Abstract
Inorganic arsenic is a known environmental toxicant and carcinogen of global public health concern. Arsenic is genotoxic and cytotoxic to human keratinocytes. However, the biological pathways perturbed in keratinocytes by low chronic dose inorganic arsenic are not completely understood. The objective of the investigation was to discover the mechanism of arsenic carcinogenicity in human epidermal keratinocytes. We hypothesize that a combined strategy of DNA microarray, qRT-PCR and gene function annotation will identify aberrantly expressed genes in HaCaT keratinocyte cell line after chronic treatment with arsenic trioxide. Microarray data analysis identified 14 up-regulated genes and 21 down-regulated genes in response to arsenic trioxide. The expression of 4 up-regulated genes and 1 down-regulated gene were confirmed by qRT-PCR. The up-regulated genes were AKR1C3 (Aldo-Keto Reductase family 1, member C3), IGFL1 (Insulin Growth Factor-Like family member 1), IL1R2 (Interleukin 1 Receptor, type 2), and TNFSF18 (Tumor Necrosis Factor [ligand] SuperFamily, member 18) and down-regulated gene was RGS2 (Regulator of G-protein Signaling 2). The observed over expression of TNFSF18 (167 fold) coupled with moderate expression of IGFL1 (3.1 fold), IL1R2 (5.9 fold) and AKR1C3 (9.2 fold) with a decreased RGS2 (2.0 fold) suggests that chronic arsenic exposure could produce sustained levels of TNF with modulation by an IL-1 analogue resulting in chronic immunologic insult. A concomitant decrease in growth inhibiting gene (RGS2) and increase in AKR1C3 may contribute to chronic inflammation leading to metaplasia, which may eventually lead to carcinogenicity in the skin keratinocytes. Also, increased expression of IGFL1 may trigger cancer development and progression in HaCaT keratinocytes.
Keywords: arsenic trioxide, chronic exposure, HaCaT cell, keratinocytes, anti-apoptosis, anti-differentiation
Introduction
Inorganic arsenic is a known environmental toxicant and carcinogen1,2 which when exposed to humans through inhalation or ingestion may result in human diseases such as intraepidermal carcinomas (Bowen disease), squamous cell carcinomas (SCC), basal cell carcinomas (BCC), and Merkel cell carcinoma (MCC), hyperkeratosis and hyperpigmentation.3 Increasing reports of arsenic (As) related cancers in different parts of the world including southeastern Michigan (USA),4 India,5 Taiwan,6 and Bangladesh,7 have raised more public health concerns about long-term exposure to arsenic through drinking water or medications.
Inorganic arsenic is genotoxic and cytotoxic to keratinocytes.8 It also causes alterations of gene expression in cultured human keratinocytes.9 Though the molecular mechanisms of action are not completely understood, it is known that arsenic can reduce DNA repair, increase growth factors, induce gene amplification, reactive oxygen production and oxidative stress, enhance cell proliferation, and alter DNA methylation and signal transduction.10 Arsenic has low mutagenic activity and can serve as a co-carcinogen.11 The intriguing property of arsenic trioxide is its ability to elicit apoptosis in some cell lines and tumorigenic in other cell lines. To explain the molecular mechanism of action of arsenic, high-throughput gene expression studies such as microarray technologies were employed to investigate multiple mechanisms together based on alterations in expression of target genes.
DNA microarrays typically consist of thousands of immobilized DNA sequences present on a miniaturized surface.12 Microarrays have been used to propose a mechanism of arsenic toxicity/carcinogenicity in skin,13 kidney,14 myeloma,15 peripheral lymphocytes,13 neural tube,16 liver17 and urogenital cells.18 Other researchers have used microarrays to investigate the effects of arsenic on keratinocytes.19,20 There is still a limited number of public domain genome-wide gene expression datasets on the effect of arsenic on epidermal cells after chronic exposure. As human exposure to arsenic occurs primarily through ingestion and skin contact, we selected HaCaT keratinocyte cell line for this in vitro study. HaCaT is the first permanent immortalized epithelial cell line from adult human skin that exhibits normal differentiation and provides a promising tool for studying regulation of keratinization in human cells.21
The objective of the investigation was to discover the mechanism of arsenic carcinogenicity in human epidermal keratinocytes. We hypothesize that a combined strategy of DNA microarray, RT-qPCR and gene function annotation will identify aberrantly expressed genes in HaCaT keratinocyte cell line after chronic treatment with arsenic trioxide. Knowledge of the biological pathways and networks of the genes that are significantly expressed or altered are pertinent to understanding the mechanism of action of arsenic carcinogenicity to HaCaT cells. Therefore, functional and pathway annotation using bioinformatics tools can help identify specific pathways of interest from the gene expression datasets.
In this article, we report the genes aberrantly expressed in HaCaT cells in response of chronic exposure to arsenic trioxide. The global transcriptomics approach identified over expression of TNFSF18 (tumor necrosis factor (ligand) superfamily, member 18), moderate expression of IL1R2 (Interleukin 1 receptor, type 2), IGFL1 (Insulin Growth Factor-Like family member 1) and AKR1C3 (Aldo-Keto Reductase family 1, member C 3) coupled with a decreased expression of RGS2 (Regulator of G-protein Signaling 2). This suggests that arsenic exposure induces immunotoxic, anti-differentiation, growth factor promotion and anti-apoptotic effects in skin keratinocytes.
Materials and Methods
Chemical and reagents
Arsenic trioxide (99.9% purity, Fisher Scientific Suwanee, GA) and fetal bovine serum (FBS, Hyclone Laboratories Logan, UT) were purchased for culturing HaCaT cell line. Cell culture supplies such as Dulbecco’s Minimum Essential Medium (DMEM), antibiotics, and phosphate buffered solution (PBS) were purchased from ATCC.
Cell culture
Our experimental design included a control (untreated HaCaT cell), and test (Treated HaCaT cell) group. The HaCaT cell line was kindly provided by Dr. N. Fusenig (German Cancer Research Center, Heidelberg, Germany). 1.5 × 105 HaCaT cells were cultured in 7.5 ml of complete DMEM containing 10% Fetal Bovine Serum (FBS) and 1% penicillin, streptomycin in T-25 culture plate. Cells were incubated in a humidified atmosphere with 5% CO2 at 37 °C. The treatment groups were exposed to 5 mg/L As2O3 (equivalent to Lethal Concentration, LC 0.5%), and passaged at 90% confluent. Chronic exposure was established by sub-culturing the treatment group up to passage 22.
RNA extraction and gene expression
Total RNA was extracted from 4 technical replicates of unexposed HaCaT cells and HaCaT cells chronically exposed to arsenic trioxide up to passage 22 using RNA STAT-60 (TEL-TEST, INC, Friendswood, TX, USA).22 A NanoDrop ND-1000 spectrophotometer (NanoDrop products, Wilmington, DE) was used to quantify the RNA by optical density reading. Also, the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) was used to determine the purity and quality of the extracted RNA. Only high quality RNA, having RNA Integrity Number (RIN) of >7.0,23 and an A260/280 absorbance ratio of >1.8, was utilized for microarray experiments.
Microarray
The Human Whole Genome OneArray™ (Phalanx Biotech, Palo Alto, CA) was used to perform DNA microarray analysis. RNA was converted to double-stranded cDNA and amplified using in vitro transcription systems that included amino-allyl UTP, and the aRNA product was subsequently conjugated with Cy5™ NHS ester (GEH Lifesciences, Pittsburgh, PA). Fragmented aRNA was hybridized at 42 °C overnight using the HybBag mixing system with 1X OneArray Hybridization Buffer (Phalanx Biotech, Palo Alto, CA), 0.01 mg/ml sheared salmon sperm DNA (Promega, Madison, WI, USA), at a concentration of 0.025 mg/ml labeled target. After hybridization, the arrays were washed according to the OneArray protocol. Raw intensity signals for each microarray were captured using a Molecular Dynamics ™ Axon 4100A scanner, measured using GenePixPro™ Software, and stored in GenePix Results (GPR) format. The data from all microarrays in each experimental set was then passed to Rosetta Resolver (Rosetta Biosoftware, Seattle, WA) for analysis.
Two-step quantitative RT-PCR
Relative quantitation using the comparative CT method24 was employed to confirm the microarray gene expression data. Untreated HaCaT cell sample was used as calibrator and Beta glucuronidase (GUSB) as endogenous control gene for normalization. Applied Biosystems (Applied Biosystems, Carlsbad, CA) standard protocol was followed. The RNA samples were reverse-transcribed for 120 min at 37 °C with High Capacity cDNA Reverse Transcription Kit. Quantitative PCR was carried out for 10 min at 95 °C, and 40 cycles of 15 sec at 95 °C, 1 min at 60 °C using 2X Power SYBR Green PCR Master Mix (Applied Biosystems, Carlsbad, CA) and 200 nM of forward and reverse primers. The primers are listed in Table 1. Triplicates of each assay were run on an Applied Biosystems 7300 Real-Time PCR system and expression fold-changes were derived using the comparative threshold (CT) method. Each replicate cycle threshold (CT) was normalized to the average CT of GUSB on a per sample basis. Applied Biosystems formula was used to calculate the relative amount of the transcripts in the arsenic treated HaCaT and the untreated sample (control), and both were normalized to the endogenous control (GUSB): ΔΔCT = ΔCT (treated) − ΔCT (control), where ΔCT is the difference in CT between the target gene and endogenous controls by subtracting the average CT of controls. The fold-change for each treated sample relative to the control sample equals 2−ΔΔCT.
Table 1.
Quantitative PCR primers for microarray validation.
| Gene | Forward | Reverse |
|---|---|---|
| AKR1C3 | GGAGAAGTGTAAGGATGCAGGATT | GTACTTGAGTCCTGGCTTGTTGAG |
| GUSB | TGATCGCTCACACCAAATCC | CCCCTTGTCTGCTGCATAGTTA |
| IGFL1 | CATCGTAGCTGTCTTTGCCATT | TGGCTGGCACAGCATCAG |
| IL1R2 | CACTACGCACCACAGTCAAGGA | ATCCATATTCCCCCCAAAACC |
| MKNK1 | CAACTCCTGTACCCCCATAACC | TGGCCTGGTCCGTGAAGA |
| PCSK1 | CCTGGAAGCAAACCCAAATC | ATCCAAATCGACTATTCACCATCA |
| PPP1R13B | GCCACACCACCTAAGAATTACCA | GAGAGGTTGAACCCGAAGGTAAA |
| RGS2 | GAATTCTGGCTGGCCTGTGA | ATGTTTATCTCTTTTGGAGCTTCCTT |
| TM4SF4 | TGGGCCTGAAGAACAATGACT | CAAGAATCCAACCACAGCAAATAT |
| TMEM70 | AAGGCATGGGATCGTTTCC | ACTCCTGGCTCAATACTGATGGA |
| TNFSF18 | AGCCCTGTATGGCTAAGTTTGG | GCCATTCTGAAGTATCTCCAGCTT |
Results
Differentially expressed genes
The criterion for selection of differentially expressed genes (DEGs) was a fold change greater than or equal to 2. Comparison of the microarray data from untreated (control) and chronically exposed HaCaT keratinocyte cell identified a total of 35 differentially expressed genes with 14 genes up-regulated (Table 2) and 21 genes down-regulated (Table 3). Genes with ≥2 fold changes and P-value ≤0.05 were considered significantly expressed and were selected for confirmation using qRT-PCR. The functional annotations including Gene Ontology for these genes were determined using the Michigan Molecular Interactions (MiMI) web tool25 (http://mimi.ncibi.org).
Table 2.
Genes up-regulated in response to chronic-dose exposure of arsenic trioxide to HaCaT keratinocyte cells.
| Gene symbol | Gene description |
|---|---|
| AGPAT4 | 1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) [ |
| AKR1C2 | aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III) |
| AKR1C3 | aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) |
| C22orf42 | Uncharacterized protein C22orf42 |
| GLT6D1 | glycosyltransferase 6 domain containing 1 |
| IGFL1 | IGF-like family member 1 |
| IL1R2 | interleukin 1 receptor, type II |
| KLHDC8A | Kelch domain-containing protein 8A |
| NQO2 | NAD(P)H dehydrogenase, quinone 2 |
| PCSK1 | proprotein convertase subtilisin/kexin type 1 |
| STRAP | STRAP serine/threonine kinase receptor associated protein |
| TMEM70 | Transmembrane protein 70 |
| TNFSF18 | tumor necrosis factor (ligand) superfamily, member 18 |
| ZFP36L1 | zinc finger protein 36, C3H type-like |
Table 3.
Genes down-regulated in response to chronic-dose exposure of arsenic trioxide to HaCaT keratinocyte cells.
| Gene symbol | Gene description |
|---|---|
| CCDC150 | coiled-coil domain containing 150 |
| CLCC1 | Chloride channel CLIC-like 1 |
| CSRP1 | Cysteine and glycine-rich protein 1 |
| FGF1 | Fibroblast growth factor 1 (acidic) |
| GCNT3 | Glucosaminyl (N-acetyl) transferase 3, mucin type |
| GDA | Guanine deaminase |
| HOXA5 | homeobox A5 |
| IMP5 | Signal peptide peptidase-like 2C Precursor (Protein SPP-like 2C) (Protein SPPL2c)(EC 3.4.23.-) (Intramembrane protease 5)(IMP5) |
| MAK | male germ cell-associated kinase |
| MKNK1 | MAP kinase interacting serine/threonine kinase 1 |
| MRC2 | mannose receptor, C type 2 |
| NP153 | nucleoporin 153 kDa |
| NT5C | 5′, 3′-nucleotidase, cytosolic |
| PHF12 | PHD finger protein 12 |
| PPP1R13B | Apoptosis-stimulating of p53 protein, protein phosphatase 1, regulatory (inhibitor) subunit 13B |
| PRKAR1B | protein kinase, cAMP-dependent, regulatory, type I, beta |
| RASL10A | RAS-like, family 10, member A |
| RGS2 | regulator of G-protein signaling 2, 24kDa |
| TM4SF4 | transmembrane 4 L six family member 4 |
| TOM1 | target of myb1 (chicken) |
| ZNF19 | zinc finger protein 19 |
Quantitative PCR confirmation of microarray data
Relative quantitation using the comparative CT method was employed to confirm the microarray gene expression data. The expression of 4 up-regulated genes and 1 down-regulated gene were confirmed by qRT-PCR for genes with a fold change of ≥2 when compared to the reference untreated control sample. The up-regulated genes were AKR1C3 (9.2 fold), IGFL1 (3.1), IL1R2 (5.9 fold), and TNFSF18 (167 fold) and down-regulated gene was RGS2 (2.0 fold). A visualization comparing the fold change obtained by microarray and qRT-PCR for 9 genes is presented in Figure 1.
Figure 1.
Comparison of fold change between microarray data and q-PCR data. Red: up-regulated; yellow: unchanged; green: down-regulated.
Biological pathway modeling
Ingenuity Pathways Analysis (IPA; Ingenuity Systems, Redwood City, CA) was used to determine models of biological pathways and networks that are significantly represented in the differentially expressed (both up and down regulated) genes.
IPA identifies networks and pathways represented in the gene lists of interest. It generates a P-value for each network and canonical pathway, which is the likelihood that a given network was identified by chance. We selected networks scoring ≥2, which have >99% confidence of not being generated by chance.26
IPA assigns biological functions to each network by using annotations from scientific literature and stored in their knowledge base. Fisher exact test is used to calculate the P-value for each biological function/disease or pathway being assigned by chance. We used Benjamini-Hochberg corrected P ≤ 0.05 to select highly significant biological functions and pathways represented in the datasets analyzed.26
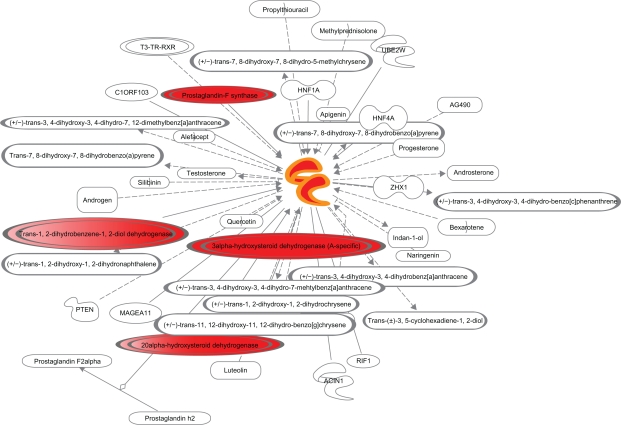
The predicted gene interaction pathways for the following up-regulated genes: IL1R2, TNFSF18 and AKR1C3 are presented in Figure 2, Figure 3 and Figure 4 respectively. Molecules in red are up-regulated while molecules in green are down-regulated in the microarray data.
Figure 2.
Interaction network of Interleukin 1 Receptor, Type II (IL1R2). Molecules in red are up-regulated while molecules in green are downregulated in the microarray analysis data.
Figure 3.
Interaction network for TNFSF18 interaction network. Molecules in red are up-regulated while molecules in green are down-regulated in the microarray data.
Figure 4.
Interaction network for AKR1C3. Molecules in red are up-regulated while molecules in green are down-regulated in the microarray data.
Discussion
In this investigation, the HaCaT keratinocyte cell line was used to pilot future investigations that will compare arsenic trioxide induced aberrantly expressed genes in a variety of human epidermal cells. The microarray experiment identified 14 up-regulated and 21 down-regulated genes with expression fold change of ≥2 in HaCaT keratinocytes exposed to chronic dose of arsenic trioxide. The criterion for selecting differentially expressed genes for qRT-PCR confirmation was a P-value ≤0.05 and fold change of ≥2. The following four up-regulated genes, AKR1C3 (9.2 fold), IGFL1 (3.1 fold), IL1R2 (5.9 fold), and TNFSF18 (167 fold) and one down-regulated gene RGS2 (2.0 fold) were selected for validation using complementary qRT-PCR approach. Subsequently, annotations for molecular function, cellular location and biological pathway were determined using bioinformatics tools. The fold change values quantified by microarray and PCR do not agree well (Fig. 1). About half of the data pairs are significantly different and sometimes totally opposite. The big variances may be due to differential transcript recognition by the two methods.27 Furthermore, different hybridization kinetics may account for genes with identical microarray values but with dissimilar qRT-PCR values (eg, IGFL1 and TMEM70). In most of the data pairs where the direction of expression is identical, the fold change was higher in the qRT-PCR as observed by other investigators.27
IL1R2 also referred to as CD121b, IL1RB, and MGC4772528 plays a vital role in immune response and it is associated with the membrane (Fig. 2). IL1R2 is a decoy receptor for inflammatory interleukin 1 (IL-1). It acts by sequestering active and inactive IL1, which in turn restricts the availability of the ligand for the functional receptor and inhibits its maturation.29–32 IL1R1 and IL1R2 are the known receptors of IL1 and cell activation which is capable of transducing the activation signal which occurs when IL1 binds to cell surface IL1R1 in conjunction with IL1R accessory protein (IL1RAP).33 IL1R2 is known as a potent, specific and natural inhibitor of IL1, but in contrast to IL1R1 it has no signaling properties when bound to IL1.29–32 Over expression of IL1R2 has been reported in human uroepithelial cell line (HUC-1) chronically exposed to arsenite.34 Our results confirmed this observation in HaCaT cell line. ILIR2 may therefore be a biomarker for chronic exposure to arsenicals. In our investigation, HaCaT cells were chronically exposed to low arsenic trioxide dose of 5 mg/L up to 22 passages. In HUC-1, IL1R2 over expression is linked with enhanced expression of Smad-interacting protein 1 (SIP-1) and reduced expression of E-cadherin.34
E-cadherin is a calcium-dependent, epithelial cell adhesion molecule, whose reduced expression has been associated with tumor dedifferentiation and increased lymph node metastasis in clinical studies involving several carcinomas.35 Furthermore, reduced expression levels of E-cadherin was associated with moderately and poorly differentiated squamous and small cell carcinoma in a limited number of patients with lung cancer.36 IL1R2 also improves cell migration34 and are suggestive of oncogenic potential of IL1R2. Gene network analysis with IL1R2 gene as illustrated in Figure 2, showed that IL1R2 interacts with IL1RAP, IL1A, IL1B and GLI1 and this agrees with previous findings.29–32 GLI1, a protein which was originally isolated from human glioblastoma37,38 is the effector of Hedgehog (Hh) signaling which critical role in carcinogenesis.39 Furthermore, GLI1 was reported to be upregulated in many tumors including basal cell carcinomas. Arsenic ingestion through drinking water has also been linked with increased risk of basal cell carcinomas (BCC).40 Thus, ILIR2 indirectly interacts with GLI1, which is the trigger for BCC indicative of contribution in skin carcinogenesis.
The cytokine TNFSF18, which can also be represented as glucocorticoid-induced tumor necrosis factor receptor-related ligand (GITRL), is a ligand for receptor TNFRSF18/AITR/GITR and it modulates T lymphocyte survival in peripheral tissues41 (Fig. 3). TNFSF18 is found in extracellular space and integral to membrane.42 We observed that TNFSF18 was significantly over expressed with fold change of more than 167. The glucocorticoid-induced tumour necrosis factor receptor-related gene (GITR) is expressed on regulatory T-cells (Treg), which are CD4+CD25+ lymphocytes. Binding of the GITR-ligand (GITRL) leads to down-regulation of the biology function of Tregs. It is believed that a defect in Tregs causes a skin condition resembling atopic dermatitis.43 Soluble forms of GITRL (sGITRL) are released by human tumor cells.44 This suggests that determination of sGITRL levels might be implemented as a tumor marker in patients.
Activated keratinocytes are known to engage intraepithelial T-cells through co-stimulatory molecules, keratinocytes express GITRL and through this important co-stimulatory molecule expressed by antigen-presenting cells (APCs).45 Furthermore, they have the potential to influence T-cell numbers in the skin via chemokine production and through a direct cell-cell effect on T-cell proliferation. This means that sustained arsenic insult could have activated the HaCaT keratinocytes, which may lead to an immunotoxigenic reaction as reported by Baumgartner-Nielsen et al.43
Aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) (AKR1C3)46 was up-regulated up to 9.2 folds in our investigation. Human AKR1C3 is an enzyme involved in steroid metabolism as illustrated in Figure 4. Elevated levels of AKR1C3 expression are implicated in leukemia cell differentiation, prostate cancer (in both androgen-dependent and androgen-independent prostate cancer),47 endometrial cancer48 and chronic inflammation.47
We observed a moderate expression (3.1 fold) of insulin growth factor-like family member 1 (IGFL1). The IGF-like (IGFL1) genes encode proteins that contain 11 conserved cysteine residues at fixed positions including two CC motif.49 The biological functions and gene interactions of IGFL1 is not very clear, however, the structure and sequence suggest that IGFL proteins are distantly related to the Insulin-like growth factors (IGF), a superfamily of growth factors. Both IGFL and IGF share gene expression patterns in many cancers.49 The human skin is the critical organ of arsenic toxicity because arsenic has a strong affinity for the keratin proteins, which are rich in the sulphur containing cysteine residues50 and potentially arsenic-binding proteins based on presence of vicinal cysteines.51 In our previous investigation,52 we observed that proteins with abundance of vicinal cysteines will increase responsiveness to arsenic-induced keratinocyte carcinogenesis. Since IGLF1 encodes proteins rich in cysteine residues, it could be playing a vital role in arsenic binding and responsiveness in keratinocytes. Further, IGFL1 is associated with embryonic tissue and was observed in libraries derived from carcinoma cell lines.49 Therefore, the increase in IGFL1 is involved in cancer development and progression and probably a marker of chronic exposure to arsenic trioxide.
The growth suppressor gene RGS2 accelerate GTPase activity of heterotrimeric G proteins, resulting in inactivation of specific signaling pathways.53 Down regulation of RGS2 occurred in human prostate tumor specimens54 as well as in recurrence and metastasis-derived colorectal cancer cell lines.55 In HaCaT keratinocyte cells, aberrant expression of RGS2 may aid in the spread of cancer or metastasis.
In conclusion, we have used a combination of microarray, gene functional annotation data and qPCR to identify genes differentially expressed in HaCaT cell line in response to chronic, low dose arsenic trioxide. Two immune response genes IL1R2 and TNFSF18 were identified which may result to chronic immunologic insult in keratinocytes. Also, the down regulation of growth inhibiting gene (RGS2) and upregulation of AKR1C3 and IGLF1 may aid chronic inflammation leading to metaplasia.
Acknowledgments
Research Centers in Minority Institutions (RCMI)—Center for Environmental Health at Jackson State University (NIH-NCRR 2G12RR013459); Mississippi NSF-EPSCoR Award (EPS-0903787); Pittsburgh Supercomputing Centre’s National Resource for Biomedical Supercomputing (T36 GM008789); National Center for Integrative Biomedical Informatics (U54DA021519); US Department of Homeland Security Science & Technology Directorate (2007-ST-104-000007; 2009-ST-062-000014; 2009-ST-104-000021). We thank Drs Robert Rice, Susan Bridges, Youping Deng and Yongqun He for their suggestions. Disclaimer: The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the funding agencies.
Footnotes
Disclosures
This manuscript has been read and approved by all authors. This paper is unique and not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.Hall M, Chen Y, Ahsan H, et al. Blood arsenic as a biomarker of arsenic exposure: results from a prospective study. Toxicology. 2006;225:225–33. doi: 10.1016/j.tox.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Tchounwou PB, Centeno JA, Patlolla AK. Arsenic toxicity, mutagenesis, and carcinogenesis—a health risk assessment and management approach. Mol Cell Biochem. 2004;255:47–55. doi: 10.1023/b:mcbi.0000007260.32981.b9. [DOI] [PubMed] [Google Scholar]
- 3.Maloney ME. Arsenic in Dermatology. Dermatol Surg. 1996;22:301–4. doi: 10.1111/j.1524-4725.1996.tb00322.x. [DOI] [PubMed] [Google Scholar]
- 4.Meliker JR, Wahl RL, Cameron LL, Nriagu JO. Arsenic in drinking water and cerebrovascular disease, diabetes mellitus, and kidney disease in Michigan: a standardized mortality ratio analysis. Environ Health. 2007;6:4. doi: 10.1186/1476-069X-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mazumder DN, Ghosh A, Majumdar KK, et al. Arsenic contamination of ground water and its health impact on population of district of nadia, west bengal, India. Indian J Community Med. 2010;35:331–8. doi: 10.4103/0970-0218.66897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng TJ, Ke DS, Guo HR. The association between arsenic exposure from drinking water and cerebrovascular disease mortality in Taiwan. Water Res. 2010 doi: 10.1016/j.watres.2010.05.040. [DOI] [PubMed] [Google Scholar]
- 7.Argos M, Kalra T, Rathouz PJ, et al. Arsenic exposure from drinking water, and all-cause and chronic-disease mortalities in Bangladesh (HEALS): a prospective cohort study. Lancet. 2010;376:252–8. doi: 10.1016/S0140-6736(10)60481-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham-Evans B, Cohly HH, Yu H, Tchounwou PB. Arsenic-induced genotoxic and cytotoxic effects in human keratinocytes, melanocytes and dendritic cells. Int J Environ Res Public Health. 2004;1:83–9. doi: 10.3390/ijerph2004020083. [DOI] [PubMed] [Google Scholar]
- 9.Rea MA, Gregg JP, Qin Q, Phillips MA, Rice RH. Global alteration of gene expression in human keratinocytes by inorganic arsenic. Carcinogenesis. 2003;24:747–56. doi: 10.1093/carcin/bgg010. [DOI] [PubMed] [Google Scholar]
- 10.Ding W, Hudson LG, Liu KJ. Inorganic arsenic compounds cause oxidative damage to DNA and protein by inducing ROS and RNS generation in human keratinocytes. Mol Cell Biochem. 2005;279:105–12. doi: 10.1007/s11010-005-8227-y. [DOI] [PubMed] [Google Scholar]
- 11.Rossman TG, Uddin AN, Burns FJ. Evidence that arsenite acts as a cocarcinogen in skin cancer. Toxicol Appl Pharmacol. 2004;198:394–404. doi: 10.1016/j.taap.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 12.Schena M. Genome analysis with gene expression microarrays. Bioessays. 1996;18:427–431. doi: 10.1002/bies.950180513. [DOI] [PubMed] [Google Scholar]
- 13.Argos M, Kibriya MG, Parvez F, et al. Gene expression profiles in peripheral lymphocytes by arsenic exposure and skin lesion status in a Bangladeshi population. Cancer Epidemiol Biomarkers Prev. 2006;15:1367–75. doi: 10.1158/1055-9965.EPI-06-0106. [DOI] [PubMed] [Google Scholar]
- 14.Sasaki A, Oshima Y, Fujimura A. An approach to elucidate potential mechanism of renal toxicity of arsenic trioxide. Exp Hematol. 2007;35:252–62. doi: 10.1016/j.exphem.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 15.Wang MC, Liu SX, Liu PB. Gene expression profile of multiple myeloma cell line treated by realgar. J Exp Clin Cancer Res. 2006;25:243–9. [PubMed] [Google Scholar]
- 16.Wlodarczyk BJ, Cabrera RM, Hill DS, et al. Arsenic-induced gene expression changes in the neural tube of folate transport defective mouse embryos. Neurotoxicology. 2006;27:547–57. doi: 10.1016/j.neuro.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Xie Y, Ducharme DM, et al. Global gene expression associated with hepatocarcinogenesis in adult male mice induced by in utero arsenic exposure. Environ Health Perspect. 2006;114:404–11. doi: 10.1289/ehp.8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su PF, Hu YJ, Ho IC, Cheng YM, Lee TC. Distinct gene expression profiles in immortalized human urothelial cells exposed to inorganic arsenite and its methylated trivalent metabolites. Environ Health Perspect. 2006;114:394–403. doi: 10.1289/ehp.8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bae DS, Hanneman WH, Yang RS, Campain JA. Characterization of gene expression changes associated with MNNG, arsenic, or metal mixture treatment in human keratinocytes: application of cDNA microarray technology. Environ Health Perspect. 2002;110(Suppl 6):931–41. doi: 10.1289/ehp.02110s6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamadeh HK, Trouba KJ, Amin RP, Afshari CA, Germolec D. Coordination of altered DNA repair and damage pathways in arsenite-exposed keratinocytes. Toxicol Sci. 2002;69:306–16. doi: 10.1093/toxsci/69.2.306. [DOI] [PubMed] [Google Scholar]
- 21.Boukamp P, Petrussevska RT, Breitkreutz D, et al. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106:761–71. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 23.Schroeder A, Mueller O, Stocker S, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Tarcea VG, Weymouth T, Ade A, et al. Michigan molecular interactions r2: from interacting proteins to pathways. Nucleic Acids Res. 2009;37:D642–6. doi: 10.1093/nar/gkn722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerling IC, Singh S, Lenchik NI, Marshall DR, Wu J. New data analysis and mining approaches identify unique proteome and transcriptome markers of susceptibility to autoimmune diabetes. Mol Cell Proteomics. 2006;5:293–305. doi: 10.1074/mcp.M500197-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Dallas PB, Gottardo NG, Firth MJ, et al. Gene expression levels assessed by oligonucleotide microarray analysis and quantitative real-time RT-PCR—how well do they correlate. BMC Genomics. 2005;6:59. doi: 10.1186/1471-2164-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dale M, Nicklin MJ. Interleukin-1 receptor cluster: gene organization of IL1R2, IL1R1, IL1RL2 (IL-1Rrp2), IL1RL1 (T1/ST2), and IL18R1 (IL-1Rrp) on human chromosome 2q. Genomics. 1999;57:177–9. doi: 10.1006/geno.1999.5767. [DOI] [PubMed] [Google Scholar]
- 29.Subramaniam S, Stansberg C, Cunningham C. The interleukin 1 receptor family. Dev Comp Immunol. 2004;28:415–28. doi: 10.1016/j.dci.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 30.Bossu P, Visconti U, Ruggiero P, et al. Transfected type II interleukin-1 receptor impairs responsiveness of human keratinocytes to interleukin-1. Am J Pathol. 1995;147:1852–61. [PMC free article] [PubMed] [Google Scholar]
- 31.Symons JA, Young PR, Duff GW. Soluble type II interleukin 1 (IL-1) receptor binds and blocks processing of IL-1 beta precursor and loses affinity for IL-1 receptor antagonist. Proc Natl Acad Sci U S A. 1995;92:1714–8. doi: 10.1073/pnas.92.5.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colotta F, Dower SK, Sims JE, Mantovani A. The type II ‘decoy’ receptor: a novel regulatory pathway for interleukin 1. Immunol Today. 1994;15:562–6. doi: 10.1016/0167-5699(94)90217-8. [DOI] [PubMed] [Google Scholar]
- 33.Dinarello CA. Therapeutic strategies to reduce IL-1 activity in treating local and systemic inflammation. Curr Opin Pharmacol. 2004;4:378–85. doi: 10.1016/j.coph.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 34.Chang SY, Su PF, Lee TC. Ectopic expression of interleukin-1 receptor type II enhances cell migration through activation of the pre-interleukin 1alpha pathway. Cytokine. 2009;45:32–8. doi: 10.1016/j.cyto.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 35.Siitonen SM, Kononen JT, Helin HJ, et al. Reduced E-cadherin expression is associated with invasiveness and unfavorable prognosis in breast cancer. Am J Clin Pathol. 1996;105:394–402. doi: 10.1093/ajcp/105.4.394. [DOI] [PubMed] [Google Scholar]
- 36.Bohm M, Totzeck B, Wieland I. Differences of E-cadherin expression levels and patterns in human lung cancer. Ann Hematol. 1994;68:81–3. doi: 10.1007/BF01715136. [DOI] [PubMed] [Google Scholar]
- 37.Kinzler KW, Bigner SH, Bigner DD, et al. Identification of an amplified, highly expressed gene in a human glioma. Science. 1987;236:70–3. doi: 10.1126/science.3563490. [DOI] [PubMed] [Google Scholar]
- 38.Saran A. Basal cell carcinoma and the carcinogenic role of aberrant Hedgehog signaling. Future Oncol. 2010;6:1003–14. doi: 10.2217/fon.10.49. [DOI] [PubMed] [Google Scholar]
- 39.Li X, Deng W, Nail CD, Bailey SK, Kraus MH, Ruppert JM, Lobo-Ruppert SM. Snail induction is an early response to Gli1 that determines the efficiency of epithelial transformation. Oncogene. 2006;25:609–621. doi: 10.1038/sj.onc.1209077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karagas MR, Stukel TA, Morris JS, et al. Skin cancer risk in relation to toenail arsenic concentrations in a US population-based case-control study. Am J Epidemiol. 2001;153:559–65. doi: 10.1093/aje/153.6.559. [DOI] [PubMed] [Google Scholar]
- 41.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–5. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gurney AL, Marsters SA, Huang RM, Pitti RM, Mark DT, Baldwin DT, Gray AM, Dowd AD, Brush AD, Heldens AD, Schow AD, Goddard AD, Wood WI, Baker KP, Godowski PJ, Ashkenazi A. Identification of a new member of the tumor necrosis factor family and its receptor, a human ortholog of mouse GITR. Curr Biol. 1999;9:215–218. doi: 10.1016/s0960-9822(99)80093-1. [DOI] [PubMed] [Google Scholar]
- 43.Baumgartner-Nielsen J, Vestergaard C, Thestrup-Pedersen K, Deleuran M, Deleuran B. Glucocorticoid-induced tumour necrosis factor receptor (GITR) and its ligand (GITRL) in atopic dermatitis. Acta Derm Venereol. 2006;86:393–8. doi: 10.2340/00015555-0118. [DOI] [PubMed] [Google Scholar]
- 44.Baltz KM, Krusch M, Baessler T, et al. Neutralization of tumor-derived soluble glucocorticoid-induced TNFR-related protein ligand increases NK cell anti-tumor reactivity. Blood. 2008;112:3735–43. doi: 10.1182/blood-2008-03-143016. [DOI] [PubMed] [Google Scholar]
- 45.Byrne AM, Goleva E, Leung DY. Identification of glucocorticoid-induced TNF receptor-related protein ligand on keratinocytes: ligation by GITR induces keratinocyte chemokine production and augments T-cell proliferation. J Invest Dermatol. 2009;129:2784–94. doi: 10.1038/jid.2009.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Azzarello JT, Lin HK, Gherezghiher A, et al. Expression of AKR1C3 in renal cell carcinoma, papillary urothelial carcinoma, and Wilms’ tumor. Int J Clin Exp Pathol. 2009;3:147–55. [PMC free article] [PubMed] [Google Scholar]
- 47.Fung KM, Samara EN, Wong C, et al. Increased expression of type 2 3alpha-hydroxysteroid dehydrogenase/type 5 17beta-hydroxysteroid dehydrogenase (AKR1C3) and its relationship with androgen receptor in prostate carcinoma. Endocr Relat Cancer. 2006;13:169–180. doi: 10.1677/erc.1.01048. [DOI] [PubMed] [Google Scholar]
- 48.Rizner TL, Smuc T, Rupreht R, Sinkovec J, Penning TM. AKR1C1 and AKR1C3 may determine progesterone and estrogen ratios in endometrial cancer. Mol Cell Endocrinol. 2006;248:126–35. doi: 10.1016/j.mce.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 49.Emtage P, Vatta P, Arterburn M, et al. IGFL: A secreted family with conserved cysteine residues and similarities to the IGF superfamily. Genomics. 2006;88:513–20. doi: 10.1016/j.ygeno.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 50.Ralph SJ. Arsenic-based antineoplastic drugs and their mechanisms of action. Met Based Drugs. 2008. 260146. [DOI] [PMC free article] [PubMed]
- 51.Kitchin KT, Wallace K. Arsenite binding to synthetic peptides based on the Zn finger region and the estrogen binding region of the human estrogen receptor-alpha. Toxicol Appl Pharmacol. 2005;206:66–72. doi: 10.1016/j.taap.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 52.Isokpehi RD, Cohly HHP, Anyanwu MN, et al. Candidate Single Nucleotide Polymorphism Markers for Arsenic Responsiveness of Protein Targets. Bioinformatics and Biology Insights. 2010;4:99–111. doi: 10.4137/BBI.S5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De VL, Zheng B, Fischer T, Elenko E, Farquhar MG. The regulator of G protein signaling family. Annu Rev Pharmacol Toxicol. 2000;40:235–71. doi: 10.1146/annurev.pharmtox.40.1.235. [DOI] [PubMed] [Google Scholar]
- 54.Cao X, Qin J, Xie Y, et al. Regulator of G-protein signaling 2 (RGS2) inhibits androgen-independent activation of androgen receptor in prostate cancer cells. Oncogene. 2006;25:3719–34. doi: 10.1038/sj.onc.1209408. [DOI] [PubMed] [Google Scholar]
- 55.Jiang Z, Wang Z, Xu Y, et al. Analysis of RGS2 expression and prognostic significance in stage II and III colorectal cancer. Biosci Rep. 2010;30:383–90. doi: 10.1042/BSR20090129. [DOI] [PubMed] [Google Scholar]