Abstract
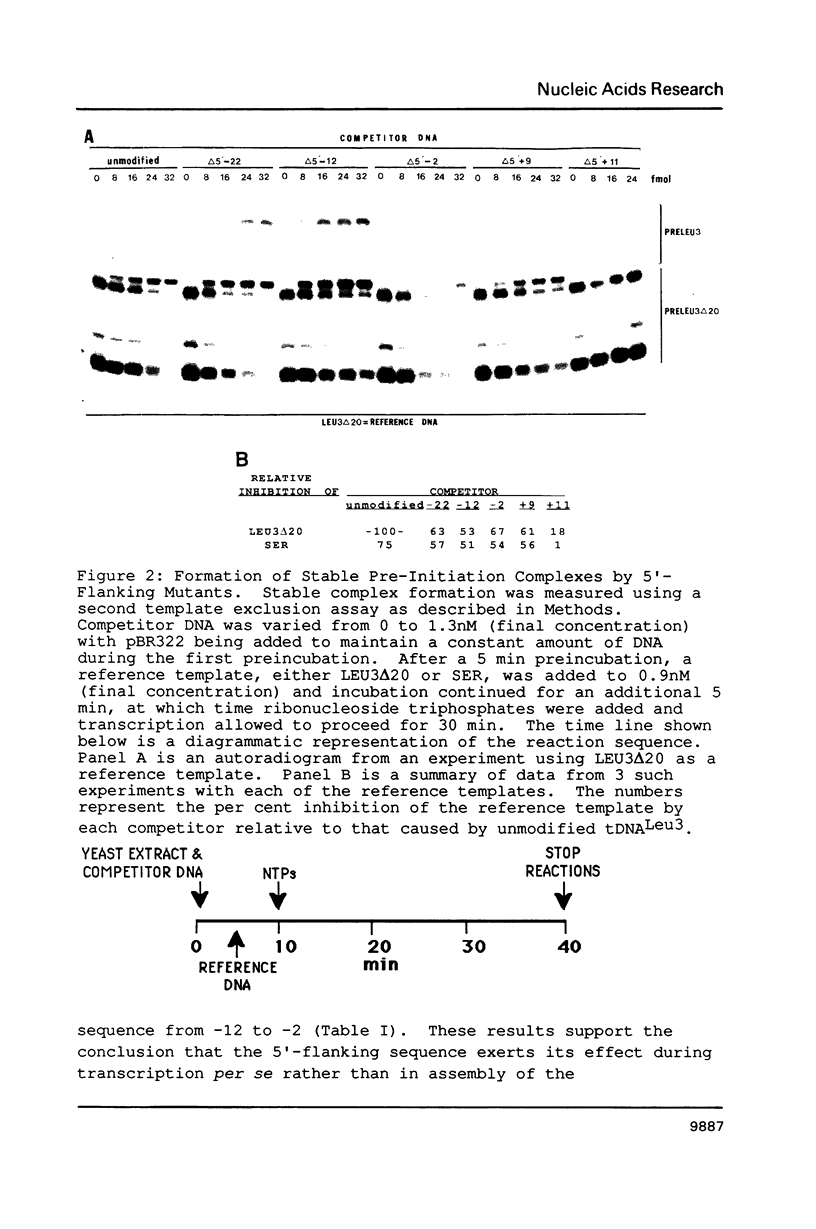
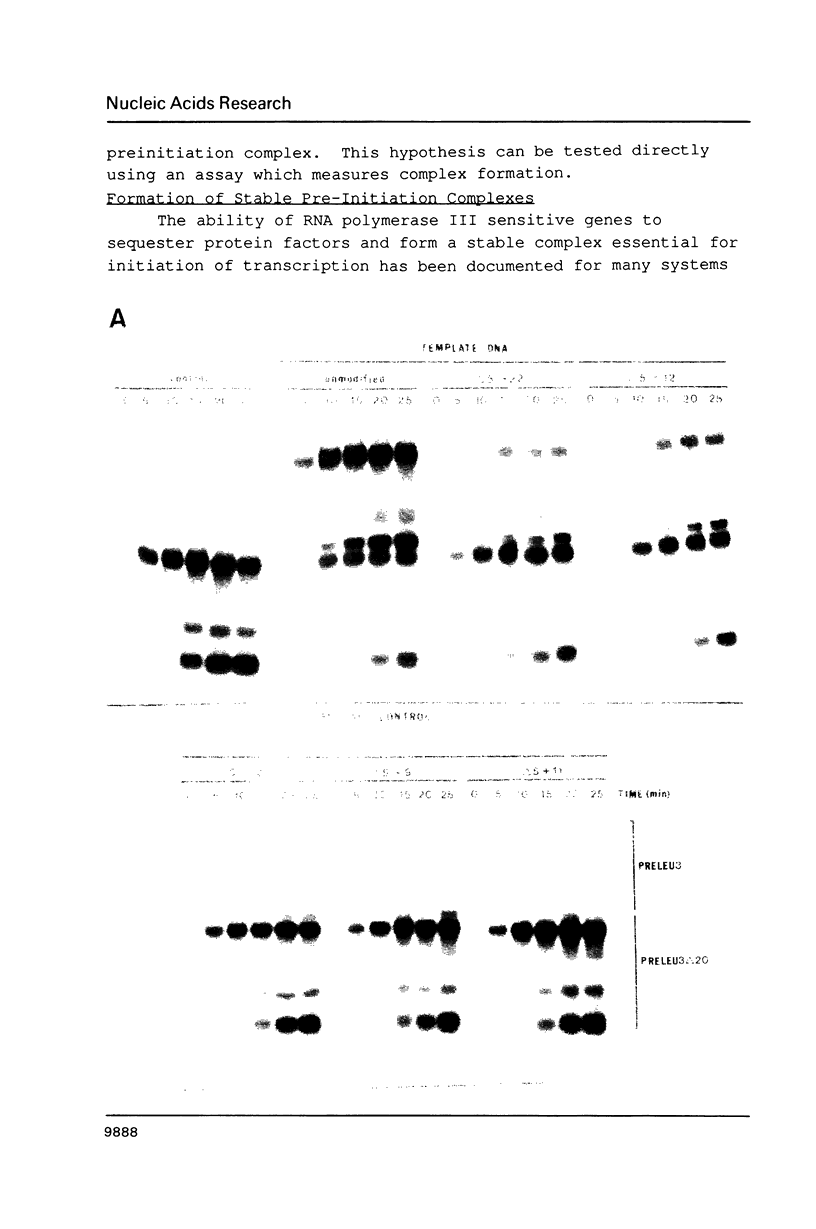
A conserved sequence that enhances transcription is present at or near the 5'-end of genes which code for several abundant yeast tRNAs, one of which is tDNA(Leu3). Mutants with alterations in this region have been compared to the parent tDNA(Leu3) and a yeast tDNA(Ser) that normally lacks the conserved sequence. The apparent KM of the transcription apparatus is insensitive to 5'-flanking sequence variation. Replacement of the normal 5'-flanking sequence does result in 3-18 fold reductions in VMAX and a 7-13 fold loss of transcription in tests with competitor genes. A second template exclusion assay revealed that sequences upstream of the conserved region effect a 2 fold change in the ability of the genes to stably sequester a component(s) of the obligatory pre-initiation complex while positions -22 to +9 have little or no effect.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison D. S., Hall B. D. Effects of alterations in the 3' flanking sequence on in vivo and in vitro expression of the yeast SUP4-o tRNATyr gene. EMBO J. 1985 Oct;4(10):2657–2664. doi: 10.1002/j.1460-2075.1985.tb03984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold G. J., Gross H. J. Unrelated leader sequences can efficiently promote human tRNA gene transcription. Gene. 1987;51(2-3):237–246. doi: 10.1016/0378-1119(87)90312-x. [DOI] [PubMed] [Google Scholar]
- Baker R. E., Gabrielsen O., Hall B. D. Effects of tRNATyr point mutations on the binding of yeast RNA polymerase III transcription factor C. J Biol Chem. 1986 Apr 25;261(12):5275–5282. [PubMed] [Google Scholar]
- Baker R. E., Hall B. D. Structural features of yeast tRNA genes which affect transcription factor binding. EMBO J. 1984 Dec 1;3(12):2793–2800. doi: 10.1002/j.1460-2075.1984.tb02211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron J. R., Loh E. Y., Davis R. W. Evidence for transposition of dispersed repetitive DNA families in yeast. Cell. 1979 Apr;16(4):739–751. doi: 10.1016/0092-8674(79)90090-4. [DOI] [PubMed] [Google Scholar]
- Chisholm G. E., Genbauffe F. S., Cooper T. G. tau, a repeated DNA sequence in yeast. Proc Natl Acad Sci U S A. 1984 May;81(10):2965–2969. doi: 10.1073/pnas.81.10.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciampi M. S., Melton D. A., Cortese R. Site-directed mutagenesis of a tRNA gene: base alterations in the coding region affect transcription. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1388–1392. doi: 10.1073/pnas.79.5.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciliberto G., Traboni C., Cortese R. Relationship between the two components of the split promoter of eukaryotic tRNA genes. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1921–1925. doi: 10.1073/pnas.79.6.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco D., Sharp S., Söll D. Identification of regulatory sequences contained in the 5'-flanking region of Drosophila lysine tRNA2 genes. J Biol Chem. 1981 Dec 10;256(23):12424–12429. [PubMed] [Google Scholar]
- Dingermann T., Burke D. J., Sharp S., Schaack J., Söll D. The 5- flanking sequences of Drosophila tRNAArg genes control their in vitro transcription in a Drosophila cell extract. J Biol Chem. 1982 Dec 25;257(24):14738–14744. [PubMed] [Google Scholar]
- Eigel A., Feldmann H. Ty1 and delta elements occur adjacent to several tRNA genes in yeast. EMBO J. 1982;1(10):1245–1250. doi: 10.1002/j.1460-2075.1982.tb00020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke D. R., Gegenheimer P., Abelson J. Nucleolytic processing of a tRNAArg-tRNAAsp dimeric precursor by a homologous component from Saccharomyces cerevisiae. J Biol Chem. 1985 Jan 25;260(2):1271–1279. [PubMed] [Google Scholar]
- Folk W. R., Hofstetter H. A detailed mutational analysis of the eucaryotic tRNAmet1 gene promoter. Cell. 1983 Jun;33(2):585–593. doi: 10.1016/0092-8674(83)90439-7. [DOI] [PubMed] [Google Scholar]
- Fuhrman S. A., Engelke D. R., Geiduschek E. P. HeLa cell RNA polymerase III transcription factors. Functional characterization of a fraction identified by its activity in a second template rescue assay. J Biol Chem. 1984 Feb 10;259(3):1934–1943. [PubMed] [Google Scholar]
- Hipskind R. A., Clarkson S. G. 5'-flanking sequences that inhibit in vitro transcription of a xenopus laevis tRNA gene. Cell. 1983 Oct;34(3):881–890. doi: 10.1016/0092-8674(83)90545-7. [DOI] [PubMed] [Google Scholar]
- Ikemura T., Ozeki H. Codon usage and transfer RNA contents: organism-specific codon-choice patterns in reference to the isoacceptor contents. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1087–1097. doi: 10.1101/sqb.1983.047.01.123. [DOI] [PubMed] [Google Scholar]
- Johnson J. D., Raymond G. J. Three regions of a yeast tRNALeu3 gene promote RNA polymerase III transcription. J Biol Chem. 1984 May 10;259(9):5990–5994. [PubMed] [Google Scholar]
- Klekamp M. S., Weil P. A. Partial purification and characterization of the Saccharomyces cerevisiae transcription factor TFIIIB. J Biol Chem. 1986 Feb 25;261(6):2819–2827. [PubMed] [Google Scholar]
- Koski R. A., Allison D. S., Worthington M., Hall B. D. An in vitro RNA polymerase III system from S. cerevisiae: effects of deletions and point mutations upon SUP4 gene transcription. Nucleic Acids Res. 1982 Dec 20;10(24):8127–8143. doi: 10.1093/nar/10.24.8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis D. S., Spiegelman G. B. Steady-state kinetic analysis of transcription of cloned tRNASer genes from Drosophila melanogaster. Eur J Biochem. 1985 Apr 15;148(2):305–313. doi: 10.1111/j.1432-1033.1985.tb08840.x. [DOI] [PubMed] [Google Scholar]
- Murphy M. H., Baralle F. E. Construction and functional analysis of a series of synthetic RNA polymerase III promoters. J Biol Chem. 1984 Aug 25;259(16):10208–10211. [PubMed] [Google Scholar]
- Nelböck P., Stucka R., Feldmann H. Different patterns of transposable elements in the vicinity of tRNA genes in yeast: a possible clue to transcriptional modulation. Biol Chem Hoppe Seyler. 1985 Nov;366(11):1041–1051. doi: 10.1515/bchm3.1985.366.2.1041. [DOI] [PubMed] [Google Scholar]
- Newman A. J., Ogden R. C., Abelson J. tRNA gene transcription in yeast: effects of specified base substitutions in the intragenic promoter. Cell. 1983 Nov;35(1):117–125. doi: 10.1016/0092-8674(83)90214-3. [DOI] [PubMed] [Google Scholar]
- Olson M. V., Page G. S., Sentenac A., Piper P. W., Worthington M., Weiss R. B., Hall B. D. Only one of two closely related yeast suppressor tRNA genes contains an intervening sequence. Nature. 1981 Jun 11;291(5815):464–469. doi: 10.1038/291464a0. [DOI] [PubMed] [Google Scholar]
- Raymond G. J., Johnson J. D. The role of non-coding DNA sequences in transcription and processing of a yeast tRNA. Nucleic Acids Res. 1983 Sep 10;11(17):5969–5988. doi: 10.1093/nar/11.17.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond K. C., Raymond G. J., Johnson J. D. In vivo modulation of yeast tRNA gene expression by 5'-flanking sequences. EMBO J. 1985 Oct;4(10):2649–2656. doi: 10.1002/j.1460-2075.1985.tb03983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeyer S. B., Olson M. V. Insertion of a repetitive element at the same position in the 5'-flanking regions of two dissimilar yeast tRNA genes. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7674–7678. doi: 10.1073/pnas.79.24.7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaack J., Sharp S., Dingermann T., Burke D. J., Cooley L., Söll D. The extent of a eukaryotic tRNA gene. 5'- and 3'-flanking sequence dependence for transcription and stable complex formation. J Biol Chem. 1984 Feb 10;259(3):1461–1467. [PubMed] [Google Scholar]
- Schaack J., Sharp S., Dingermann T., Söll D. Transcription of eukaryotic tRNA genes in vitro. II. Formation of stable complexes. J Biol Chem. 1983 Feb 25;258(4):2447–2453. [PubMed] [Google Scholar]
- Sharp S. J., Schaack J., Cooley L., Burke D. J., Söll D. Structure and transcription of eukaryotic tRNA genes. CRC Crit Rev Biochem. 1985;19(2):107–144. doi: 10.3109/10409238509082541. [DOI] [PubMed] [Google Scholar]
- Shaw K. J., Olson M. V. Effects of altered 5'-flanking sequences on the in vivo expression of a Saccharomyces cerevisiae tRNATyr gene. Mol Cell Biol. 1984 Apr;4(4):657–665. doi: 10.1128/mcb.4.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague K. U., Larson D., Morton D. 5' flanking sequence signals are required for activity of silkworm alanine tRNA genes in homologous in vitro transcription systems. Cell. 1980 Nov;22(1 Pt 1):171–178. doi: 10.1016/0092-8674(80)90165-8. [DOI] [PubMed] [Google Scholar]
- Traboni C., Ciliberto G., Cortese R. Mutations in Box B of the promoter of a eucaryotic tRNAPro gene affect rate of transcription, processing, and stability of the transcripts. Cell. 1984 Jan;36(1):179–187. doi: 10.1016/0092-8674(84)90087-4. [DOI] [PubMed] [Google Scholar]
- Wilson E. T., Larson D., Young L. S., Sprague K. U. A large region controls tRNA gene transcription. J Mol Biol. 1985 May 25;183(2):153–163. doi: 10.1016/0022-2836(85)90209-8. [DOI] [PubMed] [Google Scholar]