Abstract
The base excision repair (BER) pathway coordinates the replacement of 1 to 10 nucleotides at sites of single-base lesions. This process generates DNA substrates with various gap sizes which can alter the catalytic efficiency and fidelity of a DNA polymerase during gap-filling DNA synthesis. Here, we quantitatively determined the substrate specificity and base substitution fidelity of human DNA polymerase λ (Pol λ), an enzyme proposed to support the known BER DNA polymerase β (Pol β), as it filled 1- to 10-nucleotide gaps at 1-nucleotide intervals. Pol λ incorporated a correct nucleotide with relatively high efficiency until the gap size exceeded 9 nucleotides. Unlike Pol λ, Pol β did not have an absolute threshold on gap size as the catalytic efficiency for a correct dNTP gradually decreased as the gap size increased from 2 to 10 nucleotides and then recovered for non-gapped DNA. Surprisingly, an increase in gap size resulted in lower polymerase fidelity for Pol λ, and this downregulation of fidelity was controlled by its non-enzymatic N-terminal domains. Overall, Pol λ was up to 160-fold more error-prone than Pol β, thereby suggesting Pol λ would be more mutagenic during long gap-filling DNA synthesis. In addition, dCTP was the preferred misincorporation for Pol λ and its N-terminal domain truncation mutants. This nucleotide preference was shown to be dependent upon the identity of the adjacent 5′-template base. Our results suggested that both Pol λ and Pol β would catalyze nucleotide incorporation with the highest combination of efficiency and accuracy when the DNA substrate contains a single-nucleotide gap. Thus, Pol λ, like Pol β, is better suited to catalyze gap-filling DNA synthesis during short-patch BER in vivo, although, Pol λ may play a role in long-patch BER.
Keywords: Base excision repair, X-family DNA polymerase, DNA polymerase λ, DNA polymerase β, pre-steady state kinetics
1. INTRODUCTION
Base excision repair (BER) is the major pathway to repair single-base lesions in mammalian cells [1]. BER can proceed through the subpathways of short-patch (SP) or long-patch (LP) which result in the replacement and resynthesis of 1 or 2–10 nucleotides, respectively. For both of these subpathways, the damaged DNA base is recognized and removed by a DNA glycosylase, the DNA backbone is cleaved by apurinic/apyrimidinic endonuclease 1, nucleotide(s) is(are) incorporated by a DNA polymerase, and eventually the nicked DNA is sealed by a DNA ligase. However, these subpathways have several distinct variations: (i) one branch of LP-BER depends on the proliferating cell nuclear antigen (PCNA) [2–5], (ii) DNA polymerases β [2, 4, 6–15], δ [5, 8, 9, 16, 17], ε [5, 16], λ [18–22], ι [23–25], and θ [26, 27] are implicated in nuclear gap-filling and/or strand-displacement DNA synthesis, (iii) the 5′-deoxyribose phosphate (dRP) moiety is removed by the dRP lyase activity of a DNA polymerase in SP-BER [12, 28], and (iv) the displaced DNA strand containing the dRP group is cleaved by flap endonuclease 1 in LP-BER [9, 16]. In regards to the DNA synthesis step of BER, it remains unclear which polymerase is specifically involved in repairing each type of damaged DNA base and the preferred BER route. Clearly, each DNA polymerase functioning in BER must be able to coordinate interactions with multiple DNA repair proteins and to catalyze DNA synthesis using DNA substrates that vary in structure (e.g. gap size or DNA flap).
Pol β, a X-family DNA polymerase, is known to play a central role in SP-BER in vivo [6, 7] and is a strong candidate for functioning in LP-BER, likely independent of PCNA [15]. Pol λ, a X-family member that shares 34% sequence identity with Pol β [29], is postulated to complement or to support the in vivo function of Pol β in BER. This proposal is based on several observations: Pol λ possesses two key enzymatic activities (dRPase and gap-filling polymerase) that are required for SP-BER [18], Pol λ can repair uracil in a DNA duplex when substituted for Pol β in an in vitro BER reconstitution system [18], Pol λ is active in mediating BER when Pol β is absent or neutralized in cell extract from mouse embryonic fibroblasts [20], and Pol λ is involved in repairing oxidative DNA damage [19, 22]. Unlike Pol β, Pol λ has two non-enzymatic domains at its N-terminus: a breast cancer susceptibility gene 1 C-terminal (BRCT) domain and a proline-rich domain (Figure 1). Interestingly, the proline-rich domain has been shown to suppress polymerase activity [30], to limit strand displacement synthesis [31], and to increase polymerase fidelity on a single-nucleotide gapped DNA substrate [32]. In addition, DNA structure affects the polymerization efficiency of Pol λ [33] and Pol β [34–36]. Both enzymes prefer a single-nucleotide gapped DNA substrate with a 5′-phosphate on the downstream strand over a non-gapped primer-template DNA substrate. Based on the ternary crystal structures of Pol β [37] and a truncated form of Pol λ [38] in complex with gapped DNA and an incoming nucleotide, this substrate preference is likely due to the favorable interactions between the 5′-phosphate of the downstream strand and the dRPase domain. During BER, the DNA gap size may vary from 1–10 nucleotides with 2–4 being the most common in LP-BER [2, 9, 16, 17]. An increase in gap size would likely disrupt protein interactions with the downstream strand. Therefore, the incorporation efficiency and fidelity of a gap-filling DNA polymerase may depend on gap size. To better understand the kinetic relationship between gap size and polymerase function in BER, we have employed single-turnover kinetic assays to determine the polymerization efficiency and fidelity of Pol λ and Pol β incorporating dNTPs into DNA substrates with various gap sizes. In addition, we discerned the roles of the BRCT and proline-rich domains on gap-filling DNA synthesis catalyzed by Pol λ as well as the effects of the DNA template sequence on its nucleotide selection.
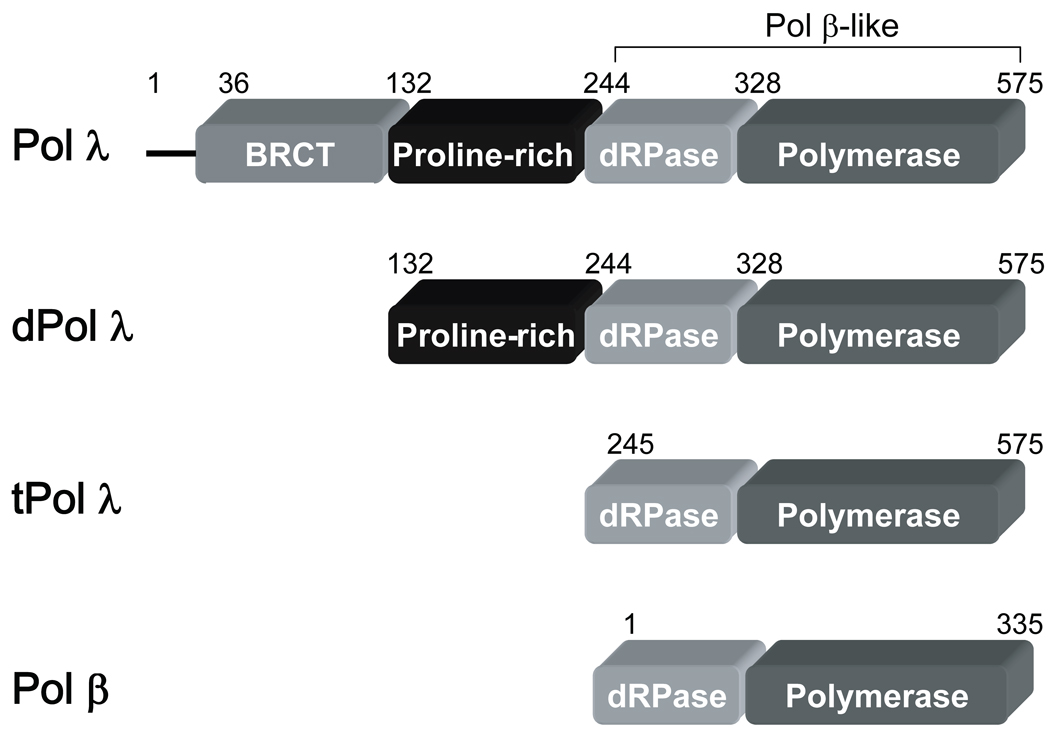
Figure 1. Domains of Pol λ, dPol λ, tPol λ, and Pol β.
Each domain is labeled in the rectangular box and the residue numbers are noted above. N-terminal residues 1–35 of Pol λ contain a nuclear localization sequence.
2. MATERIALS AND METHODS
2.1 Materials
Biochemicals and reagents were purchased from the following companies: [γ-32P]ATP, MP Biomedicals; deoxyribonucleotide-5′-triphosphates, GE Healthcare; Bio-Spin 6 columns, Bio-Rad Laboratories; OptiKinase™, USB Corporation; synthetic oligodeoxyribonucleotides 21-mer, 5′-phosphorylated 19-mers, and 41- to 50-mers, Integrated DNA Technologies.
2.2 Mutagenesis, expression and purification of DNA polymerases
Deletions, expression, and purification of wild-type human Pol λ (1–575), dPol λ (132–575), tPol λ (245–575), and human Pol β were described previously [32, 39, 40].
2.3 Single-nucleotide gapped DNA substrates
Commercially synthesized oligomers in Table 1 were purified using denaturing polyacrylamide gel electrophoresis. The 21-mer primer was radiolabeled with [γ-32P]ATP and OptiKinase™ and the unreacted [γ-32P]ATP was removed using a Bio-Spin 6 column; both steps were completed according to each of the manufacturers’s protocol. The gapped DNA substrates were prepared by mixing the 5′-[32P]-radiolabeled 21-mer, the appropriate non-radiolabeled downstream strand 19-mer, and the appropriate template (41- to 50-mer) at a 1:1.25:1.15 molar ratio, respectively. The primer-template DNA substrate was prepared by mixing 5′-[32P]-radiolabeled 21-mer and the 41-mer template at a 1:1.15 molar ratio. Then, the annealing mixture was denatured at 95 °C for 6 minutes and slowly cooled to room temperature over several hours.
Table 1.
DNA substrates*
| 21-19/41-mer |
5′-CGCAGCCGTCCAACCAACTCA pCGTCGATCCAATGCCGTCC-3′ 3′-GCGTCGGCAGGTTGGTTGAGTCGCAGCTAGGTTACGGCAGG-5′ |
| 21-19/42-mer |
5′-CGCAGCCGTCCAACCAACTCA pCGTCGATCCAATGCCGTCC-3′ 3′-GCGTCGGCAGGTTGGTTGAGTCGGCAGCTAGGTTACGGCAGG-5′ |
| 21-19/43-mer |
5′-CGCAGCCGTCCAACCAACTCA pCGTCGATCCAATGCCGTCC-3′ 3′-GCGTCGGCAGGTTGGTTGAGTCGAGCAGCTAGGTTACGGCAGG-5′ |
| 21-19/44-mer |
5′-CGCAGCCGTCCAACCAACTCA pCGTCGATCCAATGCCGTCC-3′ 3′-GCGTCGGCAGGTTGGTTGAGTCGAGGCAGCTAGGTTACGGCAGG-5′ |
| 21-19/45-mer |
5′-CGCAGCCGTCCAACCAACTCA pCGTCGATCCAATGCCGTCC-3′ 3′-GCGTCGGCAGGTTGGTTGAGTCGAGTGCAGCTAGGTTACGGCAGG-5′ |
| 21-19/46-mer |
5′-CGCAGCCGTCCAACCAACTCA pCGTCGATCCAATGCCGTCC-3′ 3′-GCGTCGGCAGGTTGGTTGAGTCGAGTGGCAGCTAGGTTACGGCAGG-5′ |
| 21-19/47-mer |
5′-CGCAGCCGTCCAACCAACTCA pCGTCGATCCAATGCCGTCC-3′ 3′-GCGTCGGCAGGTTGGTTGAGTCGAGTGCGCAGCTAGGTTACGGCAGG-5′ |
| 21-19/48-mer |
5′-CGCAGCCGTCCAACCAACTCA pCGTCGATCCAATGCCGTCC-3′ 3’-GCGTCGGCAGGTTGGTTGAGTCGAGTGCGGCAGCTAGGTTACGGCAGG-5′ |
| 21-19/49-mer |
5′-CGCAGCCGTCCAACCAACTCA pCGTCGATCCAATGCCGTCC-3′ 3′-GCGTCGGCAGGTTGGTTGAGTCGAGTGCGAGCAGCTAGGTTACGGCAGG-5′ |
| 21-19/50-mer |
5′-CGCAGCCGTCCAACCAACTCA pCGTCGATCCAATGCCGTCC-3′ 3′-GCGTCGGCAGGTTGGTTGAGTCGAGTGCGACGCAGCTAGGTTACGGCAGG-5′ |
| 21-19T/42- merCGA |
5′-CGCAGCCGTCCAACCAACTCA pTGTCGATCCAATGCCGTCC-3′ 3′-GCGTCGGCAGGTTGGTTGAGTCGACAGCTAGGTTACGGCAGG-5′ |
| 21-19/42- merCAG |
5′-CGCAGCCGTCCAACCAACTCA pCGTCGATCCAATGCCGTCC-3′ 3′-GCGTCGGCAGGTTGGTTGAGTCAGCAGCTAGGTTACGGCAGG-5′ |
| 21-19/47- merCAT |
5′-CGCAGCCGTCCAACCAACTCA pCGTCGATCCAATGCCGTCC-3′ 3′-GCGTCGGCAGGTTGGTTGAGTCATGTGCGCAGCTAGGTTACGGCAGG-5′ |
Nucleotides located in the gap are in bold, and those that were inserted to expand the gap size are underlined. p denotes the 5′-end is phosphorylated.
2.4 Measurement of the kp and Kd for single nucleotide incorporation assay
Kinetic assays were completed using optimized buffer L (50 mM Tris-Cl, pH 8.4 at 37 °C, 5 mM MgCl2, 100 mM NaCl, 0.1 mM EDTA, 5 mM DTT, 10% glycerol, and 0.1 mg/ml BSA) for Pol λ [39] and buffer B (50 mM Tris-Cl, pH 7.8 at 37 °C, 5 mM MgCl2, 50 mM NaCl, 0.1 mM EDTA, 5 mM DTT, 10% glycerol, and 0.1 mg/ml BSA) for Pol β. All kinetic experiments described herein were performed at 37 °C and the reported concentrations were final after mixing all the components. A pre-incubated solution containing Pol λ (120 nM) or Pol β (300 nM) and a DNA substrate (30 nM) was mixed with increasing concentrations (0.2–1500 µM) of nucleotide in the appropriate buffer at 37 °C. Aliquots of the reaction mixtures were quenched at various times using 0.37 M EDTA. A rapid chemical-quench flow apparatus (KinTek) was utilized for fast nucleotide incorporations. Reaction products were resolved using sequencing gel electrophoresis (17% acrylamide, 8 M urea) and quantitated with a Typhoon TRIO (GE Healthcare). The time course of product formation at each nucleotide concentration was fit to a single-exponential equation (Equation 1) using a nonlinear regression program, KaleidaGraph (Synergy Software), to yield an observed rate constant of nucleotide incorporation (kobs). The kobsvalues were then plotted as a function of nucleotide concentration and a hyperbolic equation (Equation 2) was applied to resolve the kp and apparent Kd values for nucleotide incorporation catalyzed by Pol λ or Pol β.
| Equation 1 |
| Equation 2 |
3. RESULTS
3.1 Effect of gap size on the kinetics of Pol λ
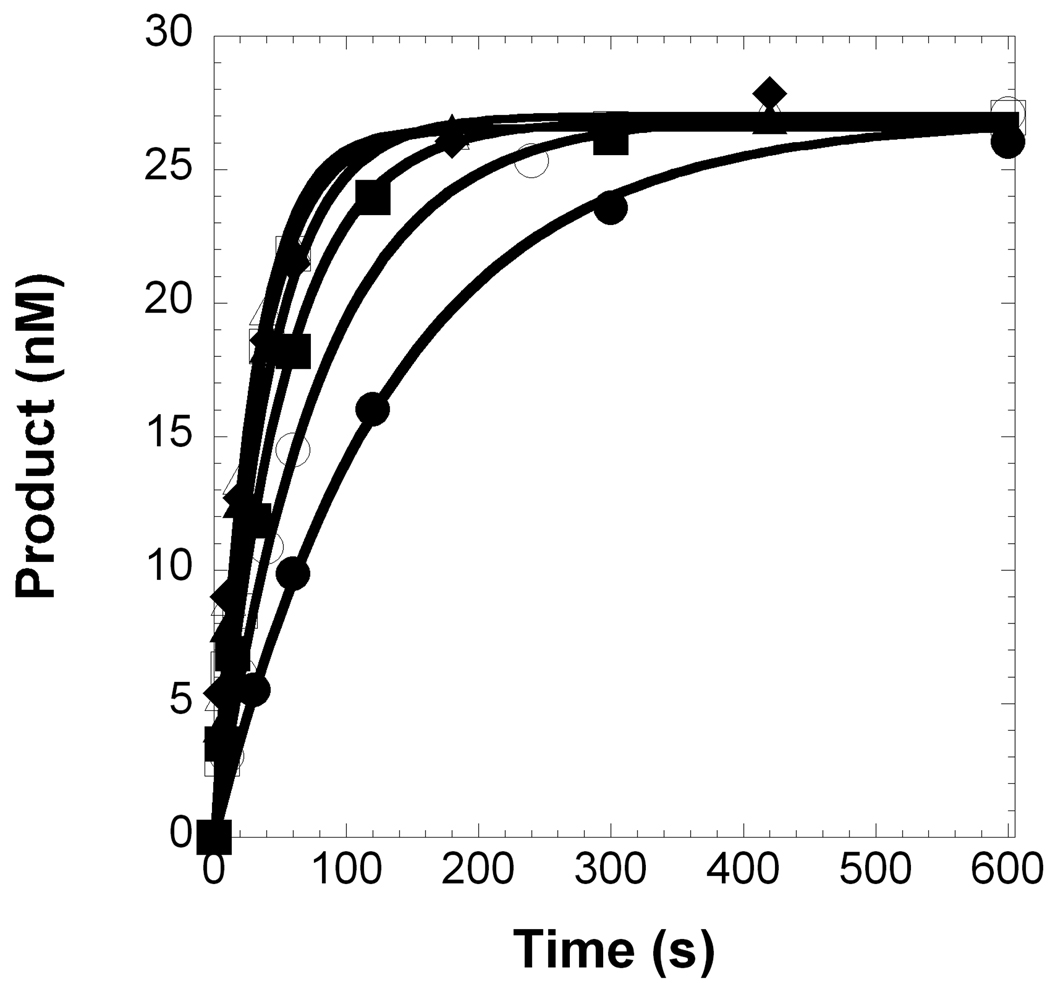
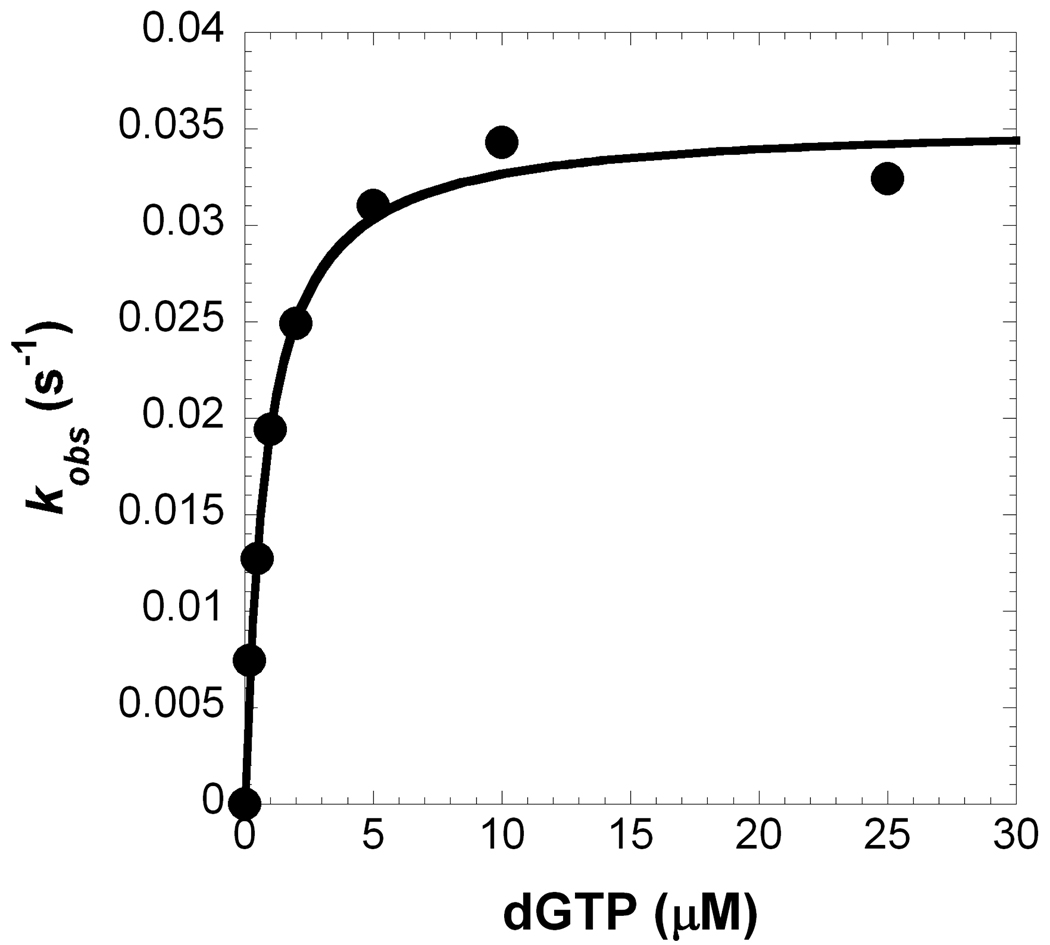
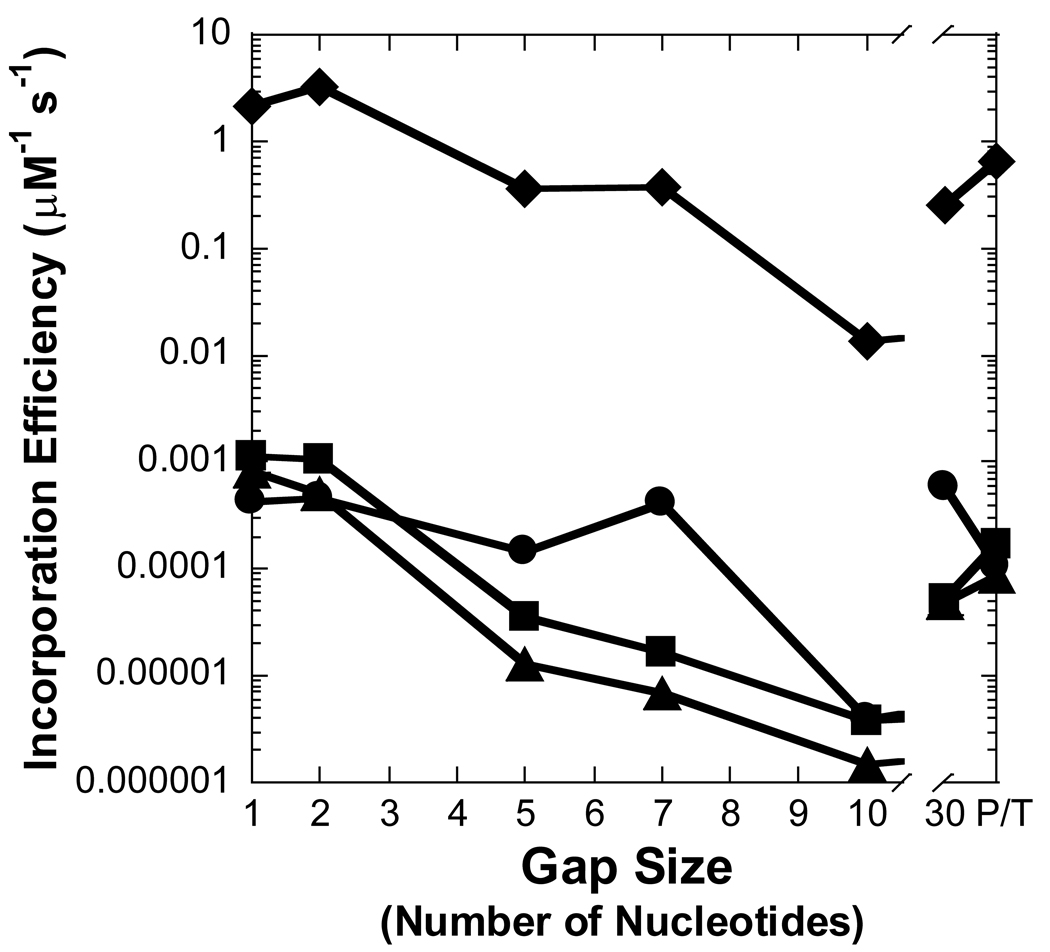
The incorporation efficiency (kp/Kd) of Pol λ inserting a dNTP opposite template dC with various DNA substrates (Table 1) was measured under single-turnover conditions (see Section 2.4). As a representative example, Pol λ catalyzed the incorporation of dGTP into a 21/41-mer DNA substrate, and a hyperbolic dependence on the observed rate constant was observed with increasing concentrations of dGTP, thereby yielding a maximum rate of nucleotide incorporation (kp) equal to 0.0353 ± 0.0008 s−1 and an apparent equilibrium dissociation constant (Kd) of the Pol λ•DNA•dGTP complex equal to 0.82 ± 0.08 µM (Figure 2 and Table 2). Similar assays were performed for correct and incorrect nucleotide incorporations into gapped and non-gapped DNA, and the kinetic parameters, incorporation efficiency, efficiency ratio relative to a gap size of 1, and base substitution fidelity are provided in Table 2. Compared to DNA with a gap size of 1, the absence of a downstream strand decreased the incorporation efficiency of a correct dGTP by 33-fold for Pol λ. This result suggested that there is a threshold on the DNA gap size for maintaining efficient catalysis. Therefore, to identify the maximum gap width for efficient gap-filling synthesis by Pol λ, the gap size was expanded by inserting nucleotides into the DNA template so that the lengths of the 21-mer primer and 19-mer strand were held constant, the template base was dC, and the first downstream base was dG (Table 1). By increasing the gap size at 1-nucleotide intervals, the maximum gap width was determined to be 9 nucleotides for a correct dNTP to be inserted relatively efficient by Pol λ, since the incorporation efficiency for a gap size of 10 was reduced by 38-fold, a level comparable to the non-gapped DNA substrate (Table 2 and Figure 3A). This kinetic effect between 1- and 10-nucleotide gapped DNA was due to a 100-fold drop in kp, since the Kd remained within 3-fold (Table 2).
Figure 2. Single-turnover kinetic parameters of nucleotide incorporation.
(A) A pre-incubated solution of Pol λ (120 nM) and 5′-[32P]-labeled 21/41-mer (30 nM) was rapidly mixed with increasing concentrations of dGTP•Mg2+ (0.2 µM, ●; 0.5 µM, ○; 1 µM, ■; 2 µM, □; 5 µM, ▲; 10 µM, △; 25 µM, ◆) for various time intervals. The solid lines are the best fits to a single-exponential equation which determined the observed rate constants, kobs. (B) The kobs values were plotted as a function of dGTP concentration. The data (●) were then fit to a hyperbolic equation, yielding a kp of 0.0353 ± 0.0008 s−1 and a Kd of 0.82 ± 0.08 µM.
Table 2.
Kinetic parameters for nucleotide incorporation into gapped or recessed DNA catalyzed by Pol λ at 37 °C.
| dNTP | kp(s−1) | Kd(µM) | kp/Kd(µM−1s−1) | Efficiency ratioa |
Fidelityb |
|---|---|---|---|---|---|
| 21-19/41-mer (1-nucleotide gap) | |||||
| dGTP | 2.7 ± 0.1 | 1.9 ± 0.2 | 1.4 | - | |
| dCTP | 0.00145 ± 0.00005 | 1.0 ± 0.1 | 1.5 × 10−3 | - | 1.0 × 10−3 |
| dATP | 0.00047 ± 0.00002 | 0.9 ± 0.1 | 5.2 × 10−4 | - | 3.7 × 10−4 |
| dTTP | 0.00135 ± 0.00009 | 2.9 ± 0.6 | 4.7 × 10−4 | - | 3.3 × 10−4 |
| 21-19/42-mer (2-nucleotide gap) | |||||
| dGTP | 1.77 ± 0.02 | 1.51 ± 0.06 | 1.2 | 1 | |
| dCTP | 0.0161 ± 0.0004 | 0.69 ± 0.08 | 2.3 × 10−2 | 16 ↑ | 2.0 × 10−2 |
| dATP | 0.00037 ± 0.00002 | 1.2 ± 0.2 | 3.1 × 10−4 | 2 ↑ | 2.6 × 10−4 |
| dTTP | 0.00070 ± 0.00003 | 3.8 ± 0.5 | 1.8 × 10−4 | 3 ↑ | 1.6 × 10−4 |
| 21-19/43-mer (3-nucleotide gap) | |||||
| dGTP | 1.99 ± 0.06 | 1.5 ± 0.2 | 1.3 | 1 | |
| dCTP | 0.0079 ± 0.0002 | 0.79 ± 0.08 | 1.0 × 10−2 | 7 ↑ | 7.5 × 10−3 |
| dATP | 0.00044 ± 0.00003 | 0.6 ± 0.2 | 7.3 × 10−4 | 1 | 5.5 × 10−4 |
| dTTP | 0.00095 ± 0.00008 | 2.8 ± 0.8 | 3.4 × 10−4 | 1 | 2.6 × 10−4 |
| 21-19/44-mer (4-nucleotide gap) | |||||
| dGTP | 1.64 ± 0.03 | 1.6 ± 0.1 | 1.0 | 1 | |
| dCTP | 0.0247 ± 0.0006 | 0.8 ± 0.1 | 3.1 × 10−2 | 21 ↑ | 2.9 × 10−2 |
| dATP | 0.00126 ± 0.00004 | 0.48 ± 0.07 | 2.6 × 10−3 | 5 ↑ | 2.6 × 10−3 |
| dTTP | 0.00129 ± 0.00007 | 2.8 ± 0.5 | 4.6 × 10−4 | 1 | 4.5 × 10−4 |
| 21-19/45-mer (5-nucleotide gap) | |||||
| dGTP | 2.15 ± 0.06 | 1.6 ± 0.2 | 1.3 | 1 | |
| dCTP | 0.053 ± 0.001 | 0.9 ± 0.1 | 5.9 × 10−2 | 41 ↑ | 4.2 × 10−2 |
| dATP | 0.0031 ± 0.0001 | 0.54 ± 0.07 | 5.7 × 10−3 | 11 ↑ | 4.3 × 10−3 |
| dTTP | 0.0118 ± 0.0006 | 2.7 ± 0.4 | 4.4 × 10−3 | 9 ↑ | 3.2 × 10−3 |
| 21-19/46-mer (6-nucleotide gap) | |||||
| dGTP | 1.53 ± 0.06 | 0.7 ± 0.1 | 2.2 | 2 ↑ | |
| dCTP | 0.0450 ± 0.0008 | 0.79 ± 0.07 | 5.7 × 10−2 | 39 ↑ | 2.5 × 10−2 |
| dATP | 0.00149 ± 0.00002 | 0.46 ± 0.03 | 3.2 × 10−3 | 6 ↑ | 1.5 × 10−3 |
| dTTP | 0.0115 ± 0.0003 | 2.0 ± 0.2 | 5.8 × 10−3 | 12 ↑ | 2.6 × 10−3 |
| 21-19/47-mer (7-nucleotide gap) | |||||
| dGTP | 1.86 ± 0.04 | 1.2 ± 0.1 | 1.6 | 1 | |
| dCTP | 0.049 ± 0.001 | 0.9 ± 0.1 | 5.4 × 10−2 | 38 ↑ | 3.4 × 10−2 |
| dATP | 0.00066 ± 0.00001 | 0.24 ± 0.02 | 2.8 × 10−3 | 5 ↑ | 1.8 × 10−3 |
| dTTP | 0.0051 ± 0.0002 | 1.1 ± 0.2 | 4.6 × 10−3 | 10 ↑ | 3.0 × 10−3 |
| 21-19/48-mer (8-nucleotide gap) | |||||
| dGTP | 1.01 ± 0.03 | 1.6 ± 0.2 | 0.6 | 2 ↓ | |
| dCTP | 0.0082 ± 0.0002 | 0.47 ± 0.06 | 1.7 × 10−2 | 12 ↑ | 2.7 × 10−2 |
| dATP | 0.00023 ± 0.00001 | 0.8 ± 0.2 | 2.9 × 10−4 | 2 ↓ | 4.6 × 10−4 |
| dTTP | 0.00072 ± 0.00002 | 5.1 ± 0.4 | 1.4 × 10−4 | 3 ↓ | 2.2 × 10−4 |
| 21-19/49-mer (9-nucleotide gap) | |||||
| dGTP | 0.83 ± 0.03 | 3.0 ± 0.3 | 0.3 | 5 ↓ | |
| dCTP | 0.00226 ± 0.00005 | 0.38 ± 0.05 | 5.9 × 10−3 | 4 ↓ | 2.1 × 10−2 |
| dATP | No incorporation | ||||
| dTTP | No incorporation | ||||
| 21-19/50-mer (10-nucleotide gap) | |||||
| dGTP | 0.026 ± 0.001 | 0.7 ± 0.1 | 3.7 × 10−2 | 38 ↓ | |
| dCTP | 0.00164 ± 0.00007 | 1.1 ± 0.2 | 1.5 × 10−3 | 1 | 3.9 × 10−2 |
| dATP | No incorporation | ||||
| dTTP | No incorporation | ||||
| 21/41-mer (no gap) | |||||
| dGTP | 0.0353 ± 0.0008 | 0.82 ± 0.08 | 4.3 × 10−2 | 33 ↓ | |
| dCTP | 0.00044 ± 0.00003 | 3.0 ± 0.7 | 1.5 × 10−4 | 10 ↓ | 3.4 × 10−3 |
| dATP | No incorporation | ||||
| dTTP | No incorporation | ||||
An upward-pointing arrow (↑) indicates the ratio was calculated as (kp/Kd)≥2-nucleotide gap/(kp/Kd)1-nucleotide gap; a downward-pointing arrow (↓) indicates the calculation used a reciprocal of the equation as follows: (kp/Kd)1-nucleotide gap/(kp/Kd)≥2-nucleotide gap.
Calculated as (kp/Kd)incorrect/[(kp/Kd)correct + (kp/Kd)incorrect].
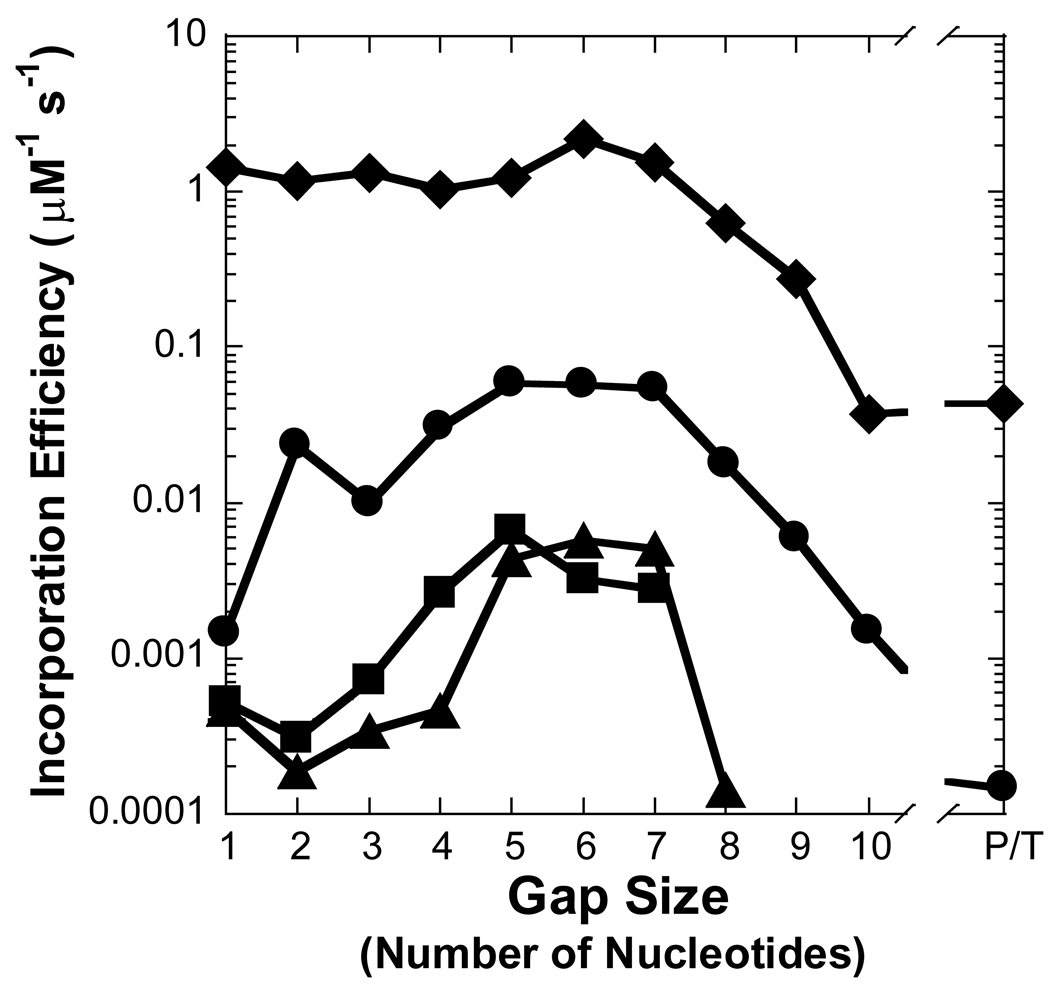
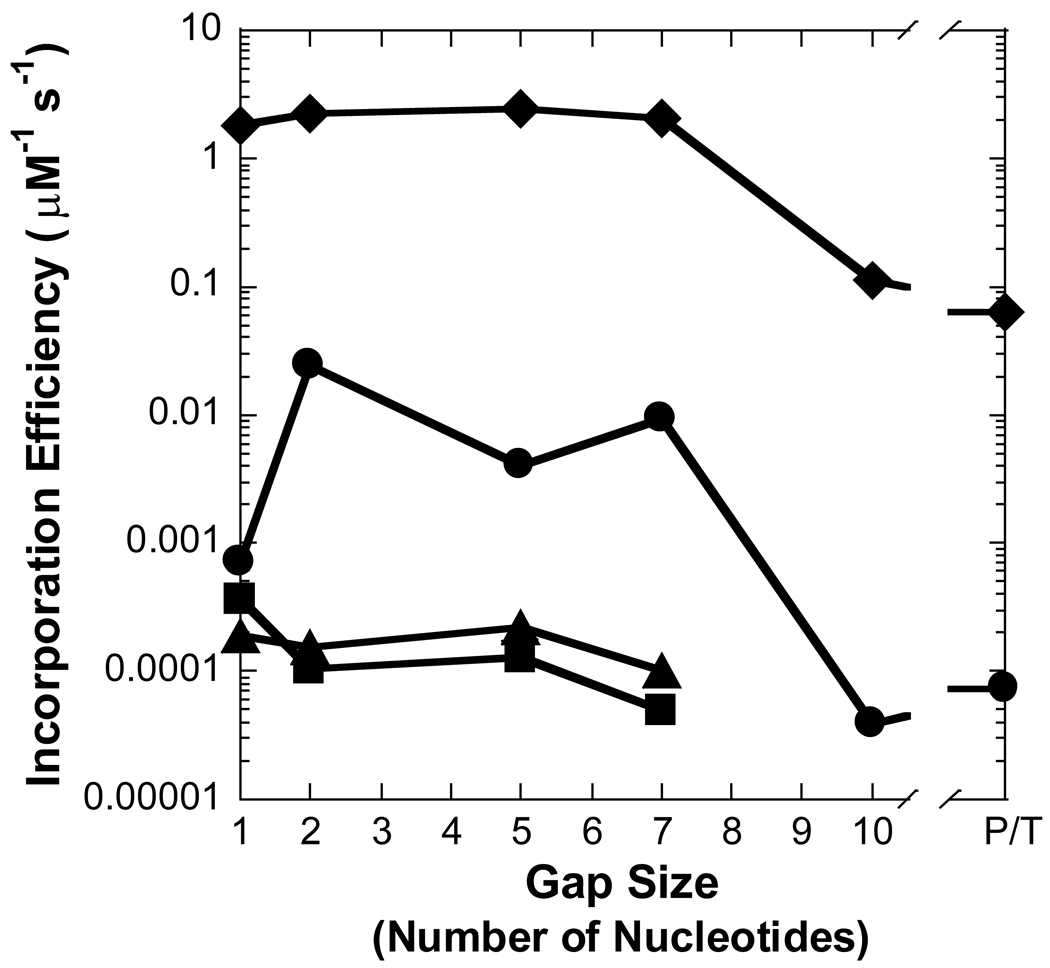
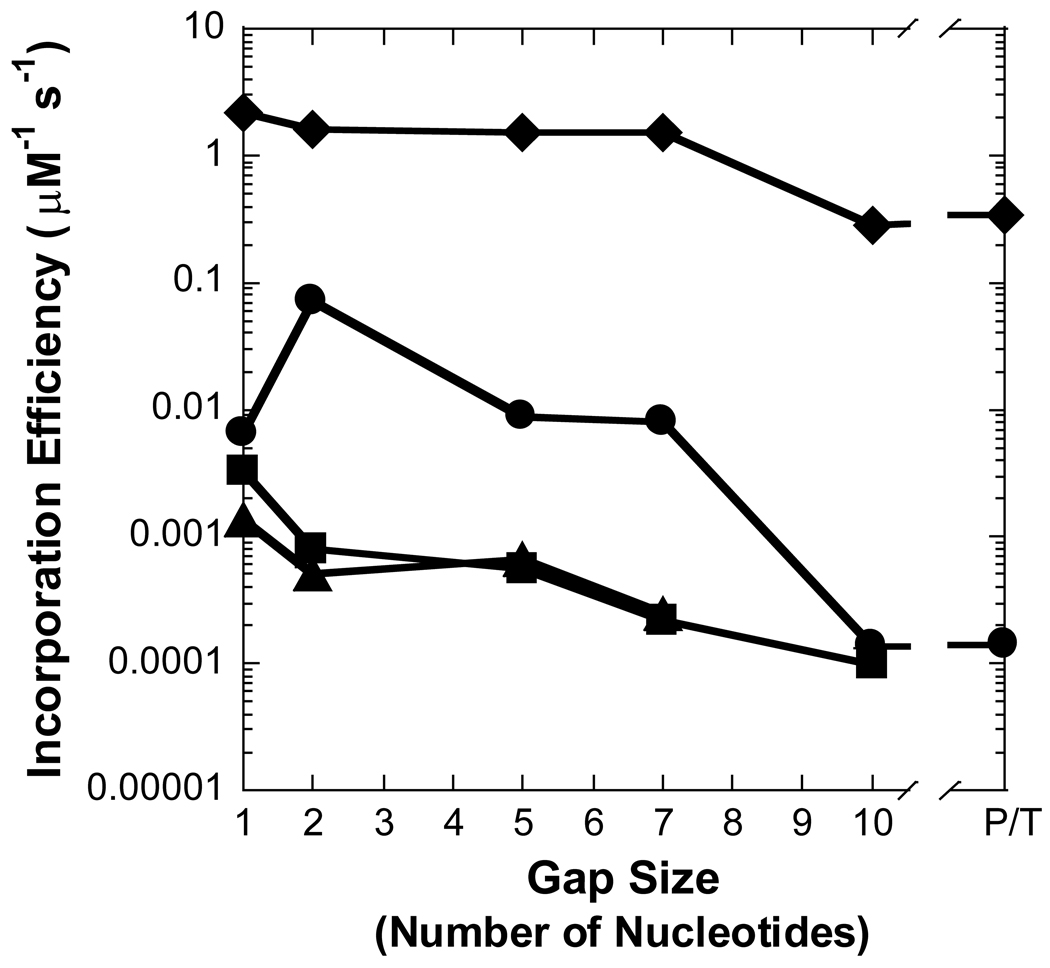
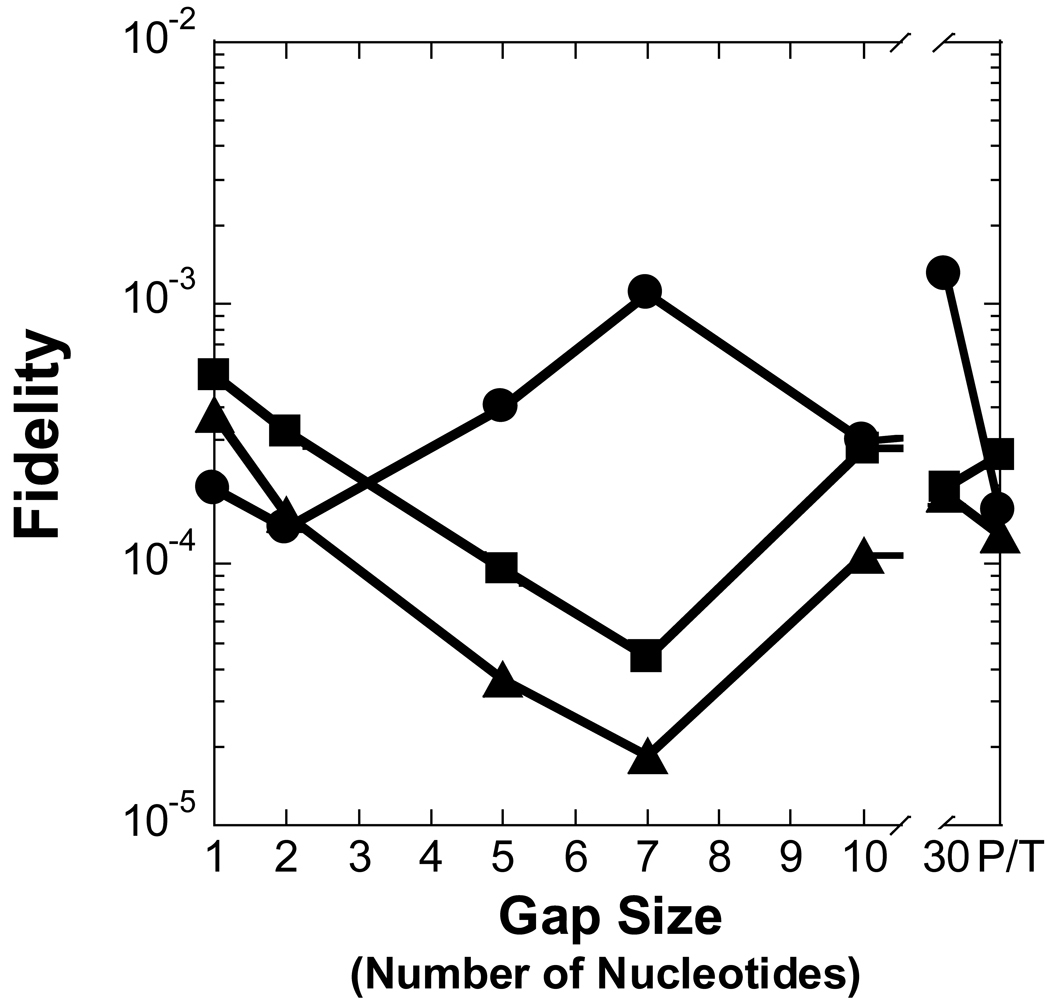
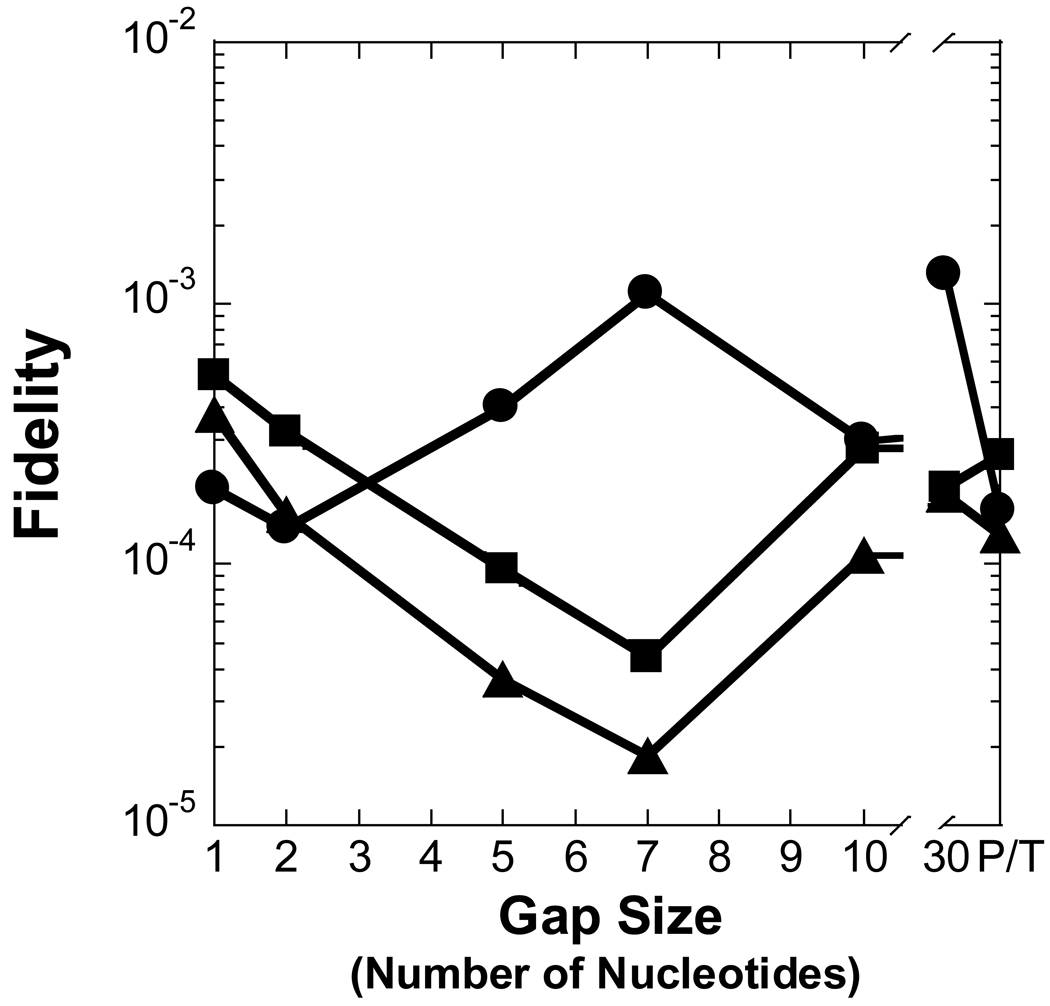
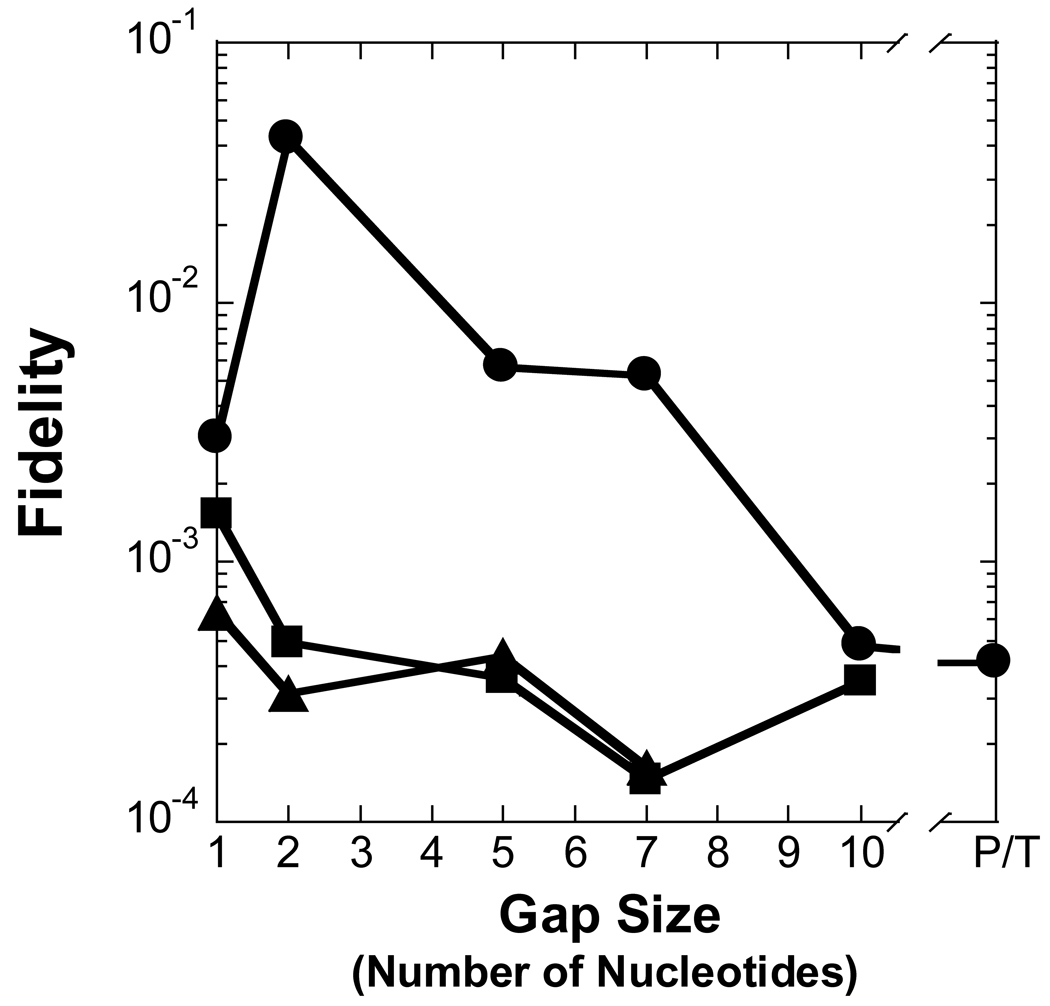
Figure 3. Effect of gap size on polymerization efficiency.
Polymerization efficiency is plotted as a function of gap size for (A) Pol λ, (B) Pol β, (C) dPol λ, and (D) tPol λ. The incoming nucleotide is represented as follows: ◆ for dGTP, ● for dCTP, ■ for dATP, and ▲ for dTTP. P/T represents data for the recessed 21/41-mer DNA substrate.
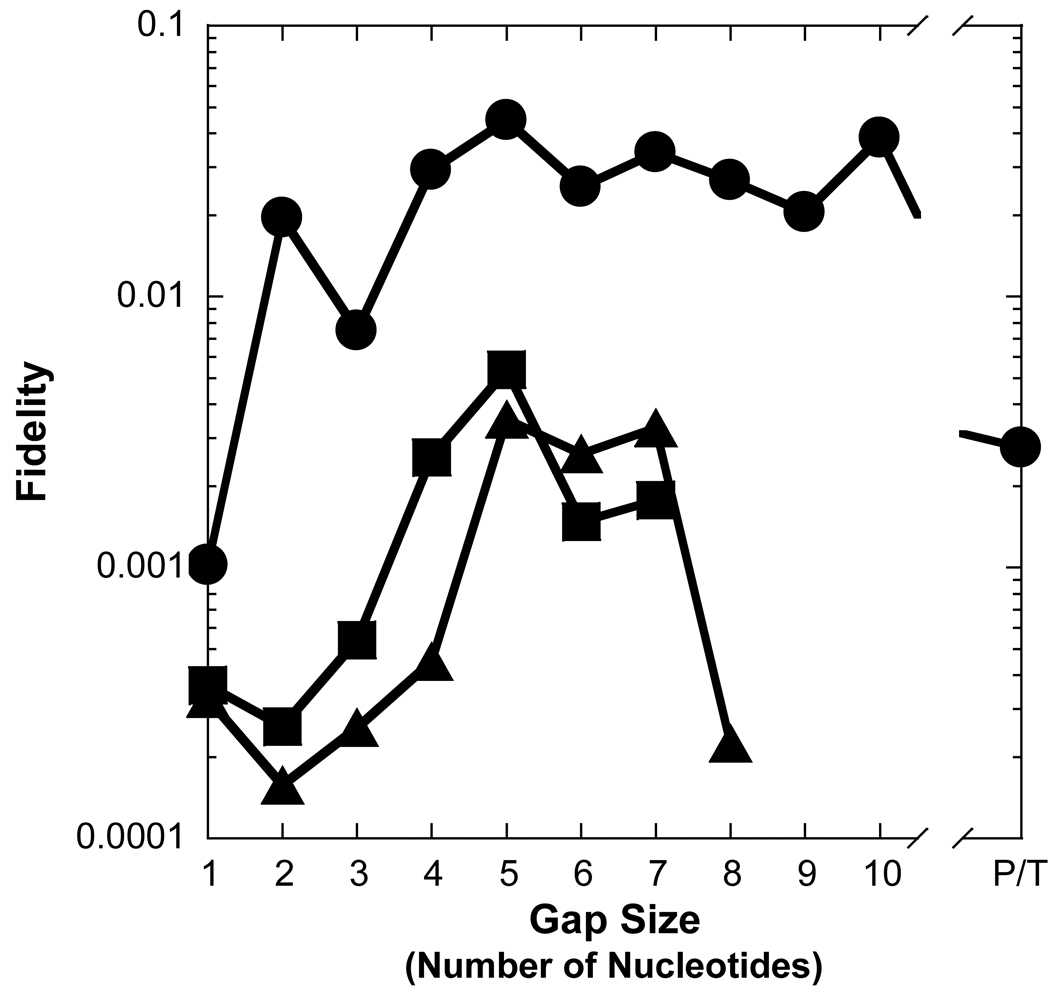
In regards to a misincorporation, the efficiency of dCTP was increased by 7- to 41-fold for gap sizes 2 to 8 when compared to a gap size of 1, thereby leading to a similar drop in polymerase fidelity (Table 2 and Figure 3A and Figure 4A). However, a different dependence on gap size was observed for incorrect dATP and dTTP. A kinetically significant increase in incorporation efficiency (5- to 12-fold) and decrease in fidelity (10−4 to 10−3) for Pol λ appeared with gap sizes of approximately 4 to 7, a narrower gap range than dCTP. The lower fidelity is due to a ∼40-fold increase in the rate of incorrect dNTP incorporation which leads to a higher incorporation efficiency. Pol λ did not incorporate dATP and dTTP for gap sizes greater than 8 nucleotides. Overall, these results suggested that the gap-filling activity of Pol λ would be efficient for gap widths of 9 nucleotides or less, although, Pol λ may be a poor candidate for LP-BER due to the higher probability of a misincorporation for gap sizes greater than 1 nucleotide. Therefore, we decided to examine the gap-filling activity of Pol β which has stronger evidence to support a biological role in LP-BER [15].
Figure 4. Effect of gap size on polymerase fidelity.
Polymerase fidelity is plotted as a function of gap size for (A) Pol λ, (B) Pol β, (C) dPol λ, and (D) tPol λ. The incoming nucleotide is represented as follows: ◆ for dGTP, ● for dCTP, ■ for dATP, and ▲ for dTTP. P/T represents data for the recessed 21/41-mer DNA substrate.
3.2 Effect of gap size on the kinetics of Pol β
The kinetic parameters were measured for Pol β inserting nucleotides into a recessed primer-template DNA substrate or gapped DNA with the selected sizes of 1, 2, 5, 7, and 10 (Table 1 and Table 3). The kinetic behavior of Pol β synthesizing DNA on the aforementioned substrates was different from Pol λ. For a correct incorporation, the catalytic efficiency of Pol β differed by a mere 3-fold for non-gapped versus single-nucleotide gapped DNA (Table 3 and Figure 3B). Surprisingly, a dramatic 160-fold reduction in the polymerase efficiency was determined with a gap size of 10 while a more modest 6-fold decrease occurred with gap sizes of 5 and 7. In contrast to Pol λ, the lower incorporation efficiency for Pol β was due to a 90-fold increase in Kdwhereas the kp was within two-fold for gap sizes of 1- and 10-nucleotides (Table 3). Like a correct dGTP incorporation, Pol β misincorporated dCTP, dATP, and dTTP with a lower catalytic efficiency as the gap size increased to 10 and then improved for non-gapped DNA. This finding suggested a “rebound effect”, whereby Pol β is capable of more efficient nucleotide incorporation when the gap size is greater than 10 nucleotides. For polymerase fidelity, a moderate 3- and 6-fold drop was observed for dCTP when the gap size was 5 and 7, respectively (Table 3 and Figure 4B). In contrast to Pol λ, the fidelity of Pol β incorporating dATP and dTTP increased up to 20-fold for gap widths greater than 5 nucleotides. Thus, these kinetic data suggested that Pol β would be less efficient but more accurate than Pol λ in LP-BER. Since different structure-function relationships were established for two similar polymerases, it was possible that the non-enzymatic BRCT and proline-rich domains may be influencing the catalytic activity of Pol λ.
Table 3.
Kinetic parameters for nucleotide incorporation into gapped or recessed DNA catalyzed by Pol β at 37 °C
| dNTP | kp(s−1) | Kd(µM) | kp/Kd(µM−1s−1) | Efficiency ratioa |
Fidelityb |
|---|---|---|---|---|---|
| 21-19/41mer (1-nucleotide gap) | |||||
| dGTP | 18.8 ± 0.4 | 8.7 ± 0.4 | 2.2 | - | |
| dCTP | 0.059 ± 0.002 | 140 ± 20 | 4.2 × 10−4 | - | 1.9 × 10−4 |
| dATP | 0.32 ± 0.02 | 280 ± 60 | 1.1 × 10−3 | - | 5.3 × 10−4 |
| dTTP | 0.27 ± 0.01 | 330 ± 40 | 8.2 × 10−4 | - | 3.7 × 10−4 |
| 21-19/42mer (2-nucleotide gap) | |||||
| dGTP | 39 ± 1 | 12 ± 2 | 3.3 | 1 | |
| dCTP | 0.0153 ± 0.0002 | 34 ± 3 | 4.5 × 10−4 | 1 | 1.4 × 10−4 |
| dATP | 0.212 ± 0.009 | 200 ± 20 | 1.1 × 10−3 | 1 | 3.3 × 10−4 |
| dTTP | 0.173 ± 0.009 | 340 ± 50 | 5.1 × 10−4 | 2 ↓ | 1.6 × 10−4 |
| 21-19/45mer (5-nucleotide gap) | |||||
| dGTP | 36 ± 2 | 100 ± 10 | 3.6 × 10−1 | 6 ↓ | |
| dCTP | 0.065 ± 0.002 | 450 ± 30 | 1.4 × 10−4 | 3 ↓ | 4.0 × 10−4 |
| dATP | 0.0188 ± 0.0006 | 540 ± 40 | 3.5 × 10−5 | 33 ↓ | 9.7 × 10−5 |
| dTTP | 0.0117 ± 0.0008 | 900 ± 100 | 1.3 × 10−5 | 63 ↓ | 3.6 × 10−5 |
| 21-19/47mer (7-nucleotide gap) | |||||
| dGTP | 37 ± 5 | 100 ± 30 | 3.7 × 10−1 | 6↓ | |
| dCTP | 0.203 ± 0.006 | 500 ± 40 | 4.1 × 10−4 | 1 | 1.1 × 10−3 |
| dATP | 0.013 ± 0.002 | 800 ± 300 | 1.6 × 10−5 | 70 ↓ | 4.4 × 10−5 |
| dTTP | 0.0096 ± 0.0005 | 1400 ± 100 | 6.9 × 10−6 | 120 ↓ | 1.9 × 10−5 |
| 21-19/50mer (10-nucleotide gap) | |||||
| dGTP | 10.5 ± 0.6 | 780 ± 90 | 1.3 × 10−2 | 160 ↓ | |
| dCTP | 0.0023 ± 0.0001 | 580 ± 60 | 4.0 × 10−6 | 110 ↓ | 2.9 × 10−4 |
| dATP | 0.00166 ± 0.00009 | 440 ± 60 | 3.8 × 10−6 | 300 ↓ | 2.8 × 10−4 |
| dTTP | 0.00090 ± 0.00006 | 620 ± 90 | 1.5 × 10−6 | 560 ↓ | 1.1 × 10−4 |
| 21/41mer (no gap) | |||||
| dGTP | 31.4 ± 0.6 | 48 ± 2 | 6.5 × 10−1 | 3 ↓ | |
| dCTP | 0.024 ± 0.001 | 230 ± 30 | 1.0 × 10−4 | 4 ↓ | 1.6 × 10−4 |
| dATP | 0.111 ± 0.007 | 650 ± 90 | 1.7 × 10−4 | 7 ↓ | 2.6 × 10−4 |
| dTTP | 0.094 ± 0.005 | 1100 ± 100 | 8.5 × 10−5 | 10 ↓ | 1.3 × 10−4 |
A downward-pointing arrow (↓) indicates the ratio was calculated as (kp/Kd)1-nucleotide gap/(kp/Kd)≥2-nucleotide gap.
Calculated as (kp/Kd)incorrect/[(kp/Kd)correct + (kp/Kd)incorrect].
3.3 Effect of non-enzymatic domains on Pol λ ’s gap-filling activity
To discern the impact of the BRCT and proline-rich domains on the catalytic activity of Pol λ, we created two truncated Pol λ mutants: dPol λ which is missing the BRCT domain and tPol λ which is missing the BRCT and proline-rich domains so that it is “Pol β-like” (Figure 1). Single-nucleotide incorporation assays were performed for dPol λ (Supplementary Table 1) and tPol λ (Supplementary Table 2) with the following set of DNA substrates: primer-template and gapped DNA with sizes of 1, 2, 5, 7, and 10 (Table 1). Like Pol λ, correct dGTP incorporation catalyzed by both Pol λ mutants remained similar for gap sizes 1, 2, 5, and 7 but decreased with a gap size of 10 or when the downstream strand was absent (Figure 3C and 3D). Interestingly, among the three Pol λ enzymes, there was a gradient for the reduced catalytic efficiency when comparing 1-nucleotide gapped DNA with non-gapped: Pol λ (33) > dPol λ (28) > tPol λ (6) (Table 2 and Supplementary Table 1 and Supplementary Table 2). Here, this gradient was closely related to the 76-, 28-, and 6-fold slower rate of dGTP incorporation for Pol λ, dPol λ, and tPol λ, respectively. Furthermore, both truncated Pol λ mutants exhibited increased incorporation efficiency (11- and 34-fold) of dCTP when the gap size was 2, thereby leading to a concomitant drop in polymerase fidelity (Figure 4C and 4D). However, beyond a gap size of 2, dPol λ continued to show improved catalytic efficiency when the gap sizes were 5 and 7 while tPol λ did not. For the dATP and dTTP misincorporations, the catalytic efficiency of both enzymes gradually decreased as the gap size increased. In general, the fidelity of both mutants for dATP increased up to 10-fold while the fidelity of dTTP remained relatively unchanged when the gap width was expanded. Together, the Pol λ results indicated that the N-terminal BRCT and proline-rich domains of Pol λ were responsible for (i) decreasing the incorporation efficiency of a correct dNTP when the gap size exceeds 9 nucleotides, (ii) enhancing misincorporations when the gap size is greater than 1 nucleotide, and (iii) downregulating polymerase fidelity when the gap size is greater than 1 nucleotide.
3.4 Effects of DNA template sequence on nucleotide preference
In general, dCTP was the preferred misincorporation for the four enzymes examined in this work. Analysis of the DNA template sequence suggested that the favorable dCTP incorporation was being instructed by the identity of the first downstream template base: dG (Table 1). For all three of the Pol λ constructs, the catalytic efficiency of dCTP misincorporation was consistently strong at a gap size of 2 (Figure 3), therefore, the 42-mer DNA template sequence was changed from 3′-CGG-5′ to 3′-CGA-5′ and 3′-CAG-5′ (Table 1). The kinetic parameters were measured for the four polymerases using the modified DNA templates (Supplementary Table 3–Supplementary Table 6). Compared to the 42-merCGG template, wild-type Pol λ and the truncated mutants displayed a reduced preference for dCTP and a greater preference for dTTP when dA was the first downstream base (Supplementary Figures 1A–C). Thus, the fidelity of Pol λ improved for dCTP but dropped for dTTP (Supplementary Figures 2A–C). Also notable is that two consecutive downstream dG bases did not strongly influence nucleotide specificities, since similar kinetic results were obtained for 42-merCGG and 42-merCGA. In regards to Pol β, the sequence-dependent effect was most pronounced at a gap size of 7 (Figure 3B and Figure 4B) so the 47-mer template was changed from 3′-CGA-5′ to 3′-CAT-5′. Like Pol λ with the 42-mers, a similar kinetic trend was observed for Pol β: dTTP misincorporation was preferred for the 47-merCAT template while dCTP was preferred for 47-merCGA (Supplementary Figure 1D and Supplementary Figure 2D). Thus, the favored misincorporation during gap-filling DNA synthesis is promoted by the base identity at the first downstream position when the gap size is 2 or greater. However, the magnitude of the misincorporation efficiency for the preferred dNTP likely depends on gap size, since Pol λ inserted dTTP with a greater catalytic efficiency into 47-merCAT than 42-merCAG (Supplementary Table 3).
4. DISCUSSION
Currently, at least six mammalian DNA polymerases (Pols β, δ, ε, λ, ι, and θ) are implicated in BER. Unfortunately, confirming their exact cellular roles in BER remains uncertain due to the functional redundancy of the polymerases, therefore, in vitro biochemical characterization has been instructive in understanding how these enzymes function in BER. This work investigated the enzymology of Pol λ and Pol β during gap-filling DNA synthesis. DNA substrates with various gap sizes were used to model DNA in SP-BER (1-nucleotide gap) and LP-BER (2- to 10-nucleotide gap). In accordance with previous studies on Pol λ and Pol β, the presence of a 5′-phosphorylated downstream strand is critical for maintaining the highest catalytic efficiency during correct nucleotide incorporation (Table 2 and Table 3) [33–36]. Examining the kinetics of nucleotide incorporation as a function of gap width at 1-nucleotide intervals for Pol λ and at key gap lengths for Pol β, dPol λ, and tPol λ revealed several important findings: (i) the gap threshold for Pol λ was 9 for a correct dNTP and as low as 7 for an incorrect dNTP, (ii) the fidelity of Pol λ dropped for gap sizes of 2 through 10 nucleotides, (iii) the BRCT and proline-rich domains downregulate the fidelity of Pol λ with the multi-nucleotide gapped DNA substrates, (iv) the preferred incorrect dNTP was influenced by the first downstream template base, and (v) for Pol β, both correct and incorrect nucleotide incorporation efficiencies dropped with an increase in gap size while its average fidelity was nearly unaffected.
4.1 Kinetic barriers for efficient nucleotide incorporation
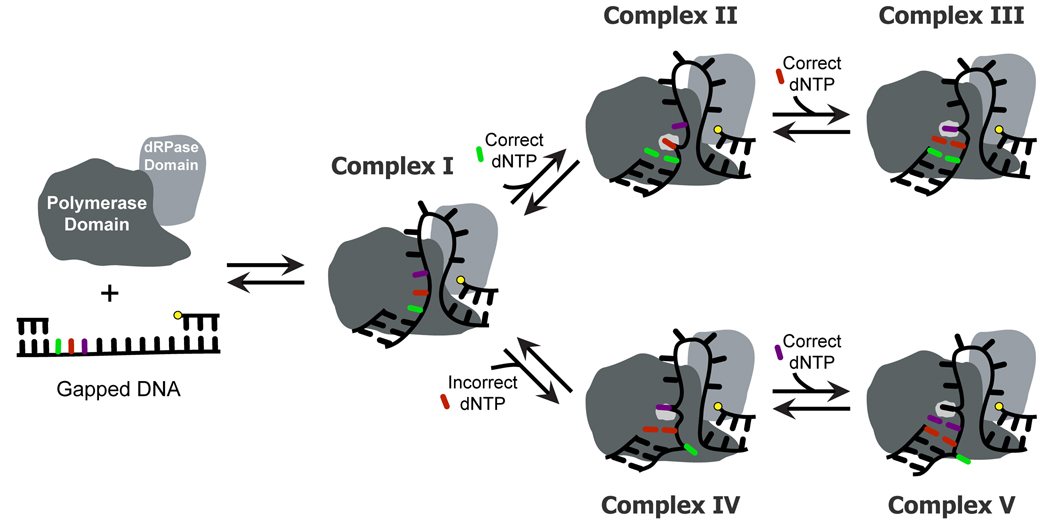
The incorporation efficiency of a correct nucleotide catalyzed by Pol λ and Pol β vary as a function of gap size (Figure 3A and 3B). Catalysis remained relatively efficient when the gap size is small, presumably due to strong interactions between the dRPase domain and the 5′-phosphorylated downstream strand based upon crystallographic evidence [37, 38]. Pol λ can remain engaged with both the 3′-hydroxyl of the upstream primer strand and 5′-phosphate of the downstream strand when the gap width is extended to 2 [41]. Interestingly, this task is achieved by a template scrunching mechanism, whereby Pol λ accommodates the first downstream template base in an extrahelical position and forms a “scrunching” binding pocket that consists of L277, H511, and R514 [41]. Since Pol λ maintained efficient catalysis of dGTP until a gap size of 10, these results supported a model in which Pol λ binds both the 3′-hydroxyl and 5′-phosphate moieties so that a DNA loop emerges between the thumb subdomain and dRPase domain when the gap is 9 nucleotides or less (Complex I in Figure 5). In this model, we speculate that the interactions between the “scrunching” binding pocket and the first downstream template base stabilize the conformation of the nascent base pair at the polymerase active site which facilitates efficient catalysis (Complex II in Figure 5). Once the intervening single-stranded DNA template surpasses 9 nucleotides, there may be an energetic penalty to accommodate such a large DNA loop, thereby preventing the polymerase domain from properly binding the 3′-hydroxyl and the nascent base pair while the dRPase domain is engaged with the 5′-phosphate. Structural studies indicate that the dRPase domain of Pol λ governs the positioning of the polymerase domain on the DNA substrate [42], therefore, sub-optimal positioning of the primer 3′-terminus at Pol λ’s active site may lead to the reduced catalytic efficiency. In addition, efficient gap-filling polymerization by Pol λ was also regulated by the non-enzymatic N-terminal domains. For a correct incorporation, the proline-rich domain was predominantly responsible for decreasing incorporation efficiency with increasing gap size (Figure 3). The crystal structure of full-length Pol λ has not been solved; therefore, it is difficult to determine how these domains alter polymerase function. Based on the dGTP incorporation efficiency differences among Pol λ, dPol λ, and tPol λ (Figure 3A, 3C, and 3D), one can postulate that there is steric interference between the protruding template loop and the N-terminal BRCT and proline-rich domains of Pol λ.
Figure 5. Model for long gap-filling DNA synthesis catalyzed by tPol λ.
Complex I shows the polymerase domain bound to the 3′-hydroxyl while the dRPase domain is bound to the 5′-phosphate (yellow dot) so that the single-stranded DNA template is looped out between the polymerase and dRPase domains. In the presence of a correct dNTP (Complex II), the first downstream template base is in the template “scrunching” binding pocket. Successive incorporations allow the downstream nucleotide that is immediately 5′ of the template base to reside in the scrunching pocket during each catalytic cycle (Complex III). In the presence of an incorrect dNTP that is complementary to the first downstream template base (Complex IV), the template base is looped out and the second downstream template base partially occupies the “scrunching” binding pocket. This complex would generate a frameshift deletion (Complex V).
Pol λ and Pol β exhibited distinct kinetic trends for the various DNA substrates which reiterated how these two X-family homologs possess different enzymatic properties [32, 43]. Pol β showed the lowest level of catalytic efficiency with a gap size of 10 but surprisingly was able to recover when the downstream strand was absent (Table 3 and Figure 3B). Previously, it has been observed that long gap-filling DNA synthesis performed by Pol β results in product accumulation at a position that corresponds to a gap size of 10 nucleotides [44]. Based on our single-turnover kinetic studies, product accumulation is likely due to the lower incorporation efficiency of Pol β. Unfortunately, there is no crystallographic evidence to show that Pol β is able to scrunch a DNA template like tPol λ. In Pol β, the analogous “scrunching” pocket residues are N37, I277, and K280, therefore, nuances likely exist in the long-patch gap-filling mechanisms of Pol β and may not follow the model for tPol λ in Figure 5. Nonetheless, Pol β may still form a binary complex similar to Complex I (Figure 5) for gapped DNA. Removing the downstream strand eliminated the steric constraints of a looped single-stranded DNA template and reduced its inhibitory effect on polymerization efficiency, thereby indicating that Pol β could achieve a more catalytically-competent ternary complex. However, such a catalytic recovery was not significant for Pol λ and its truncated mutants. Although the reason for lack of recovery is unclear, one possibility is that the absence of the interactions between the downstream primer and the dRPase domain significantly affected the conformation of the nascent base pair at Pol λ’s active site.
4.2 Pol λ was error-prone when gap size was increased
The base substitution fidelity of Pol λ dropped up to 40-fold when the gap size was increased from 1 to 10 nucleotides (Table 2). Moreover, the fidelity of Pol λ was regulated by its non-enzymatic N-terminal domains to varying degrees for each incorrect dNTP (Figure 4). For 1-nucleotide gapped DNA, the proline-rich domain of Pol λ was previously shown to increase polymerase fidelity up to 100-fold [32]. In contrast, dCTP misincorporation was not affected by the N-terminal domains at a gap size of 2, as all three Pol λ constructs showed a similar drop in fidelity (Figure 4A, C, and D). Unlike Pol λ and dPol λ, tPol λ did not sustain a lower fidelity for dCTP misinsertion into gap sizes 5 and 7, thereby indicating that the proline-rich domain was responsible for downregulating the fidelity of Pol λ during long-patch gap-filling DNA synthesis. Interestingly, both the BRCT and proline-rich domains contributed to a decrease in polymerase fidelity for dATP and dTTP misincorporations into DNA gaps with widths exceeding 2 nucleotides. Thus, deleting Pol λ’s N-terminal domains resulted in an enzyme that was kinetically more Pol β-like, i.e. the fidelity remained more constant or increased as the gap size was expanded. The relationship among polymerase fidelity, gap size, and the N-terminal domains of Pol λ can be summarized as follows: the proline-rich domain upregulates Pol λ’s fidelity during short gap-filling DNA synthesis (1-nucleotide gap) while the BRCT and proline-rich domains downregulate polymerase fidelity during long gap-filling DNA synthesis.
The incorrect nucleotide with the highest probability of being inserted by Pol λ was dCTP, and this preference was shown to be sequence dependent due to the presence of a 5′-dG adjacent to the template base dC (Figure 3A and Supplementary Figure 1A). This result is indicative of a strand misalignment which leads to the template base dC being looped out so the adjacent 5′-dG template base can base pair with dCTP. Such a dNTP-stabilized strand misalignment is supported by the fact that the preferred misincorporation was correspondingly changed from dCTP to dTTP when the first downstream template base 5′-dG was changed to 5′-dA (Supplementary Table 3) and has been observed in the crystal structures of tPol λ [45, 46]. By repositioning the first downstream template base dG, the second downstream template base may occupy the “scrunching” binding pocket to further stabilize the conformation of a misaligned dCTP:dG intermediate at the active site of Pol λ (Complex IV in Figure 5). This pathway would lead to the generation of frameshift deletions (Complex V in Figure 5), a characteristic of Pol λ. Due to multiple steric constraints in the DNA template, the second downstream template base may not be oriented properly in the “scrunching” binding pocket (Complex IV in Figure 5). In addition, the conformation of Complex IV is less optimal for catalysis than Complex II (Figure 5) because the incorporation efficiency of correct dGTP is approximately two orders of magnitude greater than incorrect dCTP (Figure 3). Assimilating these kinetic and structural findings indicates that the incoming dNTP and DNA template sequence may dictate whether Pol λ forms a slipped DNA intermediate (Complex II versus IV in Figure 5). Our results suggested that upon dCTP binding Pol λ prefers the strand misalignment route to enable dCTP:dG base pair formation, since dCTP incorporation remained enhanced for gap sizes 2 through 8 (Figure 3A). Similar effects were observed for dPol λ and tPol λ, thereby indicating the BRCT and proline-rich domains did not significantly influence template misalignment (Figures 3C and 3D). Like Pol λ, both dPol λ and tPol λ misincorporated dCTP with a greater catalytic efficiency for 2-nucleotide gapped DNA than 1-nucleotide gapped DNA. For a 1-nucleotide gap, the potential to loop out the downstream template dG was likely attenuated by the annealed downstream strand. Interestingly, Figure 3B shows that dCTP misincorporation was not preferred by Pol β with most tested gap sizes. This is consistent with published results which demonstrate that Pol λ [47] has a stronger propensity to create −1 deletions than Pol β [48].
4.3 Potential roles of Pol β and Pol λ in BER
Both Pol β and Pol λ exhibit similar efficiency and accuracy for filling single-nucleotide gapped DNA (Table 2 and Table 3). This suggests that Pol λ, like Pol β, would be competent in SP-BER in vivo. Increasing gap size led to unexpected enzyme-specific properties of Pol λ and Pol β with a single-stranded DNA break. Based on polymerase fidelity (Table 2 and Table 3), Pol β would be better than Pol λ for accurately filling larger than 1-nucleotide gaps in LP-BER. However, Pol β is not ideal for this role either since its catalytic efficiency significantly decreased with increasing gap size (Table 3 and Figure 3B). Notably, gap sizes greater than 5 nucleotides may not be physiologically relevant because most gap sizes are reported to be 2–4 nucleotides [16, 17]. In addition, the gap-filling role of Pol β and Pol λ in LP-BER may be confined to 1-nucleotide gaps generated by the alternating activities of flap endonuclease 1 and Pol β if the repair pathway proceeds via a “Hit and Run” mechanism as proposed by Wilson and coworkers [49]. Although less likely, LP-BER may also follow a tandem process in which the first insertion at the repair site is performed by Pol β or Pol λ followed by strand-displacement DNA synthesis by a replicative polymerase such as Pol δ or ε [11, 14]. In addition, the mutational spectrum in BER is predominantly single-nucleotide deletions in cell extracts [50]. Therefore, Pol λ may function in LP-BER because it has a strong propensity to create −1 deletions (see 4.2). Taken together, we cannot exclude potential roles of Pol β and Pol λ in LP-BER.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health Grant GM079403 to Z.S. J.A.B. was supported by an American Heart Association Pre-doctoral Fellowship (Grant 0815382D) and a Presidential Fellowship from The Ohio State University. L.R.P. was supported by an REU supplemental grant from the National Science Foundation Career Award to Z.S. (Grant MCB-0447899).
Abbreviations
- BER
base excision repair
- BRCT
breast cancer susceptibility gene 1 C-terminal
- flap endonuclease 1
dRP, 5′-deoxyribose phosphate
- LP
long patch
- NHEJ
non-homologous end joining
- PCNA
proliferating cell nuclear antigen
- Pol β
DNA polymerase beta
- Pol δ
DNA polymerase delta
- Pol ε
DNA polymerase epsilon
- Pol λ
DNA polymerase lambda
- SP
short patch
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 2.Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox LS, Lane DP, Abbondandolo A, Dogliotti E. Two pathways for base excision repair in mammalian cells. J Biol Chem. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- 3.Biade S, Sobol RW, Wilson SH, Matsumoto Y. Impairment of proliferating cell nuclear antigen-dependent apurinic/apyrimidinic site repair on linear DNA. J Biol Chem. 1998;273:898–902. doi: 10.1074/jbc.273.2.898. [DOI] [PubMed] [Google Scholar]
- 4.Fortini P, Pascucci B, Parlanti E, Sobol RW, Wilson SH, Dogliotti E. Different DNA polymerases are involved in the short- and long-patch base excision repair in mammalian cells. Biochemistry. 1998;37:3575–3580. doi: 10.1021/bi972999h. [DOI] [PubMed] [Google Scholar]
- 5.Stucki M, Pascucci B, Parlanti E, Fortini P, Wilson SH, Hubscher U, Dogliotti E. Mammalian base excision repair by DNA polymerases delta and epsilon. Oncogene. 1998;17:835–843. doi: 10.1038/sj.onc.1202001. [DOI] [PubMed] [Google Scholar]
- 6.Singhal RK, Prasad R, Wilson SH. DNA polymerase beta conducts the gap-filling step in uracil-initiated base excision repair in a bovine testis nuclear extract. J Biol Chem. 1995;270:949–957. doi: 10.1074/jbc.270.2.949. [DOI] [PubMed] [Google Scholar]
- 7.Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 8.Nealon K, Nicholl ID, Kenny MK. Characterization of the DNA polymerase requirement of human base excision repair. Nucleic Acids Res. 1996;24:3763–3770. doi: 10.1093/nar/24.19.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klungland A, Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1) Embo J. 1997;16:3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dianov GL, Prasad R, Wilson SH, Bohr VA. Role of DNA polymerase beta in the excision step of long patch mammalian base excision repair. J Biol Chem. 1999;274:13741–13743. doi: 10.1074/jbc.274.20.13741. [DOI] [PubMed] [Google Scholar]
- 11.Podlutsky AJ, Dianova II, Podust VN, Bohr VA, Dianov GL. Human DNA polymerase beta initiates DNA synthesis during long-patch repair of reduced AP sites in DNA. Embo J. 2001;20:1477–1482. doi: 10.1093/emboj/20.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Podlutsky AJ, Dianova II, Wilson SH, Bohr VA, Dianov GL. DNA synthesis and dRPase activities of polymerase beta are both essential for single-nucleotide patch base excision repair in mammalian cell extracts. Biochemistry. 2001;40:809–813. doi: 10.1021/bi002064s. [DOI] [PubMed] [Google Scholar]
- 13.Horton JK, Baker A, Berg BJ, Sobol RW, Wilson SH. Involvement of DNA polymerase beta in protection against the cytotoxicity of oxidative DNA damage. DNA Repair (Amst) 2002;1:317–333. doi: 10.1016/s1568-7864(02)00008-3. [DOI] [PubMed] [Google Scholar]
- 14.Parlanti E, Pascucci B, Terrados G, Blanco L, Dogliotti E. Aphidicolin-resistant and -sensitive base excision repair in wild-type and DNA polymerase beta-defective mouse cells. DNA Repair (Amst) 2004;3:703–710. doi: 10.1016/j.dnarep.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 15.Asagoshi K, Liu Y, Masaoka A, Lan L, Prasad R, Horton JK, Brown AR, Wang XH, Bdour HM, Sobol RW, Taylor JS, Yasui A, Wilson SH. DNA polymerase beta-dependent long patch base excision repair in living cells. DNA Repair (Amst) 2010;9:109–119. doi: 10.1016/j.dnarep.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pascucci B, Stucki M, Jonsson ZO, Dogliotti E, Hubscher U. Long patch base excision repair with purified human proteins. DNA ligase I as patch size mediator for DNA polymerases delta and epsilon. J Biol Chem. 1999;274:33696–33702. doi: 10.1074/jbc.274.47.33696. [DOI] [PubMed] [Google Scholar]
- 17.Matsumoto Y, Kim K, Hurwitz J, Gary R, Levin DS, Tomkinson AE, Park MS. Reconstitution of proliferating cell nuclear antigen-dependent repair of apurinic/apyrimidinic sites with purified human proteins. J Biol Chem. 1999;274:33703–33708. doi: 10.1074/jbc.274.47.33703. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Diaz M, Bebenek K, Kunkel TA, Blanco L. Identification of an intrinsic 5'-deoxyribose-5-phosphate lyase activity in human DNA polymerase lambda: a possible role in base excision repair. J Biol Chem. 2001;276:34659–34663. doi: 10.1074/jbc.M106336200. [DOI] [PubMed] [Google Scholar]
- 19.Braithwaite EK, Kedar PS, Lan L, Polosina YY, Asagoshi K, Poltoratsky VP, Horton JK, Miller H, Teebor GW, Yasui A, Wilson SH. DNA polymerase lambda protects mouse fibroblasts against oxidative DNA damage and is recruited to sites of DNA damage/repair. J Biol Chem. 2005;280:31641–31647. doi: 10.1074/jbc.C500256200. [DOI] [PubMed] [Google Scholar]
- 20.Braithwaite EK, Prasad R, Shock DD, Hou EW, Beard WA, Wilson SH. DNA polymerase lambda mediates a back-up base excision repair activity in extracts of mouse embryonic fibroblasts. J Biol Chem. 2005;280:18469–18475. doi: 10.1074/jbc.M411864200. [DOI] [PubMed] [Google Scholar]
- 21.Lebedeva NA, Rechkunova NI, Dezhurov SV, Khodyreva SN, Favre A, Blanco L, Lavrik OI. Comparison of functional properties of mammalian DNA polymerase lambda and DNA polymerase beta in reactions of DNA synthesis related to DNA repair. Biochim Biophys Acta. 2005;1751:150–158. doi: 10.1016/j.bbapap.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 22.Tano K, Nakamura J, Asagoshi K, Arakawa H, Sonoda E, Braithwaite EK, Prasad R, Buerstedde JM, Takeda S, Watanabe M, Wilson SH. Interplay between DNA polymerases beta and lambda in repair of oxidation DNA damage in chicken DT40 cells. DNA Repair (Amst) 2007;6:869–875. doi: 10.1016/j.dnarep.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bebenek K, Tissier A, Frank EG, McDonald JP, Prasad R, Wilson SH, Woodgate R, Kunkel TA. 5’-Deoxyribose phosphate lyase activity of human DNA polymerase iota in vitro. Science. 2001;291:2156–2159. doi: 10.1126/science.1058386. [DOI] [PubMed] [Google Scholar]
- 24.Prasad R, Bebenek K, Hou E, Shock DD, Beard WA, Woodgate R, Kunkel TA, Wilson SH. Localization of the deoxyribose phosphate lyase active site in human DNA polymerase iota by controlled proteolysis. J Biol Chem. 2003;278:29649–29654. doi: 10.1074/jbc.M305399200. [DOI] [PubMed] [Google Scholar]
- 25.Petta TB, Nakajima S, Zlatanou A, Despras E, Couve-Privat S, Ishchenko A, Sarasin A, Yasui A, Kannouche P. Human DNA polymerase iota protects cells against oxidative stress. Embo J. 2008;27:2883–2895. doi: 10.1038/emboj.2008.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshimura M, Kohzaki M, Nakamura J, Asagoshi K, Sonoda E, Hou E, Prasad R, Wilson SH, Tano K, Yasui A, Lan L, Seki M, Wood RD, Arakawa H, Buerstedde JM, Hochegger H, Okada T, Hiraoka M, Takeda S. Vertebrate POLQ and POLbeta cooperate in base excision repair of oxidative DNA damage. Mol Cell. 2006;24:115–125. doi: 10.1016/j.molcel.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prasad R, Longley MJ, Sharief FS, Hou EW, Copeland WC, Wilson SH. Human DNA polymerase theta possesses 5'-dRP lyase activity and functions in single-nucleotide base excision repair in vitro. Nucleic Acids Res. 2009;37:1868–1877. doi: 10.1093/nar/gkp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sobol RW, Prasad R, Evenski A, Baker A, Yang XP, Horton JK, Wilson SH. The lyase activity of the DNA repair protein beta-polymerase protects from DNA-damage-induced cytotoxicity. Nature. 2000;405:807–810. doi: 10.1038/35015598. [DOI] [PubMed] [Google Scholar]
- 29.Aoufouchi S, Flatter E, Dahan A, Faili A, Bertocci B, Storck S, Delbos F, Cocea L, Gupta N, Weill JC, Reynaud CA. Two novel human and mouse DNA polymerases of the polX family. Nucleic Acids Res. 2000;28:3684–3693. doi: 10.1093/nar/28.18.3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimazaki N, Yoshida K, Kobayashi T, Toji S, Tamai K, Koiwai O. Over-expression of human DNA polymerase lambda in E. coli and characterization of the recombinant enzyme. Genes Cells. 2002;7:639–651. doi: 10.1046/j.1365-2443.2002.00547.x. [DOI] [PubMed] [Google Scholar]
- 31.Fan W, Wu X. DNA polymerase lambda can elongate on DNA substrates mimicking non-homologous end joining and interact with XRCC4-ligase IV complex. Biochem Biophys Res Commun. 2004;323:1328–1333. doi: 10.1016/j.bbrc.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Fiala KA, Duym WW, Zhang J, Suo Z. Up-regulation of the fidelity of human DNA polymerase lambda by its non-enzymatic proline-rich domain. J Biol Chem. 2006;281:19038–19044. doi: 10.1074/jbc.M601178200. [DOI] [PubMed] [Google Scholar]
- 33.Duym WW, Fiala KA, Bhatt N, Suo Z. Kinetic effect of a downstream strand and its 5'-terminal moieties on single nucleotide gap-filling synthesis catalyzed by human DNA polymerase lambda. J Biol Chem. 2006;281:35649–35655. doi: 10.1074/jbc.M607479200. [DOI] [PubMed] [Google Scholar]
- 34.Chagovetz AM, Sweasy JB, Preston BD. Increased activity and fidelity of DNA polymerase beta on single-nucleotide gapped DNA. J Biol Chem. 1997;272:27501–27504. doi: 10.1074/jbc.272.44.27501. [DOI] [PubMed] [Google Scholar]
- 35.Ahn J, Kraynov VS, Zhong X, Werneburg BG, Tsai MD. DNA polymerase beta: effects of gapped DNA substrates on dNTP specificity, fidelity, processivity and conformational changes. Biochem J. 1998;331(Pt 1):79–87. doi: 10.1042/bj3310079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vande Berg BJ, Beard WA, Wilson SH. DNA structure and aspartate 276 influence nucleotide binding to human DNA polymerase beta. Implication for the identity of the rate-limiting conformational change. J Biol Chem. 2001;276:3408–3416. doi: 10.1074/jbc.M002884200. [DOI] [PubMed] [Google Scholar]
- 37.Sawaya MR, Prasad R, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 38.Garcia-Diaz M, Bebenek K, Krahn JM, Kunkel TA, Pedersen LC. A closed conformation for the Pol lambda catalytic cycle. Nat Struct Mol Biol. 2005;12:97–98. doi: 10.1038/nsmb876. [DOI] [PubMed] [Google Scholar]
- 39.Fiala KA, Abdel-Gawad W, Suo Z. Pre-steady-state kinetic studies of the fidelity and mechanism of polymerization catalyzed by truncated human DNA polymerase lambda. Biochemistry. 2004;43:6751–6762. doi: 10.1021/bi049975c. [DOI] [PubMed] [Google Scholar]
- 40.Brown JA, Duym WW, Fowler JD, Suo Z. Single-turnover kinetic analysis of the mutagenic potential of 8-oxo-7,8-dihydro-2'-deoxyguanosine during gap-filling synthesis catalyzed by human DNA polymerases lambda and beta. J Mol Biol. 2007;367:1258–1269. doi: 10.1016/j.jmb.2007.01.069. [DOI] [PubMed] [Google Scholar]
- 41.Garcia-Diaz M, Bebenek K, Larrea AA, Havener JM, Perera L, Krahn JM, Pedersen LC, Ramsden DA, Kunkel TA. Template strand scrunching during DNA gap repair synthesis by human polymerase lambda. Nat Struct Mol Biol. 2009;16:967–972. doi: 10.1038/nsmb.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia-Diaz M, Bebenek K, Krahn JM, Blanco L, Kunkel TA, Pedersen LC. A structural solution for the DNA polymerase lambda-dependent repair of DNA gaps with minimal homology. Mol Cell. 2004;13:561–572. doi: 10.1016/s1097-2765(04)00061-9. [DOI] [PubMed] [Google Scholar]
- 43.Garcia-Diaz M, Bebenek K, Gao G, Pedersen LC, London RE, Kunkel TA. Structure-function studies of DNA polymerase lambda. DNA Repair (Amst) 2005;4:1358–1367. doi: 10.1016/j.dnarep.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 44.Singhal RK, Wilson SH. Short gap-filling synthesis by DNA polymerase beta is processive. J Biol Chem. 1993;268:15906–15911. [PubMed] [Google Scholar]
- 45.Garcia-Diaz M, Bebenek K, Krahn JM, Pedersen LC, Kunkel TA. Structural analysis of strand misalignment during DNA synthesis by a human DNA polymerase. Cell. 2006;124:331–342. doi: 10.1016/j.cell.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 46.Bebenek K, Garcia-Diaz M, Foley MC, Pedersen LC, Schlick T, Kunkel TA. Substrate-induced DNA strand misalignment during catalytic cycling by DNA polymerase lambda. EMBO Rep. 2008;9:459–464. doi: 10.1038/embor.2008.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bebenek K, Garcia-Diaz M, Blanco L, Kunkel TA. The frameshift infidelity of human DNA polymerase lambda. Implications for function. J Biol Chem. 2003;278:34685–34690. doi: 10.1074/jbc.M305705200. [DOI] [PubMed] [Google Scholar]
- 48.Osheroff WP, Jung HK, Beard WA, Wilson SH, Kunkel TA. The fidelity of DNA polymerase beta during distributive and processive DNA synthesis. J Biol Chem. 1999;274:3642–3650. doi: 10.1074/jbc.274.6.3642. [DOI] [PubMed] [Google Scholar]
- 49.Liu Y, Beard WA, Shock DD, Prasad R, Hou EW, Wilson SH. DNA polymerase beta and flap endonuclease 1 enzymatic specificities sustain DNA synthesis for long patch base excision repair. J Biol Chem. 2005;280:3665–3674. doi: 10.1074/jbc.M412922200. [DOI] [PubMed] [Google Scholar]
- 50.Chan KK, Zhang QM, Dianov GL. Base excision repair fidelity in normal and cancer cells. Mutagenesis. 2006;21:173–178. doi: 10.1093/mutage/gel020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.