Abstract
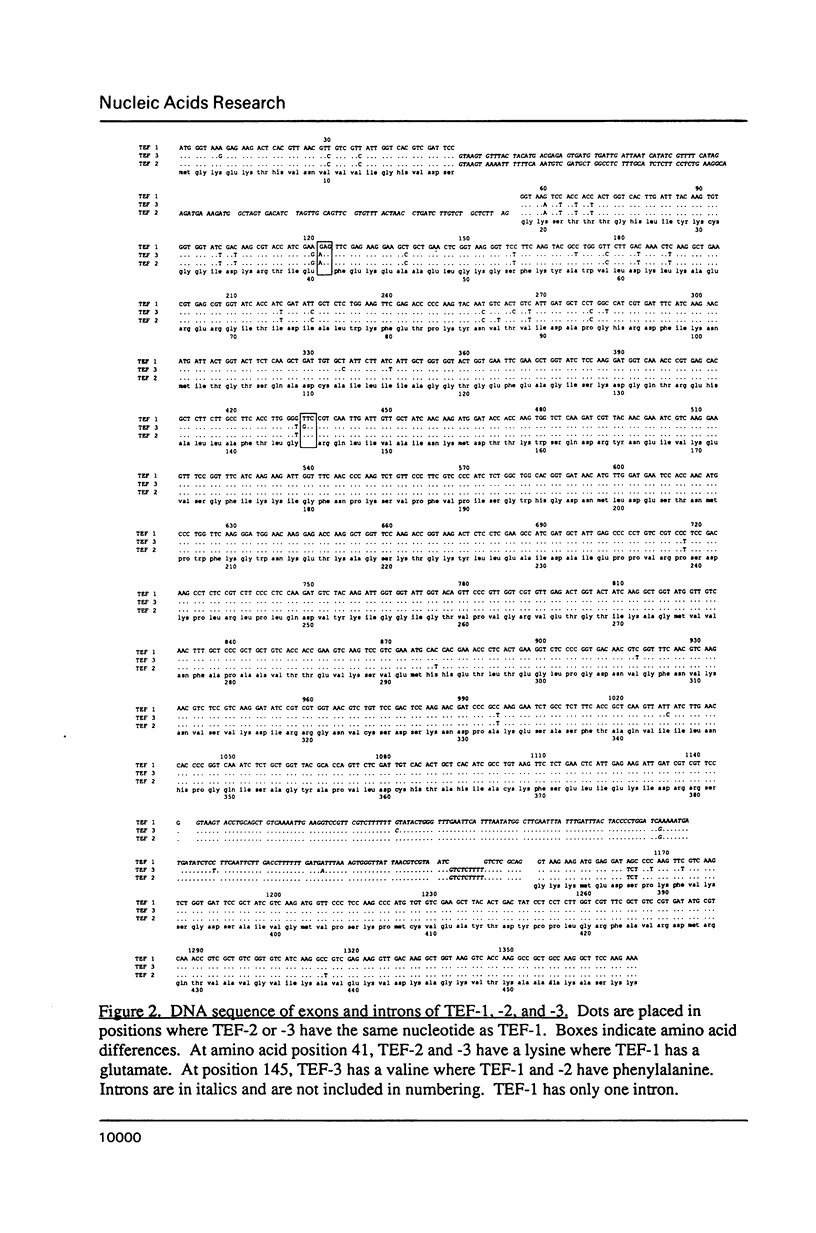
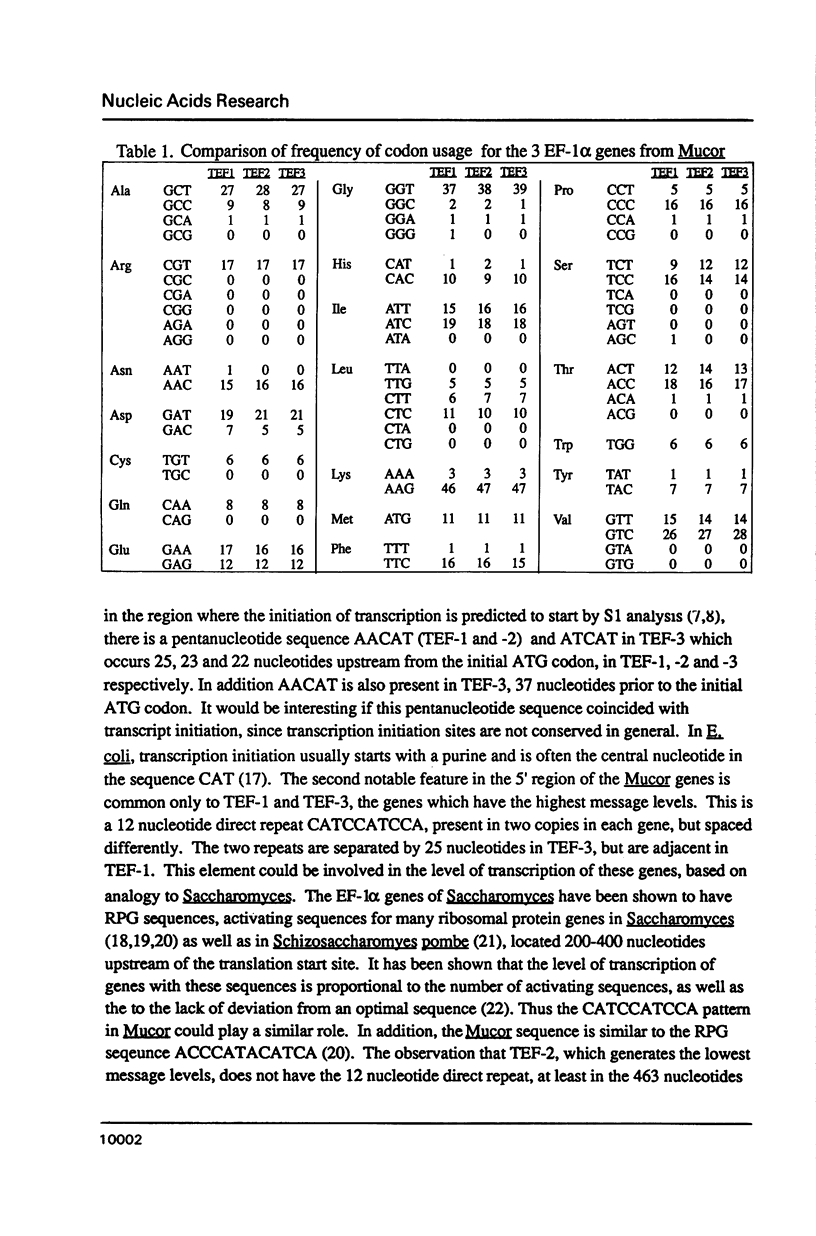
Our previous studies have shown that Mucor racemosus possesses three genes (TEF-1, -2 and -3) for EF-1 alpha, and that all three genes are transcribed. However, the level of transcription varies markedly between the three genes, with TEF-1 mRNA levels being approximately two fold higher than TEF-3 and 6 fold higher than TEF-2. We have now completed the DNA sequence of both strands of all three genes and have found that these genes are highly homologous. TEF-2 and TEF-3 are more similar to each other than they are to TEF-1. The TEF-2 and the TEF-3 coding regions differ from TEF-1 at 30 and 37 positions respectively out of 1374 nucleotides. Twenty-six of these nucleotide substitutions were common to both TEF-2 and TEF-3, and the majority of the substitutions were clustered in the 5' region of the coding sequences. While the majority of these changes were silent, TEF-2 and TEF-3 differed from TEF-1 by having a lysine instead of a glutamate at amino acid position 41. In addition, TEF-2 and -3, but not TEF-1, each have an intron located near the 5' end of the coding region, although its size and sequence is not conserved between the two genes. All three genes have a conserved intron near the 3' end of the coding region. The sequence data have been analyzed with respect to the structure and function of EF-1 alpha in protein biosynthesis.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Cottrelle P., Thiele D., Price V. L., Memet S., Micouin J. Y., Marck C., Buhler J. M., Sentenac A., Fromageot P. Cloning, nucleotide sequence, and expression of one of two genes coding for yeast elongation factor 1 alpha. J Biol Chem. 1985 Mar 10;260(5):3090–3096. [PubMed] [Google Scholar]
- Dever T. E., Glynias M. J., Merrick W. C. GTP-binding domain: three consensus sequence elements with distinct spacing. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1814–1818. doi: 10.1073/pnas.84.7.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzi W. A., Katayama C., Leathers T., Sypherd P. S. Regulation of protein synthesis factor EF-1 alpha in Mucor racemosus. Mol Cell Biol. 1985 May;5(5):1100–1103. doi: 10.1128/mcb.5.5.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hiatt W. R., Garcia R., Merrick W. C., Sypherd P. S. Methylation of elongation factor 1 alpha from the fungus Mucor. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3433–3437. doi: 10.1073/pnas.79.11.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J., Cottrelle P., Cool M., Vignais M. L., Thiele D., Marck C., Buhler J. M., Sentenac A., Fromageot P. A general upstream binding factor for genes of the yeast translational apparatus. EMBO J. 1985 Dec 16;4(13A):3539–3547. doi: 10.1002/j.1460-2075.1985.tb04114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J., Sentenac A. TUF, the yeast DNA-binding factor specific for UASrpg upstream activating sequences: identification of the protein and its DNA-binding domain. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3648–3652. doi: 10.1073/pnas.84.11.3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurnak F. Structure of the GDP domain of EF-Tu and location of the amino acids homologous to ras oncogene proteins. Science. 1985 Oct 4;230(4721):32–36. doi: 10.1126/science.3898365. [DOI] [PubMed] [Google Scholar]
- Larsen A., Sypherd P. S. Physiological control of phosphorylation ribosomal protein S6 in Mucor racemosus. J Bacteriol. 1980 Jan;141(1):20–25. doi: 10.1128/jb.141.1.20-25.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen A., Sypherd P. Ribosomal proteins of the dimorphic fungus, Mucor racemosus. Mol Gen Genet. 1979 Aug;175(1):99–109. doi: 10.1007/BF00267861. [DOI] [PubMed] [Google Scholar]
- Leer R. J., Van Raamsdonk-Duin M. M., Mager W. H., Planta R. J. Conserved sequences upstream of yeast ribosomal protein genes. Curr Genet. 1985;9(4):273–277. doi: 10.1007/BF00419955. [DOI] [PubMed] [Google Scholar]
- Linz J. E., Katayama C., Sypherd P. S. Three genes for the elongation factor EF-1 alpha in Mucor racemosus. Mol Cell Biol. 1986 Feb;6(2):593–600. doi: 10.1128/mcb.6.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linz J. E., Lira L. M., Sypherd P. S. The primary structure and the functional domains of an elongation factor-1 alpha from Mucor racemosus. J Biol Chem. 1986 Nov 15;261(32):15022–15029. [PubMed] [Google Scholar]
- Linz J. E., Sypherd P. S. Expression of three genes for elongation factor 1 alpha during morphogenesis of Mucor racemosus. Mol Cell Biol. 1987 May;7(5):1925–1932. doi: 10.1128/mcb.7.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima K., Kasai M., Nagata S., Kaziro Y. Structure of the two genes coding for polypeptide chain elongation factor 1 alpha (EF-1 alpha) from Saccharomyces cerevisiae. Gene. 1986;45(3):265–273. doi: 10.1016/0378-1119(86)90024-7. [DOI] [PubMed] [Google Scholar]
- Nischt R., Gross T., Gatermann K., Swida U., Käufer N. Sequence and regulatory responses of a ribosomal protein gene from the fission yeast Schizosaccharomyces pombe. Nucleic Acids Res. 1987 Feb 25;15(4):1477–1492. doi: 10.1093/nar/15.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlowski M., Sypherd P. S. Regulation of macromolecular synthesis during hyphal germ tube emergence from Mucor racemosus sporangiospores. J Bacteriol. 1978 Apr;134(1):76–83. doi: 10.1128/jb.134.1.76-83.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlowski M., Sypherd P. S. Regulation of translation rate during morphogenesis in the fungus Mucor. Biochemistry. 1978 Feb 21;17(4):569–575. doi: 10.1021/bi00597a002. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975 May 25;94(3):441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Teem J. L., Abovich N., Kaufer N. F., Schwindinger W. F., Warner J. R., Levy A., Woolford J., Leer R. J., van Raamsdonk-Duin M. M., Mager W. H. A comparison of yeast ribosomal protein gene DNA sequences. Nucleic Acids Res. 1984 Nov 26;12(22):8295–8312. doi: 10.1093/nar/12.22.8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]



