Abstract
TWIST1 is a transcription factor that belongs to the family of basic helix-loop-helix proteins involved in epithelial-to-mesenchymal transition and invasion processes. The TWIST1 protein possesses oncogenic, drug-resistant, angiogenic and invasive properties, and has been related with several human tumors and other pathologies. Colorectal cancer is one of the tumors in which TWIST1 is over-expressed, but its involvement in the clinical outcome of the disease is still unclear. We tested, by RT-PCR, the expression levels of TWIST1 in normal and tumor paired-sample tissues from a series of 151 colorectal cancer patients, in order to investigate its prognostic value as a tumor marker. TWIST1 expression was restricted to tumor tissues (86.1%) and correlated with lymph node metastasis (LNM). Adjusted analysis showed that the expression levels of TWIST1 correlated with overall survival (OS) and disease-free survival (DFS). Importantly, TWIST1 expression levels predicted OS specifically at stages I and II. Moreover, patients with stage II tumors and high TWIST1 levels showed even shorter survival than patients with stage III tumors. These results suggest that TWIST1 expression levels could be a tumor indicator in stage II patients and help select patients at greater risk of poor prognosis who might benefit from adjuvant chemotherapy.
Introduction
Colorectal cancer (CRC) is the second most common cause of cancer mortality in the developed countries and remains associated with a high mortality rate [1]. Metastases are the end result of tumor progression and the most common cause of death in cancer patients. The genetic bases for metastasis are beginning to be outlined [2].
Altered functions of several genes that are key players in embryonic development are related to some steps in oncogenesis, such as epithelial-mesenchymal transition (EMT). During gastrulation, certain cells from an epithelial-like structure undergo EMT and migrate to organize the mesoderm embryonic layer. The theory that tumor cells trigger EMT to allow migration and invasion has received considerable attention, since several genes involved in EMT during embryogenesis are turned up during oncogenesis [3]. Yang and co-workers, in a mouse model of breast cancer, identified genes related to each step of metastasis, particularly those involved in invasion and intravasation steps in which EMT is a necessary process [4]. In this context, the transcription factor TWIST1 was identified as an essential protein in the intravasation step. TWIST1 induces EMT in epithelial cells by activation of SNAI2 transcription [5], repression of E-cadherin-mediated cell-cell adhesion and acquisition of mesenchymal markers such as fibronectin and N-cadherin. Moreover, significant correlation was found between the expression of TWIST1 and the appearance of invasive lobular carcinomas [4]. This corroborated a previous study in which the TWIST1 promoter was much less frequently methylated in invasive lobular carcinomas than in invasive ductal carcinomas [6].
A number of studies indicate that TWIST1 possesses oncogenic [7]–[10], drug-resistant [11]–[12], angiogenic [13] and invasive [11], [14]–[17] properties. In addition, TWIST1 over-expression has been found in tumor tissues such as rhabdomyosarcoma [7], melanoma [8], pediatric osteosarcoma [18], T-cell lymphoma [10], gastric [19], prostate [11] and breast carcinoma [4], [17], [20]. Recently, it has been demonstrated that the TWIST1 protein overrides oncogene-induced senescence both in murine and human cancer cells [21].
TWIST1 over-expression in colorectal cancer is associated with gender and with a poor prognosis factor, such as nodal invasion [22], but its impact on the clinical outcome of the disease is still not clear. At the moment, pathological staging is still the most useful prognostic factor [23]. However, new molecular or clinical parameters are needed, to improve the current methods for deciding which patients could benefit from adjuvant chemotherapy and when.
We hypothesize that the expression level of TWIST1 in primary colorectal tumors determines the characteristics of the tumors, their behavior and their clinical outcome.
Materials and Methods
Patients and samples
The present study was based on a consecutive series of 151 patients undergoing surgery for colorectal cancer and included in a prospective study. Informed written consent was obtained from all participants after an explanation of the nature of the study, as approved by the Research Ethics Board of Puerta de Hierro Majadahonda University Hospital. All patients were considered sporadic cases, inasmuch as those with familial adenomatous polyposis and clinical criteria for hereditary non-polyposis colorectal cancer (Amsterdam criteria) were excluded.
Tumor and normal colon mucosa (taken at least 3 cm from the outer tumor margin) were obtained immediately after surgery, immersed in RNAlater™ (Ambion Inc, Austin, Texas), snap-frozen in liquid nitrogen and stored at −80°C until processing. All patients in the study gave written informed consent.
RNA extraction
RNA was extracted from tumor and normal samples with the RNeasy Mini Kit (Qiagen Inc., Hilden, Germany), according to the manufacturer's protocol. The RNA extracted was quantified with a NanoDrop ND-1000 Spectrophotometer (nanoDrop Technologies Inc., Wilmington, Delaware, USA).
Real-Time PCR
TWIST mRNA expression in each sample was measured as a ratio against the geometric average of three reference housekeeping genes, succinate dehydrogenase complex subunit A (SDHA), TATA binding protein (TBP) and ubiquitin C (UBC) [24]. The relative concentrations of the target and the reference genes were calculated by interpolation, using a standard curve of each gene plotted from the same serial dilution of cDNA from tumor tissue. The quantitative mRNA analysis was performed in duplicate. TWIST1 expression was only determined in tumor tissues, since normal tissues showed no expression of this gene. An arbitrary value (0.01), corresponding to half the minimum value detected in the series, was assigned to the tumors in which TWIST expression was not detected. SNAI2 expression was calculated as the ratio of its expression in tumor (T) vs its expression in normal tissue (N).
The primers used were: SDHA-5′TGGGAACAAGAGGGCATCTG 3′ forward (F) and 5′CCACCACTGCATCAAATTCATG 3′ reverse (R); TBP-5′TCTGGGATTGTACCGCAGC3′ forward (F) and 5′CGAAGTGCAATGGTCTTTAGG3′ reverse (R); UBC 5′ATTTGGGTCGCGGTTCTTG3′ forward (F) and 5′TGCCTTGACATTCTCGATGGT3′ reverse (R); TWIST1 5′ CATGTCCGCGTCCCACTAG 3′ forward (F) and 5′ TGTCCATTTTCTCCTTCTCTGG 3′ reverse (R); SNAI2 5′-GGCAAGGCGTTTTCCAGAC-3′ forward (F) and 5′-GCTCTGTTGCAGTGAGGGC-3′ reverse (R). The annealing temperature in all cases was 59°C. At the end of the PCR cycles, melting curve analyses were performed to confirm the generation of the specific expected PCR product. The PCR products were sequenced in an ABI Prism™ 377 DNA sequencer apparatus (PE Applied Biosystems). For the synthesis of cDNA, 400 ng of total RNA was retro-transcribed, using the Gold RNA PCR Core Kit (PE Biosystems, Foster City, CA). Real-time PCR was performed in a Light-Cycler apparatus (Roche Diagnostics, Mannheim, Germany), using the LightCycler-FastStartPLUS DNA Master SYBR Green I Kit (Roche Diagnostics, Mannheim, Germany).
Clinico-pathological parameters of the patients
The parameters obtained from the medical records of the 151 patients were: age, tumor location, lymph node metastases (LNM) (evaluated by optical microscopy), pathological stage (assessed by the tumor-node-metastases classification), tumor histological grade and the presence of vascular invasion in tumors.
Patients' treatment and follow-up
Colon cancer patients did not receive neo-adjuvant chemotherapy (CT). Patients with rectal carcinoma who had received preoperative treatment with CT and radiotherapy or radiotherapy alone were excluded, because of the difficulty of finding a suitable tumor for determining gene expression in these patients' surgical samples. Adjuvant treatment based on oxaliplatin (FOLFOX6, leucovorin 400 mg/m2 IV on day 1 as a 2-hour infusion, followed by 5-fluorouracil bolus of 400 mg/m2 IV on day 1, followed by 2,400 mg/m2 IV 46-hour infusion and oxaliplatin 100 mg/m2 IV as a 2-hour infusion on day 1) was administered to the 51 stage-III patients (29 colon cancer and 22 rectal cancer) without medical contra-indications who gave their written informed consent. Radiotherapy was also administered to the 22 rectal tumor cases. The median age of this subgroup of patients was 69.3 years.
Clinical follow-up after surgery and diagnosis was based on periodic visits and clinical, biochemical and imaging techniques. Ultrasonic study was performed when liver function was impaired. Overall and Disease-Free Survival (OS and DFS) were defined as the period of time from diagnosis to death and the interval between diagnosis and first recurrence, respectively.
Statistical analysis
As the distribution of the gene expression values was not normally distributed (Kolmogorov-Smirnov test), we normalized the data distribution by using log10 to carry out the statistical analysis.
The clinico-pathological parameters were contrasted with TWIST expression data in tumor tissues by the one-way ANOVA test. The General Linear Model was applied to age and stage in order to test the possible interaction between the two variables, as well as their independent value in relation to TWIST1 mRNA expression levels.
To study OS and DFS, the expression data of TWIST were divided by tertiles. The expression levels defining the three groups for the TWIST gene were 0.56 (33%) and 1.7 (66%). DFS analysis did not include patients at pathological stage IV. The relationship between the cumulative probability of OS and DFS, as well as analyzed predictors, was calculated with the Kaplan-Meier method [25], while significant differences between curves were evaluated with Mantel's log-rank test [26]. To identify factors that might be of independent significance in influencing OS and DFS, multivariate analysis (Cox proportional risk regression model) was applied [27]. Confounding and interacting variables were analyzed. The model's basic assumptions (proportional hazards) were evaluated. In all statistical tests two-tailed p values ≤ 0.05 were considered statistically significant. Statistical analyses were performed with SPSS 13.0 statistical software (SPSS Inc, Chicago, IL).
Results
TWIST1 expression is confined to tumor tissues in human colorectal cancer and is up-regulated in patients with lymph node metastasis
The analysis of TWIST1 expression levels in tumor and normal matched tissues from 151 patients with colorectal cancer showed that the expression of TWIST1 is restricted to the tumor mass. It was never detected in any normal tissues. Of 151 tumor samples tested, TWIST1 was detected in 130 cases (86.1%).
A statistical association between high levels of TWIST1 in tumor tissues and lymph node metastasis (LNM) was observed (p = 0.016), while only a trend to statistical association was found for age (p = 0.052) and tumor stage (p = 0.07) (Table 1). Though this suggests a possible interaction between the two variables, the statistical General Linear Model showed no interaction between age and tumor stage, as well as, an independent relationship for age and TWIST1 expression levels, p = 0.031. The median and range of lymph node harvesting counts were: 9 (0–34).
Table 1. Associations between the expression of TWIST gene in tumor tissues and clinico-pathological characteristics.
| Characteristics | Total (%) | Expression of TWIST | |
| Median/minimum/maximum | p a | ||
| Patients | 151 | ||
| Median age | 71 | ||
| <71 | 63 (41.72%) | 0.92/0.001/10.33 | 0.052 |
| >71 | 88 (58.28%) | 1.04/0.01/56.90 | |
| Gender | |||
| Male | 96 (63.58%) | 1.04/0.01/56.90 | NS |
| Female | 55 (36.42%) | 0.88/0.01/23.63 | |
| Tumor side | |||
| Colon | 102 (67.55%) | 0.91/0.01/56.90 | NS |
| Rectum | 49 (32.45%) | 1.67/0.01/22.78 | |
| Stage | |||
| I | 14 (9.27%) | 1.53/0.01/3.89 | 0.074 |
| II | 75 (49.67%) | 0.89/0.01/56.90 | |
| III | 51 (33.77%) | 1.16/0.01/22.78 | |
| IV | 11 (7.28%) | 1.67/0.44/23.66 | |
| Vascular invasion | |||
| Yes | 63 (41.72%) | 0.96/0.01/56.90 | NS |
| No | 88 (58.28%) | 1.00/0.01/23.63 | |
| Lymph node metastases | |||
| Positive | 62 (41.06%) | 1.19/0.01/23.63 | 0.016 |
| Negative | 89 (58.94%) | 0.94/0.01/56.90 | |
| Tumor differentiation | |||
| Good | 41 (27.15%) | 0.82/0.01/7.04 | NS |
| Moderate | 77 (50.99%) | 1.22/0.01/22.78 | |
| Poor | 33 (21.85%) | 1.67/0.01/56.90 | |
p is calculated by the ANOVA test.
NS: Not significant.
No statistical associations between TWIST1 expression levels and the other pathological variables analyzed were found (summarized in Table 1).
TWIST1 mRNA levels correlate with the transcriptional activity of the protein
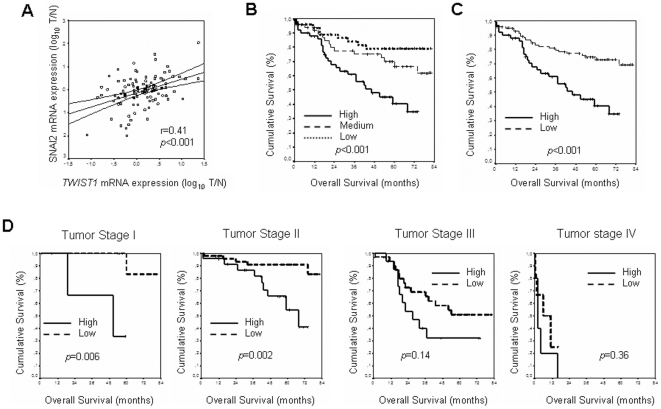
To ensure that TWIST1 mRNA levels detected in each sample correlate with protein activity, the mRNA levels of Snai2 were measured in the 130 patients in whom TWIST1 was previously detected. SNAI2 is another transcription factor involved in the EMT, whose transcription is directly regulated by TWIST1, as has been recently demonstrated [5]. As expected, a direct correlation between TWIST1 and SNAI2 mRNA levels (Pearson correlation coefficient r = 0.41, P<0.001) was found, suggesting that there is a correlation between TWIST1 mRNA levels and protein activity (Fig 1A).
Figure 1. Kaplan-Maier OS curves and TWIST1 activity.
A, Relation between expression levels of TWIST1 and SNAI2 genes, logR(T/N), in patients in which TWIST1 was previously detected. TWIST1 and SNAI2 expression directly correlates in human colon cancer, Pearson correlation coefficient r = 0.41 (P<0.001). Kaplan-Meier curves and p values for OS for TWIST1 expression levels: B, in the series distributed by tertiles: low, medium and high. C, grouping tertiles: low and medium as low. D, stratifying the series by tumor stage.
TWIST1 expression level is related to overall survival
The series was followed for a mean of five years (range of patient follow-up: 1–82 months). During this period, 54 recurrences (35.8%) were recorded and 50 patients (33.1%) died, with the five-year OS for the series at 62.4% (95% CI, 53.8%–70.98%). To carry out survival analysis, the series was divided by tertiles on the basis of TWIST1 expression levels. Thus, patients were classified with low, medium or high levels of TWIST1 expression. A statistical difference was observed in OS for the expression of TWIST1 (p<0.001): the five-year OS for each group was 79% (95% CI, 67%–91%) for those patients with low expression levels; 66.6% (95% CI, 51.9%–81.3%) for patients with medium expression levels; and 40.6% (95% CI, 24.6%–56.6%) for patients with high expression levels (Fig. 1B). Since no statistical differences were found for OS in patients with low or medium levels of TWIST1 expression and both groups behaved quite similarly (see Fig. 1B), unlike patients with high TWIST1 expression levels, these two categories were grouped. Therefore, further studies were carried out with only two categories: patients with low (the former low plus medium levels) or high expression levels of TWIST1. No changes in the correlation previously observed between OS and TWIST1 expression levels were found with this new classification. Thus, five-year OS for patients with high expression levels was the above-mentioned 40.6% (95% CI, 24.6%–56.6%), versus 72.9% (95% CI, 63.3%–82.5%) in those cases with low expression levels (p<0.001) (Fig. 1C).
Since, according to this result, the expression levels of TWIST1 in human colorectal cancer could be considered a poor prognosis factor, we were interested in the clarification of its possible prognosis value at each different colorectal tumor stage. The number of patients with low expression levels was: 9 for stage I, 51 for stage II, 34 for stage III and 6 for stage IV. Equally, the number of patients with high expression levels was: 5 for stage I, 24 for stage II, 17 for stage III and 5 for stage IV. Interestingly, the Kaplan-Meier analysis showed that the expression levels of TWIST1 correlated with OS, but only in stages I and II. Therefore, five-year OS for stage I was 33.3% (95% CI, 0%–86.65%) in patients with high levels versus 83.3% (95% CI, 53.5–100%) in patients with low expression levels (p = 0.006); for stage II, it was 54.53% (95% CI, 28.29%–80.77%) in patients with high expression levels versus 90.92% (95% CI, 82.42–99.42%) for patients with low expression levels (p = 0.002). In contrast, the Kaplan-Meier curves in stages III or IV were similar for both groups, i.e. patients with high and low TWIST1 expression levels, with no statistical differences observed (Fig. 1D).
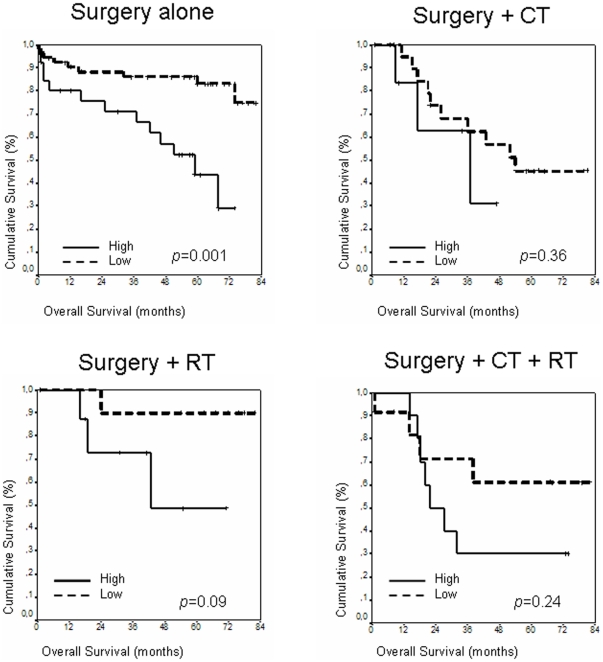
Because the different treatment protocols may affect OS and mimic the prognosis value of TWIST1, we also analyzed the prognosis value of TWIST1 at each different treatment subgroup (Fig 2). Four groups of treatment were identified. In the first group, 67 patients underwent only surgery. All of these were colon cancer cases with tumors at stages I or II (note that there were 6 patients with stage IV colon tumors and 8 patients with stage I or IV rectal tumors who also underwent only surgery, but were not considered in this analysis in order to achieve greater homogeneity for this study). Five-year OS for patients with high TWIST1 expression was 54.5% (95% CI; 81.5%–27.4%) versus 89.6% (95% CI; 99.4%–79.8%) in patients with low expression levels (p = 0.001). In a second group, 19 patients underwent radiotherapy as well as surgery: all of these were patients with stage II rectal tumors, except for two at stage I. In this group, five-year OS for patients with tumors with high expression levels was 48.6% (95% CI; 92.9%–4.3%) versus 90% (95% CI; 100%–71.6%) in patients with low expression levels (p = 0.09). No differences in OS according to TWIST1 expression levels were observed in the two remaining groups. One of these consisted of 29 patients who received chemotherapy after surgery (all were patients with stage III colon cancer tumors). The other consisted of 22 patients who received chemotherapy plus radiotherapy, as well as surgery (all of these were patients with stage III rectal cancer tumors). Again, the only clear differences in OS were found in the group considered to have good prognosis.
Figure 2. Kaplan-Maier OS curves regarding treatment protocols.
Kaplan-Meier curves and p values for OS for TWIST1 expression levels in each one of the treatment groups. CT, chemotherapy; RT, radiotherapy.
Univariate analysis was performed to determine the influence of TWIST expression and the clinico-pathological parameters in OS. Variables which could be considered statistically supported factors in OS prediction were: LNM, stage, treatment protocols and TWIST1 expression levels (Table 2). In the multivariate Cox's regression model for OS, the variables that showed an independent prognostic factor were: LNM HR 4.02 (95% CI; 2.21–7.3) (p<0.001) and TWIST1 expression levels HR 2.73 (95% CI; 1.54–4.84) (p = 0.001) (Table 2). Because LNM and tumor stage are linearly dependent covariates (tumor stages I and II are LNM negatives and tumor stage III and, probably, the vast majority at tumor stage IV are LNM positives), the variable tumor stage was not included in the multivariate analysis.
Table 2. Unadjusted and adjusted analyses of the association between TWIST expression and overall survival of colon cancer patients.
| Variable | Category | Unadjusted analysis | Adjusted analysis | ||||
| HR | (95% CI) | p Value | HR | (95% CI) | p Value | ||
| Age at diagnosis | <71 vs >71 | 0.775 | 0.44–1.37 | 0.38 | |||
| Sex of patients | Male vs female | 1.53 | 0.82–2.86 | 0.18 | |||
| Lymph node metastases | Yes vs No | 3.99 | 2.2–7.24 | <0.001 | 4.02 | 2.21–7.3 | <0.001 |
| Vascular invasion | Yes vs No | 1.66 | 0.94–2.93 | 0.079 | |||
| Stage | II vs I | 0.91 | 0.26–3.18 | 0.87 | |||
| III vs I | 3.06 | 0.92–10.2 | 0.068 | ||||
| IV vs I | 35.09 | 7.89–155.9 | <0.001 | ||||
| Histological grade | 2 vs 1 | 1.49 | 0.76–2.94 | 0.25 | |||
| 3 vs 1 | 1.02 | 0.42–2.5 | 0.96 | ||||
| Tumor side | Rectum vs colon | 1.41 | 0.79–2.50 | 0.24 | |||
| TWIST expression | High vs low | 2.72 | 1.53–4.82 | 0.001 | 2.73 | 1.5–4.84 | 0.001 |
| Treatment Protocols | CT vs surg | 2.18 | 1.08–4.39 | 0.029 | |||
| RT vs surg | 0.77 | 0.26–2.25 | 0.63 | ||||
| CT + R vs surg | 2.19 | 1.04–4.58 | 0.038 | ||||
The blank cells correspond to variables that showed no independent relationship with OS in the adjusted analysis. CT, chemotherapy; RT, radiotherapy.
Since our results suggested that TWIST1 expression levels have prognosis value at early stages I and II (both LNM negatives), we repeated the Cox's regression models, stratifying the series for their LNM status. This analysis confirmed that TWIST1 expression levels have prognosis value only in patients without lymph node metastasis (Table 3).
Table 3. Analysis of the association between TWIST expression and overall survival of colon cancer patients by lymph node metastases.
| Variable | Category | LNM positives | LNM negatives | ||||
| HR | (95% CI) | p Value | HR | (95% CI) | p Value | ||
| Age at diagnosis | <71 vs >71 | 2.28 | 0.87–5.97 | 0.09 | 0.78 | 0.38–1.62 | 0.5 |
| TWIST expression | High vs low | 6.57 | 2.31–18.7 | <0.001 | 1.84 | 0.88–3.84 | 0.1 |
TWIST1 expression levels are related to Disease-Free Survival
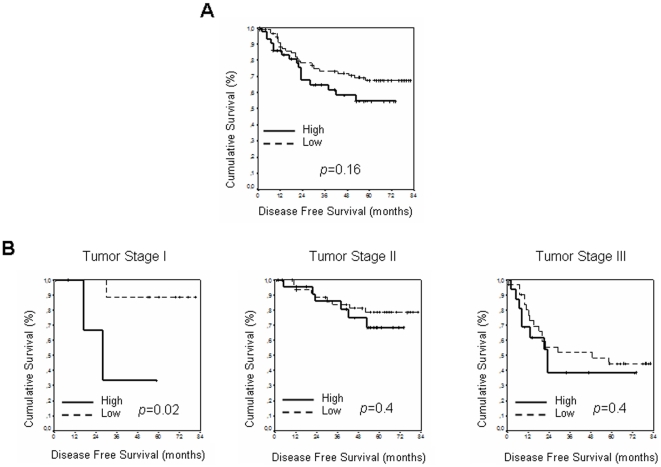
No clear difference was observed for TWIST1 expression levels and DFS after Kaplan-Meier analysis: five-year DFS for patients with low TWIST1 expression levels was 67.58% (95% CI; 57.25%–77.9%) vs 54.75% (95% CI; 39.07%–70.43%) in patients with high expression levels (Fig. 3A). However, this analysis performed in the series stratified by stage showed a correlation between TWIST1 expression levels and DFS in stage I: 88.9% (95% CI; 68.36%–100%) in patients with low expression levels vs 33.3% (95% CI; 0%–86.6%) in patients with high expression levels (p = 0.02). No correlation was found in stages II and III (Fig. 3B).
Figure 3. Kaplan-Maier DFS curves.
Kaplan-Meier curves and p values for DFS for TWIST1 expression levels: low (formed by medium and low tertiles) and high: A, in the entire series, except patients with stage IV tumor; B, at each tumor stage except stage IV.
Variables which could be considered statistically supported factors in DFS prediction, according to the Cox's model, were LNM (p<0.001) and stage (p = 0.046). However, the multivariate Cox's regression model included TWIST1 expression levels, HR 1.99 (95% CI, 1.05–3.82) (p = 0.036) and gender, HR 2.07 (95% CI; 1.04–4.15) (p = 0.038) as independent prognosis factors for DFS, as well as the variable LNM, HR 3.4 (95% CI; 1.83–6.32) (Table 4).
Table 4. Unadjusted and adjusted analyses of the association between TWIST expression and disease-free survival of colon cancer patients.
| Variable | Category | Unadjusted analysis | Adjusted analysis | ||||
| HR | (95% CI) | p Value | HR | (95% CI) | p Value | ||
| Age at diagnosis | <71 vs >71 | 0.56 | 0.3–1.07 | 0.057 | 1.68 | 0.88–3.19 | 0.112 |
| Sex of patients | Male vs female | 1.895 | 0.96–3.76 | 0.067 | 2.07 | 1.04–4.15 | 0.038 |
| Lymph node metastases | Yes vs No | 3.57 | 1.94–6.57 | <0.001 | 3.4 | 1.83–6.32 | <0.001 |
| Vascular invasion | Yes vs No | 1.69 | 0.93–3.09 | 0.087 | |||
| Stage | II vs I | 0.94 | 0.27–3.25 | 0.9 | |||
| III vs I | 3.39 | 1.02–11.28 | 0.046 | ||||
| Histological grade | 2 vs 1 | 1.91 | 0.93–3.93 | 0.08 | |||
| 3 vs 1 | 0.65 | 0.2–2.07 | 0.46 | ||||
| Tumor side | Rectum vs colon | 0.84 | 0.45–1.6 | 0.6 | |||
| TWIST expression | High vs low | 1.54 | 0.83–2.84 | 0.17 | 1.99 | 1.05–3.82 | 0.036 |
| Treatment Protocols | CT vs surg | 4.52 | 2.16–9.43 | <0.001 | |||
| RT vs surg | 1.44 | 0.51–4.03 | 0.49 | ||||
| CT + RT vs surg | 3.15 | 1.35–7.38 | 0.008 | ||||
The blank cells correspond to variables that showed no independent relationship with OS in the adjusted analysis. CT, chemotherapy; RT, radiotherapy.
TWIST1 expression in patients with stages II and III tumors and Overall Survival
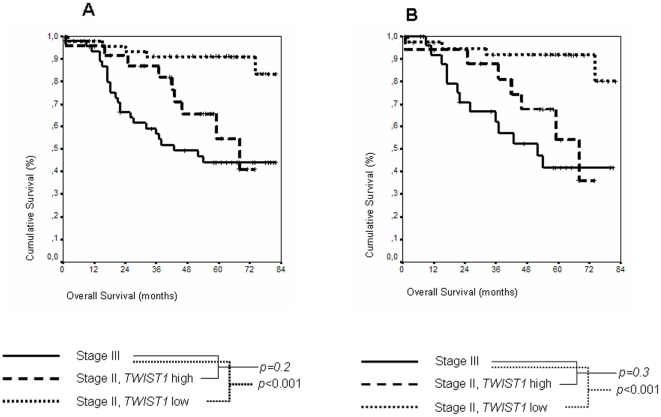
We compared the survival curves between patients with stage III tumors and patients with stage II tumors, dividing this group in two categories: those with low TWIST1 expression levels and those with high expression levels (Fig. 4A). Unlike patients with stage III tumors, none of these patients received adjuvant chemotherapy. In this analysis, we did not observe difference at five-year OS between patients at stage III, 44.04% (95% CI; 28.98%–59.09%), and patients at stage II with high TWIST1 expression levels, 54.53% (95% CI, 28.29%–80.77%). However, the difference was clear when patients in stage II expressing low TWIST1 levels were compared with stage III patients (p<0.001). Moreover, by the end of the study (82 months) cumulative survival was higher in patients with stage III tumors, 44.04% (95% CI; 28.98%–59.09%), than in patients with stage II tumors and high TWIST1 expression levels, 40.9% (95% CI; 10.52%–71.28%). This analysis was repeated, taking into account only colon cancer cases, not rectal cancer cases, in order to achieve greater homogeneity for the study. Again, the behavior of the different groups was very similar (Fig. 4B), confirming that this analysis was not affected by tumor location or treatment protocols, since all of the patients with stage III tumors were treated with chemotherapy before surgery.
Figure 4. Kaplan-Maier OS curves in stages II and III.
Kaplan-Meier curves and p values for OS at tumor stage III, tumor stage II with low TWIST1 expression levels, and tumor stage II with high TWIST1 expression levels. There is no significant difference between stage III and stage II with high TWIST1 expression levels. A, global series. B, considering only colon cancer cases.
Discussion
We examined the expression of TWIST1 in the normal and tumor tissues of a large series of 151 colorectal cancer patients. TWIST1 expression was restricted to tumor tissues, indicating that it could be a tumor marker. Moreover, its expression was associated with certain pathological parameters linked to poor prognosis, such as LNM, which corroborates a publication in this field [22].
In our view, the most significant result was that TWIST1 expression levels gave an independent prognostic factor, for both OS and DFS. Indeed, the detailed analysis of the correlation between TWIST1 expression levels and OS and DFS, at each tumor pathological stage, showed an interaction between these two variables, pathological tumor stages and TWIST1 expression. Therefore, for OS, TWIST1 was a prognostic factor only at stages I and II, losing its prognostic value in advanced stages (III and IV). In a similar way, the study showed TWIST1 as a prognostic factor for DFS only in stage I, losing this correlation in stages II and III.
The results found in our study have not been described in the literature on TWIST1 expression and patients' prognosis. Although negative results have been reported between TWIST1 expression and patient survival [28] in colon cancer cases, there are several publications that support this relationship, when analyzing the evolution of patients and mRNA TWIST1 levels in colon cancer [29] and cervix carcinoma [30]. The expression of TWIST1 and other functionally related genes, such as E-Cad, SNAIL, SLUG and HIF-1α, has also been studied in relation to survival in several tumor types, such as esophageal cancer [31], head and neck [32], [33] and bladder [34] carcinoma, with these relationships increasing when any of these genes are also overexpressed, as well as TWIST1. It is possible that a stage-by-stage analysis in some of these series would also provide results similar to ours. It would be reasonable to think that, if colon cancer develops by following steps [35], and tumor progression in these steps is caused by the accumulation of mutations in oncogenes and tumor suppressor genes, the accumulation of these at more advanced stages of disease could mimic the effects of overexpression of TWIST1 on patients' survival.
Currently, the use of adjuvant chemotherapy for all patients with stage III colon cancer after resection is part of standard treatment around the world. However, the controversy about adjuvant treatment in stage II CRC is currently unresolved. The IMPACT B2 study pooled results from five trials in Dukes B2 colon cancer patients. This study did not show any benefit in five-year overall survival: results were 80% in the control group and 82% in the 5-fluorouracil and leucovorin group [36]. Nevertheless, the four NSABP adjuvant studies showed decreased death risk with adjuvant treatment, similar to the benefit obtained in stage C [37]. Another Dutch study reported a beneficial effect of 5-fluorouracil and levamisol adjuvant therapy in stage II patients, similar to the expected benefit in stage III ones [38]. Anyway, despite the lack of data, there is growing acceptance of an informal classification system, stratifying stage II patients by risk on the basis of clinical data, as a guide for deciding whether to use adjuvant therapy. Therefore, in stage II patients with high clinico-pathological risk (intestinal obstruction, perforation, tumor adherence, poor differentiation, vascular or lymphatic invasion), adjuvant therapy can reasonably be offered.
The analysis performed to study the behavior of TWIST1 overexpression in relation to OS in subgroups of patients who received similar treatment protocols showed no significant differences, except for patients who had received no adjuvant therapy and thus patients in early stages. This supports the results found in the analysis of the complete series by stages, which showed that TWIST1 is a discriminating factor in early stages of the disease.
Although, at stage III, TWIST expression did not differ in terms of OS and DFS, it could be relevant that the OS of stage-II patients with high TWIST expression was similar to that observed in patients at stage III. Moreover, the cumulative OS at the end of the study for this group of patients was higher than the cumulative OS in patients at stage II and high expression levels of TWIST1. These preliminary observations could support the idea of TWIST1 expression as a tumor indicator at stage II, which could help select patients at greater risk who might benefit from adjuvant chemotherapy.
There is no biological explanation that justifies patients with stage I tumors and high TWIST1 expression levels showing shorter cumulative survival, 33.3% (95% CI, 0%–86.65%), than the same category with stage II tumors, 54.53% (95% CI, 28.29%–80.77%). These results obtained from Kaplan-Meier analyses at each tumor stage may seem an artefact, due to the low number of samples in a group, such as stage I. However, our conclusions are supported by the results obtained from another approach, i.e. LNM status or treatment protocols in OS, where the study was not affected by the number of samples and showed that in both cases TWIST1 mRNA expression levels have prognosis value only in early stages.
Cancer cells that are undergoing Epithelial-Mesenchymal Transition usually show deregulation of various genes. For instance, up-regulation of SNAIL1, ZEB1, ZEB2 or E12 in epithelial cells represses E-cadherin expression and induces EMT in several carcinomas [39]–[46]. Recently, the transcription factor TWIST1 has been added to the list of proteins that trigger EMT [4]. Yang et al. suggested that the expression of TWIST1 is essential in the intravasation step during the metastatic process. Expression of TWIST1 by tumor cells might enhance the intravasation steps of metastasis. In this case, tumors expressing TWIST1 would display more aggressive behavior and trigger the intravasation steps, even though no visible metastases are observed.
We show that TWIST1 expression levels may be an independent prognostic factor in patients with CRC. It may be well used in stage II to identify sub-groups of patients at high risk with a poor prognosis who might benefit from adjuvant chemotherapy.
Acknowledgments
We thank M. Eaude for help with the English manuscript, as well as I. Millan and F. García for biostatistical assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by grants from: Foundation AECC, S-GEN/0266/2006, RD06/0020/0020, SAF2010-20750 and RD06/0020/0040. JMG is supported by ISCIII-CP08/00217. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Tenesa A, Dunlop MG. New insights into the aetiology of colorectal cancer from genome-wide association studies. Nat Rev Genet. 2009;10:353–358. doi: 10.1038/nrg2574. [DOI] [PubMed] [Google Scholar]
- 2.Nguyen DX, Massagué J. Genetic determinants of cancer metastasis. Nat Rev Genet. 2007;8:341–352. doi: 10.1038/nrg2101. [DOI] [PubMed] [Google Scholar]
- 3.Kang Y, Massagué J. Epithelial-Mesenchymal transitions: Twist in development and metastasis. Cell. 2004;118:277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 4.Yang J, Mani SA, Donaher JL Ramaswamy S, Itzykson RA, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Casas E, Kim J, Bendesky A, Ohno-Machado L, Wolfe CJ, et al. Snail2 is an essential mediator of Twist1-induced epithelial mesenchymal transition and metastasis. Cancer Res. 2011;71:245–254. doi: 10.1158/0008-5472.CAN-10-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fackler MJ, McVeigh M, Evron E, Garrett E, Mehrotra J, et al. DNA methylation of RASSF1A, HIN-1, RAR-beta, Cyclin D2 and Twist in situ and invasive lobular breast carcinoma. Int J Cancer. 2003;107:970–975. doi: 10.1002/ijc.11508. [DOI] [PubMed] [Google Scholar]
- 7.Maestro R, Dei Tos AP, Hammamori Y, Krasnokutsky S, Sartorelli V, et al. Twist is a potential oncogene that inhibits apoptosis. Genes Dev. 1999;13:2207–2217. doi: 10.1101/gad.13.17.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoek K, Rimm DL, Williams KR, Zhao H, Ariyan S, et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 2004;64:5270–5280. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- 9.Valsesia-Wittmann S, Magdeleine M, Dupasquier S, Garin E, Jallas AC, et al. Oncogenic cooperation between H-Twist and N-Myc overrides failsafe programs in cancer cells. Cancer Cell. 2004;6:625–630. doi: 10.1016/j.ccr.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 10.van Doorn R, Dijkman R, Vermeer MH, Out-Luiting JJ, van der Raaij-Helmer EM, et al. Aberrant expression of the tyrosine kinase receptor EphA4 and the transcription factor twist in Sezary syndrome identified by gene expression analysis. Cancer Res. 2004;64:5578–5586. doi: 10.1158/0008-5472.CAN-04-1253. [DOI] [PubMed] [Google Scholar]
- 11.Kwok WK, Ling MT, Lee TW, Lau TC, Zhou C, et al. Up-regulation of TWIST in prostate cancer and its implication as a therapeutic target. Cancer Res. 2005;65:5153–5162. doi: 10.1158/0008-5472.CAN-04-3785. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Ling MT, Guan XY, Tsao SW, Cheung HW, et al. Identification of a novel function of TWIST, a bHLH protein, in the development of acquired Taxol resistance in human cancer cells. Oncogene. 2004;23:474–482. doi: 10.1038/sj.onc.1207128. [DOI] [PubMed] [Google Scholar]
- 13.Mironchik Y, Winnard PT, Jr, Vesuna F, Kato Y, Wildes F, et al. Twist overexpression induces in vivo angiogenesis and correlates with chromosomal instability in breast cancer. Cancer Res. 2005;65:10801–10809. doi: 10.1158/0008-5472.CAN-05-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elias MC, Tozer KR, Silber JR, Mikheeva S, Deng M, et al. TWIST is expressed in human gliomas and promotes invasion. Neoplasia. 2005:824–837. doi: 10.1593/neo.04352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee TK, Poon RT, Yuen AP, Ling MT, Kwok WK, et al. Twist overexpression correlates with hepatocellular carcinoma metastasis through induction of epithelial-mesenchymal transition. Clin Cancer Res. 2006;12:5369–5376. doi: 10.1158/1078-0432.CCR-05-2722. [DOI] [PubMed] [Google Scholar]
- 16.Horikawa T, Yang J, Kondo S, Yoshizaki T, Joab I, et al. Twist and epithelial-mesenchymal transition are induced by the EBV oncoprotein Latent Membrane Protein 1 and are associated with metastatic nasopharyngeal carcinoma. Cancer Res. 2007;67:1970–1978. doi: 10.1158/0008-5472.CAN-06-3933. [DOI] [PubMed] [Google Scholar]
- 17.Cheng GZ, Chan J, Wang Q, Zhang W, Sun CD, et al. Twist transcriptionally up-regulates AKT2 in breast cancer cells leading to increased migration, invasion, and resistance to Paclitaxel. Cancer Res. 2007;67:1979–1987. doi: 10.1158/0008-5472.CAN-06-1479. [DOI] [PubMed] [Google Scholar]
- 18.Entz-Werle N, Stoetzel C, Berard-Marec P, Kalifa C, Brugiere L, et al. Frequent genomic abnormalities at TWIST in human pediatric osteosarcomas. Int J Cancer. 2005;117:349–355. doi: 10.1002/ijc.21068. [DOI] [PubMed] [Google Scholar]
- 19.Rosivatz E, Becker I, Specht K, Fricke E, Luber B, et al. Differential expression of the epithelial-mesenchymal transition regulators snail, SIP1, and twist in gastric cancer. Am J Pathol. 2002;161:1881–1891. doi: 10.1016/S0002-9440(10)64464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watanabe O, Imamura H, Shimizu T, Kinoshita J, Okabe T, et al. Expression of twist and wnt in human breast cancer. Anticancer Res. 2004;24:3851–3856. [PubMed] [Google Scholar]
- 21.Ansieau S, Bastid J, Doreau A, Morel AP, Bouchet BP, et al. Induction of EMT by Twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14:79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 22.Valdés_Mora F, Gómez del Pulgar T, Bandrés E, Cejas P, Ramírez de Molina A, et al. TWIST overexpression is associated with nodal invasion and male sex in primary colorectal cancer. Ann Surg Oncol. 2009;16:78–87. doi: 10.1245/s10434-008-0166-x. [DOI] [PubMed] [Google Scholar]
- 23.Deans GT, Parks TG, Rowlands BJ, Spence RA. Prognostic factors in colorectal cancer. Br J Surg. 1992;79:608–613. doi: 10.1002/bjs.1800790706. [DOI] [PubMed] [Google Scholar]
- 24.Vandesompele J, de Preter K, Pattyn F, Poppe B, Van RN, et al. Accurate normalization or real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034 (on-line access) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaplan EL, Meier P. Non-parametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 26.Peto R, Pike MC, Armitage P, Breslow NE, Cox DR, et al. Design and analysis of randomized clinical trials requiring prolonged observations of each patient: II. Analysis and examples. Br J Cancer. 1977;35:1–39. doi: 10.1038/bjc.1977.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomita N, Jiang W, Hibshoosh H, Warburton D, Kahn SM, et al. Isolation and characterization of a highly malignant variant of the SW480 human colon cancer cell line. Cancer Res. 1992;52:6840–6847. [PubMed] [Google Scholar]
- 28.Hong R, Lim SC. Overexpression of Twist in colorectal adenocarcinoma. Basic and Applied Pathology. 2009;2:15–20. [Google Scholar]
- 29.Okada T, Suehiro Y, Ueno K, Mitomori S, Kaneko S, et al. TWIST1 hypermethylation is observed frequently in colorectal tumors and its overexpression is associated with unfavorable outcomes in patients with colorectal cancer. Genes Chromosomes Cancer. 2010;49:452–462. doi: 10.1002/gcc.20755. [DOI] [PubMed] [Google Scholar]
- 30.Shibata K, Kajiyama K, Ino M, Terauchi E, Yamamoto A, et al. Twist expression in patients with cervical cancer is associated with poor disease outcome. Ann Oncol. 2008;19:81–85. doi: 10.1093/annonc/mdm344. [DOI] [PubMed] [Google Scholar]
- 31.Sasaki K, Natsugoe S, Ishigami S, Matsumoto M, Okumura H, et al. Significance of Twist expression and its association with E-cadherin in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2009 doi: 10.1186/1756-9966-28-158. Doi: 10.1186/1756-9966-28-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, et al. Direct regulation of TWIST by HIF-1α promotes metastasis. Nat Cell Biol. 2008;10:295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 33.Yang MH, Hsu DSS, Wang HW, Wang HJ, Lan HY, et al. Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nat Cell Biol. 2010;12:982–992. doi: 10.1038/ncb2099. [DOI] [PubMed] [Google Scholar]
- 34.Yu Q, Zhang K, Wang X, Liu X, Zhang Z. Expression of transcription factors sanil, slug, and twist in human bladder carcinoma. J Exp Clin Cancer Res. 2010 doi: 10.1186/1756-9966-29-119. Doi: 10.1186/1756-9966-29-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 36.International Multicentrere Pooled Analysis of B2 Colon Cancer Trials (IMPACT B2) investigators. Efficacy of adjuvant fluorouracil and folinic acid in B2 colon cancer. J Clin Oncol. 1999;17:1356–1363. [PubMed] [Google Scholar]
- 37.Mamounas E, Wieand S, Wolmark N, Bear HD, Atkins JN, et al. Comparative efficacy of adjuvant chemotherapy in patients with Dukes'B versus Dukes'C colon cancer: Results from four National Surgical Adjuvant Breast and Bowel Project adjuvant Studies (C-01, C-02, C-03, and C-04). J Clin Oncol. 1999;17:1349–1355. doi: 10.1200/JCO.1999.17.5.1349. [DOI] [PubMed] [Google Scholar]
- 38.Taal BG, Tinteren H, Zoetmulder FA. Adjuvant 5FU plus levamisole in colonic or rectal cancer: improved survival in stage II and III. Br J cancer. 2001;85:1437–1443. doi: 10.1054/bjoc.2001.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Batlle E, Sancho E, Francí C, Domínguez D, Monfar M, et al. The transcription factor Snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 40.Cano A, Pérez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, et al. The transcription factor Snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 41.Grooteclaes ML, Frisch SM. Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene. 2000;19:3823–3828. doi: 10.1038/sj.onc.1203721. [DOI] [PubMed] [Google Scholar]
- 42.Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, et al. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24:2375–2385. doi: 10.1038/sj.onc.1208429. [DOI] [PubMed] [Google Scholar]
- 43.Guaita S, Puig I, Francí C, Garrido M, Domínguez D, et al. Snail induction of epithelial mesenchymal transition in tumor cells is accompanied by MUC1 repression and ZEB1 expression. J Biol Chem. 2002;277:39209–39216. doi: 10.1074/jbc.M206400200. [DOI] [PubMed] [Google Scholar]
- 44.Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, et al. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7:1267–1278. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- 45.Pérez-Moreno MA, Locascio A, Rodrigo I, Dhondt G, Portillo F, et al. A new role for E12/E47 in the repression of E-cadherin expression and epithelial-mesenchymal transitions. J Biol Chem. 2001;276:27424–27431. doi: 10.1074/jbc.M100827200. [DOI] [PubMed] [Google Scholar]
- 46.Peinado H, Olmeda D, CanoA Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]