Abstract
During the last several years, intracellular lipid droplets have become the focus of intense study. No longer an inert bystander, the lipid droplet is now known as a dynamic organelle contributing lipids to many cellular events. However, while the dynamics of cholesterol efflux from both the plasma membrane and lipid droplets have been studied, less is known regarding the efflux of sphingomyelin from these membranes. In order to address this issue, sphingomyelin efflux kinetics and binding affinities from different intracellular pools were examined. When compared to the plasma membrane, lipid droplets had a smaller exchangeable sphingomyelin efflux pool and the time required to efflux that pool was significantly shorter. Fluorescence binding assays revealed that proteins in the plasma membrane and lipid droplet pool bound sphingomyelin with high affinity. Further characterization identified adipose differentiation-related protein (ADRP) as one of the sphingomyelin binding proteins in the lipid droplet fraction and revealed that ADRP demonstrated saturable binding to 6-((N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino)-hexanoyl)sphingosyl-phosphocholine (NBD-sphingomyelin) and also 2-(6-(7-nitrobenz-2-oxa-1, 3-diazol-4-yl)amino)hexanoyl-1-hexadecanoyl-sn-glycero-3-phosphocholine (NBD-phosphatidylcholine) with binding affinities in the nanomolar range. Taken together, these results suggest that lipid droplet associated proteins such as ADRP may play a significant role in regulating the intracellular distribution of phospholipids and lipids in general. Overall, insights from the present work suggest new and important roles for lipid droplets and ADRP in phospholipid metabolism.
Keywords: Lipid droplets, Sphingomyelin, Efflux, ADRP
Introduction
As defined by the body mass index, greater than one-third of the American adult population is considered to be obese, with a greater risk of developing cardiovascular disease and diabetes [1–3]. When chronic over-nutrition leading to obesity occurs, excess lipids are stored in intracellular organelles called lipid droplets. Once thought to be simple storage vesicles for neutral lipids, it is now clear that lipid droplets serve as reservoirs for lipids and proteins involved in many cellular processes. However, despite growing awareness of the health problems associated with excessive lipids in tissues and cells, details on the molecular mechanism of intracellular lipid storage remain unclear.
Lipid droplets have been the focus of increasing scrutiny in the last decade and the subject of several reviews [4–6]. Lipid droplets are composed of a neutral lipid core surrounded by a phospholipid monolayer, within which proteins reside [7, 8]. Initially, the only known proteins were the perilipins [7], adipose differentiation-related protein (ADRP) [9, 10], P200 capsule protein [11, 12], and vimentin [11]. Proteomic analysis later revealed associations with proteins involved in cell signaling, cholesterol/fatty acid metabolism, and membrane trafficking [13–15]. Caveolin proteins, normally found in flask-like invaginations of the plasma membrane called caveolae, were also present. However, while the function of plasma membrane caveolins in binding and facilitating the transport of cholesterol and fatty acids is well-documented [16–19], it remains unclear what role these proteins play in lipid droplet function.
In cells, the bulk of cholesterol and sphingolipids such as sphingomyelin are in the plasma membrane, a lipid bilayer where the deposition of cholesterol is defined by its high affinity for the saturated acyl chains of sphingolipids. Cholesterol and sphingomyelin are essential membrane components, yet, excess levels are toxic and increase the risk for arteriosclerosis. Therefore, the levels are strictly regulated through several processes, including efflux, to remove excess lipids from the cell and the body. As lipid reservoirs, the regulation of cholesterol and sphingolipid levels within lipid droplets is important to maintain lipid homeostasis. However, while the dynamics of cholesterol efflux from the plasma membrane and lipid droplets have been studied [20], less is known regarding the efflux of sphingomyelin from membranes. The present work was undertaken to examine the efflux kinetics and binding affinity of different intracellular sphingomyelin pools. The results presented herein establish, for the first time, ADRP as a phospholipid (sphingomyelin, phosphatidylcholine) binding protein and define the affinity and capacity of NBD-sphingomyelin for lipid droplet and plasma membrane proteins.
Materials and Methods
Materials
Silica Gel G and Silica Gel 60 thin layer chromatography (TLC) plates were from Analtech (Newark, DE) and EM Industries, Inc. (Darmstadt, Germany), respectively. Neutral lipid standards were purchased from Nu-Chek Prep, Inc. (Elysian, MN), while phospholipid standards were obtained from Avanti (Alabaster, AL). Rabbit polyclonal antiserum to mouse ADRP was prepared as described [21]. Rabbit anti-human caveolin-1 (cat. no. 610059) was purchased from BD Transduction Laboratories (Palo Alto, CA). Rabbit anti-human caveolin-2 (cat. no. NB100-79911) and rabbit anti-human Na+ K+-ATPase (cat. no. NB100-80005) were purchased from Novus Biologicals (Littleton, CO). Mouse anti-human cytochrome c oxidase (clone AS70, cat. no. C9229) was purchased from Sigma (St. Louis, MO). Mouse anti-human transferrin receptor (CD71, clone H68.4, cat. no. 13-6800), NBD-sphingomyelin, Nile red, and NBD-phosphatidylcholine were purchased from Invitrogen (Camarillo, CA). Primary antibodies were diluted 1:1,000 for Western blot analysis. Human high-density lipoprotein (HDL) was from Calbiochem (San Diego, CA). All reagents and solvents used were of the highest grade available and were cell culture tested.
L-Cell Culture
Murine L-cells (L arpt−tk−) were grown to confluency in Higuchi medium [22] supplemented with 10% fetal bovine serum (Hyclone, Logan, UT), as described in [23]. For fluorescence imaging and efflux experiments, cells were seeded at a density of 50,000 cells/chamber onto Lab-Tek Chamber Coverglass slides (Nunc, Naperville, IL). To ensure a monolayer, samples were examined within 20 h of seeding. Lipid droplets were isolated from L-cells grown to confluency on culture trays (245 × 245 × 25 mm, Nunc, Naperville, IL), as described previously [8].
Laser Scanning Confocal Microscopy
Confocal and co-localized images were acquired on a Bio-Rad (Hercules, CA) MRC-1024 Laser Scanning Confocal Imaging System using multiple photomultiplier tubes under the control of LaserSharp v.3.2 software (Bio-Rad). For probe excitation, the MRC-1024 system utilized a 15-mW krypton-argon laser (American Laser Corp., Salt Lake City, UT) with a 5-mW output. Cells cultured on Lab-Tek Chamber Coverglass slides were placed on the stage of a Zeiss Axiovert 135 inverted epifluorescence microscope (Zeiss, Thornwood, NY) and examined with a 63× oil-immersion, infinity objective (numerical aperture 1.4). For co-localization experiments, cells were incubated with NBD-sphingomyelin (0.2 µM) and Nile red (0.1 µM) for 10 min in a 37°C, 5% CO2 incubator. After incubation, cells were washed with phosphate-buffered saline (PBS) buffer and then sequentially imaged for NBD-sphingomyelin (488 nm excitation, 540/30 emission, green channel) and Nile red (568 nm excitation, HQ598/40 emission, red channel). The confocal images from the green and red channels were merged and appeared yellow where superimposition occurred (red and green are additive and yield yellow to orange in the RGB color space). NBD-sphingomyelin was mainly localized to lipid droplets and the plasma membrane, with other internal staining evident after 20–30 min of incubation. Pixel fluorograms were constructed and correlation coefficients used to estimate the extent of probe overlap were determined from the fluorograms as described previously [20]. Samples were exposed to the light source for minimal time periods to minimize photobleaching effects. The gain and black levels of each photomultiplier tube were optimized to minimize the levels of the autofluorescence signal and to increase the dynamic range. Residual auto fluorescence remaining was subtracted from the data. Image files were analyzed using either Metamorph software (West Chester, PA) or NIH Image, a program written by W. Rasband and available by anonymous FTP from http://zippy.nimh.nih.gov.
NBD-Sphingomyelin Stability
In experiments performed to assess the stability of the NBD-sphingomyelin probe, L-cells were grown to confluency on 35-mm tissue culture dishes as described above. Cells were incubated with NBD-sphingomyelin (0.5 µM) in PBS for 3 min at 37°C, after which the buffer was removed and the cells washed with PBS to remove unincorporated NBD-labeled probe. Thereafter, the cells were incubated in PBS at 37°C for various timed intervals (0–60 min). At the indicated time, the buffer was removed, the cells were washed once with PBS before freezing, and then scraped into n-hexane-2-propanol extraction solvent 3:2 (v/v) [24]. Polar lipids (including NBD-sphingomyelin) were separated from neutral lipids using silicic acid columns washed with chloroform–methanol 59:1 (v/v), followed by methanol. The polar lipids were resolved on Silica Gel 60 TLC plates using chloroform:methanol:water:acetic acid (150:112.5:6:10.5, v/v/v/v). NBD-labeled lipids were visualized on the TLC plates under UV light at 360 nm using an Alpha Innotech FluorChem Imager (San Leandro, CA). Levels of NBD-sphingomyelin remaining at each time point were compared to NBD-sphingomyelin at time zero (initial concentration) to obtain the percentage of NBD-sphingomyelin remaining in the cells.
NBD-Sphingomyelin Efflux
In order to perform the HDL-mediated NBD-sphingomyelin efflux experiments, cells were loaded with NBD-sphingomyelin (0.2 µM) and Nile red (0.1 µM) in PBS buffer for 10 min in a 37°C, 5% CO2 incubator. After incubation, the cells were washed with PBS buffer and then imaged sequentially for NBD-sphingomyelin (488 nm excitation, 540/30 emission) and Nile red (568 nm excitation, HQ598/40 emission). Lipid droplets were identified by morphology and confirmed by Nile red and NBD-sphingomyelin co-localization data. Once a suitable field of cells was chosen, a medial section passing through 5–10 cells was selected, and the section was scanned for the initial fluorescence intensity of NBD-sphingomyelin. HDL (10 µg HDL/mL) was added to start the efflux experiment. Since stability studies indicated that greater than 85% of the NBD-sphingomyelin probe remained intact after 40 min incubation, all experiments were completed within 40 min. Fluorescence intensity in the total area of lipid droplet and plasma membrane was measured over time and was used to gauge the extent of NBD-sphingomyelin efflux from each cell compartment. Cells were imaged and analyzed by Metamorph software to graphically determine the separate contributions of the efflux process. Data points were fitted (R2 = 0.996) to the following three-parameter, sigmoidal equation: y = A/(1 − e−((t − t0)/b)), where y is the percentage of HDL-mediated NBD-sphingomyelin efflux at time t, A indicates the percentage of sphingolipid pool available for efflux, 1/b is an apparent rate constant (k), and t0 is the time required to reach 50% of the maximal efflux, representing the half-time (t1/2) of the exchangeable pool. Half-time and k values were apparent values due to dependence on HDL acceptor concentrations. Using a nonlinear least squares routine, all parameters were obtained simultaneously from the fitted data. Unique fittings were obtained individually for each time course involving individual cells. Statistical methods involving the paired t-test (GraphPad Prism, San Diego, CA) were used to compare the parameters derived from lipid droplet and plasma membrane efflux curves. Experiments were performed in triplicate with n = 50–60 lipid droplets from n = 25 cells. Values with P < 0.05 were considered to be statistically significant.
Western Blot Analysis
Expression levels of lipid droplet (ADRP, caveolin-1, caveolin-2), plasma membrane (Na+, K+-ATPase, transferrin), and mitochondrial (cytochrome c oxidase) protein markers in homogenate and lipid droplet fractions isolated from L-cells [8] were determined by Western blot analysis. In brief, cell homogenate (5 µg/lane) and lipid droplet (10 µg protein/lane) proteins were subjected to SDS-PAGE using 12% tricine gels before transferring to 0.45-µm nitrocellulose paper (Sigma Chemical Co., St. Louis, MO) by electroblotting in a continuous buffer system at 0.8 mA/cm2 for 2 h. After transfer, blots were blocked in 3% gelatin in TBST (10 mM Tris–HCL, pH 8, 100 mM NaCl, 0.05% Tween-20) before incubation with the primary antibodies. Alkaline-phosphatase conjugates of goat anti-rabbit IgG and Sigma Fast 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolim tablets (Sigma, St. Louis, MO) were used to visualize bands of interest. Proteins were quantitated by densitometric analysis after image acquisition using a single-chip CCD (charge-coupled device) video camera and a computer workstation (IS-500 system from Alpha Innotech, San Leandro, CA). Image files were analyzed (mean 8-bit grayscale density) using NIH Image, available by anonymous FTP from http://zippy.nimh.nih.gov.
Lipid Analyses
Lipid droplets were isolated from L-cell fibroblasts as described in [8]. Lipids from lipid droplets were extracted with n-hexane-2-propanol 3:2 (v/v) and resolved into individual lipid classes using Silica Gel G TLC plates developed in petroleum ether:diethyl ether:methanol:acetic acid 90:7:2:0.5 [8]. Total cholesterol, free fatty acid, triacylglyceride, cholesteryl ester, and phospholipid content were determined by the method of Marzo et al. [25]. Individual phospholipids, including phosphatidic acid (PA), ethanolamine glycerophospholipid (PE), phosphatidylinositol (PI), phosphatidylserine (PS), choline glycerophospholipid (PC), and sphingomyelin (SM), were resolved from total phospholipid (PL) samples in the following manner: PL were eluted from the Silica Gel G TLC plates using chloroform:methanol:HCL (100:50:0.375, v/v/v) and applied to Silica Gel 60 TLC plates, where individual phospholipids were resolved using chloroform: methanol:water:acetic acid (150:112.5:6:10.5, v/v/v/v). Individual phospholipids were quantitated densiometrically as described previously [26, 27]. Protein concentration was determined by the method of Bradford from the dried protein extract residue digested overnight in 0.2 M KOH [28]. All glassware was washed with sulfuric acid-chromate before use.
NBD-Sphingomyelin Binding Affinity
Lipid droplet and plasma membrane fractions were isolated as described previously [8, 26]. To measure the sphingomyelin and choline glycerophospholipid (phosphatidylcholine) binding affinities of membranes (lipid droplet and plasma membrane) and lipid binding proteins (ADRP, ACBP), a fluorescent ligand, saturation binding assay was performed with NBD-sphingomyelin and NBD-phosphatidylcholine using a steady-state photon-counting fluorimeter [29]. Briefly, the NBD-labeled lipid (in 100 µM stock) was added incrementally (0.2–4.0 µL) to membrane (50 µg), ADRP (180 nM), or ACBP (180 nM) protein aliquots equilibrated in phosphate buffer (10 mM, pH 7.4). After each addition, the samples were allowed to mix at 25°C for 3–4 min before exciting the NBD-labeled probe at 480 nM. Fluorescence emission spectra was recorded from 490 to 600 nm and integrated after each aliquot addition. Corrections were made for blank (without lipid droplet or plasma membrane present) and background emission. The data were fitted using a simple, single binding site model as in [30] to determine binding capacities (Bmax) and binding affinities from the Kd values. Since Bmax was protein-mass-dependant, values were normalized by the protein mass for each sample.
Results
Intracellular Localization of NBD-Sphingomyelin
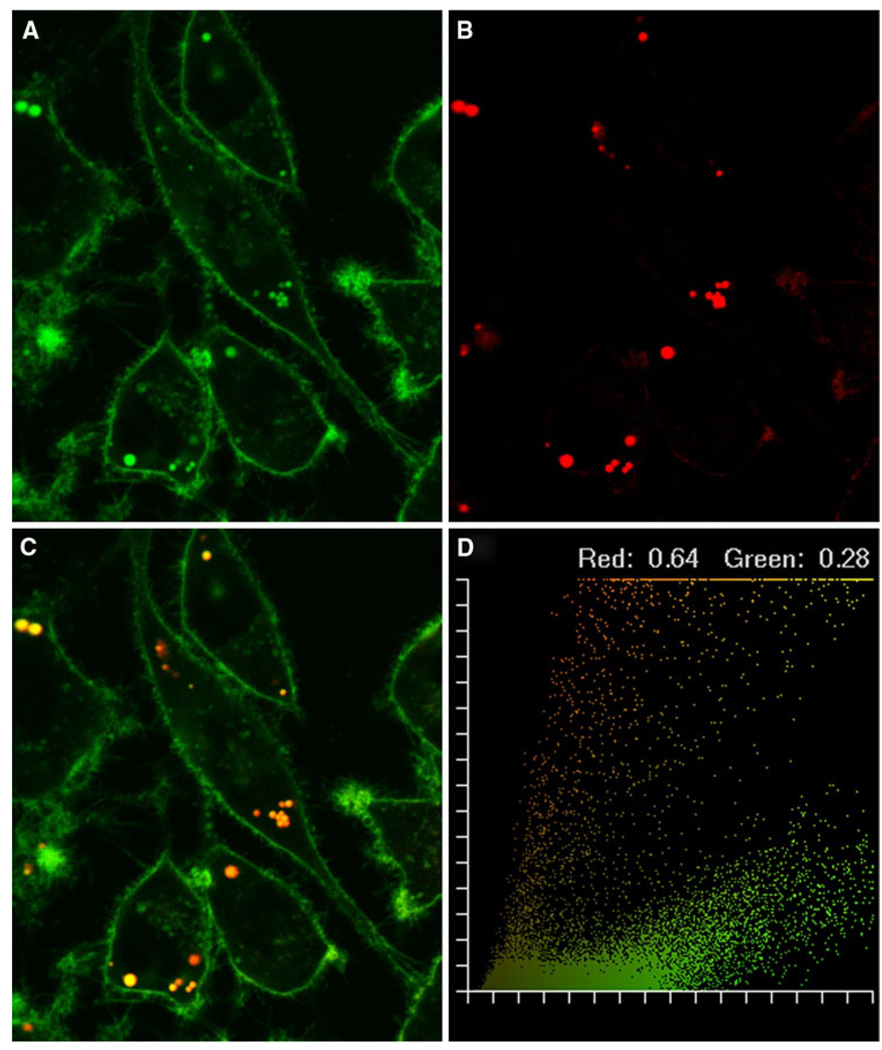
While it has been shown that NBD-labeled cholesterol and filipin (a cholesterol analog) localize to lipid droplets in living cells [8, 31], it was not known whether NBD-labeled sphingomyelin targeted to the lipid droplet phospholipid monolayer. Therefore, a series of experiments was performed with NBD-sphingomyelin and Nile red, a lipid droplet marker, to establish the intracellular localization of sphingomyelin in living cells. The NBD-sphingomyelin probe was initially observed in the plasma membrane, followed by lipid droplets after 5–10 min (Fig. 1a). After 20 min, the presence of NBD-sphingomyelin was observed in areas resembling the endoplasmic reticulum (ER) and Golgi apparatus. Nile red co-staining confirmed NBD-sphingomyelin targeted to lipid droplets (Fig. 1b). Overlap of Nile red with NBD-sphingomyelin was observed as yellow to red pixels, indicating the co-localization of probes (Fig. 1c). The extent of co-localization was quantified in pixel fluorograms where coefficients of overlap indicated that 28% of the NBD-label co-localized with the lipid stain (Nile red), while 64% of the Nile red was co-localized with NBD-sphingomyelin (Fig. 1d). These results were in agreement with NBD-sphingomyelin targeting to both the lipid droplet and plasma membrane compartments.
Fig. 1.
Intracellular distribution of NBD-sphingomyelin and Nile red in L-cell fibroblasts. Co-localization patterns of NBD-sphingomyelin and Nile red were shown using pseudo-coloring derived from confocal image acquisition from red- and green-specific PMT channels. A 24-bit RGB image was created from the red plus green plus blue (null) channels. Red and green are additive in the RGB color space, yielding yellow-orange. L-cell fibroblasts stained with NBD-sphingomyelin (a) and Nile red (b) were combined to yield yellow-orange (c) where co-localization occurred. Superimposition of the probes was graphically demonstrated in a pixel fluorogram (d). The correlation coefficients corresponding to red and green were proportional to the degree of fluorescence probe co-localizing in each component of the image relative to the total fluorescence and indicated the extent of probe overlap. The cells were examined using the Bio-Rad MRC-1024 confocal system. Objective, 63×
HDL-Mediated NBD-Sphingomyelin Efflux from Lipid Droplets and the Plasma Membrane
Since there are at least three different metabolic pathways for sphingomyelin catabolism in cultured fibroblasts, including: (1) degradation by sphingomyelinase in the lysosome; (2) hydrolysis by cellular sphingomyelinase in the plasma membrane to form ceramide and phosphorylcholine; or (3) hydrolysis of sphingomyelin to form lecithin (review in [32]), it was necessary to determine the stability of the NBD-labeled probe in culture. Stability studies were performed as described in the “Materials and Methods” to ensure that the NBD-labeled probe was intact during the time frame of the efflux experiments. After 40 min incubation of NBD-sphingomyelin in culture, more than 85% of the probe remained (data not shown). Therefore, all experiments were kept under 40 min to ensure minimal probe degradation.
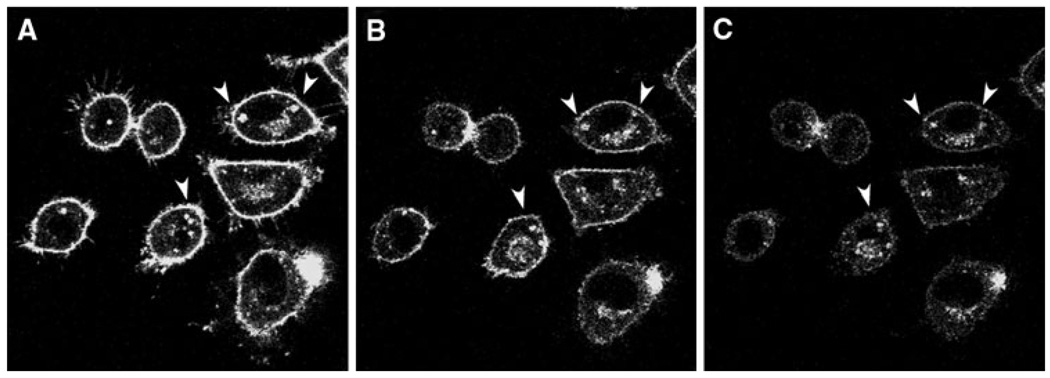
HDL-mediated NBD-sphingomyelin efflux from lipid droplets and the plasma membrane was next examined. The existence of mobile pools of cholesterol in lipid droplets was previously reported in [20], where the analysis of data from HDL-mediated NBD-cholesterol efflux curves revealed that lipid droplets contained two kinetic sterol domains, one with t1/2 = 10–20 min and the other with t1/2 near 1 min, suggesting vesicular- and protein-mediated sterol transfer, respectively [20]. However, examining sterol domains within the plasma membrane was not possible with NBD-cholesterol since the probe was not observed in the plasma membrane [20]. In contrast, NBD-sphingomyelin exhibited a high fluorescence intensity in both plasma membrane and lipid droplets (Figs. 1 and 2). Therefore, the relative distribution of fluorescence in lipid droplets and the plasma membrane during HDL-mediated sphingomyelin efflux could be observed as in Fig. 2 at: 0 min (Fig. 2a), 20 min (Fig. 2b), and 30 min (Fig. 2c) from the point of HDL addition.
Fig. 2.
HDL-mediated NBD-sphingomyelin efflux in L-cell fibroblasts. Cells were labeled with NBD-sphingomyelin (0.2 µM) and incubated with HDL (10 µg HDL/mL) to begin the efflux process. a 0 min, b 20 min, and c 30 min after the addition of HDL. The arrows indicate high-retention lipid droplets during the time course. Cells were examined with the Bio-Rad MRC-1024 confocal imaging system. Objective, 63×
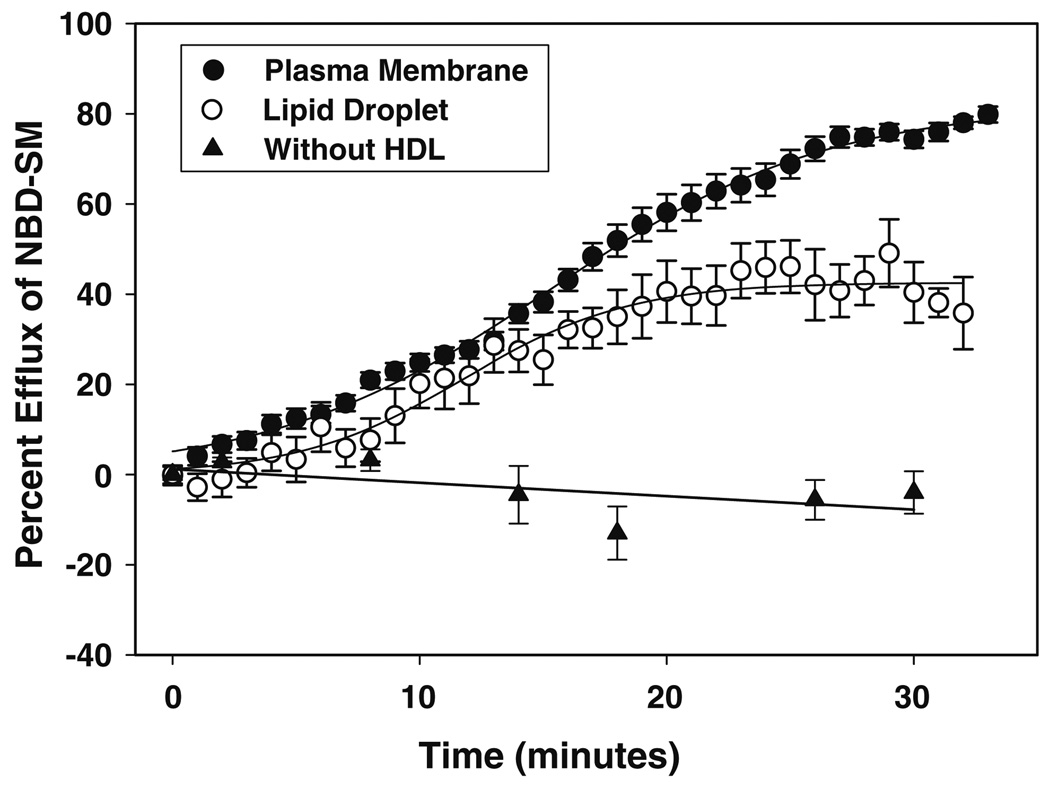
Next, cells were imaged and graphically partitioned to separate the lipid droplet contribution from the plasma membrane component of the cell as described in the “Materials and Methods” section. Kinetic analysis of NBD-sphingomyelin fluorescence revealed that efflux from the lipid droplet (Fig. 3, open circles) and the plasma membrane (Fig. 3, closed circles) best fit to a multi-parameter, sigmoidal equation (R2 = 0.996). Further examination revealed that the maximal HDL-mediated efflux from lipid droplets was less than that from the plasma membrane, suggesting that lipid droplets contained a smaller exchangeable sphingomyelin efflux pool. Parameters derived from the lipid droplet and plasma membrane efflux curves included the available NBD-sphingomyelin pool size (A), rate constant (k), and time required to reach 50% of maximal efflux (T0). A comparison of the available sphingomyelin pool size (Table 1) between lipid droplets and plasma membrane revealed not only that lipid droplets had a smaller exchangeable pool size (42.6 ± 1.1 vs. 81.9 ± 1.2), but also that the time required to efflux that pool was 22% less in lipid droplets, in keeping with a significantly faster lipid droplet versus plasma membrane efflux rate constant (0.3 ± 0.03 vs. 0.18 ± 0.06, P < 0.04).
Fig. 3.
Percent efflux of NBD-sphingomyelin. The percentage of HDL-mediated NBD-sphingomyelin efflux from the plasma membrane and lipid droplet compartments of living cells was determined as described in the “Materials and Methods” section. Data analysis of NBD-sphingomyelin fluorescence showed that efflux from the lipid droplet (open circles) and the plasma membrane (closed circles) best fit the following sigmoidal equation: y = A/(1 − e−((t− −t0)/b)), where y is the percentage of HDL-mediated NBD-sphingomyelin efflux at time t, A indicates the percentage of sphingolipid pool available for efflux, 1/b is an apparent rate constant (k), and t0 is the time required to reach 50% of the maximal efflux, representing the half-time (t1/2) of the exchangeable pool. Percentage efflux in the presence of medium without HDL present is indicated by closed triangles
Table 1.
Kinetic parameters of HDL-mediated NBD-sphingomyelin efflux from lipid droplets and plasma membrane
| Cell compartment | A | k(min−1) | T0 (min) |
|---|---|---|---|
| Lipid droplet | 42.6 ± 1.1 | 0.3 ± 0.03 | 11.8 ± 0.4 |
| Plasma membrane | 81.9 ± 1.2* | 0.18 ± 0.06* | 15.2 ± 0.3* |
The percentage of exchangeable sphingolipid pool size (A), rate constant (k), and time required to reach 50% of the maximal efflux (T0) was determined for lipid droplet and plasma membrane cell compartments within living cells as described in the “Materials and Methods” section. Values represent the mean ± SEM, with n = 25 cells, 50–60 lipid droplets
Indicates significance, P < 0.003 as compared to lipid droplets
The slower plasma membrane efflux parameters may also, in part, be attributed to the efflux of NBD-sphingomyelin from lipid droplets and other compartments cycling through the plasma membrane [33]. These contributions were included in the efflux measurements to ensure that the natural dynamics of the cell were represented. Thus, the kinetic domains of NBD-labeled sphingomyelin within lipid droplets versus the plasma membrane exhibited significant differences.
NBD-Sphingomyelin Binding Affinity for Proteins in Isolated Membrane Fractions
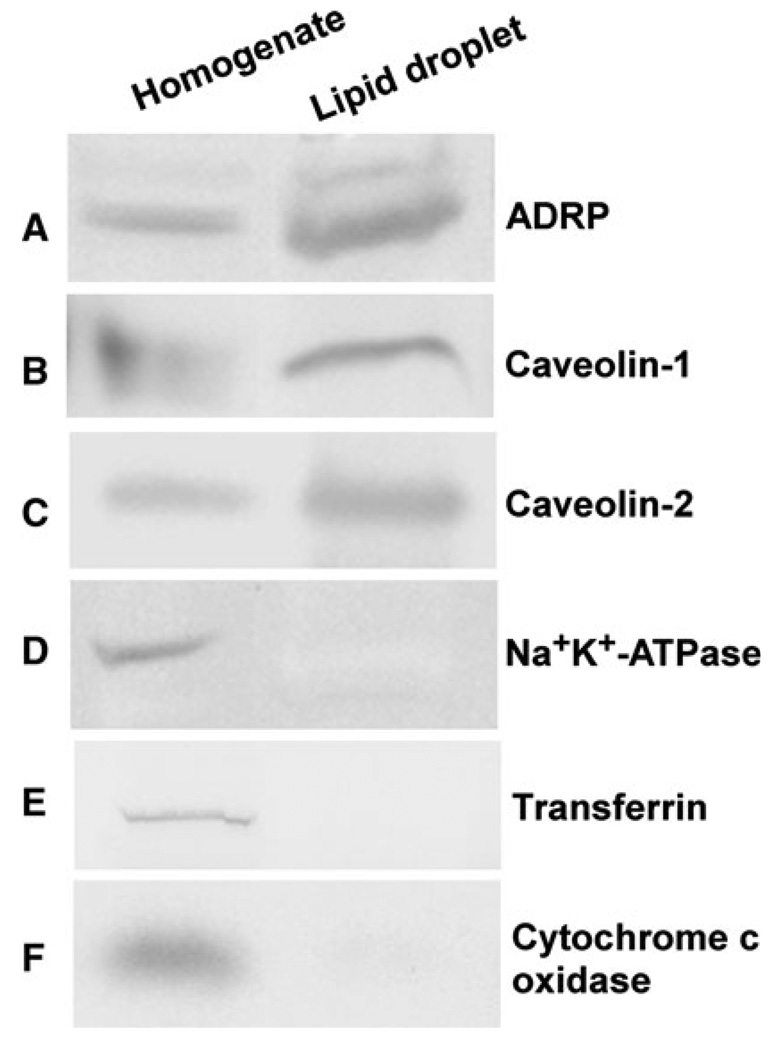
In order to determine the affinity of proteins associated with lipid droplets and the plasma membrane for sphingomyelin, fluorescence binding assays were performed. First, the purity and enrichment of isolated lipid droplets and plasma membranes were determined by Western blot analysis as described in [8, 26]. Protein expression of the lipid droplet marker proteins ADRP (Fig. 4a), caveolin-1 (Fig. 4b), and caveolin-2 (Fig. 4c) was detected in L-cell homogenates (lane 1) and purified lipid droplets (lane 2). The plasma membrane protein markers Na+, K+-ATPase (Fig. 4d), and transferrin (Fig. 4e), while present in the L-cell homogenate (lane 1), were not detected in the lipid droplet enriched fraction (lane 2), indicating that there was little to no plasma membrane present in the purified lipid droplet fractions. Similarly, the mitochondrial protein marker, cytochrome c oxidase (Fig. 4f), while observed in the homogenate (Fig. 4, lane 1), was not present in the lipid droplet (Fig. 4, lane 2) fraction.
Fig. 4.
Western blot analysis of lipid droplet proteins isolated from L-cell fibroblasts. Western blots on cell homogenate (lane 1) and lipid droplets (lane 2) were probed with the following affinity-purified antibodies as described in the “Materials and Methods” section: a anti-ADRP, b anti-caveolin-1, c anti-caveolin-2, d anti-Na+, K+-ATPase, e transferrin, and f anti-cytochrome c oxidase
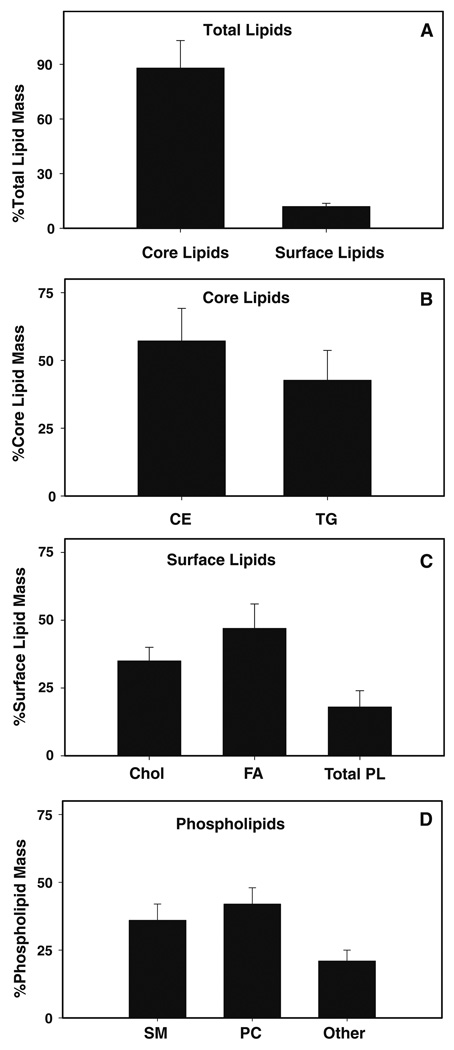
Second, the enrichment of neutral lipids and the presence of sphingomyelin in the isolated lipid droplet fraction were verified by lipid analysis. There has been some conjecture as to whether lipid droplets contain sphingomyelin in the phospholipid monolayer [34]. However, numerous studies on lipid droplets isolated from different cell types, including those from alveolar macrophages [35], Niemann–Pick cells [36], mammary tissue [37], and HepG2 cells [38], record the presence of sphingomyelin in lipid droplet preparations. It should be noted that the lack of sphingomyelin, PS, and PA observed in the former study [34] may be based on the specific cell type or from the isolation and extraction procedure. In the present work, lipid droplet lipids were extracted and the lipid profile resolved as described in the “Materials and Methods” section. The lipid droplet fraction showed an almost exclusive presence of core lipids (cholesteryl ester and triacylglyceride) over surface lipids (sphingomyelin, cholesterol, phospholipids, and fatty acids), with the core lipids present at almost equal levels in L-cell lipid droplets (Fig. 5), further confirming the purity and enrichment of the lipid droplet preparation since lipid droplets are the only organelle composed mostly of cholesteryl esters and triglycerides. Analysis of the individual phospholipid classes revealed the following lipid profile (nmol/mg protein): sphingomyelin, 1.8 ± 0.3; PC, 2.1 ± 0.3; PS/PI, 0.7 ± 0.1; and PE, 0.35 ± 0.2. PA was below the level of detection. These results confirmed the enrichment of neutral lipids and the presence of sphingomyelin (among other lipids) in the isolated lipid droplet fraction.
Fig. 5.
Percentage lipid mass of lipid droplets isolated from L-cell fibroblasts. Percentage lipid mass of total lipids (a), core lipids (b), surface lipids (c), and phospholipids (d) were determined from lipid droplets isolated from L-cells. Core lipids include triacylglycerol (TG) and cholesteryl esters (CE). Surface lipids include cholesterol (Chol), phospholipids (PL), and free fatty acids (FA). Phospholipids include sphingomyelin (SM), choline glycerophospholipid (PC), phosphatidylserine (PS), phosphatidylinositol (PI), ethanolamine glycerophospholipid (PE), and phosphatidic acid (PA)
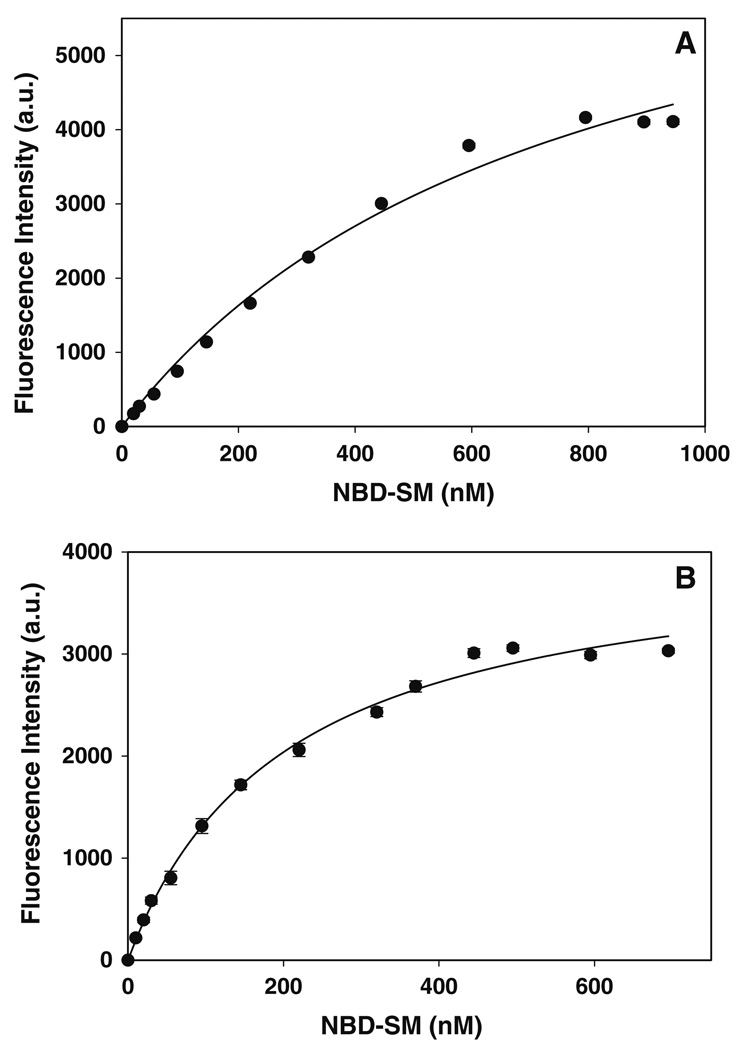
Third, the ability of proteins associated with the plasma membrane and lipid droplets to bind NBD-labeled sphingomyelin and phosphatidylcholine was established through a fluorescent ligand, saturation binding assay using a steadystate photon-counting fluorimeter [29]. The interaction of plasma membrane and lipid droplet associated proteins with NBD-labeled lipids was monitored as increased fluorescence intensity at 530 nm, consistent with localization of the ligand in a more hydrophobic environment. Plasma membrane (Fig. 6a) and lipid droplet (Fig. 6b) fractions exhibited saturation binding for NBD-sphingomyelin. From the saturation curves, fitted to a simple, single binding site model as in [29], binding affinities (Kd) and binding capacities (Bmax) were determined (Table 2). Analysis of the binding data revealed that the association of NBD-sphingomyelin for proteins in the lipid droplet fraction was 3.8-fold greater (P < 0.05, n = 3) than that for plasma membrane proteins (Kd 201 ± 19 nM vs. Kd 758 ± 118 nM). When NBD-sphingomyelin binding studies were performed with recombinant ADRP (Fig. 7a), it was shown that ADRP exhibited similar binding affinities to that of the purified lipid droplet fraction (Kd 201 ± 19 nM vs. Kd 257 ± 64 nM). Differences were also observed in the NBD-sphingomyelin binding capacities (Bmax). Under the experimental conditions presented herein, differences in Bmax indicated differences in the number of lipid binding proteins and lipid binding sites. The NBD-sphingomyelin binding capacity of lipid droplet proteins was 1.9-fold lower (P < 0.005, n = 3) than that observed with plasma membranes (Table 2), suggesting that the plasma membrane contained more sphingomyelin binding proteins. However, there was no significant difference between the NBD-sphingomyelin binding capacity of the lipid droplet protein fraction and ADRP. Thus, fluorescence binding assays revealed the presence of sphingomyelin binding proteins in the purified lipid droplet fraction. Further characterization identified ADRP as one of the sphingomyelin binding proteins in the lipid droplet fraction.
Fig. 6.
Titration of plasma membrane and lipid droplet fractions with NBD-sphingomyelin. The titration of plasma membranes (a) and lipid droplet (b) fractions with NBD-sphingomyelin was followed by an increase in fluorescence intensity as described in the “Materials and Methods” section. The fluorescent intensity data, representing the mean ± SE of three independent measurements, were analyzed to determine the dissociation constant Kd and binding capacity Bmax
Table 2.
Binding affinity and capacity of plasma membrane and lipid droplet proteins, ADRP, and ACBP for NBD-labeled sphingomyelin and phosphatidylcholine
| Protein | NBD label | Kd (nM) | Bmax/µg protein |
|---|---|---|---|
| PM | NBD-SM | 758 ± 119 | 156 ± 13 |
| LD | NBD-SM | 201 ± 19a | 82 ± 3a |
| ADRP | NBD-SM | 257 ± 64a | 111 ± 12 |
| ADRP | NBD-PC | 15 ± 3a,b,c | 148 ± 8 |
| ACBP | NBD-SM | NDa,b,c | NDa,b,c |
The binding affinity and capacity of plasma membrane (PM) and lipid droplet (LD) proteins and ADRP and ACBP for NBD-labeled sphingomyelin (SM) and phosphatidylcholine (PC) was measured by a fluorescent lipid binding assay as described in the “Materials and Methods” section. Values represent the mean ± SEM, n = 3
Indicates significance, P < 0.02 as compared to PM binding of NBD-SM
Indicates significance, P < 0.0001 as compared to LD binding of NBD-SM
Indicates significance, P < 0.02 as compared to ADRP binding of NBD-SM
ND indicates not detected
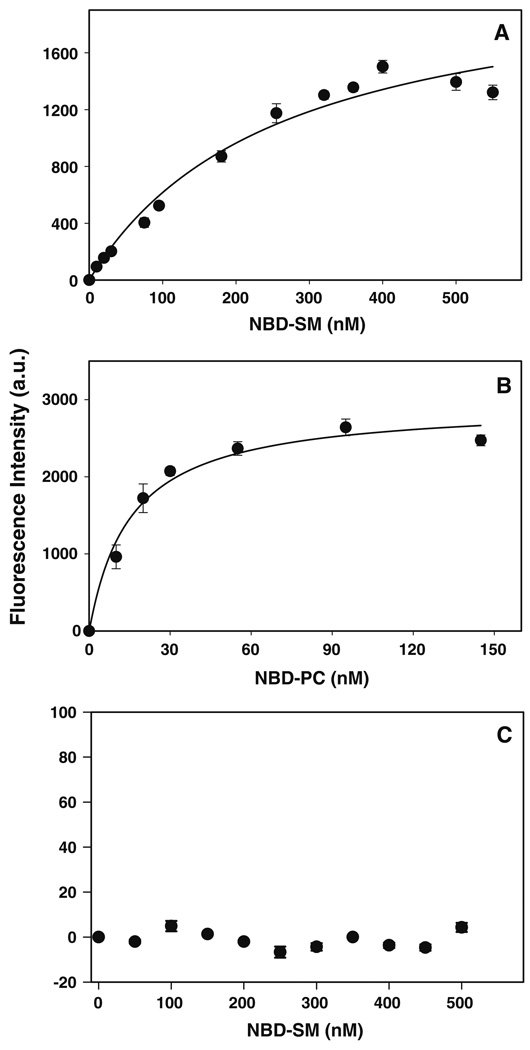
Fig. 7.
Titration of ADRP and ACBP with NBD-labeled lipids. The titration of ADRP (a, b) or ACBP (c) with NBD-sphingomyelin (a, c) or NBD-phosphatidylcholine (b) was followed by an increase in fluorescence intensity as described in the “Materials and Methods” section. The fluorescent intensity data, representing the mean ± SE of three independent measurements, were analyzed to determine the dissociation constant Kd and binding capacity Bmax
The ability of ADRP to interact with another NBD-labeled phospholipid (NBD-phosphatidylcholine) was also examined. Upon titration of ADRP with increasing concentrations of NBD-phosphatidylcholine, an increase in fluorescence intensity was observed as before with NBD-sphingomyelin, indicating binding (Fig. 7b). Analysis of the data revealed that ADRP bound NBD-phosphatidylcholine with high affinity (Kd = 15 ± 3 nM), 17.1-fold greater than that observed for NBD-sphingomyelin (Table 2). Despite this, there were no significant differences between the Bmax of NBD-labeled sphingomyelin and phosphatidylcholine indicating that, regardless of the stronger binding affinity of ADRP for NBD-phosphatidylcholine, ADRP contained the same number of sphingomyelin and phosphatidylcholine binding sites.
The specificity of ADRP binding sphingomyelin was examined next. The NBD-sphingomyelin fluorescence binding assay was performed with an unrelated protein, acyl-CoA binding protein (ACBP), a cytosolic lipid binding protein that exclusively binds long-chain fatty acyl-CoA [39]. Increased fluorescence was not observed when ACBP was titered with increasing concentrations of NBD-sphingomyelin (Fig. 7c). A plot of the maximal intensity of NBD-sphingomyelin emission at 530 nm versus increasing NBD-sphingomyelin concentration yielded a line with scattered points, not the saturation curve observed previously with ADRP binding of sphingomyelin and phosphatidylcholine. These results suggested that NBD-sphingomyelin did not enter the ACBP binding pocket and indicated that ACBP had little to no sphingomyelin binding affinity in this assay.
In summary, fluorescence binding assays indicated that the purified lipid droplet and plasma membrane protein fractions contained sphingomyelin binding proteins. The lipid droplet associated protein ADRP was identified as a phospholipid binding protein with nanomolar affinity for sphingomyelin and phosphatidylcholine.
Discussion
Although there is growing interest in the role that lipid droplets play in lipid transport, cell signaling, immune function, and membrane trafficking (review in [40, 41]), questions remain regarding the means by which lipid droplet proteins and lipids interact to regulate cellular dynamics. Cholesterol and sphingomyelin are essential membrane components, and when levels are high, the ability to remove excess lipids through storage or efflux remains an effective method for cells to regulate otherwise harmful lipids. Therefore, the regulation of cholesterol and sphingomyelin within lipid reservoirs such as lipid droplets is important to maintain lipid homeostasis. However, while the dynamics of cholesterol efflux from lipid droplets and the plasma membrane has been studied [20], less is known regarding the efflux of sphingomyelin from these membranes. The present work was undertaken to examine the efflux kinetics and binding affinities of different intracellular sphingomyelin pools in lipid droplets versus the plasma membrane.
In preparation for efflux studies, NBD-sphingomyelin targeting to lipid droplets in living cells was established. It should be noted that, while there are some reports that NBD-labeled lipids do not distribute to the same organelles as the natural lipids [42], it has been shown in other work that NBD-labeled probes are metabolized similarly [29] and traffic by similar uptake, intracellular, and secretory pathways [29, 43]. In CHO-K1 cells, NBD-sphingomyelin internalized from the plasma membrane to vesicles and other internal sites, and then transported back to the plasma membrane [33, 44]. In a similar fashion, NBD-sphingomyelin in primary cultures of rat astrocytes [45] and in L-cells (present work) were endocytosed and sorted to several intracellular sites, including lipid droplets (results herein). Furthermore, with other NBD-labeled probes such as NBD-cholesterol, there is substantial evidence to suggest that NBD-cholesterol undergoes metabolism and utilizes the same intracellular trafficking pathways as the endogenous lipid [20, 29]. For example, in hamster intestinal enterocytes, CaCO2 cells, and L-cells [29, 43, 46], NBD-cholesterol was esterified similarly to cholesterol and dehydroergosterol, a natural cholesterol analog [47], while in CaCO2 cells and L-cells, NBD-cholesterol was shown to traffic by similar pathways [29, 43, 46]. Taken together, these studies suggest that, while the behaviors of NBD-labeled probes are not identical to their natural lipid counterparts, they can be useful to examine lipid trafficking and metabolism in living cells. Therefore, NBD-sphingomyelin was used to show sphingomyelin targeting to lipid droplets in living cells. Next, NBD-sphingomyelin was used to perform efflux studies where HDL-mediated NBD-sphingomyelin efflux from lipid droplets and the plasma membrane was graphically delineated. Kinetic analysis of the NBD-sphingomyelin efflux curves from lipid droplets revealed the presence of resolvable sphingolipid pools that exhibited properties which were different from those found in the plasma membrane (Table 1). The exchangeable sphingomyeslin pool (A) in lipid droplets (42.6 ± 1.1) was smaller than the sphingomyelin pool associated with plasma membranes (81.9 ±1.2). Despite this, the rate of sphingomyelin efflux was faster in lipid droplets and the half-time was significantly less. Sphingomyelin in lipid droplets is constrained to the surface, bound and unbound to resident proteins. Consistent with the data in the present work, at least two sphingomyelin pools were observed with lipid droplets, one smaller, dynamic pool with a half-time of 11.8 min, and another larger, relatively inert pool with a half-time on the order of hours to days. It should be noted that HDL-mediated NBD-cholesterol [20] and NBD-sphingomyelin efflux rates are comparable, yet faster and reach equilibrium quicker (minutes) than that observed in similar studies with [3H]-labeled lipids (hours) [20]. These results may be due to the higher aqueous solubility of NBD-labeled lipids versus the natural lipid counterparts [48]. Nevertheless, the rapid turnover of NBD-sphingomyelin in the current study is in keeping with the fact that L-cells endocytose the equivalent of their entire cell surface membrane within about 2 h [49], similar to the activity of macrophages [50].
In contrast to lipid droplets, sphingomyelin in the plasma membrane exhibited a small, inert pool (18% of total sphingolipid) and a large, exchangeable pool comprising 82% of the available sphingomyelin (half-time of 15.2 min) was observed. Sphingomyelin from this large, dynamic plasma membrane pool is readily available for participation in at least three different sphingomyelin metabolic pathways, including the degradation of sphingomyelin by sphingomyelinases in the lysosome, hydrolysis of sphingomyelin to form lecithin, and the sphingomyelin cycle [32, 51]. In cells, most of the sphingomyelin is located in the outer leaflet of the plasma membrane (review in [52]). The sphingomyelin cycle begins with the activation of sphingomyelinases located in the plasma membrane which hydrolyze sphingomyelin to form phosphorylcholine and ceramide. Ceramide acts as a second messenger with effects in multiple cellular processes, including the activation of specific kinases and phosphatases, cell cycle arrest, induction of cell differentiation, and apoptosis [53]. The presence of ceramide generated from activated sphingomyelinases also leads to the clustering of CD95 and CD40 receptors [54] and the activation of a number of signaling agents, including tumor necrosis factor α, γ-interferon, and arachidonate [55–57]. Sphingomyelin is resynthesized by the addition of a phosphorylcholine headgroup from phosphatidylcholine through the action of sphingomyelin synthase to complete the sphingomyelin cycle. However, while evidence for the above events occurring in lipid droplets is minimal, the presence of a small dynamic sphingomyelin pool in the lipid droplet monolayer is evocative of new roles for the storage organelle in lipid signaling.
The extent of specific organelle protein–lipid interactions in lipid droplets was next examined using a NBD-sphingomyelin fluorescence binding assay as described in the “Materials and Methods” section. While both lipid droplet and plasma membrane proteins exhibited saturable binding to NBD-labeled sphingomyelin, binding affinities of NBD-sphingomyelin for lipid droplet proteins (Kd 201 ± 19 nM) were higher than for plasma membrane proteins (Kd 758 ± 119 nM). In agreement with the binding studies, lipid droplets exhibited a larger inert sphingomyelin pool with strong binding affinity for sphingomyelin in comparison to the plasma membrane, which had a small inert sphingomyelin pool with weak sphingomyelin binding affinity. Thus, not only did the kinetic domains of NBD-labeled sphingomyelin within lipid droplets and the plasma membrane exhibit significant differences, the results presented herein show, for the first time, that collective proteins associated with lipid droplets versus plasma membrane show differential binding affinity to NBD-sphingomyelin.
While little is known regarding the identity or binding affinities of sphingomyelin-binding proteins within lipid droplets, the surface coat protein ADRP [10, 20, 29] was a likely choice based on its ability to bind other lipid molecules. ADRP was shown to bind hydrophobic lipids such as NBD-stearic acid (Kd 145 nM) and NBD-cholesterol (Kd 2.0 nM) in a 1:1 stoichiometry with high affinity [20, 29], and to also increase fatty acid uptake [9]. As shown in the present work, ADRP demonstrated saturable binding to NBD-sphingomyelin and also NBD-phosphatidylcholine with binding affinities in the nanomolar range. However, there was no significant difference in NBD-sphingomyelin binding capacity between the lipid droplet protein fraction and ADRP. Thus, fluorescence binding assays revealed the presence of sphingomyelin binding proteins in the purified lipid droplet fraction. Further examination confirmed that ADRP contributed to this pool and bound sphingomyelin and phosphatidylcholine with high affinity. The specificity of ADRP binding of sphingomyelin was also examined by determining whether an unrelated protein, ACBP, would bind sphingomyelin. A plot of the maximal intensity of NBD-sphingomyelin emission versus increasing NBD-sphingomyelin concentration yielded a line with scattered points, indicating that ACBP had little to no sphingomyelin binding affinity in this assay. Thus, while ADRP exhibited high affinity for sphingomyelin and phosphatidylcholine, performing the same assay with ACBP revealed little to no binding of sphingomyelin.
While this work presents the first evidence of a lipid droplet associated protein exhibiting phospholipid binding ability, there are other phospholipid binding proteins with similar binding affinities. Sterol carrier protein-2 (SCP-2) bound sphingomyelin with affinity near 173 nM [58], an affinity slightly higher than that observed with ADRP (Kd = 257 nM). Moreover, SCP-2 exhibited high binding affinities for other sphingolipids, including ceramides, GM1, and globosides [58]. In addition, surface plasmon resonance measurements of lysenin, a 41-kDa protein that causes contraction of the rat vascular smooth muscle, bound to sphingomyelin membranes with Kd = 5.3 nM [59]. However, while there are other proteins that bind sphingomyelin with similar affinity, it is ADRP’s position on the surface of the lipid droplet that makes it uniquely qualified to provide phospholipids to meet the needs of the cell during membrane synthesis, signaling, and cellular metabolism. Indeed, the ability of ADRP to bind with high affinity the two major phospholipids in cell membranes (sphingomyelin, phosphatidylcholine) suggests that ADRP may play an important role in intracellular phospholipid trafficking and lipid metabolism in general.
In summary, the results presented herein confirmed, through confocal fluorescence imaging and fluorescence binding studies, the existence of exchangeable sphingolipid domains in lipid droplets. In addition, the present investigation shows, for the first time, that ADRP binds phospholipids with high affinity. More importantly, ADRP exhibited similar binding affinities to other known phospholipid binding proteins. These results suggest that ADRP may play a significant role in regulating the intracellular distribution of phospholipids and lipids in general. Overall, insights from the present work suggest new and important roles for lipid droplets in phospholipid and lipid metabolism.
Acknowledgments
The helpful technical assistance of Bonnie M. McCann, Jessica Z. Howsley, Meredith Dixon, and Anas M. Fathalla was much appreciated. This work was supported in part by the USPHS National Institutes of Health grant DK70965 (BPA).
Abbreviations
- ADRP
Adipose differentiation-related protein
- ACBP
Acyl-CoA binding protein
- NBD-sphingomyelin
6-((N-(7-nitrobenz-2-oxa-1, 3-diazol-4-yl)amino)-hexanoyl)sphingosyl-phosphocholine
- NBD-phosphatidylcholine
2-(6-(7-nitrobenz-2-oxa-1, 3-diazol-4-yl)amino)hexanoyl-1-hexadecanoyl-sn-glycero-3-phosphocholine
- Chol
Cholesterol
- SM
Sphingomyelin
- PL
Phospholipid
- TG
Triacylglycerides
- CE
Cholesteryl ester
- FFA
Free fatty acid
- PE
Ethanolamine glycerophospholipid
- PI
Phosphatidylinositol
- PS
Phosphatidylserine
- PC
Choline glycerophospholipid or phosphatidylcholine
- PA
Phosphatidic acid
Contributor Information
Avery L. McIntosh, Department of Physiology and Pharmacology, Texas A&M University, TVMC, College Station, TX 77843, USA
Stephen M. Storey, Department of Physiology and Pharmacology, Texas A&M University, TVMC, College Station, TX 77843, USA
Barbara P. Atshaves, Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, MI 48824, USA, atshaves@msu.edu
References
- 1.Rexrode KM, Manson JE, Hennekens CH. Obesity and cardiovascular disease. Curr Opin Cardiol. 1996;11:490–495. doi: 10.1097/00001573-199609000-00007. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz MW, Brunzell JD. Regulation of body adiposity and the problem of obesity. Arterioscler Thromb Vasc Biol. 1997;17:223–238. doi: 10.1161/01.atv.17.2.233. [DOI] [PubMed] [Google Scholar]
- 3.Ogden CL, Carroll MD, Flegal KM. Epidemiologic trends in overweight and obesity. Endocrinol Metab Clin North Am. 2003;32:741–760. doi: 10.1016/s0889-8529(03)00074-4. [DOI] [PubMed] [Google Scholar]
- 4.Bickel PE, Tansey JT, Welte MA. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim Biophys Acta. 2009;1791:419–440. doi: 10.1016/j.bbalip.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujimoto T, Ohsaki Y, Cheng J, Suzuki M, Shinohara Y. Lipid droplets: a classic organelle with new outfits. Histochem Cell Biol. 2008;130:263–279. doi: 10.1007/s00418-008-0449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- 7.Londos C, Brasaemle DL, Schultz CJ, Segrest JP, Kimmel AR. Perilipins, ADRP, and other proteins that associate with intracellular neutral lipid droplets in animal cells. Semin Cell Dev Biol. 1999;10:51–58. doi: 10.1006/scdb.1998.0275. [DOI] [PubMed] [Google Scholar]
- 8.Atshaves BP, Storey SM, McIntosh AL, Petrescu AD, Lyuksyutova OI, Greenberg AS, Schroeder F. Sterol carrier protein-2 expression modulates protein and lipid composition of lipid droplets. J Biol Chem. 2001;276:25324–25335. doi: 10.1074/jbc.M100560200. [DOI] [PubMed] [Google Scholar]
- 9.Gao J, Serrero G. Adipose differentiation related protein (ADRP) expressed in transfected COS-7 cells selectively stimulates long chain fatty acid uptake. J Biol Chem. 1999;274:16825–16830. doi: 10.1074/jbc.274.24.16825. [DOI] [PubMed] [Google Scholar]
- 10.Brasaemle DL, Barber T, Wolins NE, Serrero G, Blanchette-Mackie EJ, Londos C. Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet-associated protein. J Lipid Res. 1997;38:2249–2263. [PubMed] [Google Scholar]
- 11.Wang SM, Fong TH, Hsu SY, Chien CL, Wu JC. Reorganization of a novel vimentin-associated protein in 3T3-L1 cells during adipose conversion. J Cell Biochem. 1997;67:84–91. [PubMed] [Google Scholar]
- 12.Fong TH, Wang SM, Lin HS. Immunocytochemical demonstration of a lipid droplet-specific capsule in cultured Leydig cells of the golden hamsters. J Cell Biochem. 1996;63:366–373. doi: 10.1002/(sici)1097-4644(19961201)63:3<366::aid-jcb12>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 13.Liu P, Ying Y, Zhao Y, Mundy DI, Zhu M, Anderson RGW. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J Biol Chem. 2004;279:3787–3792. doi: 10.1074/jbc.M311945200. [DOI] [PubMed] [Google Scholar]
- 14.Brasaemle DL, Dolios G, Shapiro L, Wang R. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J Biol Chem. 2004;279:46835–46842. doi: 10.1074/jbc.M409340200. [DOI] [PubMed] [Google Scholar]
- 15.Fujimoto Y, Itabe H, Sakai J, Makita M, Noda J, Mori M, Higashi Y, Kojima S, Takano T. Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim Biophys Acta. 2004;1644:47–59. doi: 10.1016/j.bbamcr.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 16.Trigatti BL, Anderson RGW, Gerber GE. Identification of caveolin-1 as a fatty acid binding protein. Biochem Biophys Res Commun. 1999;255:34–39. doi: 10.1006/bbrc.1998.0123. [DOI] [PubMed] [Google Scholar]
- 17.Uittenbogaard A, Ying YS, Smart EJ. Characterization of a cytosolic heat-shock protein-caveolin chaperone complex. Involvement in cholesterol trafficking. J Biol Chem. 1998;273:6525–6532. doi: 10.1074/jbc.273.11.6525. [DOI] [PubMed] [Google Scholar]
- 18.Uittenbogaard A, Everson WV, Matveev SV, Smart EJ. Cholesteryl ester is transported from caveolae to internal membranes as part of a caveolin-annexin II lipid–protein complex. J Biol Chem. 2002;277:4925–4931. doi: 10.1074/jbc.M109278200. [DOI] [PubMed] [Google Scholar]
- 19.Murata M, Peränen J, Schreiner R, Wieland F, Kurzchalia TV, Simons K. VIP21/caveolin is a cholesterol-binding protein. Proc Natl Acad Sci USA. 1995;92:10339–10343. doi: 10.1073/pnas.92.22.10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atshaves BP, Starodub O, McIntosh AL, Petrescu A, Roths JB, Kier AB, Schroeder F. Sterol carrier protein-2 alters high density lipoprotein-mediated cholesterol efflux. J Biol Chem. 2000;275:36852–36861. doi: 10.1074/jbc.M003434200. [DOI] [PubMed] [Google Scholar]
- 21.Atshaves BP, Petrescu AD, Starodub O, Roths JB, Kier AB, Schroeder F. Expression and intracellular processing of the 58 kDa sterol carrier protein-2/3-oxoacyl-CoA thiolase in transfected mouse L-cell fibroblasts. J Lipid Res. 1999;40:610–622. [PubMed] [Google Scholar]
- 22.Higuchi K. An improved chemically defined culture medium for strain L mouse cells based on growth responses to graded levels of nutrients including iron and zinc ions. J Cell Physiol. 1970;75:65–72. doi: 10.1002/jcp.1040750108. [DOI] [PubMed] [Google Scholar]
- 23.Atshaves BP, Storey SM, Petrescu AD, Greenberg CC, Lyuksyutova OI, Smith RI, 3rd, Schroeder F. Expression of fatty acid binding proteins inhibits lipid accumulation and alters toxicity in L cell fibroblasts. Am J Physiol Cell Physiol. 2002;283:C688–C703. doi: 10.1152/ajpcell.00586.2001. [DOI] [PubMed] [Google Scholar]
- 24.Hara A, Radin NS. Lipid extraction of tissues with a low-toxicity solvent. Anal Biochem. 1978;90:420–426. doi: 10.1016/0003-2697(78)90046-5. [DOI] [PubMed] [Google Scholar]
- 25.Marzo A, Ghirardi P, Sardini D, Meroni G. Simplified measurement of monoglycerides, diglycerides, triglycerides, and free fatty acids in biological samples. Clin Chem. 1971;17:145–147. [PubMed] [Google Scholar]
- 26.Atshaves BP, Gallegos AM, McIntosh AL, Kier AB, Schroeder F. Sterol carrier protein-2 selectively alters lipid composition and cholesterol dynamics of caveolae/lipid raft vs nonraft domains in L-cell fibroblast plasma membranes. Biochemistry. 2003;42:14583–14598. doi: 10.1021/bi034966+. [DOI] [PubMed] [Google Scholar]
- 27.Eckert GP, Igbavboa U, Müller WE, Wood WG. Lipid rafts of purified mouse brain synaptosomes prepared with or without detergent reveal different lipid and protein domains. Brain Res. 2003;962:144–150. doi: 10.1016/s0006-8993(02)03986-0. [DOI] [PubMed] [Google Scholar]
- 28.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 29.Frolov A, Petrescu A, Atshaves BP, So PTC, Gratton E, Serrero G, Schroeder F. High density lipoprotein-mediated cholesterol uptake and targeting to lipid droplets in intact L-cell fibroblasts. A single- and multiphoton fluorescence approach. J Biol Chem. 2000;275:12769–12780. doi: 10.1074/jbc.275.17.12769. [DOI] [PubMed] [Google Scholar]
- 30.Schroeder F, Myers-Payne SC, Billheimer JT, Wood WG. Probing the ligand binding sites of fatty acid and sterol carrier proteins: effects of ethanol. Biochemistry. 1995;34:11919–11927. doi: 10.1021/bi00037a033. [DOI] [PubMed] [Google Scholar]
- 31.Prattes S, Hörl G, Hammer A, Blaschitz A, Graier WF, Sattler W, Zechner R, Steyrer E. Intracellular distribution and mobilization of unesterified cholesterol in adipocytes: triglyceride droplets are surrounded by cholesterol-rich ER-like surface layer structures. J Cell Sci. 2000;113:2977–2989. doi: 10.1242/jcs.113.17.2977. [DOI] [PubMed] [Google Scholar]
- 32.Spence MW, Clarke JTR, Cook HW. Pathways of sphingomyelin metabolism in cultured fibroblasts from normal and sphingomyelin lipidosis subjects. J Biol Chem. 1983;258:8595–8600. [PubMed] [Google Scholar]
- 33.Lipsky NG, Pagano RE. Intracellular translocation of fluorescent sphingolipids in cultured fibroblasts: endogenously synthesized sphingomyelin and glucocerebroside analogues pass through the Golgi apparatus en route to the plasma membrane. J Cell Biol. 1985;100:27–34. doi: 10.1083/jcb.100.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartz R, Li WH, Venables B, Zehmer JK, Roth MR, Welti R, Anderson RGW, Liu P, Chapman KD. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J Lipid Res. 2007;48:837–847. doi: 10.1194/jlr.M600413-JLR200. [DOI] [PubMed] [Google Scholar]
- 35.Ishii I, Onozaki R, Takahashi E, Takahashi S, Fujio N, Harada T, Morisaki N, Shirai K, Saito Y, Hirose S. Regulation of neutral cholesterol esterase activity by phospholipids containing negative charges in substrate liposome. J Lipid Res. 1995;36:2303–2310. [PubMed] [Google Scholar]
- 36.Knudson AG., Jr Inborn errors of sphingolipid metabolism. Am J Clin Nutr. 1961;9:55–62. doi: 10.1093/ajcn/9.1.55. [DOI] [PubMed] [Google Scholar]
- 37.Hood LF, Patton S. Isolation and characterization of intracellular lipid droplets from bovine mammary tissue. J Dairy Sci. 1973;56:858–863. doi: 10.3168/jds.S0022-0302(73)85267-1. [DOI] [PubMed] [Google Scholar]
- 38.Tauchi-Sato K, Ozeki S, Houjou T, Taguchi R, Fujimoto T. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J Biol Chem. 2002;277:44507–44512. doi: 10.1074/jbc.M207712200. [DOI] [PubMed] [Google Scholar]
- 39.Frolov A, Schroeder F. Acyl coenzyme A binding protein. Conformational sensitivity to long chain fatty acyl-CoA. J Biol Chem. 1998;273:11049–11055. doi: 10.1074/jbc.273.18.11049. [DOI] [PubMed] [Google Scholar]
- 40.Simons K, Ikonen E. How cells handle cholesterol. Science. 2000;290:1721–1726. doi: 10.1126/science.290.5497.1721. [DOI] [PubMed] [Google Scholar]
- 41.Murphy DJ. The biogenesis and functions of lipid bodies in animals, plants, and microorganisms. Prog Lipid Res. 2001;40:325–438. doi: 10.1016/s0163-7827(01)00013-3. [DOI] [PubMed] [Google Scholar]
- 42.Mukherjee S, Zha X, Tabas I, Maxfield FR. Cholesterol distribution in living cells: fluorescence imaging using dehydroergosterol as a fluorescent cholesterol analog. Biophys J. 1998;75:1915–1925. doi: 10.1016/S0006-3495(98)77632-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sparrow CP, Patel S, Baffic J, Chao Y-S, Hernandez M, Lam M-H, Montenegro J, Wright SD, Detmers PA. A fluorescent cholesterol analog traces cholesterol absorption in hamsters and is esterified in vivo and in vitro. J Lipid Res. 1999;40:1747–1757. [PubMed] [Google Scholar]
- 44.Koval M, Pagano RE. Lipid recycling between the plasma membrane and intracellular compartments: transport and metabolism of fluorescent sphingomyelin analogues in cultured fibroblasts. J Cell Biol. 1989;108:2169–2181. doi: 10.1083/jcb.108.6.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomás M, Durán JM, Lázaro-Diéguez F, Babiá T, Renau-Piqueras J, Egea G. Fluorescent analogues of plasma membrane sphingolipids are sorted to different intracellular compartments in astrocytes; Harmful effects of chronic ethanol exposure on sphingolipid trafficking and metabolism. FEBS Lett. 2004;563:59–65. doi: 10.1016/S0014-5793(04)00245-5. [DOI] [PubMed] [Google Scholar]
- 46.Reaven E, Tsai L, Azhar S. Intracellular events in the “selective“ transport of lipoprotein-derived cholesteryl esters. J Biol Chem. 1996;271:16208–16217. doi: 10.1074/jbc.271.27.16208. [DOI] [PubMed] [Google Scholar]
- 47.McIntosh AL, Gallegos AM, Atshaves BP, Storey SM, Kannoju D, Schroeder F. Fluorescence and multiphoton imaging resolve unique structural forms of sterol in membranes of living cells. J Biol Chem. 2003;278:6384–6403. doi: 10.1074/jbc.M205472200. [DOI] [PubMed] [Google Scholar]
- 48.Avdulov NA, Chochina SV, Igbavboa U, Warden CS, Schroeder F, Wood WG. Lipid binding to sterol carrier protein-2 is inhibited by ethanol. Biochim Biophys Acta. 1999;1437:37–45. doi: 10.1016/s0005-2760(98)00178-7. [DOI] [PubMed] [Google Scholar]
- 49.Schroeder F, Kinden DA. Measurement of phagocytosis using fluorescent latex beads. J Biochem Biophys Methods. 1983;8:15–27. doi: 10.1016/0165-022x(83)90017-9. [DOI] [PubMed] [Google Scholar]
- 50.Tangirala RK, Jerome WG, Jones NL, Small DM, Johnson WJ, Glick JM, Mahlberg FH, Rothblat GH. Formation of cholesterol monohydrate crystals in macrophage-derived foam cells. J Lipid Res. 1994;35:93–104. [PubMed] [Google Scholar]
- 51.Perry DK, Hannun YA. The role of ceramide in cell signaling. Biochim Biophys Acta. 1998;1436:233–243. doi: 10.1016/s0005-2760(98)00145-3. [DOI] [PubMed] [Google Scholar]
- 52.Schroeder F, Atshaves BP, Gallegos AM, McIntosh AL, Liu JC, Kier AB, Huang H, Ball JM. Lipid rafts and caveolae organization. In: Lisanti MP, Frank PG, editors. Advances in molecular and cell biology. Boca Raton: CRC Press; 2004. [Google Scholar]
- 53.Ghosh S, Strum JC, Bell RM. Lipid biochemistry: functions of glycerolipids and sphingolipids in cellular signaling. FASEB J. 1997;11:45–50. doi: 10.1096/fasebj.11.1.9034165. [DOI] [PubMed] [Google Scholar]
- 54.Grassme H, Jekle A, Riehle A, Schwarz H, Berger J, Sandhoff K, Kolesnick R, Gulbins E. CD95 signaling via ceramide-rich membrane rafts. J Biol Chem. 2001;276:20589–20596. doi: 10.1074/jbc.M101207200. [DOI] [PubMed] [Google Scholar]
- 55.Okazaki T, Bielawska A, Bell RM, Hannun YA. Role of ceramide as a lipid mediator of 1 alpha,25-dihydroxyvitamin D3-induced HL-60 cell differentiation. J Biol Chem. 1990;265:15823–15831. [PubMed] [Google Scholar]
- 56.Kim MY, Linardic C, Obeid L, Hannun Y. Identification of sphingomyelin turnover as an effector mechanism for the action of tumor necrosis factor alpha and gamma-interferon. Specific role in cell differentiation. J Biol Chem. 1991;266:484–489. [PubMed] [Google Scholar]
- 57.Jayadev S, Linardic CM, Hannun YA. Identification of arachidonic acid as a mediator of sphingomyelin hydrolysis in response to tumor necrosis factor alpha. J Biol Chem. 1994;269:5757–5763. [PubMed] [Google Scholar]
- 58.Atshaves BP, Jefferson JR, McIntosh AL, Gallegos AM, McCann BM, Landrock KK, Kier AB, Schroeder F. Effect of sterol carrier protein-2 expression on sphingolipid distribution in plasma membrane lipid rafts/caveolae. Lipids. 2007;42:871–884. doi: 10.1007/s11745-007-3091-z. [DOI] [PubMed] [Google Scholar]
- 59.Yamaji A, Sekizawa Y, Emoto K, Sakuraba H, Inoue K, Kobayashi H, Umeda M. Lysenin, a novel sphingomyelin-specific binding protein. J Biol Chem. 1998;273:5300–5306. doi: 10.1074/jbc.273.9.5300. [DOI] [PubMed] [Google Scholar]