the relationship between the survival curve and the underlying distribution.
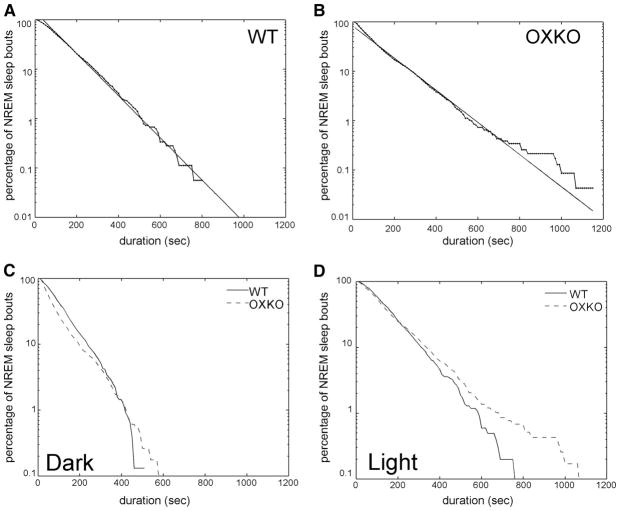
Survival analysis of NREM sleep bouts
We analyzed the effects of orexins on the maintenance of sleep by comparing NREM sleep-bout durations in WT and OXKO mice (Fig. 4). As in the analysis of wake-bout durations, both individual data and data pooled by genotype were considered. WT and OXKO mice had an average of 191 ± 27 and 276 ± 22 NREM sleep bouts, respectively. NREM sleep-bout durations in both groups followed an exponential distribution (r2 = 0.99 in WT and OXKO mice), consistent with previous reports (Blumberg et al. 2007; Lo et al. 2004). Thus although fragmentation of NREM sleep increased the total number of NREM bouts in OXKO mice, the survival of NREM bouts was similar between WT and OXKO mice. Surprisingly, maximal bout durations were longer in OXKO mice (P < 0.01), resulting in a characteristic timescale τ that was slightly larger in OXKO mice compared with that of WT mice. Despite longer maximal bout durations, the increased fragmentation of NREM sleep in OXKO mice resulted in shorter mean bout durations compared with those of WT mice, consistent with previous work (Mochizuki et al. 2004). The exponential distribution of NREM bouts persisted in the dark and light periods. Similar exponential distributions for survival of NREM bouts in WT and OXKO mice suggest that the mechanism that terminates NREM bouts is intact in OXKO mice.
FIG. 4.
OXKO and WT mice have similar distributions of nonrapid eye movement (NREM) sleep bouts. A and B: in both groups, NREM-bout durations over 24 h follow an exponential distribution as shown by linear regression of semilog data (r2 = 0.99 in each group). C and D: this exponential pattern is preserved in separate analyses of dark- and light-period NREM sleep (r2 > 0.96 for each group).
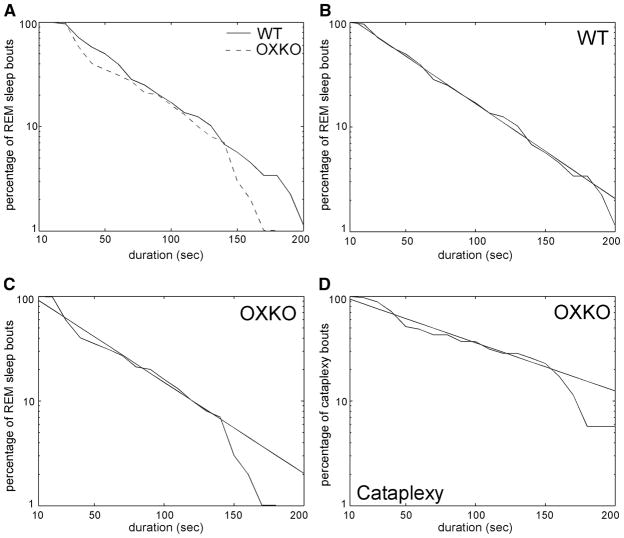
Survival analysis of REM sleep and cataplexy bouts
To investigate orexin-dependent differences in REM sleep, we compared REM-bout durations between WT and OXKO mice (Fig. 5). We did not analyze light- and dark-period data separately because REM sleep is rare during the dark period in WT mice. WT and OXKO mice had an average of 74 ± 9 and 98 ± 15 REM sleep bouts, respectively. In both groups, the distribution of bout durations <150 s was exponential with similar characteristic timescale τ (r2 > 0.98 in WT and OXKO mice). In OXKO mice, the fall-off in survival of REM-bout durations after 150 s was steeper than predicted by the exponential, but this was not observed in WT mice. This difference in the tail of the distributions resulted in a significant difference between survival curves (log-rank and Wilcoxon statistical tests, P < 0.01).
FIG. 5.
Survival of rapid eye movement (REM) sleep and cataplexy bouts is similar in OXKO and WT mice. A: the survival of REM bouts <150 s is similar for both groups, but OXKO mice have a more rapid fall-off in the right-hand tail region of the survival curve (log-rank and Wilcoxon tests, P < 0.01). B and C: in both groups, survival of REM bouts <150 s is exponential with similar timescales as shown by linear regression of semilog data (r2 = 0.98 in each group). D: in OXKO mice, the survival of cataplexy bouts also follows an exponential distribution, but like REM sleep in these animals, few bouts last longer than 150 s (r2 = 0.96).
OXKO mice had an average of 23 ± 10 cataplexy bouts. These episodes were scored as a distinct state and never occurred in WT mice. The survival of cataplexy-bout durations <150 s followed an exponential distribution (r2 = 0.96). Compared with REM sleep, the fall-off in bout durations in cataplexy was slower as reflected by a longer characteristic timescale than that associated with REM sleep (Fig. 5). Interestingly, as with REM-bout durations in OXKO mice, the survival of cataplexy durations showed a steep fall-off for bouts >150 s.
Implementation of orexin signaling in the model changes network dynamics
This survival analysis strongly suggested that orexins play an essential role in the maintenance of intermediate and long bouts of wakefulness. Wake bouts lasting <60 s did not show orexin-related differences. Based on this observation and neuroanatomic evidence, we modeled orexin effects [gOX(tW)] as a time-dependent increase in the strength of inhibition from the wake- to the sleep-promoting populations in the model network. By including time dependence in our implementation, we focused the orexin effect on intermediate and long wake bouts while leaving short bouts unaffected.
We selected the time course of orexin onset, ρ = 0.7 (min), based on the survival analyses showing that orexin effects are minimal during the first 60 s of wakefulness. A range of ρ values produced similar results, although when ρ was sufficiently large, the activation of gOX(tW) was nearly instantaneous and the temporally targeted effects of the function were lost. Fast activation of gOX(tW) is analogous to a fixed increase in the maximal strength of inhibition from wake- to sleep-promoting populations. Although such an increase extends the mean duration of sustained wake bouts, consistent with the state-consolidating role of orexins, it also inappropriately increases the total amount of wakefulness.
The key to the implementation was the differential effect of gOX(tW) on simulated brief and sustained wake bouts. In our model, separate mechanisms produce brief and sustained awakenings: sustained wake bouts are mainly limited by homeostatic sleep drive, but brief wake bouts are generated by intrinsic excitability in the wake-promoting populations (Diniz Behn et al. 2007). To generate OXKO mouse–like data, it was necessary to choose ρ such that gOX(tW) was minimal during brief awakenings but active during sustained wake bouts. This allowed the delicate mechanism underlying the production of brief wake bouts to function normally and resulted in realistic simulation of the fragmented sleep/wake behavior of OXKO mice: wake bouts became shorter; brief awakenings became more frequent; and the total amounts of wakefulness, NREM sleep, and REM sleep did not change. The general structure of the model was robust to variation of parameters (for details see Diniz Behn et al. 2007).
Simulating behavior of an orexin knockout mouse
To quantify the narcolepsy-like behavior generated by changes to OXS, we simulated behavior of WT and OXKO mice by setting the parameter reflecting the magnitude of orexin effects, gmax, to 1 and 0, respectively. By simply changing gmax and holding all other parameters fixed, we captured most differences in sleep/wake architecture between genotypes. Although WT and OXKO mice spend similar amounts of time in each state, OXKO mice have more frequent and shorter bouts of wakefulness and NREM sleep (Fig. 6). The simulated sleep/wake data reproduced these changes.
FIG. 6.
Model simulations replicate the differences in sleep/wake behavior between WT and OXKO mice. A and B: WT and OXKO mice have similar daily percentages of wakefulness, NREM sleep, and REM sleep. C and D: compared with WT mice, OXKO mice have more wake and NREM sleep bouts. E and F: the mean durations of wake and NREM bouts are shorter in OXKO mice. All of these differences are reflected in the simulated behavior.
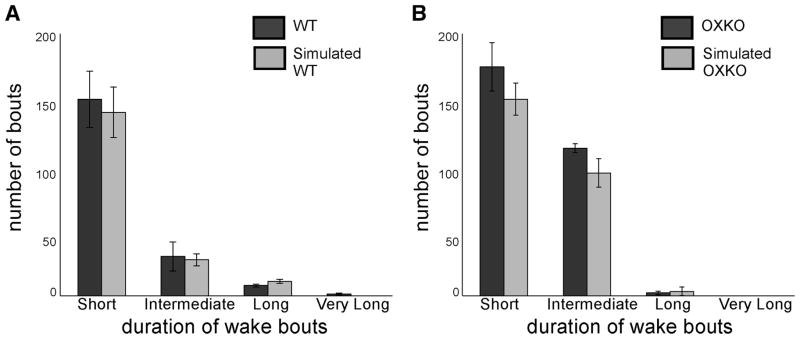
The oscillatory mechanism underlying brief wake bouts in the model resulted in a normal distribution of brief wake-bout durations inconsistent with the observed power-law behavior, so we could not perform conventional survival analysis of simulated wake bouts. Therefore to evaluate the maintenance of sleep/wake states in simulated WT and OXKO mouse data, we compared the distributions of bout durations for experimental and simulated data (Fig. 7). We considered short (<60 s), intermediate (60 –1,000 s), long (1,000 – 6,000 s), and very long (>6,000 s) bout durations. Compared with WT mice, OXKO mice had more short and intermediate wake bouts and fewer long wake bouts. In addition, OXKO mice did not produce very long bouts of wakefulness. The simulated sleep/wake behavior reproduces this shift toward shorter bouts.
FIG. 7.
In OXKO mice, time spent in wakefulness occurs in short- and intermediate-length bouts rather than long bouts. This shift as shown by histograms with bins corresponding to short (<60 s), intermediate (60 –1,000 s), long (1,000 – 6,000 s), and very long (>6,000 s) wake bouts. A: WT mice spend much of their waking time in long and very long wake bouts. B: OXKO mice show reduced numbers of long and very long wake bouts balanced by increased numbers of short- and intermediate-length bouts. The absence of bouts >6,000 s in OXKO mouse data highlights the inability of these animals to maintain very long bouts.
In the validation of our original model of sleep/wake behavior in WT mice, we compared probabilities of transitioning from one behavioral state to another (given that a state transition occurred) in experimental and simulated data (Diniz Behn et al. 2007). Earlier reports showed that raw numbers of state transitions are higher in OXKO mice than in WT mice, but state-transition probabilities are similar between the two genotypes (Mochizuki et al. 2004). Our analysis was consistent with these results with two exceptions: 1) if brief wake bouts and sustained wake bouts are considered as separate states, then OXKO mice were more likely to transition from REM sleep to sustained wakefulness than WT mice; and 2) the probability of transitioning from wakefulness to cataplexy in OXKO mice was almost 0.2, resulting in a lower probability of transitioning from wakefulness to NREM sleep than that seen in WT mice. With the exception of cataplexy-related transitions, the transition probabilities in the simulated data were similar to those observed experimentally (for details see Supplemental Table S1).1
DISCUSSION
Using survival analysis, we found that orexin deficiency has little impact on wake bouts <1 min. This suggests a slow onset of functional orexin effects, which may arise from delayed activation of the orexin neurons and their targets. In contrast, the survival of NREM and REM sleep bouts was very similar between the genotypes, consistent with the wake-active profile of orexin neurons (Lee et al. 2005; Mileykovskiy et al. 2005). We incorporated these findings into a mathematical model of the mouse sleep/wake network by modeling orexin effects as a time-dependent increase in the strength of inhibition from wake- to sleep-promoting populations. The model accurately reflects the fragmented sleep/wake behavior of narcolepsy and leads to several predictions. By integrating neurophysiology of the sleep/wake network with emergent properties of behavioral data, our model provides a novel framework for exploring network dynamics and mechanisms associated with normal and pathologic sleep/wake behavior.
Limitations of the current model
Our model accurately reflects many aspects of sleep/wake physiology and makes several useful predictions, but it includes some simplifications that limit its ability to fully replicate sleep/wake behavior. First, although the oscillatory mechanism underlying brief awakenings in our model generates brief awakenings consistent with observed sleep architecture, it does not permit replication of the power-law behavior observed in experimental data. Furthermore, the ON– OFF dichotomy of activity in our populations neglects probable differences in population activity within a given state. For example, VLPO neurons fire slowly in light NREM sleep and much faster in sustained, deep NREM sleep (Szymusiak et al. 1998). Little is currently known about temporal variations in the activity of state-regulatory populations, but these patterns will need to be integrated into future models of sleep/wake behavior.
Second, this model does not include circadian variations in sleep/wake behavior. Recent studies have identified anatomic pathways through which circadian signals may modulate sleep/wake behavior (Saper et al. 2005), but the physiologic and dynamic aspects of this control are complex and probably bidirectional (Aston-Jones et al. 2001; Deboer et al. 2003; Lu et al. 2000; Mistlberger 2005; Saint-Mleux et al. 2004, 2007). Thus we focused on how orexin helps produce long bouts of wakefulness, but future modeling efforts should incorporate circadian modulation of sleep/wake behavior.
Another limitation of our model is that it only partially reflects the complexity of the orexin system. We focus on projections from orexin neurons to wake-promoting monoaminergic and cholinergic populations, but orexin neurons innervate many brain regions and may influence sleep/wake behavior through other pathways such as projections to midline thalamus and cortex (Peyron et al. 1998). In addition, by modeling orexin signaling as a modulatory effect rather than the output of an independent population, we were not able to investigate factors affecting orexin neuron activity. For example, consideration of feedback to the orexin system from wake-active populations (Eriksson et al. 2001; Grivel et al. 2004; Li and van den Pol 2005; Muraki et al. 2004) may be necessary to model the role of orexin under altered conditions such as sleep deprivation.
Last, our model assumes coordinated activity within and between neuronal populations with similar state-dependent firing profiles, but in vivo, these neuronal populations fire with complex patterns of activity that probably influence network dynamics. For example, LC neurons are most active during periods of high attention and have tonic and phasic modes of activity (Aston-Jones and Bloom 1981; Aston-Jones et al. 1991). By assuming that wake-promoting populations behave in a coordinated way, we cannot account for cataplexy, a state in which the LC and DR fall silent, whereas the TMN remains active (John et al. 2004; Wu et al. 1999, 2004). Future work in which each neuronal population is modeled separately should permit the expression of simulated cataplexy and perhaps other dissociated states.
Despite these limitations, our model reflects the fragmented sleep/wake behavior of narcolepsy more accurately than prior efforts. Early recordings of OXKO mice reported an apparent increase in REM sleep, especially during the dark period when REM sleep is typically absent (Chemelli et al. 1999; Hara et al. 2001). Therefore Tamakawa and colleagues (2006) created a detailed physiologic model in which the absence of orexins increased REM sleep. However, more recent analyses of OXKO mice have shown that this apparent increase in REM sleep during the night can be attributed to cataplexy-like behaviors (Mochizuki et al. 2004; Willie et al. 2003) and, as discussed earlier, additional work is needed to investigate the neuronal mechanisms underlying cataplexy. Phillips and Robinson (2007) developed a model of the sleep/wake network in which orexins inhibit the VLPO. Although this increased state transitions in their model, their results were limited by an inability to replicate the severe state fragmentation characteristic of narcolepsy. The unique ability of our model to describe both brief and sustained wake bouts (based on different mechanisms) results in a novel and physiologically based framework in which to explore the state instability of narcolepsy.
Integration with current neurophysiology
During wakefulness, the orexin neurons are hypothesized to promote arousal mainly via excitation of monoaminergic and cholinergic neurons (Burlet et al. 2002; Eggermann et al. 2001; Eriksson et al. 2001; Hagan et al. 1999; Huang et al. 2001; Liu et al. 2002). In part, these systems drive wakefulness by increasing activity in the cortex, basal forebrain, and thalamus, but NE, 5-HT, and ACh also inhibit neurons in the VLPO (Gallopin et al. 2000; Osaka and Matsumura 1995). Our modeling demonstrates that normal consolidation of wakefulness and sleep may be achieved through orexin-mediated inhibition of sleep-promoting neurons. Orexins have no direct effect on VLPO neurons, but the VLPO region contains numerous orexin fibers (Chou et al. 2002). We speculate that orexins act presynaptically in the VLPO region to promote release of monoamines or ACh as has been demonstrated with NE release in the cortex (Hirota et al. 2001). Alternatively, orexins may enhance monoaminergic inhibition of the VLPO by reducing reuptake of monoamines. Further experiments are needed to help define the critical sites at which orexins inhibit sleep.
Recent studies raise interesting questions about whether normal orexin activity causes awakenings from sleep or helps maintain wakefulness. Adamantidis and colleagues (2007) showed that photostimulation of orexin neurons increased the probability of awakening from either NREM sleep or REM sleep. Consistent with our model, wakefulness often followed the photostimulation by about 25 s, perhaps because of a delayed functional effect of orexin neuron activity. The experiments show that robust orexin neuron activity can produce awakenings, but in vivo, orexin neurons may not reach similar high levels of firing until the animal is physically active, such as occurs during long wake bouts (Lee et al. 2005; Mileykovskiy et al. 2005). We predict that if used during wakefulness, even low levels of photostimulation of orexin neurons would be very effective in producing sustained wakefulness. Future studies that mimic normal patterns of orexin neuron firing may clarify the effects of orexin on maintenance of wakefulness.
Implications of the survival analysis
Survival analysis of bouts of wakefulness and sleep provides new perspectives on the dynamics of sleep/wake behavior. Consistent with previous work (Blumberg et al. 2007; Lo et al. 2004), we found that NREM sleep-bout durations follow an exponential distribution. However, our survival analysis of wake bouts identified several differences between WT and OXKO mice, suggesting that different neural mechanisms may govern the termination of short, intermediate, and long wake bouts. First, the shortest 90% of wake bouts in WT and OXKO mice will obey a power-law distribution, consistent with previous work (Blumberg et al. 2007; Lo et al. 2004). Lo and colleagues (2004) suggest that short-term nonperiodic fluctuations in the interactions of sleep/wake-promoting populations, such as intrinsic excitability, might produce brief awakenings (Taddese and Bean 2002). Our new observations indicate that this process persists for wake bouts lasting ≤600 s in OXKO mice, but whether this is due to changes in intrinsic excitability or to unmasking of power-law behavior in intermediate-length bouts is unknown. These predictions could be studied by survival analysis of wake bouts in mice with changes in sleep/wake-promoting populations and their interactions, such as VLPO-lesioned or histamine receptor knockout mice (Huang et al. 2006; Lu et al. 2000).
Second, in both WT and OXKO mice, very long wake bouts follow an exponential structure. The tails of the wake-bout distributions differ most between genotypes during the dark (active) period, probably because high orexin tone during the night helps maintain long wake bouts (Fujiki et al. 2001; Zeitzer et al. 2003). Survival in the tail of the distribution is probably governed by homeostatic sleep drive, and future experiments examining the effects of sleep deprivation and adenosine antagonists may improve our understanding of these very long wake bouts.
Third, and of most relevance to this project, survival curves of wake bouts in OXKO mice lack the plateau region seen in WT mice data. Between 100 and 1,000 s, very few bouts of wakefulness terminate in WT mice, yet this plateau is absent in OXKO mice, suggesting that orexins play an essential role in maintaining wakefulness during this period. Saper and colleagues (2001) proposed that orexins act like a “finger on the flip-flop sleep/wake switch,” preventing inappropriate state transitions. Our analysis indicates that in the first minute of wakefulness, transitions back to sleep occur easily. After this interval, orexins almost completely prevent transitions into sleep for almost 1,000 s, at which point the buildup of homeostatic sleep drive may again trigger transitions into sleep. In the absence of orexins, transitions into sleep may occur at any time, resulting in the poor consolidation of wakefulness observed in OXKO mice. These differences were not reported in a prior study of 21-day-old mice (Blumberg et al. 2007), most likely because of relatively late development of the orexin system (prepro-orexin levels are low before day 20; de Lecea et al. 1998) and maturation of sleep/wake behavior in general.
Modeling supports a delayed functional role of orexins
Based on our survival analysis and preliminary modeling, we implemented orexin effects as a time-dependent increase in inhibition from wake- to sleep-promoting populations. Our model produces simulated sleep/wake behavior very similar to that of WT and OXKO mice, suggesting that despite widespread projections of orexin neurons, the indirect inhibition of VLPO neurons by orexins is sufficient for normal consolidation of wakefulness and sleep. In the absence of orexins, reduced inhibition of the sleep-promoting population destabilizes the model’s flip-flop mechanism, thus providing more opportunities for bouts of sleep interrupted by brief bouts of wakefulness. In addition, the interaction between the time course of orexin effects and brief wake bouts contributes to the stabilizing role of orexins; minimizing modeled orexin effects during brief awakenings allows orexins to consolidate sleep/wake behavior without altering the total amounts of time spent in each state.
Several mechanisms may explain the delay in functional effects of orexins until 1 to 2 min after the onset of wake. Delayed firing by the orexin neurons appears unlikely because extracellular recordings show that these cells can begin firing several seconds before wakefulness begins (Lee et al. 2005; Mileykovskiy et al. 2005). One possibility is that behavioral effects are apparent only with relatively high levels of orexin neuron activity, similar to the observations in photostimulation experiments (Adamantidis et al. 2007). Alternatively, Li and van den Pol (2006) suggest that the delay may arise from interactions of colocalized neuropeptides in the orexin neurons. Orexin neurons contain both orexin and dynorphin, a generally inhibitory neuropeptide (Chou et al. 2001). Orexin indirectly excites orexin neurons, whereas dynorphin directly inhibits them (Li and van den Pol 2006; Li et al. 2002). However, coapplication of orexin and dynorphin initially reduces but then increases spike frequency in orexin neurons (Li and van den Pol 2006), representing a shift from autoinhibition to autoexcitation. This shift may be caused by desensitization of dynorphin receptors because the inhibition of orexin neurons by dynorphin is attenuated after about 90 s. Other wake-promoting targets may undergo similar shifts from inhibition to excitation over the first 1 to 2 min of orexin neuron firing, delaying the onset of orexin-mediated excitation as suggested by our survival analysis. Additional experiments are needed to determine just how the neurotransmitters of the orexin neurons interact to influence the sleep/wake network.
Supplementary Material
Acknowledgments
We thank R. Torgovitsky for help with survival analyses. OXKO mice were a kind gift from T. Sakurai of Kanazawa University.
GRANTS
This work was supported by National Institutes of Health Grants HL-007901, NS-055367, HL-60292, and DP1OD003646.
APPENDIX
As described in the text, orexin effects are modulated by the function gOX(tW) given by
where gmax = 0.033, ρ = 0.7 (min), and tW is a variable tracking time elapsed since the onset of the wake bout.
The sleep/wake network model includes 12 variables: the activity of wake-, sleep-, and REM sleep-promoting populations is described, respectively, by vW, vS, and vR; recovery variables for these populations are uW, uS, and uR; tw and ts measure time elapsed since the onset of the current wake or sleep bout, respectively; and homeostatic NREM sleep drive is described by h; homeostatic REM sleep drive effects are described by eV and r = rf + rs.
The activity and recovery of each population are modeled by a two-dimensional system of relaxation oscillator equations
where the ci represents coupling terms in which parameters OXW and OXS appear:
The saturating functions describing the onset of inhibition from sleep-to wake-promoting populations, respectively, are given by the following equations
with constant parameters gSmax, gWmax, ρS, and ρW. The variables ts and tw measure time elapsed since the onset of the current wake or sleep bout and are governed by the equations
The NREM homeostatic sleep drive h is governed by an equation in which OXh appears
where τhW and τhS are time constants controlling the growth and decay of h during wakefulness and sleep, respectively, and nh is a noise term. To obtain the appropriate reduction in inhibition from wake- to sleep-promoting populations, gW(tW) is multiplied by (1 − h) in cW; hence, this strength of inhibition varies inversely with h.
For the simulation results presented here, OXW = 0 and OXh = 0. We set OXS = 1 in the WT case and OXS = 0 in the OXKO case.
Homeostatic REM sleep effects include a gating mechanism based on reduced activity in wake-active populations (ev) and a drive (r) composed of two processes (rf and rs) acting on different timescales (Franken 2002). The variable ev is governed by the equation
where τev is a parameter controlling the rate of growth of ev; the occurrence of a REM bout resets ev to 0.
The fast process (rf) is involved in timing REM sleep within a sleep bout and is governed by
where r∞ and τrf are parameters describing the maximal value and time constant of rf, respectively; rf is reset to zero by the occurrence of a REM bout.
The slow process (rs) regulates the daily amount of REM sleep and is governed by the equation
where parameters rsup and rsdwn control the rate of growth and decay of rs.
The parameter values used in the simulations and additional equation details are given in Diniz Behn et al. (2007).
Footnotes
The online version of this article contains supplemental data.
References
- Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de Lecea L. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature. 2007;450:420–424. doi: 10.1038/nature06310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison PD. Survival Analysis Using SAS: A Practical Guide. Cary, NC: SAS Institute; 1995. [Google Scholar]
- Aston-Jones G, Bloom FE. Activity of norepinephrine-containing locus coeruleus neurons in behaving rats anticipates fluctuations in the sleep-waking cycle. J Neurosci. 1981;1:876–886. doi: 10.1523/JNEUROSCI.01-08-00876.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aston-Jones G, Chen S, Zhu Y, Oshinsky M. A neural circuit for circadian regulation of arousal. Nat Neurosci. 2001;4:732–738. doi: 10.1038/89522. [DOI] [PubMed] [Google Scholar]
- Aston-Jones G, Chiang C, Alexinsky T. Discharge of noradrenergic locus coeruleus neurons in behaving rats and monkeys suggests a role in vigilance. Prog Brain Res. 1991;88:501–520. doi: 10.1016/s0079-6123(08)63830-3. [DOI] [PubMed] [Google Scholar]
- Beuckmann CT, Sinton CM, Williams SC, Richardson JA, Hammer RE, Sakurai T, Yanagisawa M. Expression of a poly-glutamine-ataxin-3 transgene in orexin neurons induces narcolepsy-cataplexy in the rat. J Neurosci. 2004;24:4469–4477. doi: 10.1523/JNEUROSCI.5560-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg MS, Coleman CM, Johnson ED, Shaw C. Developmental divergence of sleep-wake patterns in orexin knockout and wild-type mice. Eur J Neurosci. 2007;25:512–518. doi: 10.1111/j.1460-9568.2006.05292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg MS, Seelke AM, Lowen SB, Karlsson KA. Dynamics of sleep-wake cyclicity in developing rats. Proc Natl Acad Sci USA. 2005;102:14860–14864. doi: 10.1073/pnas.0506340102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burlet S, Tyler CJ, Leonard CS. Direct and indirect excitation of laterodorsal tegmental neurons by hypocretin/orexin peptides: implications for wakefulness and narcolepsy. J Neurosci. 2002;22:2862–2872. doi: 10.1523/JNEUROSCI.22-07-02862.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemelli RM, Willie JT, Sinton CM, Elmquist JK, Scammell T, Lee C, Richardson JA, Williams SC, Xiong Y, Kisanuki Y, Fitch TE, Nakazato M, Hammer RE, Saper CB, Yanagisawa M. Narcolepsy in orexin knockout mice: molecular genetics of sleep regulation. Cell. 1999;98:437–451. doi: 10.1016/s0092-8674(00)81973-x. [DOI] [PubMed] [Google Scholar]
- Chou TC, Bjorkum AA, Gaus SE, Lu J, Scammell TE, Saper CB. Afferents to the ventrolateral preoptic nucleus. J Neurosci. 2002;22:977–990. doi: 10.1523/JNEUROSCI.22-03-00977.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TC, Lee CE, Lu J, Elmquist JK, Hara J, Willie JT, Beuckmann CT, Chemelli RM, Sakurai T, Yanagisawa M, Saper CB, Scammell TE. Orexin (hypocretin) neurons contain dynorphin. J Neurosci. 2001;21:RC168. doi: 10.1523/JNEUROSCI.21-19-j0003.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Siwek DF. Single cell activity patterns of pedunculopontine tegmentum neurons across the sleep-wake cycle in the freely moving rats. J Neurosci Res. 2002;70:611–621. doi: 10.1002/jnr.10405. [DOI] [PubMed] [Google Scholar]
- Dauvilliers Y, Arnulf I, Mignot E. Narcolepsy with cataplexy. Lancet. 2007;369:499–511. doi: 10.1016/S0140-6736(07)60237-2. [DOI] [PubMed] [Google Scholar]
- de Lecea L, Kilduff TS, Peyron C, Gao X, Foye PE, Danielson PE, Fukuhara C, Battenberg EL, Gautvik VT, Bartlett FS, 2nd, Frankel WN, van den Pol AN, Bloom FE, Gautvik KM, Sutcliffe JG. The hypocretins: hypothalamus-specific peptides with neuroexcitatory activity. Proc Natl Acad Sci USA. 1998;95:322–327. doi: 10.1073/pnas.95.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deboer T, Vansteensel MJ, Detari L, Meijer JH. Sleep states alter activity of suprachiasmatic nucleus neurons. Nat Neurosci. 2003;6:1086–1090. doi: 10.1038/nn1122. [DOI] [PubMed] [Google Scholar]
- Diniz Behn CG, Brown EN, Scammell TE, Kopell NJ. A mathematical model of network dynamics governing mouse sleep-wake behavior. J Neurophysiol. 2007;97:3828–3840. doi: 10.1152/jn.01184.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggermann E, Serafin M, Bayer L, Machard D, Saint-Mleux B, Jones BE, Muhlethaler M. Orexins/hypocretins excite basal forebrain cholinergic neurones. Neuroscience. 2001;108:177–181. doi: 10.1016/s0306-4522(01)00512-7. [DOI] [PubMed] [Google Scholar]
- Eriksson KS, Sergeeva O, Brown RE, Haas HL. Orexin/hypocretin excites the histaminergic neurons of the tuberomammillary nucleus. J Neurosci. 2001;21:9273–9279. doi: 10.1523/JNEUROSCI.21-23-09273.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- España RA, Baldo BA, Kelley AE, Berridge CW. Wake-promoting and sleep-suppressing actions of hypocretin (orexin): basal forebrain sites of action. Neuroscience. 2001;106:699–715. doi: 10.1016/s0306-4522(01)00319-0. [DOI] [PubMed] [Google Scholar]
- Fadel J, Frederick-Duus D. Orexin/hypocretin modulation of the basal forebrain cholinergic system: insights from in vivo microdialysis studies. Pharmacol Biochem Behav. 2008 January 19; doi: 10.1016/j.pbb.2008.01.008. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Franken P. Long-term vs. short-term processes regulating REM sleep. J Sleep Res. 2002;11:17–28. doi: 10.1046/j.1365-2869.2002.00275.x. [DOI] [PubMed] [Google Scholar]
- Fujiki N, Cheng T, Yoshino F, Nishino S. Specificity of direct transitions from wake to REM sleep in orexin/ataxin-3 narcoleptic mice (Abstract) Sleep. 2006;29:A225. doi: 10.1016/j.expneurol.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki N, Yoshida Y, Ripley B, Honda K, Mignot E, Nishino S. Changes in CSF hypocretin-1 (orexin A) levels in rats across 24 hours and in response to food deprivation. Neuroreport. 2001;12:993–997. doi: 10.1097/00001756-200104170-00026. [DOI] [PubMed] [Google Scholar]
- Gallopin T, Fort P, Eggermann E, Cauli B, Luppi PH, Rossier J, Audinat E, Muhlethaler M, Serafin M. Identification of sleep-promoting neurons in vitro. Nature. 2000;404:992–995. doi: 10.1038/35010109. [DOI] [PubMed] [Google Scholar]
- Grivel J, Cvetkovic V, Bayer L, Machard D, Tobler I, Mühlethaler M, Serafin M. The wake-promoting hypocretin/orexin neurons change their response to noradrenaline after sleep deprivation. J Neurosci. 2005;25:4127–4130. doi: 10.1523/JNEUROSCI.0666-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan J, Leslie R, Patel S, Evans M, Wattam T, Holmes S, Benham C, Taylor S, Routledge C, Hemmati P, Munton R, Ashmeade T, Shah A, Hatcher J, Hatcher P, Jones D, Smith M, Piper D, Hunter A, Porter R, Upton N. Orexin A activates locus coeruleus cell firing and increases arousal in the rat. Proc Natl Acad Sci USA. 1999;96:10911–10916. doi: 10.1073/pnas.96.19.10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halasz P, Terzano M, Parrino L, Bodizs R. The nature of arousal in sleep. J Sleep Res. 2004;13:1–23. doi: 10.1111/j.1365-2869.2004.00388.x. [DOI] [PubMed] [Google Scholar]
- Hara J, Beuckmann CT, Nambu T, Willie JT, Chemelli RM, Sinton CM, Sugiyama F, Yagami K, Goto K, Yanagisawa M, Sakurai T. Genetic ablation of orexin neurons in mice results in narcolepsy, hypophagia, and obesity. Neuron. 2001;30:345–354. doi: 10.1016/s0896-6273(01)00293-8. [DOI] [PubMed] [Google Scholar]
- Hirota K, Kushikata T, Kudo M, Kudo T, Lambert DG, Matsuki A. Orexin A and B evoke noradrenaline release from rat cerebrocortical slices. Br J Pharmacol. 2001;134:1461–1466. doi: 10.1038/sj.bjp.0704409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath T, Peyron C, Diano S, Ivanov A, Aston-Jones G, Kilduff T, van Den Pol A. Hypocretin (orexin) activation and synaptic innervation of the locus coeruleus noradrenergic system. J Comp Neurol. 1999;415:145–159. [PubMed] [Google Scholar]
- Huang ZL, Mochizuki T, Qu WM, Hong ZY, Watanabe T, Urade Y, Hayaishi O. Altered sleep-wake characteristics and lack of arousal response to H3 receptor antagonist in histamine H1 receptor knockout mice. Proc Natl Acad Sci USA. 2006;103:4687–4692. doi: 10.1073/pnas.0600451103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZL, Qu WM, Li WD, Mochizuki T, Eguchi N, Watanabe T, Urade Y, Hayaishi O. Arousal effect of orexin A depends on activation of the histaminergic system. Proc Natl Acad Sci USA. 2001;98:9965–9970. doi: 10.1073/pnas.181330998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hungs M, Mignot E. Hypocretin/orexin, sleep and narcolepsy. Bioessays. 2001;23:397–408. doi: 10.1002/bies.1058. [DOI] [PubMed] [Google Scholar]
- John J, Wu MF, Boehmer LN, Siegel JM. Cataplexy-active neurons in the hypothalamus: implications for the role of histamine in sleep and waking behavior. Neuron. 2004;42:619–634. doi: 10.1016/s0896-6273(04)00247-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Hassani OK, Jones BE. Discharge of identified orexin/hypocretin neurons across the sleep-waking cycle. J Neurosci. 2005;25:6716–6720. doi: 10.1523/JNEUROSCI.1887-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Gao XB, Sakurai T, van den Pol AN. Hypocretin/orexin excites hypocretin neurons via a local glutamate neuron-A potential mechanism for orchestrating the hypothalamic arousal system. Neuron. 2002;36:1169–1181. doi: 10.1016/s0896-6273(02)01132-7. [DOI] [PubMed] [Google Scholar]
- Li Y, van den Pol AN. Direct and indirect inhibition by catecholamines of hypocretin/orexin neurons. J Neurosci. 2005;25:173–183. doi: 10.1523/JNEUROSCI.4015-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, van den Pol AN. Differential target-dependent actions of coexpressed inhibitory dynorphin and excitatory hypocretin/orexin neuropeptides. J Neurosci. 2006;26:13037–13047. doi: 10.1523/JNEUROSCI.3380-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu RJ, van den Pol AN, Aghajanian GK. Hypocretins (orexins) regulate serotonin neurons in the dorsal raphe nucleus by excitatory direct and inhibitory indirect actions. J Neurosci. 2002;22:9453–9464. doi: 10.1523/JNEUROSCI.22-21-09453.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CC, Chou T, Penzel T, Scammell TE, Strecker RE, Stanley HE, Ivanov P. Common scale-invariant patterns of sleep-wake transitions across mammalian species. Proc Natl Acad Sci USA. 2004;101:17545–17548. doi: 10.1073/pnas.0408242101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Bjorkum AA, Xu M, Gaus SE, Shiromani PJ, Saper CB. Selective activation of the extended ventrolateral preoptic nucleus during rapid eye movement sleep. J Neurosci. 2002;22:4568–4576. doi: 10.1523/JNEUROSCI.22-11-04568.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Greco MA, Shiromani P, Saper CB. Effect of lesions of the ventrolateral preoptic nucleus on NREM and REM sleep. J Neurosci. 2000;20:3830–3842. doi: 10.1523/JNEUROSCI.20-10-03830.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileykovskiy BY, Kiyashchenko LI, Siegel JM. Behavioral correlates of activity in identified hypocretin/orexin neurons. Neuron. 2005;46:787–798. doi: 10.1016/j.neuron.2005.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistlberger RE. Circadian regulation of sleep in mammals: role of the suprachiasmatic nucleus. Brain Res. 2005;49:429–454. doi: 10.1016/j.brainresrev.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Mochizuki T, Crocker A, McCormack S, Yanagisawa M, Sakurai T, Scammell TE. Behavioral state instability in orexin knock-out mice. J Neurosci. 2004;24:6291–6300. doi: 10.1523/JNEUROSCI.0586-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraki Y, Yamanaka A, Tsujino N, Kilduff TS, Goto K, Sakurai T. Serotonergic regulation of the orexin/hypocretin neurons through the 5-HT1A receptor. J Neurosci. 2004;24:7159–7166. doi: 10.1523/JNEUROSCI.1027-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka T, Matsumura H. Noradrenaline inhibits preoptic sleep-active neurons through alpha 2-receptors in the rat. Neurosci Res. 1995;21:323–330. doi: 10.1016/0168-0102(94)00871-c. [DOI] [PubMed] [Google Scholar]
- Peyron C, Tighe DK, van den Pol AN, de Lecea L, Heller HC, Sutcliffe JG, Kilduff TS. Neurons containing hypocretin (orexin) project to multiple neuronal systems. J Neurosci. 1998;18:9996–10015. doi: 10.1523/JNEUROSCI.18-23-09996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AJ, Robinson PA. A quantitative model of sleep-wake dynamics based on the physiology of the brainstem ascending arousal system. J Biol Rhythms. 2007;22:167–179. doi: 10.1177/0748730406297512. [DOI] [PubMed] [Google Scholar]
- Saint-Mleux B, Bayer L, Eggermann E, Jones BE, Muhlethaler M, Serafin M. Suprachiasmatic modulation of noradrenaline release in the ventrolateral preoptic nucleus. J Neurosci. 2007;27:6412–6416. doi: 10.1523/JNEUROSCI.1432-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Mleux B, Eggermann E, Bisetti A, Bayer L, Machard D, Jones BE, Muhlethaler M, Serafin M. Nicotinic enhancement of the noradrenergic inhibition of sleep-promoting neurons in the ventrolateral preoptic area. J Neurosci. 2004;24:63–67. doi: 10.1523/JNEUROSCI.0232-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saper CB, Chou TC, Scammell TE. The sleep switch: hypothalamic control of sleep and wakefulness. Trends Neurosci. 2001;24:726–731. doi: 10.1016/s0166-2236(00)02002-6. [DOI] [PubMed] [Google Scholar]
- Saper CB, Scammell TE, Lu J. Hypothalamic regulation of sleep and circadian rhythms. Nature. 2005;437:1257–1263. doi: 10.1038/nature04284. [DOI] [PubMed] [Google Scholar]
- Scammell TE. The neurobiology, diagnosis, and treatment of narcolepsy. Ann Neurol. 2003;53:154–166. doi: 10.1002/ana.10444. [DOI] [PubMed] [Google Scholar]
- Strecker RE, Morairty S, Thakkar MM, Porkka-Heiskanen T, Basheer R, Dauphin LJ, Rainnie DG, Portas CM, Greene RW, McCarley RW. Adenosinergic modulation of basal forebrain and preoptic/anterior hypothalamic neuronal activity in the control of behavioral state. Behav Brain Res. 2000;115:183–204. doi: 10.1016/s0166-4328(00)00258-8. [DOI] [PubMed] [Google Scholar]
- Szymusiak R, Alam N, McGinty D. Discharge patterns of neurons in cholinergic regions of the basal forebrain during waking and sleep. Behav Brain Res. 2000;115:171–182. doi: 10.1016/s0166-4328(00)00257-6. [DOI] [PubMed] [Google Scholar]
- Szymusiak R, Alam N, Steininger TL, McGinty D. Sleep-waking discharge patterns of ventrolateral preoptic/anterior hypothalamic neurons in rats. Brain Res. 1998;803:178–188. doi: 10.1016/s0006-8993(98)00631-3. [DOI] [PubMed] [Google Scholar]
- Taddese A, Bean BP. Subthreshold sodium current from rapidly inactivating sodium channels drives spontaneous firing of tuberomammillary neurons. Neuron. 2002;33:587–600. doi: 10.1016/s0896-6273(02)00574-3. [DOI] [PubMed] [Google Scholar]
- Tamakawa Y, Karashima A, Koyama Y, Katayama N, Nakao M. A quartet neural system model orchestrating sleep and wakefulness mechanisms. J Neurophysiol. 2006;95:2055–2069. doi: 10.1152/jn.00575.2005. [DOI] [PubMed] [Google Scholar]
- Willie JT, Chemelli RM, Sinton CM, Tokita S, Williams SC, Kisanuki YY, Marcus JN, Lee C, Elmquist JK, Kohlmeier KA, Leonard CS, Richardson JA, Hammer RE, Yanagisawa M. Distinct narcolepsy syndromes in orexin receptor-2 and orexin null mice: molecular genetic dissection of non-REM and REM sleep regulatory processes. Neuron. 2003;38:715–730. doi: 10.1016/s0896-6273(03)00330-1. [DOI] [PubMed] [Google Scholar]
- Wu MF, Gulyani SA, Yau E, Mignot E, Phan B, Siegel JM. Locus coeruleus neurons: cessation of activity during cataplexy. Neuroscience. 1999;91:1389–1399. doi: 10.1016/s0306-4522(98)00600-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MF, John J, Boehmer LN, Yau D, Nguyen GB, Siegel JM. Activity of dorsal raphe cells across the sleep-waking cycle and during cataplexy in narcoleptic dogs. J Physiol. 2004;554:202–215. doi: 10.1113/jphysiol.2003.052134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitzer JM, Buckmaster CL, Parker KJ, Hauck CM, Lyons DM, Mignot E. Circadian and homeostatic regulation of hypocretin in a primate model: implications for the consolidation of wakefulness. J Neurosci. 2003;23:3555–3560. doi: 10.1523/JNEUROSCI.23-08-03555.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.